Abstract
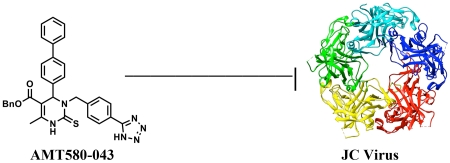
Human polyomaviruses are generally latent but can be reactivated in patients whose immune systems are suppressed. Unfortunately, current therapeutics for diseases associated with polyomaviruses are non-specific, have undefined mechanisms of action, or exacerbate the disease. We previously reported on a class of dihydropyrimidinones that specifically target a polyomavirus-encoded protein, T antigen, and/or inhibit a cellular chaperone, Hsp70, that is required for virus replication. To improve the antiviral activity of the existing class of compounds, we performed Biginelli and modified multi-component reactions to obtain new 3,4-dihydropyrimidin-2(1H)-ones and -thiones for biological evaluation. We also compared how substituents at the N-1 versus N-3 position in the pyrimidine affect activity. We discovered that AMT580-043, a N-3 alkylated dihydropyrimidin-2(1H)-thione, inhibits the replication of a disease-causing polyomavirus in cell culture more potently than an existing drug, cidofovir.
Graphical abstract
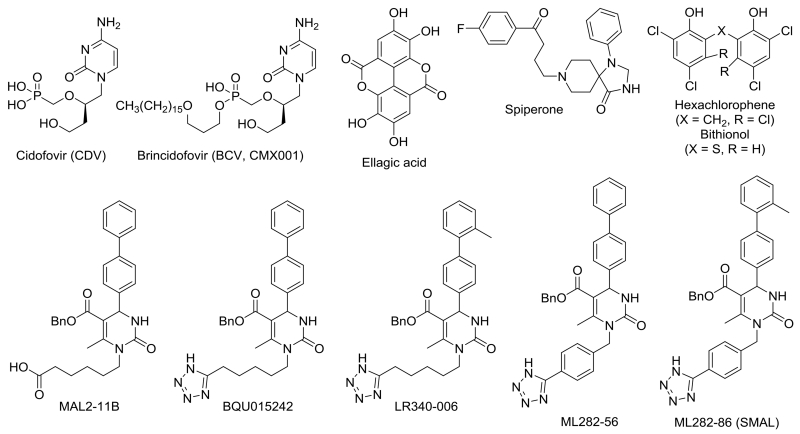
Polyomaviruses are double stranded DNA viruses that are endemic in the human population but are usually not disease-causing. However, in individuals who are immune-compromised, select members of this virus family can be reactivated and cause serious ailments.1,2 For example, BK virus reactivation in renal transplant patients undergoing immunosuppressive therapies leads to BK virus associated nephropathy (BKVAN), which is observed in 5-10% of all kidney transplant recipients. More than half of these individuals will ultimately lose the donated organ. BK virus reactivation is also evident in cancer patients who undergo bone marrow transplants and are given immunosuppressants, leading to hemorrhagic cystitis. Similarly, 5-10% of HIV-infected individuals ultimately succumb to progressive multifocal leukoencephalopathy (PML), which arises from JC virus reactivation. Due to recent improvements in virus identification and sequencing techniques, the number of known polyomaviruses has increased, such that there are now 12 human polyomavirus types. Several of these new polyomavirus family members have also been linked to disease.3 Unfortunately, existing therapeutics to treat polyomavirus infections are non-specific and/or exhibit unwanted side-effects.4 Cidofovir (CDV), an FDA approved treatment for cytomegalovirus (CMV) retinitis in AIDS patients, is an acyclic dCMP analog that inhibits DNA polymerase and is commonly used off-label to treat polyomavirus infections (Figure 1).4 Brincidofovir (BCV, CMX001) is a prodrug of CDV and shares its mechanism of action.5 A cell based high-throughput screen for simian virus 40 (SV40) and polyomavirus (BK and JC) inhibitors also detected activity with ellagic acid and spiperone.6
Figure 1.
Inhibitors of BK polyomavirus replication and propagation.
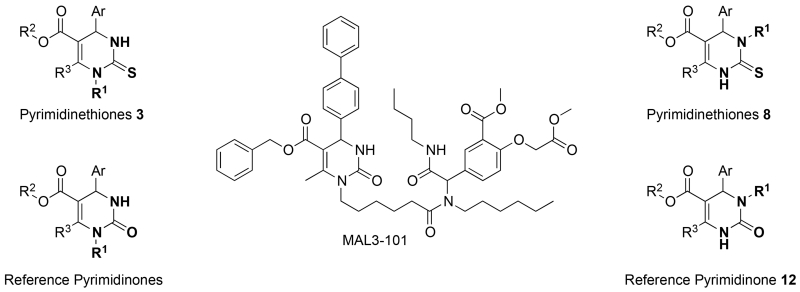
We previously reported on the synthesis, screening, and preliminary structure-activity relationship (SAR) studies of multi-component reaction-derived dihydropyrimidinones that inhibit the growth of polyomaviruses, in particular MAL2-11B, BQU015242, LR340-006, ML282-56, and ML282-86 (SMAL) (Figure 1); we also identified hexachlorophene and bithionol as polyomavirus inhibitors.7-11 The most specific dihydropyrimidinone compounds were identified based on their ability to inhibit the ATPase activity of T antigen, which is a polyomavirus-encoded factor. T antigen lacks human homologs and is required to catalyze replication of the polyomavirus genome.12 Some of our polyomavirus inhibitors also compromised the ability of T antigen, which contains a region with homology to Hsp40 chaperones, to activate the ATPase activity of the Hsp70 molecular chaperone. Hsp70 is required for T antigen-catalyzed polyomavirus replication.13,14 Our most potent compound, SMAL,7 inhibited T antigen ATPase activity with an IC50 of 5 μM. However, SMAL was as effective in cell-based viral replication assays as compounds that were less potent in the in vitro ATPase assay. Therefore, in order to identify improved inhibitors of polyomavirus replication, we now synthesized and characterized a class of SMAL variants that bear a thiourea group in place of the urea moiety in the dihydropyrimidinones (Figure 2). This modification allowed us to investigate the influence of electronic and physicochemical inhibitor properties on antiviral activity, such as: (1) the greater electron-density and polarizability of the thiocarbonyl compared to the carbonyl group; (2) the modification of H-bond donor (HBD) and H-bond acceptor (HBA) properties in the thiourea vs urea, and (3) the increased acidity of the thiourea NH group vs the amide NH.15,16 In addition, we switched key moieties at the N-1 and N-3 positions in the heterocycle to determine whether altered placement of active side chains affects T antigen and Hsp70 inhibition.
Figure 2.
Structures of heterocyclic cores probed in this work and compound MAL3-101, a reference pyrimidinone along with 12 (AMT628-003), MAL2-11B, BQU015242, LR340-006, ML282-56, and ML282-86 (SMAL). See Supplemental Materials for an overview table that lists, in alphabetical order, all compound codes and the corresponding structures for all assayed analogs, as well as their sequential compound numbers from the synthetic schemes and their corresponding UPCMLD codes.
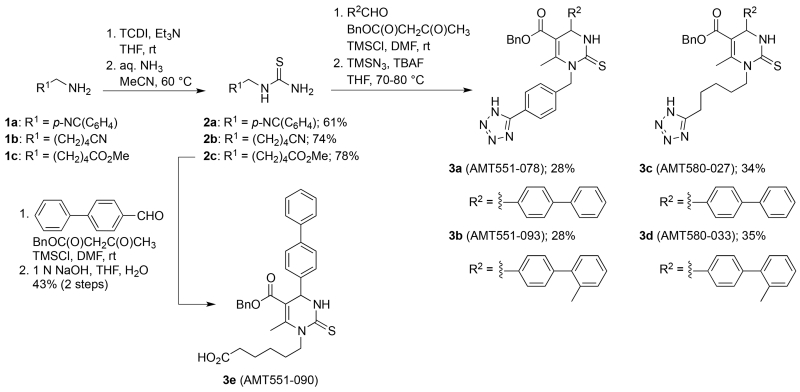
The synthesis of N-1 substituted 3,4-dihydropyrimidine-2(1H)-thiones was achieved through a series of Biginelli reactions (Scheme 1).17-19 Thioureas 2a-c were obtained in 61-78% yield by condensation of amines 1 with thiocarbonyldiimidazole (TCDI) in the presence of trimethylamine, followed by aminolysis with aqueous ammonia. According to a procedure described by Tolmachev and co-workers,20 2c was sonicated in the presence of 1,1′-biphenyl]-4-carbaldehyde, benzyl 3-oxobutanoate and TMSCl for 1 h and subsequently stirred at rt for several days, followed by saponification with 1 N NaOH to yield the desired Biginelli acid 3e (AMT551-090). In an analogous fashion, thioureas 2a and 2b were converted to the heterocycle-linked nitriles which were further modified with trimethylsilylazide and TBAF to give the corresponding tetrazoles 3a-d.
Scheme 1.
Preparation of N-1 substituted 3,4-dihydropyrimidine-2(1H)-thiones.
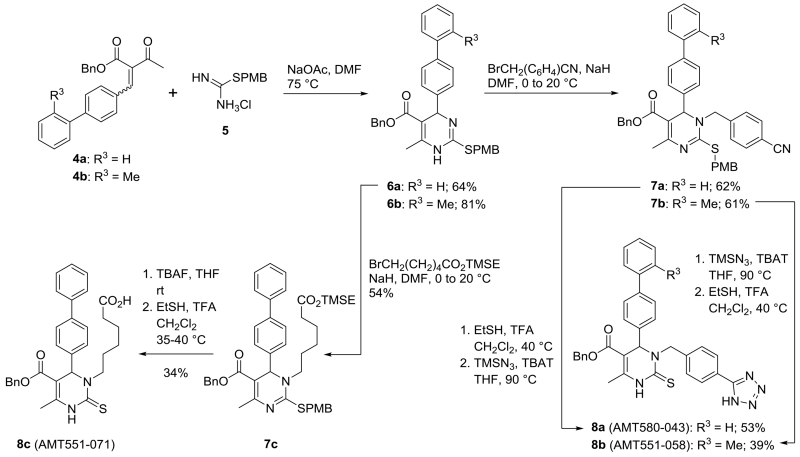
The synthesis of the regioisomeric N-3 alkylated analogs of dihydropyrimidinethiones 3 used the Atwal modification of the Biginelli reaction (Scheme 2). Enones 4 were condensed with 2-(4-methoxybenzyl)isothiouronium chloride (5) in the presence of sodium acetate in DMF to give the desired isothioureas 6 in 64-81% yield. Regioselective N-3 alkylation21 with bromonitrile and bromoester electrophiles gave the dihydropyrimidines 7a-c in 54-62% yield. Nitriles 7a and 7b were converted to tetrazoles 8a and 8b, respectively, whereby tetrazole formation with TMS-azide and tetrabutylammonium triphenyldifluorosilicate (TBAT) could proceed or follow the S-debenzylation with ethanethiol and trifluoroacetic acid (TFA). The 2-(trimethylsilyl)ethyl (TMSE) ester 7c was desilylated with tetrabutylammonium fluoride (TBAF) before cleavage of the isothiourea with ethanethiol and TFA to give acid 8c.
Scheme 2.
Preparation of N-3 substituted 3,4-dihydropyrimidine-2(1H)-thiones.
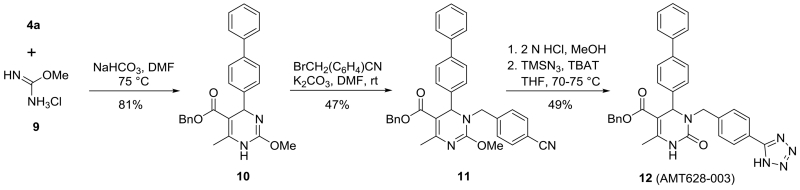
Finally, in order to compare the effect of N-3 vs N-1 substitution on the dihydropyrimidinone in the carbonyl series, we also prepared tetrazole 12 (Scheme 3). The enone 4a was reacted with methyl isourea hydrochloride 9 in the presence of NaHCO3 in DMF to give methoxy pyrimidine 10, which was selectively alkylated with 4-(bromomethyl)benzonitrile to give 11 in 38% overall yield from 9. After hydrolysis of the isourea moiety, tetrazole formation with TMS-azide provided dihydropyrimidinone 12, which differed from dihydropyrimidinethione 8a solely by the presence of a carbonyl function in place of the thiocarbonyl group at C-2 of the 6-membered heterocycle.
Scheme 3.
Preparation of an N-3 substituted 3,4-dihydropyrimidine-2(1H)-one.
Previously, we had found that SMAL and certain SMAL analogs inhibited both the ATPase activity of T antigen and the ability of T antigen’s homologous Hsp40 domain to stimulate Hsp70 ATPase activity;7 therefore, we assessed all new compounds and several control molecules for their ability to affect the ATPase activity of human Hsp70 in the presence or absence of a human Hsp40 homolog, Hdj2 (see Supplemental Materials). Consistent with previous data,10 SMAL and a SMAL analog, BQU015242, modestly inhibited the endogenous Hsp70 ATPase activity but had a more pronounced effect on Hsp40-activated Hsp70 activity (4.3- and 1.3-fold, respectively; Table 1). Most striking was AMT580-043, which inhibited the co-chaperone stimulated activity by 5.3-fold. Because AMT551-078 was somewhat less active (4.5-fold inhibition), we concluded that substitution at the N-3 position vs N-1 appears to augment the inhibitory activity. Furthermore, the AMT580-043 dihydropyrimidinethione was slightly more active than the AMT628-003 dihydropyrimidinone (5.3-fold versus 5.0-fold inhibition). In each of these compounds, replacing the flexible linker that ties the tetrazole to the dihydropyrimidine with the more rigid phenyl group in the side chain increased activity. Derivatives containing a carboxylic acid in place of the tetrazole (e.g. MAL2-11B or AMT551-090) either lacked inhibitory activity or modesty stimulated Hsp40-dependent activation of Hsp70. We also examined the ability of each compound to inhibit the ATPase activity of purified T antigen, as previously published.7 As shown in Table I, several compounds exhibited robust inhibition of activity. First, consistent with the reported EC50 for SMAL (~5 μM),7 this compound significantly reduced the ATPase activity of T antigen when applied at a final concentration of 10 μM. Second, an analysis of the remaining compounds extends the structure-activity relationship obtained in our previous work.7 We find that the thiourea moiety in combination with a tetrazole in the side chain is necessary but not sufficient for inhibition of T antigen activity. Although not without exception (e.g., AMT580-027), the compounds that contain both of these motifs are the most active inhibitors. One possibility is that the active compound complex with zinc or interfere with the zinc-finger domain in T antigen, which might disrupt the structure of this hexameric ATPase. This hypothesis will be examined in future studies.
Table 1.
Fold change differences in ATPase activity of Hsp70 in the presence or absence of an Hsp40, Hdj2, and the indicated compounds (at a final concentration of 100 μM). The activity of purified T antigen was examined in the presence of a final concentration of 10 μM compound.
| Cmpd | Hsp70 | Hsp70 + Hdj2 |
T antigen |
|---|---|---|---|
| SMAL | −1.2 | −4.3 | −4.3 |
| BQU015242 | −1.2 | −1.3 | −1.4 |
| AMT551-093 (3b) | NC | −3.7 | −7.7 |
| ML282-56 | NC | −5.0 | −2.0 |
| AMT551-078 (3a) | −1.4 | −4.5 | −5.5 |
| AMT551-058 (8b) | −1.3 | −4.8 | −7.5 |
| AMT628-003 (12) | −1.2 | −5.0 | −2.3 |
| AMT580-043 (8a) | −1.7 | −5.3 | −4.5 |
| LR340-006 | −1.4 | −3.0 | −2.1 |
| AMT580-033 (3d) | NC | −2.5 | −4.7 |
| AMT580-027 (3c) | NC | −1.7 | −2.1 |
| MAL2-11B | NC | +1.5 | −1.5 |
| AMT551-090 (3e) | NC | NC | −1.5 |
| AMT551-071 (8c) | NC | NC | −1.6 |
NC, no statistically significant change in activity. Data were normalized to activities in reactions lacking compound but containing an equivalent amount of DMSO. Reactions contained a 1:0.5 molar ratio of Hsp70:Hsp40. Data represent the means of three independent experiments.
The ultimate goal of our program is to identify more potent and efficacious compounds that inhibit the replication of polyomaviruses. Therefore, we next tested whether representative compounds inhibit the growth of human cells in culture. Many cancer cell lines rely on Hsp70 for proliferation,22 and to begin to determine whether members of this new compound series were cytotoxic, we measured the 50% growth inhibition (GI50) values of select compounds when incubated with a rhabdomyosarcoma cell line (RMS13) as well as with a breast cancer cell line, MCF7 (see Supplemental Material). MAL3-101, the parent lead structure for SMAL and all of the compounds reported herein, was used as a positive control as this dihydropyrimidinone inhibits the growth of cancer cells both in vitro and in vivo.8,23,24 As anticipated, MAL3-101 inhibited the growth of the two cancer lines with a GI50 of 6.0-13 μM (Table 2). In contrast, all of the newly prepared compounds were significantly less toxic, with GI50 values of 62-124 μM.
Table 2.
GI50 (μM) values for the indicated compounds against RMS13 and MCF7 cells
| Cmpd | RMS13 | MCF7 | Cmpd | RMS13 | MCF7 |
|---|---|---|---|---|---|
| MAL3-101 | 13 ± 9 | 6.0 ± 1 | LR340-006 | 100 ± 7 | 72 ± 1 |
| SMAL | 73 ± 4 | 140 ± 6 | AMT580-033 | 100 ± 8 | ND |
| AMT551-093 | 68 ± 1 | 160 ± 18 | AMT580-027 | 110 ± 0.0 | ND |
| ML282-56 | 96 ± 5 | 160 ± 9 | MAL2-11B | 120 ± 6 | ND |
| AMT551-078 | 87 ± 5 | 140 ± 7 | AMT551-090 | 100 ± 3 | ND |
| AMT551-058 | 62 ± 7 | ND | AMT551-071 | 110 ± 3 | ND |
| AMT580-043 | 72 ± 6 | 110 ± 4 |
Fifty-percent growth inhibition of the indicated cell lines in the presence of the compounds was calculated as described in the Supplemental Materials. Data represent the means of 2 independent experiments each run in triplicate, ± SD. ND, not determined
Because the novel dihydropyrimidines were less toxic than MAL3-101, and because some of the compounds were significantly more potent inhibitors than MAL3-101 in ATPase assays, we next measured the replication of BK virus and JC virus, which, respectively, lead to BKVAN and PML in immunocompromised patients (see above). Specifically, we examined the activity of AMT580-043 in polyomavirus infected cells because this was the most potent compound in the Hsp40-Hsp70 ATPase assay (Table 1). We then compared the activity of AMT580-043 with BQU015242,10 SMAL,7 and cidofovir. As shown in Table 3, each of these four compounds inhibited the replication of BKV in human foreskin fibroblasts with similar potency, and EC50 values ranged from 1.1 μM (cidofovir) to 3.9 μM (BQU015242). However, cidofovir was significantly less toxic than the dihydropyrimidines. In contrast, when the replication of JCV was assessed in green monkey kidney cells, AMT580-043 was by far the most potent compound, with an EC50 of 0.75 μM versus an EC50 of 2.0 μM for cidofovir. It is also noteworthy that AMT580-043 is one of the most potent inhibitors of T antigen ATPase activity examined in this study (Table I). Moreover, the specificity indices (CC50/EC50) for AMT580-043 and cidofovir were essentially identical (23 and 24, respectively). These results indicate that AMT580-043 should be further optimized to offset the catastrophic consequences of PML, a disease that has a 3-month mortality rate of 20-50%.24 While some of the physicochemical features of AMT580-043 (MW 572.7, logP 6.4) are still in need of improvement, other parameters (logD 4.8, HBA 4, HBD 2, number of rotatable bonds <10, passes Veber filter25) are attractive, making this compound a suitable lead structure for further medicinal chemistry optimization.
Table 3.
Antiviral Activity of select pyrimidines against BK Virus in human foreskin fibroblasts
| Cmpd | EC50 (μM)a | CC50 (μM)a |
|---|---|---|
| BQU015242 | 3.9 ± 4.0b | >67 ± 46b |
| SMAL | 1.7 ± 0.76 | 20 ± 17.8 |
| AMT580-043 | 1.8 ± 0.86 | 17 ± 1.7 |
| Cidofovir | 1.1 ± 1.2 | >100 ± 0 |
Values shown represent the average EC50 value and CC50 value from three independent experiments ±SD.
Values shown are from two independent experiments. EC50: effective concentration at which 50% activity is evident; CC50: cytotoxic concentration to cause death in 50% of viable cells.
Supplementary Material
Table 4.
Antiviral Activity of select pyrimidines against JC Virus in COS7 monkey fibroblast cells
| Cmpd | EC50 (μM)a | CC50 (μM)a |
|---|---|---|
| BQU015242 | 3.5 ± 2.6 | 24 ± 3.1 |
| SMAL | 4.2 ± 2.5 | 21 ± 1.0 |
| AMT580-043 | 0.75 ± 0.6 | 18 ± 0.17 |
| Cidofovir | 2.0 ± 1.6 | 49 ± 0.1 |
Values shown represent the average EC50 value and CC50 value from two independent experiments.
Acknowledgments
This work was supported by National Institutes of Health grant DK079307 (The Pittsburgh Center for Kidney Research) and a Howard Hughes Collaborative Innovation Award (JLB), an American Australian Association Merck Company Foundation Fellowship (AM-T), and National Institutes of Health grant GM067082 (PW). MNP was funded in whole or in part by the National Institutes of Allergy and Infectious Diseases, National Institutes of Health, under contract HHSN272201100016I. The authors also thank Annette N. Chiang, Paul Cantolupo, Jim Pipas, Peter G. Chambers, Michael Lyons, Bettina Quade, Lynn Resnick, Mary Liang, and Taber Lewis for reagents, discussions, analyses, and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- [1].Kean JM, Rao S, Wang M, Garcea RL. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pinto M, Dobson S. J. Infect. 2014;68:S2. doi: 10.1016/j.jinf.2013.09.009. [DOI] [PubMed] [Google Scholar]
- [3].DeCaprio JA, Garcea RL. Nat. Rev. Microbiol. 2013;11:264. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Andrei G, Topalis D, De Schutter T, Snoeck R. Antivir. Res. 2015;114:21. doi: 10.1016/j.antiviral.2014.10.012. [DOI] [PubMed] [Google Scholar]
- [5].Tylden GD, Hirsch HH, Rinaldo CH. Antimicrob. Agents Chemother. 2015;59:3306. doi: 10.1128/AAC.00238-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goodwin EC, Atwood WJ, DiMaio D. J. Virol. 2009;83:5630. doi: 10.1128/JVI.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ireland AW, Gobillot TA, Gupta T, Seguin SP, Liang M, Resnick L, Goldberg MT, Manos-Turvey A, Pipas JM, Wipf P, Brodsky JL. Bioorg. Med. Chem. 2014;22:6490. doi: 10.1016/j.bmc.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Adam C, Baeurle A, Brodsky JL, Wipf P, Schrama D, Becker JC, Houben R. PLoS ONE. 2014;9:e92041. doi: 10.1371/journal.pone.0092041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Seguin SP, Ireland AW, Gupta T, Wright CM, Miyata Y, Wipf P, Pipas JM, Gestwicki JE, Brodsky JL. Antivir. Res. 2012;96:70. doi: 10.1016/j.antiviral.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huryn DM, Brodsky JL, Brummond KM, Chambers PG, Eyer B, Ireland AW, Kawasumi M, LaPorte MG, Lloyd K, Manteau B, Nghiem P, Quade B, Seguin SP, Wipf P. Proc. Natl. Acad. Sci. USA. 2011;108:6757. doi: 10.1073/pnas.1015251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wright CM, Seguin SP, Fewell SW, Zhang H, Ishwad C, Vats A, Lingwood CA, Wipf P, Fanning E, Pipas JM, Brodsky JL. Virus Res. 2009;141:71. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Sem. Cancer Biol. 2009;19:218. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fewell SW, Pipas JM, Brodsky JL. Proc. Natl. Acad. Sci. USA. 2002;99:2002. doi: 10.1073/pnas.042670999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sullivan CS, Pipas JM. Microbiol. Mol. Biol. Rev. 2002;66:179. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nelli YR, Antunes S, Salaün A, Thinon E, Massip S, Kauffmann B, Douat C, Guichard G. Isosteric Substitutions of Urea to Thiourea and Selenourea in Aliphatic Oligourea Foldamers: Site-Specific Perturbation of the Helix Geometry, Chemistry – A European Journal. 2015;21:2870. doi: 10.1002/chem.201405792. [DOI] [PubMed] [Google Scholar]
- [16].Bordwell FG, Branca JC, Hughes DL, Olmstead WN. J. Org. Chem. 1980;45:3305. [Google Scholar]
- [17].Werner S, Turner DM, Lyon MA, Huryn DM, Wipf P. Synlett. 2006;2006:2334. [Google Scholar]
- [18].Arnold DM, LaPorte MG, Anderson SM, Wipf P. Tetrahedron. 2013;69:7719. doi: 10.1016/j.tet.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suresh, Sandhu JS. ARKIVOC. 2012;1:66. [Google Scholar]
- [20].Ryabukhin SV, Plaskon AS, Ostapchuk EN, Volochnyuk DM, Tolmachev AA. Synthesis. 2007;3:417. [Google Scholar]
- [21].Atwal KS, Rovnyak GC, O’Reilly BC, Schwartz J. J. Org. Chem. 1989;54:5898. [Google Scholar]
- [22].Murphy ME. Carcinogenesis. 2013;34:1181. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Braunstein MJ, Scott SS, Scott CM, Behrman S, Walter P, Wipf P, Coplan JD, Chrico W, Joseph D, Brodsky JL, Batuman O. J. Oncol. 2011;2011:232037. doi: 10.1155/2011/232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brew BJ, Davies NWS, Cinque P, Clifford DB, Nath A. Nat. Rev. Neurol. 2010;6:667. doi: 10.1038/nrneurol.2010.164. [DOI] [PubMed] [Google Scholar]
- [25].Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. J. Med. Chem. 2002;45:2615. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.