Abstract
Small heterodimer partner (SHP, NR0B2) is a nuclear orphan receptor without endogenous ligands. Due to its crucial inhibitory role in liver cancer, it is of importance to identify small molecule agonists of SHP. As such, we initiated a probe discovery effort to identify compounds capable of modulating SHP function. First, we performed binding assays using small molecule microarrays (SMMs) and discovered 5-(diethylsulfamoyl)-3-hydroxynaphthalene-2-carboxylic acid (DSHN) as a novel activator of SHP. DSHN transcriptionally activated Shp mRNA, but also stabilized SHP protein by preventing its ubiquitination and degradation. Second, we identified Ccl2 as a new SHP target gene by RNA-seq. We showed that activation of SHP by DSHN repressed Ccl2 expression and secretion by inhibiting p65 activation of CCL2 promoter activity, as demonstrated in vivo in Shp−/− mice and in vitro in HCC cells with SHP overexpression and knockdown. Third, we elucidated a strong inhibitory effect of SHP and DSHN on HCC cell migration and invasion by antagonizing the effect of CCL2. Lastly, by interrogating a publically available database to retrieve SHP expression profiles from multiple types of human cancers, we established a negative association of SHP expression with human cancer metastasis and patient survival. In summary, the discovery of a novel small molecule activator of SHP provides a therapeutic perspective for future translational and preclinical studies to inhibit HCC metastasis by blocking Ccl2 signaling.
Keywords: nuclear receptor, small molecule probe, cell migration and invasion, liver cancer, gene therapy
Introduction
Chemokines have been implicated to play an important role in many aspects of tumor cell biology, including regulation of tumor cell growth, metastasis, and host immune response (1). Chemokine (C-C motif) ligand 2 (CCL2), also known as monocyte chemoattractant protein-1 (MCP-1), recruits and activates monocytes during the inflammatory response. Recent studies showed that patients with high levels of tumor associated CCL2 expression had a significantly shorter survival (2). CCL2 association with the epithelial mesenchymal transition (EMT) and metastases was also reported (3). Targeting tumor-infiltrating macrophages via CCL2/CCR2 signaling was examined as a therapeutic strategy against hepatocellular carcinoma (4). Using a small molecule inhibitor to modulate CCL2 production may serve as a mean to suppress hepatocellular carcinoma (HCC) progression (5).
Small heterodimer partner (SHP, NR0B2) is a unique orphan nuclear receptor that acts as a transcriptional repressor (6) and inhibits transcription of its downstream target genes (7). SHP plays a crucial role in various hepatic metabolic pathways to control bile acid and lipid metabolism, homocysteine homeostasis and circadian rhythm (8–15). Although the SHP protein contains a ligand-like binding domain (16), it remains unsolved whether SHP has endogenous ligands (17).
Our previous studies revealed a role of SHP in the negative regulation of HCC cell proliferation (18). The expression of SHP was markedly diminished in human HCC due to epigenetic silencing (19) and SHP repressed DNA methylation via suppressing DNA methyltransferase (Dnmt) expression (20, 21). Activation of SHP by its agonists retinoid 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid (AHPN) and 4-[3′-(1-adamantyl)-4′-hydroxyphenyl]-3-chlorocinnamic acid (3-Cl-AHPC) induced HCC cell apoptosis by modulating Bcl2 function (22, 23). SHP interacted with p53 and Mdm2 proteins to control their ubiquitination and stability (24, 25). These studies suggest strongly that SHP functions as a tumor suppressor in HCC.
The above findings prompted us to further explore whether there is a connection between the role of SHP and HCC metastasis. In the present study, we investigated the effect of SHP in HCC cell invasion and migration using a novel chemical probe. First, we discovered a small molecule compound 5-(diethylsulfamoyl)-3-hydroxynaphthalene-2-carboxylic acid (namely, DSHN) that functions as a transcriptional activator of SHP using small molecule microarrays (SMMs) (26). Second, we identified Ccl2 as a downstream target gene of SHP. Third, we showed that activation of SHP by its novel agonist inhibited HCC cell invasion and migration by suppressing Ccl2 expression. Lastly, we established a negative correlation between SHP expression and human cancer metastasis and patient survival. Because extrahepatic metastasis of HCC is a significant clinical problem, our studies shed lights on the perspective of using small molecule activator of SHP to inhibit HCC metastasis by blocking Ccl2 signaling.
Materials and Methods
Animals, Cell lines, Plasmids and Reagents
Wild-type (wt) and Shp−/− (sko), non-transgenic control (nc) and hepatocyte specific Shp transgenic (stg) mice were described previously (9, 22). Protocols for animal use were approved by the Institutional Animal Care Committee at the University of Utah and University of Connecticut. Nmuli, Hepa1, HEK293T, MHCC97H and MHCC97L, Hep3B and Huh7 cells were purchased from ATCC in 2010 and were made aliquots and stored in liquid nitrogen tank immediately after the 1st passage. All cell lines were last confirmed by short tandem repeat analysis of cellular DNA (PowerPlex 1.2 Kit, Promega) in 2015. When cells were recovered from liquid nitrogen in 2015 they were found to be free of mycoplasma (e-Myco Kit, Boca Scientific). The cell lines were passaged for less than 6 months when used for experiments. The CCL2 promoter was generated from Dr. Teizo Yoshimura’s laboratory from National Cancer Institute. Adenovirus Shp and siSHP were described previously (9). The recombinant human CCL2 protein was purchased from BioLegend.
Small-Molecule Microarray Screens
SMMs were manufactured as described previously using isocyanate-mediate capture (27). In total, 21,600 printed features were screened, including 9,152 products of diversity-oriented synthesis, 9,152 commercial compounds that included natural products and drug-like compounds, 772 known bioactive compounds, 144 compound controls, and 2,380 DMSO solvent controls. Printed microarrays were screened using three replicates and incubated with 1 μg/mL of purified SHP N-terminally tagged with His6 in TBST buffer for 60 minutes at 4 °C with gentle agitation. The protein was purchased from Panomics (#RP1030) and supplied at 0.1 mg/mL in a storage buffer of 50 mM Tris-HCl (pH 8.0), 138 mM NaCl, 27 mM KCl, and 1 mM DTT. Slides were briefly rinsed in TBST buffer and then incubated with an AlexaFluor-647-labeled anti-His5 antibody (Qiagen #34600) for 60 minutes at 4 °C with gentle agitation. The slides were washed in TBST buffer for 1 minute followed by a wash in TBS buffer 1 minute and dried by centrifugation. Dried slides were scanned for fluorescence at 635 nm using a Genepix 4000B microarray scanner. Control arrays were probed with antibody only. GenePix Pro 6.0 software was used to identify fluorescence intensity values for a set 300 μm diameter centered over each feature. Each SMM spot intensity was scored by its deviation from a population of vehicle-control spots on the same slide as described previously (28). Three replicate measurements were combined as weighted averages of deviations, normalized by the variance of corresponding vehicle-control distributions and measurement uncertainties. We called a positive ‘hit’ any compound whose normalized score for protein binding exceeded the expected score for the most extreme acceptable vehicle-control outlier at a fixed statistical significance (family-wise error rate p < 0.05 by Holm-Bonferroni method) (28), indicating a greater likelihood that the compound was a member of a putative ‘hit’ distribution than of the vehicle-control distribution.
Serum Shock
Huh7 cells were grown to confluence in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were starved in the same medium with no FBS for 18 hr, 50% horse serum was added for 2 hr, and then the medium was changed back to starvation medium. Cells were harvested at 4 hr intervals for analyses (29).
ELISA Assay
The level of CCL 2 was measured using CCL 2 ELISA Ready-SET-Go! kit (eBioscience) following manufacturer’s specifications. In brief, cell culture media were collected and centrifuged at 5,000 g for 10 min and stored at −80°C until use. For mouse samples, liver tissues were homogenized in lysis buffer (50 mM Tris-HCl, 2 mM EDTA, pH 7.4 and 1x protease inhibitors) (Thermo Scientific) on ice. Samples were centrifuged at 13,000 g for 10 min and stored at −80°C until use. Mouse serum was centrifuged and stored at −80°C. Experiments were performed in triplicate wells and the data were averaged.
Cell Migration and Invasion Assays
The cell invasion and migration assays were described previously (30, 31). Briefly, cells were infected with the indicated adenovirus for 48 hr. The cells were then serum starved for 24 hr and 5×104 cells were seeded on Transwell inserts (8 μm pore size; Culterx 96 well cell migration assay, cat# 3465-095-K). Cells were allowed to migrate for 16 hr, and the non-migratory cells were removed from the insert with a cotton swab. The migrated cells were fixed for 10 min (3.7% v/v formaldehyde in PBS) before staining with 0.1% crystal violet for 15 min, followed by washing with PBS. Pictures were taken with Microfire/Qcam CCD Olympus 1×81 microscope. Crystal violet stained cells were counted and then lysed with 1% SDS for 30 min and absorbance was measured at 595 nm. Cell invasion assays were performed similarly as migration assays, except that cells were seeded on pre-coated inserts with BME solution (Cultrex 96 well BME cell invasion Assay, cat# 3455-096-K). Invaded cells were treated with Cell Dissociation Solution/Calcein-AM for 1 hr, and fluorescence was measured at 485 nm excitation and 520 nm emission.
qPCR, Transient Transfection and Luciferase Assay
In brief, cells were transfected with the plasmids as indicated in the Figure legends. Transfection was carried out using Lipofectamine 2000 (Invitrogen). Luciferase activities were measured and normalized against Renilla activities (Promega). Experiments were done in three independent triplicate transfection assays. Detailed methods were described previously (8, 9). Mammalian two hybrid assay was described previously (16).
Statistical analysis
All the experiments were performed in triplicate and repeated at least two times. The data presented as the mean values ± standard error of the mean (SE). Statistical analysis was carried out using the Student t test for unpaired data to compare the values between the two groups. P<0.01 was considered statistically significant.
Results
Discovering a small molecule that functions as a transcriptional activator of SHP
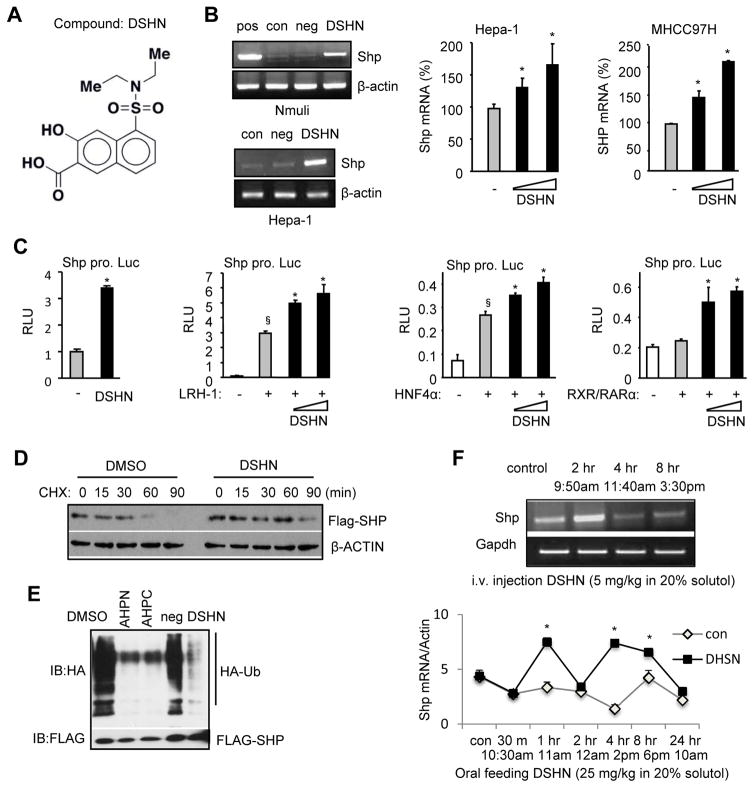
We used SMMs for ligand discovery (32), which generated 20 hits (Supp Fig. S1). We tested the ability of the positive compounds to alter SHP expression and identified compound #29, i.e. 5-(diethylsulfamoyl)-3-hydroxynaphthalene-2-carboxylic acid (namely, DSHN) as an assay positive that bound SHP protein applied to the microarray. The chemical structure and detailed information of DSHN is shown in Fig. 1A and Supp Fig. S2. Our initial test observed that treatment of HCC cells (MHCC97H, Hepa1, Huh7) with high concentrations of DSHN did not affect cell viability as determined by MTT assay (Supp Fig. S3). Further analysis showed that DSHN markedly reactivated Shp expression in both mouse (Nmuli, Hepa-1) and human (MHCC97H) hepatic cells in which Shp expression was lost (Fig. 1B). In addition, DSHN activated Shp promoter luciferase reporter (Fig. 1C, 1st panel) and augmented the transactivation of several known Shp activators dose-dependently, including live receptor homolog 1 (LRH-1), hepatocyte nuclear factor 4α (HNF4α), and retinoid X receptor/retinoic acid receptor α (RXR/RARα) (Fig. 1C, 2nd to 4th panels). DSHN also increased SHP protein half-life (30 min in DMSO group vs. 90 min in DSHN group) as determined by inhibiting protein synthesis using cycloheximide (CHX) (Fig. 1D). The increased stability of SHP protein by DSHN was associated with the ability of DSHN to prevent SHP protein ubiquitination and degradation (Fig. 1E). AHPN and AHPC were reported to bind to SHP (33) and inhibit SHP protein ubiquitination (22), thus both served as positive controls. On the other hand, DSHN did not alter the interaction of SHP with LRH-1 and HNF4α (Supp Fig. S4A–4B), as determined by mammalian two-hybrid approach.
Figure 1. Identifying a small molecule that activates SHP gene transcription.
(A) The structure of compound DSHN, which was identified by small molecule microarray (SMM).
(B) Left: Semi-quantitative PCR of Shp mRNA in normal mouse hepatocyte Nmuli (upper panel) and hepatoma Hepa-1 (lower panel) cells treated with DSHN (20 μM). pos: positive control, cells transfected with a mouse Shp (mShp) expression plasmid; con, control, DMSO; neg: negative control, cells treated with an unrelated compound that does not induce Shp. Right: qPCR of Shp mRNA in mouse Hepa-1 (left) and human MHCC97H (right) cells treated with DSHN (20, 30 μM). Data is represented as mean ± SE (*p<0.01 vs. DMSO, grey bar).
(C) Relative luciferase (luc) activity (RLU) of mShp promoter (pro.) without (first) or with co-transfected LRH-1 (second), HNF4α (third), and RXR/RARα (fourth) in the absence (-, DMSO) or presence of DSHN (50, 100 μm). Data is represented as mean ± SE of triplicate assays (§p<0.01 vs. white bar; *p<0.01 vs. gray bar).
(D) Western blot of SHP protein in Hepa-1 cells treated with DSHN (20 μM) and protein synthesis inhibitor cycloheximide (CHX) to determine SHP protein half-life. The cells were transfected Flag-Shp plasmid (1 μg/well) for 24 hrs before harvested at indicated time point.
(E) Ubiquitination analysis of SHP protein. HEK 293T cells were cotransfected with Flag-Shp (1 μg/well) and HA-Ub (2 μg/well) for 24 hrs before the treatment with Shp agonists (AHPN, AHPC), an unrelated compound (neg) and DSHN (20 μM).
(F) Semi-quantitative PCR (upper) and qPCR (bottom) of Shp mRNA expression in mice treated with DSHN (upper: 5 mg/kg for i.v. injection or lower: 25 mg/25 kg for oral feeding). Data is represented as mean ± SE (*p<0.01 vs. solvent treated group). Mice of two month-old were used (n=5). ALL data are representative of two or more experiments.
To test whether DSHN induces Shp expression in vivo in mouse liver, we treated mice with DSHN using two methods. Because Shp is an integral part of the liver circadian clock network and its expression shows a rhythmic variation (9), we collected samples at multiple time points. First, DSHN was given to mice via i.v. injection and livers were collected before injection and 2 hr, 4 hr, and 8 hr after injection. Shp mRNA exhibited a peak induction 2 hr after DSHN treatment (Fig. 1F, upper panel). Next, mice were treated with DSHN by oral feeding and livers were collected over a longer time period for 24 hr. Shp mRNA showed a clear rhythm with peak inductions at 1 hr and 4 hr after DSHN treatment (Fig. 1F, lower panel). To further confirm a direct effect of DSHN on SHP expression, we conducted serum shock experiment to synchronize Huh7 cells (29). The expression of SHP mRNA exhibited two peaks; both were highly induced upon DSHN treatment (Supp Fig. S4C). Overall, compound DSHN is a potent activator of SHP through both transcriptional and post-transcriptional regulation.
Ccl2 is a direct transcriptional target of SHP
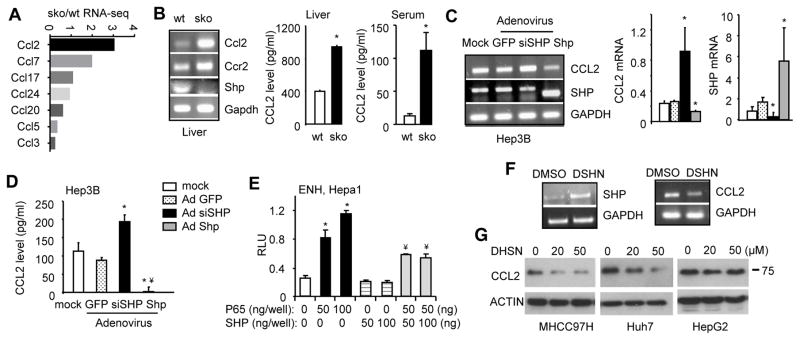
A recent study established a role of SHP in TLR-dependent inflammatory response (34). From our RNA-sequencing (RNA-seq) datasets (GEO accession no: GSE43893) (13), we found an upregulation in the expression of Ccl family members in SHP−/− (sko) vs. wild-type (wt) liver (Fig. 2A). In particular, Ccl2 showed the highest induction. Due to the importance of CCL2 in both inflammation and cancer metastasis, we examined Ccl2 expression regulation by SHP. The upregulation of Ccl2 and its receptor Ccr2 mRNA in sko vs. wt liver was confirmed by RT-PCR (Fig. 2B, left). This was accompanied by the elevated levels of CCL2 production in sko liver and serum (Fig. 2B, middle and right). In contrast, Ccl2 mRNA was reduced in hepatocyte Shp-overexpressed transgenics (stg) (20) as compared to the non-transgenic control (nc) (Supp Fig. S5A).
Figure 2. SHP functions as a transcriptional suppressor of Ccl2.
(A) RNA-seq of relative expression levels of Ccl family in sko vs wt liver.
(B) Left: Semi-quantitative PCR of hepatic Ccl2, Ccr2 and Shp mRNA in wildtype (wt) and Shp−/− (sko). Middle and Right: ELISA of CCL2 levels in liver and serum of wt and sko. Data is represented as mean ± SE of triplicate assays (*p<0.01 vs. white bar).
(C) Semi-quantitative PCR (left) and qPCR (middle and right) of CCL2 and SHP mRNA in Hep3B cells with Shp overexpression or knockdown (siSHP) using adenovirus (100 MOI). Lower band: mouse Shp (Ade-Shp); higher band: endogenous human SHP. Data is represented as mean ± SE of triplicate assays (*p<0.01 vs. white bar).
(D) ELISA of CCL2 protein levels in culture medium of Hep3B cells infected with indicated adenovirus (Ad, 100 MOI). Data is represented as mean ± SE of triplicate assays (*p<0.01 vs. white bar; ¥p<0.01 vs. siSHP, black bar).
(E) Promoter luciferase report assay. Hepa1 cells were cotransfected with the indicated plasmids for 48 hrs. Data is represented as mean ± SE of triplicate assays (*p<0.01 vs. white bar; ¥p<0.01 vs. 2nd bar).
(F) Semi-quantitative PCR of SHP (left) and CCL2 (right) mRNA in Huh7 cells treated with DSHN for 24 hrs (10 μm).
(G) Western blot of CCL2 protein in HCC cells treated with different doses of DSHN for 24 hr (MHCC97H, Huh7) or 48 hr (HepG2). Data are representative of two or more experiments.
To determine a direct cell autonomous inhibition of Ccl2 by SHP, we used in vitro cell culture system. Hep3B cells expressed high basal level of SHP thus were used to examine the effect of SHP knockdown on CCL2 expression. CCL2 mRNA (Fig. 2C), as well as CCL2 production (Fig. 2D), were markedly increased by SHP knockdown (siSHP) but decreased by Shp overexpression (Ade-Shp). CCL2 secretion was also largely diminished by Ade-Shp in Huh7 cells (Supp Fig. S5B).
We analyzed Ccl2 promoter and identified NF-κB binding sites (Supp. Fig. S5C). SHP was reported to interact with NF-κB subunit p65 (RelA) to mediate cell apoptosis (35). Thus we examined the effect of SHP on p65-meidated Ccl2 promoter activity. Promoter luciferase reporter assays revealed that p65 dose-dependently activated Ccl2 promoter (both ENH and LNG), which was markedly suppressed by Shp (Fig. 2E and Supp. Fig. S5D). In addition, DSHN treatment diminished CCL2 mRNA expression that correlated with SHP activation (Fig. 2F). Furthermore, CCL2 protein was dose-dependently diminished by DSHN in multiple HCC cells (Fig. 2G). Interestingly, a stronger inhibition was observed in MHCC97H and Huh7 (high migration potential) as compared to HepG2 (low migration potential), suggesting DSHN may be more effective in invasive cells. Taken together, our results demonstrate that Ccl2 is a direct transcriptional target of Shp and its expression can be inhibited by DSHN.
Activation of SHP inhibits Ccl2-mediated HCC cell migration and invasion
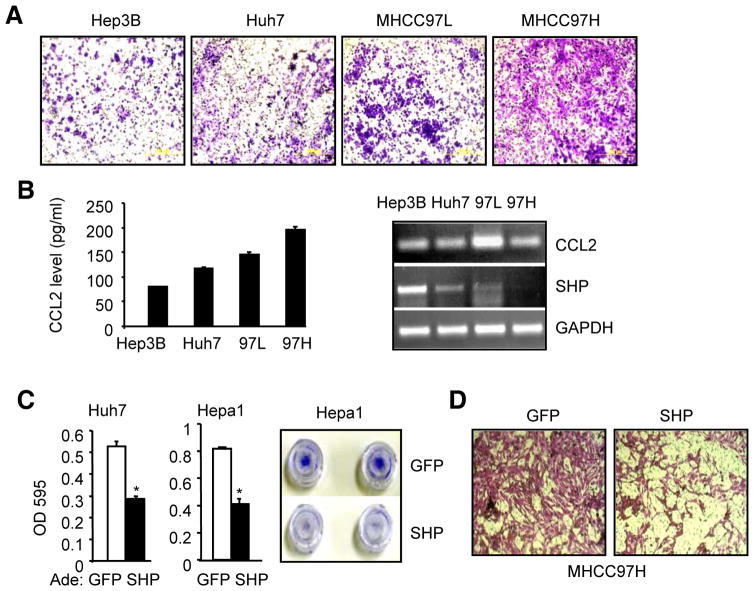
SHP served as a tumor suppressor in HCC by inhibiting cell proliferation and activing apoptosis (18, 22), however, the role of SHP in cell migration remains unknown. By comparing different HCC cell lines, MHCC97H exhibited the highest migration potential as compared to Hep3B, Huh7 and MHCC97L (Fig. 3A). Interestingly, CCL2 production and its expression were positively correlated with the migration potential of HCC cells (Fig. 3B), whereas SHP expression showed an inverse correlation. We determined the effect of SHP in inhibiting HCC cell migration and invasion by examining multiple HCC cell lines. Cell migration assay revealed that overexpression of SHP markedly suppressed the potential of Huh7 and Hepa1 (Fig. 3C) and MHCC97H cells (Fig. 3D) to migrate through the Transwell membrane.
Figure 3. Overexpression of SHP inhibits HCC cell invasion and migration.
(A) Comparing the migration potential in Hep3B, Huh7, HMCC97L and MHCC97H cells. Migrated cells were visualized by the staining of crystal violet.
(B) Left: The levels of secreted CCL2 protein in culture medium as examined by ELISA. Right: Semi-quantitative PCR of CCL2 mRNA in various HCC cells.
(C) HCC cell migration assay. Huh7 (left) and Hepa1 (middle) cells were overexpressed Ade-SHP (adenovirus, 50 MOI) for 48 hrs. Data is represented as mean ± SE of triplicate assays (*p<0.01 vs. white bar). Right: Images of invaded cells stained with crystal violet.
(D) HCC cell migration assay. MHCC97H cells were transduced with Ade-SHP (50 MOI) for 48 hrs and migrated cells were visualized by the staining of crystal violet. Data are representative of two or more experiments.
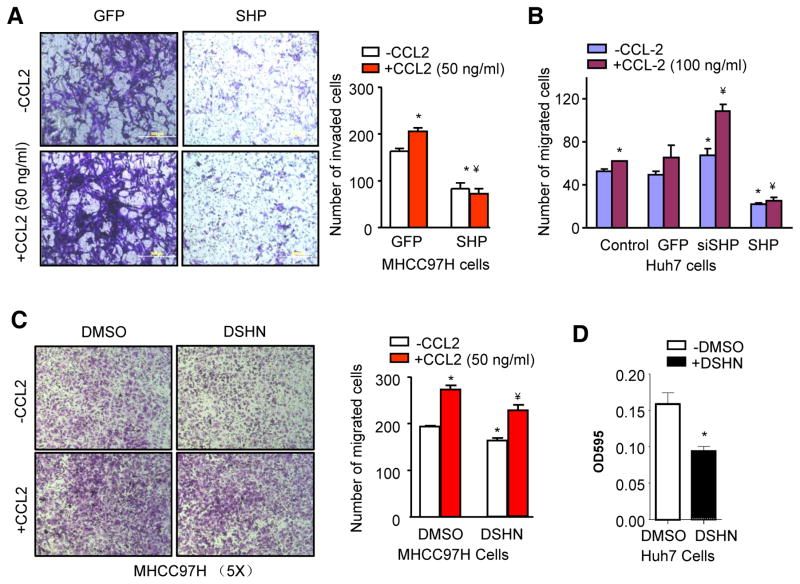
Next, we determined the direct effect of SHP on CCL2-mediated 97H cell invasion. Treatment of recombinant CCL2 protein dramatically stimulated 97H cell invasion, whereas overexpression of SHP antagonized the ability of CCL2 to promote cell invasion (Fig. 4A). Huh7 cells expressed higher basal level of SHP than 97H thus were used to examine the effect of SHP knockdown. As expected, siSHP markedly augmented the effect of CCL2 to stimulate Huh7 cell migration (Fig. 4B). In addition, treatment of 97H cells with DSHN significantly diminished CCL2-induced cell migration (Fig. 4C). The cell migration potential of Huh7 cells were also inhibited by DSHN (Fig. 4D). It was noted that the inhibitory effect of DSHN was less strikingly than that of Ade-SHP (Fig. 4A), which could be due to the fact that the level of SHP was much highly expressed and induced by AdeSHP than by DSHN. Overall, the results suggest that SHP inhibition of HCC cell invasion and migration is at least in part mediated by its ability to suppress CCL2 expression and function.
Figure 4. Activation of SHP inhibits CCl2-mediated HCC cell invasion and migration.
(A) HCC invasion assay. MHCC97H cells were transduced with Ade-SHP for 48 hrs before subjected to invasion assay in the absence or presence of CCL2 (50 ng/ml). The invaded cells were stained with crystal violet and counted. Quantitative results on the right: *p<0.01 vs. GFP (-CCL2); ¥p<0.01 vs. GFP (+CCL2).
(B) HCC migration assay. Huh7 cells were transduced with indicated adenovirus for 48 hrs before subjected to migration assay in the absence or presence of CCL2 (100 ng/ml). The migrated cells were stained with crystal violet and counted. *p<0.01 vs. GFP (-CCL2); ¥p<0.01 vs. GFP (+CCL2).
(C) HCC migration assay. DSHN group: 50 μM for 48 hrs; CCL2 group: 50 ng/ml for 24 hrs. The migrated cells were stained with crystal violet and counted. Quantitative results on the right: *p<0.05 vs. DMSO (-CCL2); ¥p<0.01 vs. DMSO (+CCL2) and DSHN (-CCL2).
(D) HCC migration assay in Huh7 cells treated with DSHN (50 μM). *p<0.01 vs. DMSO. Data are representative of two or more experiments.
SHP expression is negatively associated with cancer metastasis and patient survival
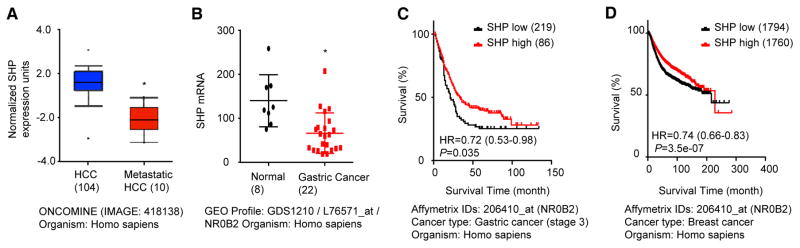
To further establish the clinical relevance of SHP in human cancer metastasis and survival, we interrogated publically available database to retrieve SHP gene expression profile from multiple types of human cancers. The DNA copy number of SHP was noticeable reduced in HCC patients based on The Cancer Genome Atlas (TCGA) database (Supp. Fig. S6). As expected, SHP expression was further decreased in human metastatic HCC vs. HCC (Fig. 5A). SHP expression was also markedly decreased in human gastric cancer relative to normal (Fig. 5B) (36).
Figure 5. The expression of SHP is negatively associated with cancer metastasis.
(A) The expression of SHP in human HCC specimens. The data is derived from ONCOMINE (IMAGE: 418138).
(B) The expression of SHP in human gastric cancer. The data is derived from Profile: GDS1210 / L76571_at / NR0B2.
(C) Kaplan-Meier survival plots. The gastric cancer dataset was divided into two groups according to the expression levels of SHP from 305 patients. Grouped datasets were subjected to Kaplan-Meier survival analysis.
(D) Kaplan-Meier survival plots. The breast cancer dataset was divided into two groups according to the expression levels of SHP from 3554 patients. Grouped datasets were subjected to Kaplan-Meier survival analysis.
The Kaplan Meier plotter is capable to assess the effect of 54,675 / 22,277 genes on survival using 10,188 cancer samples. These include 4,142 breast, 1,648 ovarian, 2,437 lung and 1,065 gastric cancer patients with a mean follow-up of 69 / 40 / 49 / 33 months (37). Kaplan Meier plotter analysis revealed a significant association between SHP expression and patient’s survival time. The high expression of SHP were associated significantly with better patient survival in human gastric cancer (Fig. 5C) and breast cancer (Fig. 5D). SHP expression was also negatively associated with patients’ survival time in ovarian cancer and lung cancer. However, the p value did not reach significance (Supp Fig. S7A–B). Nonetheless, SHP expression was significantly decreased in lung cancer compared to normal epithelium (Supp Fig. S7C). Together, these clinical data analyses clearly indicate that SHP may function as an important survival factor and its expression may serve as a negative prognostic factor associated with high rates of metastasis and decreased disease-free survival in a wide-range of human cancers.
Discussion
Small-molecule microarrays (SMMs) have been used previously to identify small molecules that bind and modulate specific protein targets in a cellular context, including various transcription factors (26, 28, 38). Using the SMM based approach, we identified DSHN as a putative binder and novel agonist of SHP. Our results demonstrate a dual-function of DSHN; it not only activates Shp promoter to induce Shp gene transcription but also prevents proteasome mediated SHP degradation to increase SHP protein stability. More importantly, DSHN may serve as a potential therapeutic agent to inhibit HCC metastasis via SHP-mediated repression of Ccl2 signaling.
SHP possesses the essential characteristics as a tumor suppressor gene, based on its ability to inhibit HCC cell proliferation (18), activate apoptosis (22, 23), repress DNA methylation (20, 21), suppress HCC cell migration and invasion (the present study), its downregulation in HCC (19), and its inhibition of oncogenic non-coding RNAs (39, 40). In particular, SHP expression is negatively correlated with patient survival in several types of aggressive and highly metastatic human cancers, including gastric cancer and breast cancer, suggesting it may be an attractive target for cancer therapy that is not limited to HCC. It was of considerable interest to note that SHP was identified as a single gene prognostic lung cancer biomarker (41). The regulatory function of SHP in other cancers remain to be investigated.
One major obstacle to explore the possibility of targeting SHP in HCC therapeutics is that SHP is an orphan receptor and endogenous ligands for SHP have not yet been identified. Significant efforts were made in the past, which identified AHPN and 3-Cl-AHPC as the first exogenous SHP agonists (33) that were potent apoptosis inducer (22). Our study opens possible new avenues for treatment of liver cancer by restoring the lost SHP expression in HCC using DSHN. Of equal importance, identifying Ccl2 as a SHP target gene allows future mechanistic studies on the action of DSHN beyond liver cancer. DSHN might be tested in preclinical models to see whether it can induce SHP expression and inhibit tumorigenesis or malignant behavior. In summary, our study highlights the potential use of SHP as a therapeutically tractable gene in future translational and preclinical studies.
Supplementary Material
Acknowledgments
Grant Support: L. Wang is supported by NIH R01ES025909, R01DK080440, R01DK104656, R21AA022482, R21AA024935, VA Merit Award 1I01BX002634, P30 DK34989 (Yale Liver Center) and the National Natural Scientific Foundation of China (Grant No. 81572443). A.N. Koehler is supported by N01-CO-12400 and R01CA160860.
Abbreviations
- CCL2
chemokine (C-C motif) ligand 2
- MCP-1
monocyte chemoattractant protein-1
- EMT
epithelial mesenchymal transition
- HCC
hepatocellular carcinoma
- SHP
small heterodimer partner
- AHPN
6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid
- 3-Cl-AHPC
4-[3′-(1-adamantyl)-4′-hydroxyphenyl]-3-chlorocinnamic acid
- Bcl2
B-cell lymphoma 2
- MDM2
mouse double minute 2 homolog
- DSHN
5-(diethylsulfamoyl)-3-hydroxynaphthalene-2-carboxylic acid
- SMMs
small molecule microarrays
- wt
wild type
- sko
Shp−/−
- stg
Shp-overexpressed transgenic mice
- LRH-1
live receptor homolog 1
- HNF4α
hepatocyte nuclear factor 4α
- RXR/RARα
retinoid X receptor/retinoic acid receptor α
- CHX
cycloheximide
- TLRs
toll-like receptors
- ade
adenovirus
- ENH
enhancer
- LNG
long enhancer
- TCGA
The Cancer Genome Atlas
Footnotes
No conflict of interest to disclose for all authors.
References
- 1.Conti I, Rollins BJ. CCL2 (monocyte chemoattractant protein-1) and cancer. Semin Cancer Biol. 2004;14:149–54. doi: 10.1016/j.semcancer.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, et al. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–9. [PubMed] [Google Scholar]
- 3.Lee CC, Ho HC, Su YC, Lee MS, Hung SK, Lin CH. MCP1-Induced Epithelial-Mesenchymal Transition in Head and Neck Cancer by AKT Activation. Anticancer Res. 2015;35:3299–306. [PubMed] [Google Scholar]
- 4.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2015 doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 5.Chen TA, Wang JL, Hung SW, Chu CL, Cheng YC, Liang SM. Recombinant VP1, an Akt inhibitor, suppresses progression of hepatocellular carcinoma by inducing apoptosis and modulation of CCL2 production. PLoS One. 2011;6:e23317. doi: 10.1371/journal.pone.0023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudraiah S, Zhang X, Wang L. Nuclear Receptors as Therapeutic Targets in Liver Disease: Are We There Yet? Annu Rev Pharmacol Toxicol. 2016;56:605–26. doi: 10.1146/annurev-pharmtox-010715-103209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchiya H, da Costa KA, Lee S, Renga B, Jaeschke H, Yang Z, et al. Interactions Between Nuclear Receptor SHP and FOXA1 Maintain Oscillatory Homocysteine Homeostasis in Mice. Gastroenterology. 2015;148:1012–23. e14. doi: 10.1053/j.gastro.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SM, Zhang Y, Tsuchiya H, Smalling R, Jetten AM, Wang L. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology. 2015;61:497–505. doi: 10.1002/hep.27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta S, Wang L, Moore DD, Osborne TF. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter by nuclear receptors liver receptor homologue-1 and small heterodimer partner: a mechanism for differential regulation of cholesterol synthesis and uptake. J Biol Chem. 2006;281:807–12. doi: 10.1074/jbc.M511050200. [DOI] [PubMed] [Google Scholar]
- 11.Suh JH, Huang J, Park YY, Seong HA, Kim D, Shong M, et al. Orphan nuclear receptor small heterodimer partner inhibits transforming growth factor-beta signaling by repressing SMAD3 transactivation. J Biol Chem. 2006;281:39169–78. doi: 10.1074/jbc.M605947200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Xu N, Xu J, Kong B, Copple B, Guo GL, et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr-1/SHP/EID1 network. Hepatology. 2014;60:919–30. doi: 10.1002/hep.27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalling RL, Delker DA, Zhang Y, Nieto N, McGuiness MS, Liu S, et al. Genome-wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease. American journal of physiology Gastrointestinal and liver physiology. 2013;305:G364–74. doi: 10.1152/ajpgi.00077.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Bonzo JA, Gonzalez FJ, Wang L. Diurnal regulation of the early growth response 1 (Egr-1) protein expression by hepatocyte nuclear factor 4alpha (HNF4alpha) and small heterodimer partner (SHP) cross-talk in liver fibrosis. J Biol Chem. 2011;286:29635–43. doi: 10.1074/jbc.M111.253039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabbi-Anneni I, Cooksey R, Gunda V, Liu S, Mueller A, Song G, et al. Overexpression of nuclear receptor SHP in adipose tissues affects diet-induced obesity and adaptive thermogenesis. American journal of physiology Endocrinology and metabolism. 2010;298:E961–70. doi: 10.1152/ajpendo.00655.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, et al. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem. 2010;285:24871–81. doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cellanetti M, Gunda V, Wang L, Macchiarulo A, Pellicciari R. Insights into the binding mode and mechanism of action of some atypical retinoids as ligands of the small heterodimer partner (SHP) Journal of computer-aided molecular design. 2010;24:943–56. doi: 10.1007/s10822-010-9386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–98. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Andrews GK, Wang L. Zinc-induced Dnmt1 expression involves antagonism between MTF-1 and nuclear receptor SHP. Nucleic acids research. 2012;40:4850–60. doi: 10.1093/nar/gks159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Wang L. Nuclear receptor SHP inhibition of Dnmt1 expression via ERRgamma. FEBS Lett. 2011;585:1269–75. doi: 10.1016/j.febslet.2011.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, et al. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Molecular and cellular biology. 2010;30:1341–56. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang L. Characterization of the mitochondrial localization of the nuclear receptor SHP and regulation of its subcellular distribution by interaction with Bcl2 and HNF4alpha. PLoS One. 2013;8:e68491. doi: 10.1371/journal.pone.0068491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Zhang Y, Kemper JK, Wang L. Cross-regulation of protein stability by p53 and nuclear receptor SHP. PLoS One. 2012;7:e39789. doi: 10.1371/journal.pone.0039789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Wang L. An autoregulatory feedback loop between Mdm2 and SHP that fine tunes Mdm2 and SHP stability. FEBS Lett. 2012;586:1135–40. doi: 10.1016/j.febslet.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. Journal of the American Chemical Society. 2003;125:8420–1. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 27.Bradner JE, McPherson OM, Mazitschek R, Barnes-Seeman D, Shen JP, Dhaliwal J, et al. A robust small-molecule microarray platform for screening cell lysates. Chemistry & biology. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Clemons PA, Bodycombe NE, Carrinski HA, Wilson JA, Shamji AF, Wagner BK, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18787–92. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell metabolism. 2010;12:174–86. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893–9. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA-127 and TGFbeta/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013;8:e65256. doi: 10.1371/journal.pone.0065256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vegas AJ, Fuller JH, Koehler AN. Small-molecule microarrays as tools in ligand discovery. Chemical Society reviews. 2008;37:1385–94. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farhana L, Dawson MI, Leid M, Wang L, Moore DD, Liu G, et al. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer research. 2007;67:318–25. doi: 10.1158/0008-5472.CAN-06-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW, Jin HS, et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nature immunology. 2011;12:742–51. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 35.Murshed F, Farhana L, Dawson MI, Fontana JA. NF-kappaB p65 recruited SHP regulates PDCD5-mediated apoptosis in cancer cells. Apoptosis : an international journal on programmed cell death. 2014;19:506–17. doi: 10.1007/s10495-013-0939-y. [DOI] [PubMed] [Google Scholar]
- 36.Oncomine Research Edition. cited 2015. Available from: https://www.oncomine.org/
- 37.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pop MS, Stransky N, Garvie CW, Theurillat JP, Hartman EC, Lewis TA, et al. A small molecule that binds and inhibits the ETV1 transcription factor oncoprotein. Molecular cancer therapeutics. 2014;13:1492–502. doi: 10.1158/1535-7163.MCT-13-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Scientific reports. 2016;6:20559. doi: 10.1038/srep20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Yang Z, Whitby R, Wang L. Regulation of miR-200c by nuclear receptors PPARalpha, LRH-1 and SHP. Biochemical and biophysical research communications. 2011;416:135–9. doi: 10.1016/j.bbrc.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong Y, Xie Y, Xiao G, Behrens C, Girard L, Wistuba II, et al. Nuclear receptor expression defines a set of prognostic biomarkers for lung cancer. PLoS medicine. 2010;7:e1000378. doi: 10.1371/journal.pmed.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.