Abstract
Macromolecular reagents can be targeted to tumors through active and passive mechanisms. ‘Active’ targeting involves moieties, such as receptor ligands, to direct tumor cell binding, while ‘passive’ targeting relies on long reagent circulating half-life, abnormal tumor vasculature, and poor lymphatic drainage for tumor entrapment. Here we sought to study the impact of reagent circulating half-life on ‘active’ and ‘passive’ tumor uptake. The humanized PSMA-targeting antibody, HuJ591, was utilized as the ‘active’ targeting agent. HuJ591 was labeled with a Near Infrared (NIR) dye and its circulating half-life was modified by conjugation to high-molecular-weight Polyethylene Glycol (PEG). PEGylation did not negatively impact PSMA binding specificity. ‘Active’ and ‘passive’ tumor targeting of intravenously injected antibody conjugates were then quantified by NIR fluorescent imaging of immunocompromised mice bearing bilateral isogenic PSMA-positive and PSMA-negative human tumor xenografts. Two isogenic tumor pairs were applied, PC3 +/- PSMA (PC3-PIP/PC3-Flu) or LMD-MDA-MB-231 +/- PSMA (LMD-PSMA/LMD). This study provided a unique model system to simultaneously observe ‘active’ and ‘passive’ tumor targeting within a single animal. ‘Passive’ targeting was observed in all PSMA-negative tumors, and was not enhanced by enhanced size or extended circulating half-life. Interestingly, ‘active’ targeting was only successful in some situations. Both PSMA-positive tumor models could be actively targeted with J591-IR800 and J591-PEG10K. However, the larger J591-PEG30K enhanced ‘active’ targeting in the PC-3 tumor models, but inhibited ‘active’ targeting the LMD-MDA-MB-231 tumor model. Successful ‘active’ targeting was associated with higher PSMA expression. These results support the potential for ‘active’ targeting to enhance overall macromolecular reagent uptake within tumors.
Keywords: Targeted Cancer Therapeutics, Enhanced Permeability and Retention (EPR), Passive Targeting, Active Targeting, Nanomaterials, Prostate Specific Membrane Antigen (PSMA)
Introduction
The concept of a tumor-targeted reagent may be viewed differently, or defined differently, among various specialties in the medical research community. In the macromolecule and nanoparticle community, tumor targeting seeks to maximize reagent delivery to the tumor and to reduce reagent exposure at non-specific sites (1, 2). Macromolecular reagents can accumulate within tumors following intravenous (i.v.) injection by both ‘active’ (receptor-targeted) and ‘passive’ (non-receptor targeted) mechanisms. ‘Passive targeting’ does not involve reagent interaction with defined cell-surface receptors or tumor markers, but rather occurs inertly through the common leakiness of tumor vasculature, the poor lymphatic drainage of tumors, and the long blood circulating half-life of larger macromolecular reagents such as polymers and nanoparticles. This phenomenon is commonly known as the Enhanced Permeability and Retention (EPR) effect (3-6). The EPR effect produces a high local concentration of therapeutics within the tumor that generally increases with reagent size and blood circulating half-life. Second generation actively targeted reagents differ by being engineered to bind cell surface markers, such as ligand-specific receptors. Targeting ligands, including antibodies or small molecules, can be conjugated directly to reagent surfaces to achieve this cell-specific ‘active’ targeting (7-10).
The impact of ‘active’ and ‘passive’ targeting are debatable, and they have been challenging to study (11, 12). The conjugation of ‘active’ targeting ligands to a reagent can result in unpredictable changes in size, shape and surface charge. Because of this, it is often unsuitable to compare the tumor uptake of an actively-targeted reagent to the original ‘naked’ reagent (13, 14). For example, ‘active’ targeting studies could be inaccurately interpreted as positive based on non-receptor-mediated tumor accumulation of more negatively charged reagents. Similarly, the artificial expression (or knock-down) of an actively-targeted receptor may change the tumor microenvironment and consequentially the ‘passive’ uptake of reagents through changes in cell-surface charge, tumor interstitial pressure, vascularity, extracellular matrix components, phagocyotosis or macropinocytosis rates (15-18). For example, targeted reagents may be falsely concluded as unsuccessful based on reduced tumor vascularity within receptor-positive tumors. In light of these challenges, there is a significant need for further analyses of ‘active’ and ‘passive’ tumor targeting in well controlled model systems.
The goal of this study was to develop a carefully controlled model system where ‘active’ and ‘passive’ tumor targeting could be evaluated simultaneously in a single animal model. With this system, we sought to analyze the effects of blood circulating half-life on the ‘active’ and ‘passive’ targeting of a macromolecular reagent. These results provide a novel and temporal view of ‘active’ and ‘passive’ tumor targeting with three different sized reagents, of varying blood circulating half-life, over a five day period.
Materials and Methods
Materials
Murine J591 and humanized HuJ591 were obtained from Neil H. Bander (Weil Cornell Medical College, New York, NY). PEG reagents were purchased from Laysan Bio Inc. (Arab, AL). IRDye® 800CW NHS Ester was purchased from (LI-COR, Lincoln, NE). All other reagents/chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless mentioned otherwise. Antibodies used for IHC include goat anti-human IgG (H+L) cross adsorbed antibody (Thermo Scientific, Rockford, IL), rabbit monoclonal anti-PSMA antibody (Cell signaling Technology Inc., Denver, MA), Alexa Fluor donkey 568 anti-goat IgG (Life Technologies Inc., Fredrick, MD) and PowerVision Poly-HRP anti-rabbit IgG (Leica Biosystem, Australia).
Cell Lines
Human PSMA-expressing PC-3 prostate cancer cells (PC3-PIP) and isogenic PSMA-negative control cells (PC3-Flu) were obtained from Dr. Warren Heston (Cleveland Clinic, Cleveland, OH) (19). Human PSMA-expressing LMD-MDA-MB-231 breast cancer cells (LMD-PSMA) and isogenic PSMA-negative control cells (LMD) were previously developed (20). Cells were cultured in RPMI 1640 (Corning Cellgro, Manassas, VA) plus 10% FBS (Sigma-Aldrich, St. Louis, MO), 50 μg/ml Gentamicin (Quality Biological, Gaithersburg, MD) and 5 μg/ml Ciprofloxacin (US Biological, Salem, MA). Cells were maintained at 37°C, 5% CO2 in a humidified incubator. Cells were authenticated by STR profiling (Johns Hopkins GRCF DNA services; November 2015) and confirmed > 90% homology with parental ATCC cell lines. Further mycoplasma monitoring occurs semiannually.
J591 NIR fluorescent conjugates
100 μL of 3 mM IRDye® 800CW NHS Ester (∼10 molar fold excess in DMSO) was added to 1 mL of J591 antibody (5 mg/mL in PBS) and kept 1 h at ambient temperature. Unreacted dye was removed by Zebra desalting column (Thermo Scientific, Rockford IL). J591-IR800 was PEGylated with 10-fold molar excess of Methoxy-poly(ethyleneglycol) succinimidyl valerate (mPEG-SVA). mPEG10000-SVA (28 mg/mL) and mPEG30000-SVA (84 mg/mL) stocks were made in anhydrous DMSO. 40 μL of each stock was added to 400 μL of J591-IR800 in a glass vial and stirred at ambient temperature for 3 h. Unreacted reagents were separated by 40k MWCO Zebra desalting column. Conjugate concentrations were determined by BCA protein Assay (Thermo Scientific).
J591 conjugate characterization
Dye conjugation efficiency and stoichiometry was confirmed by absorbance at 280/745 nm. Conjugate sizes were compared by 10% PAGE (BioRad Lab Inc., Hercules, CA; 200 ng of each J591-conjugates), visualized by LICOR Odyssey imaging system (LI-COR Biosciences, Lincoln, NE). Sizes were estimated by primary amines and 75% conjugation efficiency. Sizes of PEGylated conjugates were determined by Transmission Electron Microscopy (TEM). Bright-field images were taken on an FEI Tecnai 12 TWIN TEM equipped with 16 bit 2K x2K FEI Eagle bottom mount camera and SIS Megaview III wide-angle camera. TEM grids were prepared by drying 10 μL of sample on copper grids with holey carbon films overnight (grids from Ted Pella). Particle size was determined using ImageJ, where at least 23 particles were analyzed per sample.
Western Blot and PSMA quantification
Immunoblots were performed as previously reported (21). Cell-lysates were separated by SDS–PAGE, transferred to nitrocellulose membranes, blocked with Odyssey Blocking buffer (LI-COR Biosciences, Lincoln, NE) and incubated with muJ591/β-actin antibody for 1 h at room temperature. Bands were detected through anti-mouse-LICOR conjugated antibody (45 min). Bands were imaged and quantified by LI-COR odyssey imaging system.
PSMA specific cell binding assay
PSMA-positive (LMD-PSMA, PC3-PIP) and PSMA-negative (LMD, PC3-Flu) cells were plated in quadruplicate in 96 well format at equal density. The next day cells were fixed (4% para-formaldehyde), rinsed, and blocked with 3% BSA. Following PBS wash, J591-IR800 conjugates (in 50 μL PBS) were added for 1 h at 37° C. After washing (3 x 100 μL PBS-T followed by 1 x 100 μL of PBS) plates were quantified by LI-COR odyssey imaging system.
Competitive binding studies
PSMA positive (LMD-PSMA) and PSMA negative (LMD) cells were plated, fixed and blocked in a 96 well plate format. 100 ng of J591-conjugates were mixed with increasing concentrations of unlabeled J591 (1 μM to 1 nM) in 200 μL of PBS. PBS was utilized as a negative control. 50 μL was added per well, 37° C for 1 h. After washing, the plates were imaged and quantified by LI-COR odyssey imaging system. The percent binding was calculated according to the following formula
Blood circulating half-life
Animal studies were performed according to protocols approved by the Animal Care and Use Committee at Johns Hopkins University. 12-16 weeks old athymic Nu/Nu mice were injected with ∼ 10 μg (in 100 μL of PBS) of J591-800, J591-PEG10k or J591-PEG30k (n= 5 for each group) via tail vein. At various time points 15-20 μL tail-veins were nicked (25G needle) and blood was collected in heparinized capillary tubes (Polymer technology Systems Inc., Indianapolis, IN). Fluorescence intensity was measured by imaging capillaries with the LI-COR odyssey imaging system. The first time point (blood drawn < 1 h after the injection) was considered at 0 h (T0). The percent ID was calculated using the following formula:
Near Infrared animal imaging and signal quantification
A 2:1 suspension of 1.2 x 106 LMD or PC3-Flu tumor cells and Matrigel™ (BD Biosciences, Billerica, MA) was subcutaneously injected into the upper flanks of Athymic Nu/Nu mice, while an equivalent suspension of 1.5 x 106 LMD-PSMA or PC3-PIP was injected into the contralateral flank. When the tumors were ∼0.2 cc, 15 micrograms of J591-800, J591-PEG10k, J591-PEG30k was injected intravenously via tail vein (n=5 in each group). Animals were anaesthetized with isoflurane, and serial images of the dorsal, left, right and ventral surfaces were obtained using the LI-COR Pearl NIR Imager at 24, 48, 72 and 120 h post-injection. Images were acquired and processed using Pearl Cam Software (LI-COR Biosciences, Lincoln, NE) with 800 nm and visual channels. Regions of interest (ROIs) were drawn around tumor or background (muscle) for quantification. All images were normalized by matching the minimum and maximum signal and the tumor signal was calculated using the following formula:
The PSMA specific targeting was calculated using the following formula:
Ex vivo tumor analysis
120 h post injection the mice were euthanized and tumor, liver, spleen and kidneys were removed. Organs were placed on a non-fluorescent petri dish and imaged with LI-COR odyssey imaging system at 800 nm. For performing the quantitative analysis, ROIs were drawn on each corresponding tumors/organ and normalized by size. See Supplementary Figure S1 for details of ex vivo tumor lysis methods.
Immunohistochemistry
The resected tumor was fixed using formalin and embedded in paraffin. Adjacent sections were cut from the embedded tumor and slides were made for histological analysis. For detection of J591 conjugates, the slides were deparaffinized, treated with xylene, rehydrated and incubated with Goat Anti-human Ab followed by Alexa-Fluor 568 Anti-goat IgG. Similarly, for PSMA IHC, slides were first incubated with rabbit anti-PSMA Ab followed by Poly-HRP anti- Rabbit IgG. PSMA images were obtained in Nikon Eclipse E400 optical microscope and all other slides were imaged using Zeiss Axiocam inverted fluorescence laser scanning microscope equipped with 40X lens.
Statistical Analysis
Statistical analysis was performed using either Microsoft Excel (Microsoft, Seattle, WA) or Prism software (GraphPad Software, Inc., La Jolla, CA). The IC50 values of the conjugates were determined by non-linear curve fitting (one site- Fit IC50 model) using GraphPad Prism and group comparisons were evaluated by Two-Way ANOVA with Bonferroni posttests. Similar non-linear curve fitting (One phase decay model) was performed to determine the blood-half-life using GraphPad. Pearson r value was determined by analyzing Positive correlations using GraphPad (For Supplementary Figure S1). For statistical significance, unpaired, two-tailed t test was used, and P values of less than 0.05 were considered significant.
Results
Development and characterization of targeting agents and tumor models
Prostate Specific Membrane Antigen (PSMA) was selected as the cell surface marker for ‘active’ tumor targeting. PSMA is an integral membrane protein that is over-expressed on the surface of aggressive prostate cancer (PCa) cells (19, 22). Moreover, it is a well-characterized target for PCa imaging and therapeutic agents (23, 24). The humanized J591 (HuJ591) monoclonal antibody (mAb) is an established PSMA-specific targeting reagent with proven clinical targeting ability in human metastatic PCa (25). J591 has also had success as a targeting ligand for nanoparticles (21, 26). We therefore selected this antibody as the ‘active’ targeting reagent.
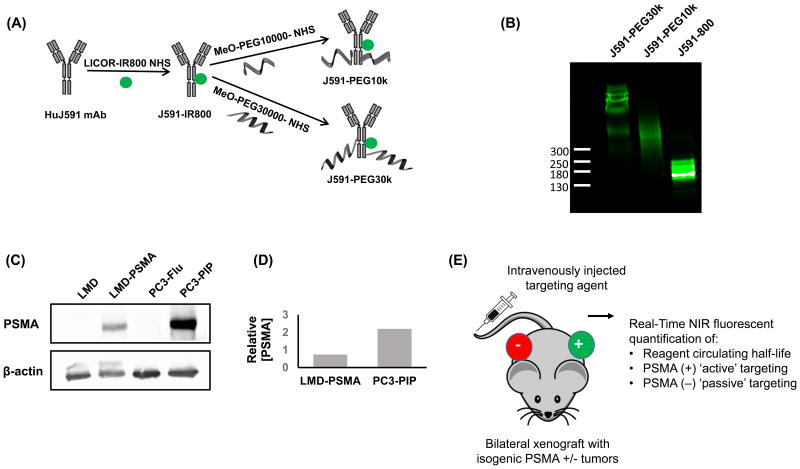
The HuJ591 mAb was first labelled with the NIR dye, IRDye® 800CW, through primary amine conjugation (Figure 1A). Dyes with this wavelength emission have minimal interference with blood and other tissues, making them detectable by in vivo NIR fluorescent imaging at depths of several centimeters (27, 28). The resulting J591-IR800 was then divided and further modified through conjugation with 10,000 or 30,000 molecular weight PEG, at high stoichiometric ratios, in order to increase antibody size, blood circulating half-life, and potential for ‘passive’ tumor uptake (3). The three resulting conjugates were termed J591-IR800, J591-PEG10k and J591-PEG30k. Because each antibody conjugate initiated from the same lot of J591-IR800, each product has equal antibody NIR-dye labeling density, but differing molecular weight.
Figure 1. An isogenic model system for evaluating ‘active’ and ‘passive’ targeting.
A, Left, schematic of HuJ591 IR800 labeling and PEGylation to generate three PSMA-targeted agents with increasing molecular weight. B, Analysis of the three resulting J591-conjugate sizes by denaturing polyacrylamide gel electrophoresis. Protein MW markers indicated on left. Image obtained by IR800 NIR gel imaging. C, Analysis of PSMA expression by Western Blot. β-actin was applied as a loading control. D, Quantification of relative, β-actin normalized, PSMA expression from Western Blot. E, Schematic of in vivo model. PSMA-positive and PSMA-negative isogenic cell lines pairs were applied to generate bilateral subcutaneous xenograft tumors. Mice with established tumors were then intravenously injected with each J591 targeting agent. NIR fluorescent imaging was then applied to quantify ‘active’ targeting in PSMA-positive tumors and ‘passive’ targeting in PSMA-negative tumors.
Denaturing polyacrylamide gel electrophoresis was applied to visualize the conjugation products. The results demonstrate three different antibody-conjugates of increasing molecular weight (Figure 1B). The relative molecular weights of each conjugate cannot be accurately determined by comparison to standard protein markers; therefore, sizes were estimated as 180, 255 and 405 kDa based on PEG molecular weight and the number of potential conjugation sites (Table 1). NIR-dye labeling density was determined for each conjugate by spectrophotometric absorbance at 280/745 nm. The results confirm a consistent dye-labeling density of approximately 1.3 Dye molecules per antibody (Table 1). Analysis by Transmission electron microscopy (TEM) measured an average diameter of 28.5 nm for J591-PEG10k, and 43.9 nm for J591-PEG30k conjugates (Table 1). Further characterization of the conjugates by Dynamic Light Scattering was unsuccessful because of NIR fluorescent dye interference.
Table 1. Characteristics of J591-conjugates.
| Conjugate | Estimated MW(kDa) | Average Size (nm) | Dye per Antibody (280/745 nm) | Relative immuno-reactivity | Log IC50 | half-life in blood-circulation (h) |
|---|---|---|---|---|---|---|
| J591-800 | ∼180 | N.D. | 1.29 | 100% | -7.8 | 8.8 |
| J591-PEG10k | ∼255 | 28.5 | 1.32 | ∼58% | -8.6 | 15.5 |
| J591-PEG30k | ∼405 | 43.9 | 1.33 | ∼35% | -8.8 | 25.2 |
Two previously established isogenic PSMA-positive and PSMA-negative human cancer cell line pairs were applied to study ‘active’ versus ‘passive’ targeting, respectively. The LMD and LMD-PSMA cell line pair was previously derived from a PSMA-negative breast cancer cell line, LMD-MDA-MB-231, following transduction with a PSMA-expressing lentivirus (20). Similarly, the PC3-PIP (PSMA-positive) and PC3-Flu (PSMA-negative) cell line pair were previously derived from retroviral transduction of the PSMA-negative PCa cell line, PC3, with PSMA-expressing or negative control vectors (19). Western blotting demonstrates the relative PSMA expression between these two cell line pairs (Figure 1C). Quantitative analysis indicates that PC3-PIP cells express ∼2 fold higher levels of PSMA when compared to LMD-PSMA cells (Figure 1D). The schematic in Figure 1E summarizes how these cell lines were applied to generate the ‘active’ and ‘passive’ targeting models. Bilateral isogenic subcutaneous xenograft tumor pairs, one PSMA-positive and one PSMA-negative, were established in immunocompromised mice. Once tumors were established, the three different J591-conjugates were intravenously injected and NIR-fluorescent imaging was applied to quantify the PSMA-specific ‘active’ targeting and PSMA-negative ‘passive’ targeting, respectively, over a five day period. This bilateral tumor imaging approach is commonly applied to study receptor-specific targeting in vivo (29-31). To our knowledge, this is the first application of such a system to evaluate ‘active’ and ‘passive’ tumor targeting simultaneously.
J591-conjugate PSMA binding specificity and affinity
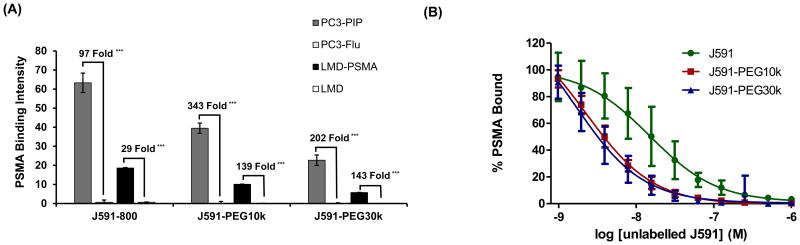
The PSMA-binding specificity of each antibody conjugate was evaluated by NIR fluorescent cell binding assay. Both isogenic cell models were plated to equal density, fixed the following day, and probed with an equal mass of each J591 conjugate (100 ng). NIR fluorescent image quantification confirmed the PSMA binding specificity of all three conjugates in both isogenic cell models (Figure 2A). The fold PSMA binding for each isogenic cell line pair and antibody-conjugate indicate a 2-5 fold increase in PSMA binding specificity following PEGylation (from 97 to 202 fold, and from 29 to 143 fold PSMA binding specificity). The overall binding level, or immunoreactivity of the conjugates, was inversely associated with conjugate size (Figure 2A, Table 1), which is expected given the lower fluorescent specific activity of the higher molecular weight conjugates.
Figure 2. Binding specificity and affinity of J591-PEG conjugates.
A, NIR immunofluorescent cell binding (1 h) by J591-IR800, J591-PEG10k and J591-PEG30k (100 ng, each) in LMD and PC3 isogenic cell models. PSMA Binding Intensity represents average NIR fluorescence intensity per well (N=4). The relative fold increase in PSMA-specific binding is indicated per cell model. *** indicates statistical significance (p < 0.001) as determined by paired two-tailed T-test. B, Competitive inhibition of J591-IR800, J591-PEG10k and J591-PEG30k LMD-PSMA cell binding (100 ng) by free unlabeled J591 at varying concentrations (4 log dilution series). Results are relative to non-blocked controls (N=4). Error bars represent standard deviation of the mean.
To interrogate the PSMA binding affinity of the conjugates, a competitive binding assay was performed on LMD-PSMA cells. Equal mass of each NIR-conjugate (100 ng) was mixed with increasing amounts of competing unlabeled J591. LMD-PSMA cell binding was quantified by NIR fluorescence and presented as percent PSMA bound, relative to unblocked control. Competitive inhibition produced the expected sigmoid curve and reflected the nanomolar binding affinity of each conjugate (Figure 2B). The Log IC50 results indicate a moderate but statistically significant (p<0.01, Two-way ANOVA) reduction in PSMA binding affinity following PEG conjugation (Table 1). However, there were no significant differences in binding affinity between J591-PEG10k and J591-PEG30k (p =0.91, Two-way ANOVA).
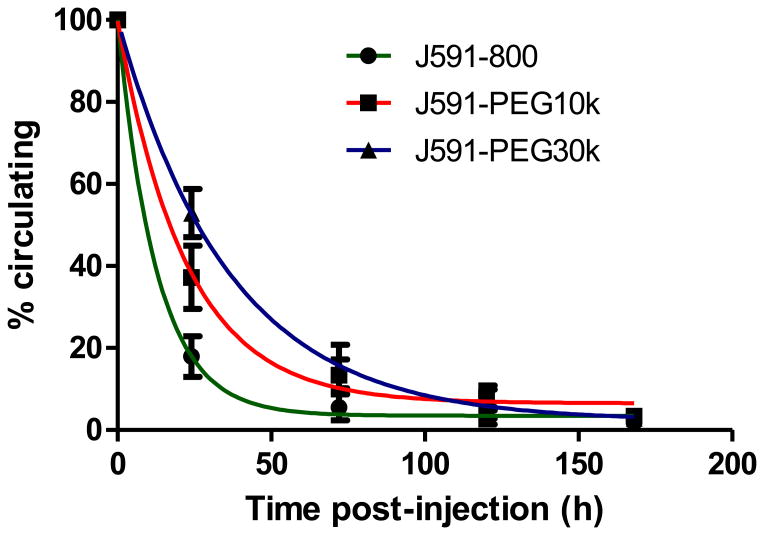
Extension of conjugate circulating half-life by PEGylation
The blood circulation half-lives of the three J591 conjugates were measured at predetermined time points following i.v. injection. Samples were isolated from tail vein nicks by glass capillary tube and conjugate levels were quantified by NIR fluorescent imaging. The results demonstrate that high molecular weight PEGylation significantly extended the blood-circulation time of each conjugate (Figure 3). The calculated half-lives of J591-IR800, J591-PEG10k and J591-PEG30k were 8.8, 15.5 and 25.2 hours, respectively (Table 1).
Figure 3. J591-conjugate circulating half-life.
15 μg of each of the three J591-conjugates were injected via tail vein (N=5 per group). Blood samples were collected by glass microcapillary at indicated times and quantified by 800 nm NIR fluorescent imaging. Results represent percent injected dose remaining in circulation, relative to the first time point. Error bars indicate standard deviation of the mean.
Characterization of real-time quantitative NIR tumor imaging
This model applies live NIR fluorescent animal imaging to follow ligand targeting over time. Live imaging offers a non-end point analysis that can be performed at multiple time points within a single animal, thus minimizing the number of experimental animals required and the potential for error. Previous studies have supported the use of NIR-Dye conjugates for real-time analysis of reagent biodistribution and tumor uptake (32). To investigate the integrity of real time imaging in our model, animals bearing established LMD and LMD-PSMA bilateral tumors were i.v. injected with J591-IR800. NIR fluorescent signal was obtained from live tumor imaging (signal/area), and results were compared to NIR-Dye fluorescent signal obtained from the same tumors following necropsy and lysis (% injected dose/g). A statistically significant correlation was observed between in vivo imaging and ex vivo NIR ligand quantification (r value >0.7 and P<0.001, Supplementary Figure S1), supporting the use of real-time animal imaging in this model. Ex vivo imaging was also applied to verify that comparable levels of J591-conjugate were injected (Supplementary Figure S2A), which could also be queried by in vivo imaging of other tissues, such as muscle (Supplementary Figure S2B).
Additional studies were performed to determine whether PSMA gene expression altered the tumor microenvironment. Tumor vascular permeability of PSMA-positive and PSMA-negative tumors was assessed by the Evans Blue Dye extravasation assay, following intravenous injection of the Evans Blue Dye (33). The results show that all four tumor models have comparable vascular permeability or leakiness (Supplementary Figure S3), indicating that ‘passive’ uptake should be similar between PSMA-positive and PSMA-negative tumor models. To further study the effects of PSMA expression on tumor vascularity and leakiness, we evaluated the uptake of a non-targeted PEG-IR800 fluorescent dye, which lacked any antibody components, in established LMD and LMD-PSMA tumors (Supplementary Figure S4). Similar levels of ‘passive’ PEG-IR800 uptake were observed in PSMA-positive and PSMA-negative tumors. These results support the integrity of the tumor models to study ‘active’ and ‘passive’ tumor targeting in real time.
‘Active’ and ‘Passive’ targeting in LMD-PSMA expressing isogenic tumors
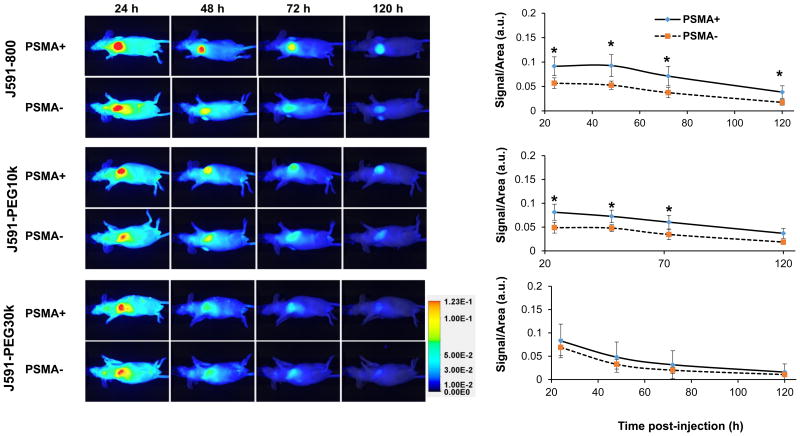
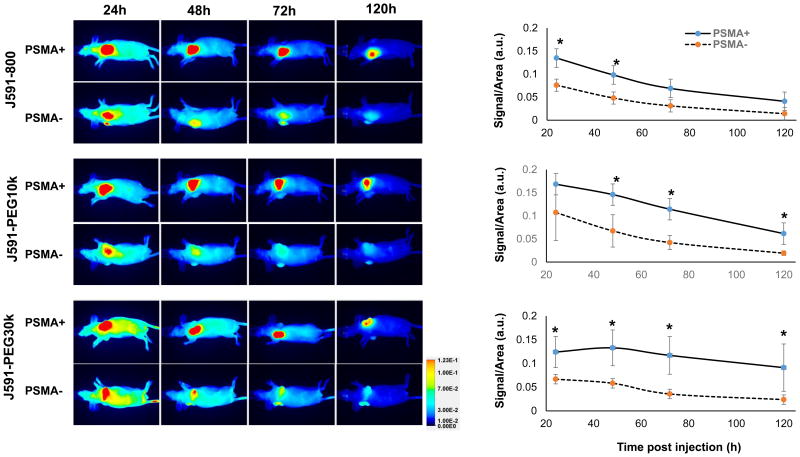
‘Active’ and ‘passive’ conjugate targeting were first compared in the isogenic LMD and LMD-PSMA xenograft model, which expresses relatively lower PSMA levels when compared to PC3-PIP cells (Figure 2D). Mice bearing established PSMA-positive LMD-PSMA tumors on the left flank, and PSMA-negative LMD tumors on the right flank, were i.v. injected with each of the three antibody conjugates (N=5 per group) and whole body prone and supine imaging was performed by LiCOR Pearl NIR fluorescence at 24, 48, 72 and 120 h post-injection (Figure 4). The tumors were similar in size and weight for each injected group (Supplementary Table S1). The NIR signal was quantified per tumor area and normalized to muscle for each animal and time point. Significant NIR fluorescent signal was detected in all tumors, regardless of PSMA expression status, indicating consistent ‘passive’ targeting by the antibody and antibody-PEG conjugates. The overall signal and kinetics of ‘passive’ targeting were similar among the three conjugates, indicating that extended circulation time did not impact ‘passive’ targeting in this model (Figure 4, PSMA-). The relative degree of ‘active targeting’ for each conjugate can be determined by comparing the level of NIR signal in PSMA positive versus PSMA negative tumors. The maximum relative level of ‘active’ targeting for this tumor model was observed with the smallest conjugate, J591-IR1800 (Figure 4). The PEGylated J591-PEG10k demonstrated similar and significant PSMA-targeting specificity at 24, 48, and 72 h post injection. Surprisingly no significant ‘active’ PSMA-targeting was observable with PEG30k at any time point, despite the known targeting activity of the conjugate in vitro (Figure 2). Tumors were further analyzed ex vivo by NIR fluorescent imaging, after the last time point, and the results were comparable to those observed in vivo (Supplementary Figures S5). Ex vivo analysis of conjugate biodistribution to non-tumor sites at necropsy indicated no significant difference in liver uptake between the reagents. However, the larger molecules (J591-PEG10k and J591-PEG30k) had notably lower kidney and spleen uptake, when compared to J591-IR800 (Supplementary Figure S6).
Figure 4. Near-infrared fluorescent imaging and quantification of J591-conjugate biodistribution in LMD ± PSMA xenografts.
Mice (5 animal/group) were i.v. injected with 15 μg each of J591-IR800, J591-PEG10k or J591-PEG30k and imaged after 24, 48, 72 and 120 h by LICOR PERAL optical imaging. Images of a representative animal from each group are provided. Graphs represent average signal/area (N=5) in arbitrary unit (a.u.), as described in materials and methods, over time for each tumor. Error bars represent standard deviation of the mean. Asterisk indicates statistical significance (t-test; p<0.05) between PSMA positive and negative groups at specified time points.
‘Active’ and ‘Passive’ targeting in PC-3 PSMA expressing isogenic tumors
The uptake of the same J591 NIR conjugates was then evaluated in the higher-PSMA expressing PC3-PIP/PC3-Flu isogenic tumor model. Mice bearing established PC3-PIP (PSMA+) tumors on left flank and contralateral PC3-Flu (PSMA-) tumors on the right flank were i.v. injected with each of the three antibody conjugates (N=5 per group) and whole body NIR fluorescent imaging was performed as described above. ‘Passive’ PSMA-negative tumor uptake and tumor clearance rates were similar to those observed in the LMD ± PSMA model (Figure 5). Thus, in both models, extended circulation half-life was not associated with increased ‘passive’ tumor uptake. Notably, in this high PSMA-expressing PC3 tumor model, all three J591-conjugates demonstrated significant ‘active’ targeting (Figure 5). Importantly, the same lot of J591-PEG30K that was previously observed to have no ‘active’ targeting to LMD-PSMA tumors (Figure 4), was the most effective ‘active’ targeting reagent in the PC3±PSMA model (Figure 5). Here, the separation between ‘active’ and ‘passive’ targeting increased with conjugate molecular weight, particularly at later time points (Figure 5). Tumors were analyzed ex-vivo, after the last imaging time point (120 h), and the results support the trends observed in vivo (Supplementary Figure S7).
Figure 5. Near-infrared fluorescent imaging and quantification of J591-conjugate biodistribution in PC3 ± PSMA xenografts.
Mice (5 animal/group) were i.v. injected with 15 μg each of J591-IR800, J591-PEG10k or J591-PEG30k and imaged after 24, 48, 72 and 120 h by LICOR PERAL optical imaging. Images of a representative animal from each group are provided. Graphs represent average signal/area (N=5) over time for each tumor. Error bares represent standard deviation of the mean. Asterisk indicates statistical significance (t-test, p<0.05) between PSMA positive and negative groups at specified time points.
Collectively, these results indicate that the extended circulating half-life primarily benefited ‘active’ targeting, rather than ‘passive’ targeting. Further, these results imply that the higher PSMA-expression of PC3-PIP tumors contributed to improved ‘active’ targeting, when compared to the same reagents in the lower PSMA-expressing LMD tumor model. However, additional differences between the PC3 and LMD tumor models, unrelated to PSMA expression, may have also contributed to these results.
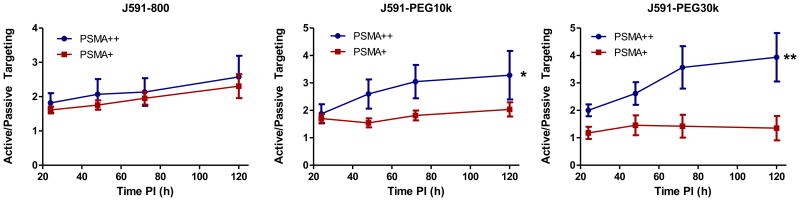
Kinetics of ‘active’ tumor targeting
To further evaluate the kinetics ‘active’ targeting for each conjugate, the ratio of active/passive targeting for low PSMA-expressing LMD-PSMA tumors (PSMA+) and high PSMA-expressing PC3-PIP tumors (PSMA++) was plotted over time (Figure 6). In the lower PSMA-expressing LMD-PSMA model, the relative levels of ‘active’ targeting decreased with higher molecular weight reagents, and the kinetics of binding became relatively flat over time. On the other hand, the extended circulating half-life was beneficial for the higher PSMA-expressing PC3 tumor model, where ‘active’ targeting progressively increased with reagent molecular weight and circulating half-life. Collectively, the data presents a general increase in ‘active’ targeting throughout the time course of the experiment, indicating that ‘active’ targeting may be better evaluated at later time points. It is also notable that the relative level of ‘active’ targeting begins to plateau for J591-PEG10k and J591-PEG30k in PC3-PIP tumors, suggesting that ‘active’ targeting may be approaching equilibrium at later time points.
Figure 6. Kinetics of ‘active’ targeting.
Relative active/passive targeting analysis of J591-conjugates over time as calculated from whole animal imaging. Active targeting signal represents fold PSMA specific targeting (average, N=5)). Error bars represent standard error and * Indicates p< 0.05, ** indicates p<0.01 as determined by unpaired two-tailed t-test.
Pathologic analysis of tumor targeting
The tumor models and tumor targeting were further studied at the microscopic level. To achieve this, systemic J591-IR800 and J591-PEG30k injections were repeated and tumors were harvested 24 hours after intravenous injection. PSMA immunohistochemistry (IHC) confirmed the specificity of PSMA expression in both isogenic models and verified the relatively higher PSMA expression level within PC3-PIP tumors, when compared to LMD-PSMA tumors (Supplementary Figure S8A). Importantly, PSMA-positive cells were not detected in PSMA-negative tumors, demonstrating that PSMA-positive cells did not migrate or become established in contralateral PSMA negative tumors. Further, PSMA expression was not detected in the murine tumors despite the known expression of PSMA in human neovasculature (19). These results indicate that J591 does not recognize murine endothelial cells or murine neovasculature, and that the uptake of J591-conjugates in PSMA-negative tumors was truly ‘passive’ and independent of PSMA expression.
HuJ591 is a humanized antibody; therefore the injected conjugates could be detected by IHC or immunofluorescence (IF). Anti-human IgG IF-microscopy was performed to visualize conjugate binding and intratumoral distribution. The relative level of J591-conjugate staining by IF microscopy was consistent with targeting levels observed in previous NIR-imaging studies (Supplementary Figure 8B). Specifically, the highest level of IF staining (red) was observed with J591-PEG30k in PC3-PIP tumors. Conjugate staining in PSMA-positive tumors was generally homogeneous and not suggestive of any negative effects of reagent molecular weight on tumor penetration or biodistribution. PSMA negative tumors demonstrated a generally weaker conjugate staining with a more heterogeneous distribution pattern that was not consistent with PSMA staining. Staining in both tumor models was too diffuse to associate J591-conjugate ‘passive’ targeting with any specific cell type or compartment.
Discussion
Macromolecular reagents, including nanoparticles, hold great promise for improved drug delivery and enhanced therapeutic indices. The first generation of cancer nanomedicines relied on ‘passive’ targeting, for example with the approved liposomal doxorubicin delivery agent Doxil® (34). Additional passively targeted drug-formulations were subsequently approved by the FDA, including Daunoxome® for HIV related Kaposi's sarcoma and Abraxane® for metastatic breast cancer (35). These formulations reduce the adverse side effects of the chemotherapeutic agent and moderately improve the overall survival for some cancers. Although, the benefits of these formulations may primarily be through altered pharmacokinetics (36). These successes have inspired the development of next-generation ‘actively’ targeted reagents. While significant progress has been made with ‘actively’ targeted nanomaterials, including a few agents that have entered clinical trials, many hurdles remain (11). The actual benefit of ‘active’ targeting has not yet been unambiguously proven in human solid tumors (37). ‘Active’ targeting is a stochastic process that involves multiple interactions as the whole reagent is in transit, before it is even exposed to the desired target (38). Thus, the transition from in vitro targeting affinity and specificity to in vivo delivery is not always straightforward.
The concept of ‘passive’ targeting is commonly applied to nanoparticles; however, this phenomenon was originally described with polymeric drugs and chemically modified proteins (3). Polymers with a molecular size larger than 40 kDa, or proteins larger than 48 kDa, have the potential for EPR-mediated tumor uptake through reduced renal clearance and extended circulating half-lives (5). The largest molecule described in the original was a murine immunoglobulin (150 kDa), which had similar tumor accumulation to other high molecular weight molecules such as bovine serum albumin (69 kDa) (3). Seymour and colleagues evaluated even larger macromolecules of N-(2-hydroxypropyl)methacrylamide (HPMA), ranging from 22 to 778 kDa (39). They confirmed the predicted 40 kDa cut off for renal clearance and also observed enhanced, but comparable, tumor uptake of larger molecules (39). These results indicate a potential saturation point for EPR-mediated tumor uptake with molecules of high molecular weight and extended plasma circulating half-life. On the other hand, enhanced ‘passive’ tumor uptake has been observed with larger and longer circulating block copolymer micelle (BCM) nanoparticles, which already began with long circulation half-lives and ‘passive’ targeting abilities (40). Therefore, the saturation point for EPR-mediated tumor uptake differ among particles or materials. Here, conjugate size and circulation half-life (Table 1) did not result in enhanced ‘passive targeting’ (Figures 4-5). These results indicate that high molecular weight PEGylation of whole, intact antibodies may not further enhance their passive uptake into the tumor microenvironment.
Historically, antibody PEGylation has been employed to reduce immunogenicity (41). This has become less necessary with the development of humanized antibodies; although it is still commonly applied to increase the plasma half-life of antibody fragments. Each PEG subunit is predicted to associate with 2-3 water molecules, resulting in enhanced size, reduced renal clearance, and reduced uptake by resident macrophage in the liver and spleen (42, 43). Indeed, PEGylation significantly reduced J591 accumulation within the kidney and spleen; however no differences were observed in the liver (Supplementary Figure S6). The effects of random PEGylation on antibody affinity are mixed, and can result in minor to significant effects on antibody binding capabilities (44, 45). Here we applied PEG molecules that are significantly larger than those typically utilized for altered immunogenicity. HuJ591PEGylation resulted in relatively minor effects on PSMA binding affinity, and no loss of binding specificity (Figure 2). Rather, the fold PSMA binding actually increased.
The current study provided a unique model to simultaneously observe ‘active’ and ‘passive’ tumor targeting within a single animal. Interestingly, ‘active’ targeting was only successful in some situations. Both PSMA-positive tumor models could be actively targeted with J591-IR800 and J591-PEG10K. However, higher molecular weight PEGylation enhanced ‘active’ targeting in the PC-3 tumor models, but inhibited ‘active’ targeting the LMD-MDA-MB-231 tumor model. One notable difference between these tumor models was PSMA expression level.
It has been previously reported that ‘active’ tumor targeting by BCM nanoparticles was only successful in tumors with high ligand expression (40). This is consistent with our observations of J591-PEG30K targeting to PC3-PIP, but not LMD-PSMA, tumors. In fact antibody targeting, under saturating conditions, is directly dependent upon the number of available ligand binding sites (46). However, the concept of ‘active’ targeting for nanoparticles and macromolecular reagents has not been as straightforward (11, 47). For example, two well-known studies found that ‘active’ targeting did not enhance the total amount of nanoparticle accumulation within the tumor, but rather only enhanced the uptake of particles into the tumor cells (48, 49). Thus, it remains unclear whether ‘active’ or ‘passive’ targeting forces are superior.
Here, ‘active’ targeting clearly resulted in overall higher levels of J591-IR800, J591-PEG10K and J591-PEG30K in PSMA-positive tumors. These differences cannot be attributed to contrasting vascular permeability between tumors, by evidence of Evans Blue Dye extravasation analysis (Supplementary Figure S3), or by potential PSMA-mediated effects on passive macromolecule uptake (Supplementary Figure S4). Further, the loss of ‘active’ targeting in LMD-PSMA tumors cannot be explained by loss of J591-PEG30K affinity, because these experiments were performed with the same lot of reagent and the LMD-PSMA studies were performed before PC3-PIP tumor studies. In summary, these results support the potential for ‘active’ macromolecular targeting in some tumor models. While these reagents may not be considered nanoparticles, their size is comparable to some particle formulations (Table 1).
While the present study is novel and informative, it is important to discuss its limitations. These data embody targeting through a single cancer cell antigen that, while clinically validated, may not be representative of other commonly targeted cancer antigens. These studies also applied a modified antibody that may not be representative of other targeting ligands or reagents, such as larger or more complex nanoparticles. Future studies involving alternate materials, targeting reagents or targeted receptors are needed to confirm or negate these observations.
Supplementary Material
Acknowledgments
We thank Torga Gonzalo for technical help in fluorescent microscopy, Munizia Uddin for help with IHC, and Catherine Foss and Ben Copeland for help in animal imaging.
Financial Support: National Cancer Institute CCNE (U54CA151838) for M.G. Pomper, P.C. Searson and pilot for S.E. Lupold, (CA134675) for M.G. Pomper, the Prostate Cancer Foundation and the Safeway Foundation for S.E. Lupold and A. Mukherjee.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose
References
- 1.Strebhardt K, Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nature reviews Cancer. 2008;8:473–80. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 2.Gerber DE. Targeted therapies: a new generation of cancer treatments. American family physician. 2008;77:311–9. [PubMed] [Google Scholar]
- 3.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer research. 1986;46:6387–92. [PubMed] [Google Scholar]
- 4.Fang J, Nakamura H, Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Advanced drug delivery reviews. 2011;63:136–51. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Maeda H. Toward a full understanding of the EPR effect in primary and metastatic tumors as well as issues related to its heterogeneity. Advanced drug delivery reviews. 2015;91:3–6. doi: 10.1016/j.addr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Wong AD, Ye M, Ulmschneider MB, Searson PC. Quantitative Analysis of the Enhanced Permeation and Retention (EPR) Effect. PloS one. 2015;10:e0123461. doi: 10.1371/journal.pone.0123461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nature reviews Cancer. 2002;2:750–63. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo LY, Theek B, Storm G, Kiessling F, Lammers T. Recent progress in nanomedicine: therapeutic, diagnostic and theranostic applications. Current opinion in biotechnology. 2013;24:1159–66. doi: 10.1016/j.copbio.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stylianopoulos T, Jain RK. Design considerations for nanotherapeutics in oncology. Nanomedicine : nanotechnology, biology, and medicine. 2015 doi: 10.1016/j.nano.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Ho W, Zhang X, Bertrand N, Farokhzad O. Cancer nanomedicine: from targeted delivery to combination therapy. Trends in molecular medicine. 2015;21:223–32. doi: 10.1016/j.molmed.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirollo KF, Chang EH. Does a targeting ligand influence nanoparticle tumor localization or uptake? Trends in biotechnology. 2008;26:552–8. doi: 10.1016/j.tibtech.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Kwon IK, Lee SC, Han B, Park K. Analysis on the current status of targeted drug delivery to tumors. Journal of controlled release : official journal of the Controlled Release Society. 2012;164:108–14. doi: 10.1016/j.jconrel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albanese A, Tang PS, Chan WC. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annual review of biomedical engineering. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 14.Ma N, Ma C, Li C, Wang T, Tang Y, Wang H, et al. Influence of nanoparticle shape, size, and surface functionalization on cellular uptake. Journal of nanoscience and nanotechnology. 2013;13:6485–98. doi: 10.1166/jnn.2013.7525. [DOI] [PubMed] [Google Scholar]
- 15.Pluen A, Boucher Y, Ramanujan S, McKee TD, Gohongi T, di Tomaso E, et al. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4628–33. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong RT, Boucher Y, Kozin SV, Winkler F, Hicklin DJ, Jain RK. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer research. 2004;64:3731–6. doi: 10.1158/0008-5472.CAN-04-0074. [DOI] [PubMed] [Google Scholar]
- 17.Goodman TT, Olive PL, Pun SH. Increased nanoparticle penetration in collagenase-treated multicellular spheroids. International journal of nanomedicine. 2007;2:265–74. [PMC free article] [PubMed] [Google Scholar]
- 18.Ernsting MJ, Murakami M, Roy A, Li SD. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:782–94. doi: 10.1016/j.jconrel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB. Five different anti-prostate-specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor-associated neovasculature. Cancer research. 1999;59:3192–8. [PubMed] [Google Scholar]
- 20.Neuman BP, Eifler JB, Castanares M, Chowdhury WH, Chen Y, Mease RC, et al. Real-time, near-infrared fluorescence imaging with an optimized dye/light source/camera combination for surgical guidance of prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:771–80. doi: 10.1158/1078-0432.CCR-14-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee A, Darlington T, Baldwin R, Holz C, Olson S, Kulkarni P, et al. Development and screening of a series of antibody-conjugated and silica-coated iron oxide nanoparticles for targeting the prostate-specific membrane antigen. ChemMedChem. 2014;9:1356–60. doi: 10.1002/cmdc.201300549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perner S, Hofer MD, Kim R, Shah RB, Li H, Moller P, et al. Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Human pathology. 2007;38:696–701. doi: 10.1016/j.humpath.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Mease RC, Foss CA, Pomper MG. PET imaging in prostate cancer: focus on prostate-specific membrane antigen. Current topics in medicinal chemistry. 2013;13:951–62. doi: 10.2174/1568026611313080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ristau BT, O'Keefe DS, Bacich DJ. The prostate-specific membrane antigen: lessons and current clinical implications from 20 years of research. Urologic oncology. 2014;32:272–9. doi: 10.1016/j.urolonc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandit-Taskar N, O'Donoghue JA, Durack JC, Lyashchenko SK, Cheal SM, Beylergil V, et al. A Phase I/II Study for Analytic Validation of 89Zr-J591 ImmunoPET as a Molecular Imaging Agent for Metastatic Prostate Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-15-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milowsky MI, Nanus DM, Kostakoglu L, Sheehan CE, Vallabhajosula S, Goldsmith SJ, et al. Vascular targeted therapy with anti-prostate-specific membrane antigen monoclonal antibody J591 in advanced solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:540–7. doi: 10.1200/JCO.2006.07.8097. [DOI] [PubMed] [Google Scholar]
- 27.Frangioni JV. In vivo near-infrared fluorescence imaging. Current opinion in chemical biology. 2003;7:626–34. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Vahrmeijer AL, Hutteman M, van der Vorst JR, van de Velde CJ, Frangioni JV. Image-guided cancer surgery using near-infrared fluorescence. Nature reviews Clinical oncology. 2013;10:507–18. doi: 10.1038/nrclinonc.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mease RC, Dusich CL, Foss CA, Ravert HT, Dannals RF, Seidel J, et al. N-[N-[(S)-1,3-Dicarboxypropyl]carbamoyl]-4-[18F]fluorobenzyl-L-cysteine, [18F]DCFBC: a new imaging probe for prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:3036–43. doi: 10.1158/1078-0432.CCR-07-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Penet MF, Nimmagadda S, Li C, Banerjee SR, Winnard PT, Jr, et al. PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS nano. 2012;6:7752–62. doi: 10.1021/nn301725w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs AV, Tse BW, Pearce AK, Yeh MC, Fletcher NL, Huang SS, et al. Evaluation of Polymeric Nanomedicines Targeted to PSMA: Effect of Ligand on Targeting Efficiency. Biomacromolecules. 2015;16:3235–47. doi: 10.1021/acs.biomac.5b00913. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira S, Cohen R, Walsum MS, van Dongen GA, Elias SG, van Diest PJ, et al. A novel method to quantify IRDye800CW fluorescent antibody probes ex vivo in tissue distribution studies. EJNMMI research. 2012;2:50. doi: 10.1186/2191-219X-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunjachan S, Pola R, Gremse F, Theek B, Ehling J, Moeckel D, et al. Passive versus active tumor targeting using RGD- and NGR-modified polymeric nanomedicines. Nano letters. 2014;14:972–81. doi: 10.1021/nl404391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uziely B, Jeffers S, Isacson R, Kutsch K, Wei-Tsao D, Yehoshua Z, et al. Liposomal doxorubicin: antitumor activity and unique toxicities during two complementary phase I studies. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1995;13:1777–85. doi: 10.1200/JCO.1995.13.7.1777. [DOI] [PubMed] [Google Scholar]
- 35.Dawidczyk CM, Kim C, Park JH, Russell LM, Lee KH, Pomper MG, et al. State-of-the-art in design rules for drug delivery platforms: lessons learned from FDA-approved nanomedicines. Journal of controlled release : official journal of the Controlled Release Society. 2014;187:133–44. doi: 10.1016/j.jconrel.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nature reviews Clinical oncology. 2010;7:653–64. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Meel R, Vehmeijer LJ, Kok RJ, Storm G, van Gaal EV. Ligand-targeted particulate nanomedicines undergoing clinical evaluation: current status. Advanced drug delivery reviews. 2013;65:1284–98. doi: 10.1016/j.addr.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Florence AT. “Targeting” nanoparticles: the constraints of physical laws and physical barriers. Journal of controlled release : official journal of the Controlled Release Society. 2012;164:115–24. doi: 10.1016/j.jconrel.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 39.Seymour LW, Miyamoto Y, Maeda H, Brereton M, Strohalm J, Ulbrich K, et al. Influence of molecular weight on passive tumour accumulation of a soluble macromolecular drug carrier. European journal of cancer. 1995;31A:766–70. doi: 10.1016/0959-8049(94)00514-6. [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Fonge H, Hoang B, Reilly RM, Allen C. The effects of particle size and molecular targeting on the intratumoral and subcellular distribution of polymeric nanoparticles. Molecular pharmaceutics. 2010;7:1195–208. doi: 10.1021/mp100038h. [DOI] [PubMed] [Google Scholar]
- 41.Chapman AP. PEGylated antibodies and antibody fragments for improved therapy: a review. Advanced drug delivery reviews. 2002;54:531–45. doi: 10.1016/s0169-409x(02)00026-1. [DOI] [PubMed] [Google Scholar]
- 42.Swierczewska M, Lee KC, Lee S. What is the future of PEGylated therapies? Expert opinion on emerging drugs. 2015;20:531–6. doi: 10.1517/14728214.2015.1113254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moghimi SM, Szebeni J. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Progress in lipid research. 2003;42:463–78. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 44.Kitamura K, Takahashi T, Yamaguchi T, Noguchi A, Noguchi A, Takashina K, et al. Chemical engineering of the monoclonal antibody A7 by polyethylene glycol for targeting cancer chemotherapy. Cancer research. 1991;51:4310–5. [PubMed] [Google Scholar]
- 45.Koumenis IL, Shahrokh Z, Leong S, Hsei V, Deforge L, Zapata G. Modulating pharmacokinetics of an anti-interleukin-8 F(ab')(2) by amine-specific PEGylation with preserved bioactivity. International journal of pharmaceutics. 2000;198:83–95. doi: 10.1016/s0378-5173(99)00458-5. [DOI] [PubMed] [Google Scholar]
- 46.Thurber GM, Weissleder R. Quantitating antibody uptake in vivo: conditional dependence on antigen expression levels. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2011;13:623–32. doi: 10.1007/s11307-010-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lammers T, Kiessling F, Hennink WE, Storm G. Drug targeting to tumors: principles, pitfalls and (pre-) clinical progress. Journal of controlled release : official journal of the Controlled Release Society. 2012;161:175–87. doi: 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 48.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15549–54. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer research. 2006;66:6732–40. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.