Abstract
The programmed cell death protein 1 (PD-1) limits effector T-cell functions in peripheral tissues and its inhibition leads to clinical benefit in different cancers. To better understand how PD-1 blockade therapy modulates the tumor-host interactions, we evaluated three syngeneic murine tumor models, the BRAFV600E-driven YUMM1.1 and YUMM2.1 melanomas, and the carcinogen-induced murine colon adenocarcinoma MC38. The YUMM cell lines were established from mice with melanocyte-specific BRAFV600E mutation and PTEN loss (BRAFV600E/PTEN-/-). Anti–PD-1 or anti–PD-L1 therapy engendered strong antitumor activity against MC38 and YUMM2.1, but not YUMM1.1. PD-L1 expression did not differ between the three models at baseline or upon interferon stimulation. Whereas mutational load was high in MC38, it was lower in both YUMM models. In YUMM2.1, the antitumor activity of PD-1 blockade had a critical requirement for both CD4 and CD8 T cells, as well as CD28 and CD80/86 costimulation, with an increase in CD11c+CD11b+MHC-IIhigh dendritic cells and tumor associated macrophages in the tumors after PD-1 blockade. Compared to YUMM1.1, YUMM2.1 exhibited a more inflammatory profile by RNA sequencing analysis, with an increase in expression from chemokine-trafficking genes that are related to immune cell recruitment and T-cell priming. In conclusion, response to PD-1 blockade therapy in tumor models requires CD4 and CD8 T cells and costimulation that is mediated by dendritic cells and macrophages.
Keywords: melanoma, programmed cell death-1 blockade, BRAF mutant, mutational load, costimulation
Introduction
The development of inhibitors of the programmed cell death protein 1 (PD-1) or its ligand (PD-L1) represents a paradigm shift in the treatment of advanced cancers, with significant clinical benefits demonstrated in patients with several different histologies (1-4). Tumor responses are associated with a higher number of pretreatment PD-L1–expressing tumor and myeloid cells (5, 6), a high mutational load leading to increase in antigen-specific T-cell recognition (7, 8), the ability of PD-1/PD-L1 blockade to increase antigen presentation (9, 10) and modulate the tumor microenvironment (10, 11), and pre-existing CD8 T-cell infiltration (5, 12). A higher tumor mutational load induced by carcinogens such as ultraviolet light for melanoma (13) or cigarette smoking for lung carcinomas (14), would allow T cells to better differentiate between cancer and normal cells, thereby leading to immune recognition that could be unleashed by PD-1 blockade therapy.
Despite these advances, a better understanding is needed of the tumor-host interactions and how anti–PD-1 agents modulate cellular and molecular characteristics of each individual microenvironment. It is widely accepted that PD-1 blockade agents regulate T-cell activity in peripheral tissues in the context of infection or in tumors where PD-1/L1 checkpoint is the dominant inhibitory pathway. However, anti–PD-1 interacts earlier with T cells positively regulated by B7-CD28 costimulation (15) and this interaction is less well characterized (16-18).
In this study, we analyzed different tumor-host characteristics that might influence the effects of PD-1 blockade in murine models with a fully functional immune system. We conclude that T-cell priming and costimulation are required for anti–PD-1 therapy response to be effective in the melanoma tumor models in vivo.
Materials and Methods
Mice, cell lines and reagents
C57BL/6 mice, B6.Cg-Braftm1MmcmPtentm1HwuTg(Tyr-cre/ERT2)13Bos/BosJ, B6.129S2-Cd28tm1Mak/J and B6.129S4-Cd80tm1Shr Cd86tm2Shr/J mice (Jackson Laboratories, Bar Harbor, ME) were bred and kept under defined-flora pathogen-free conditions at the AALAC-approved animal facility of the Division of Experimental Radiation Oncology, UCLA, and used under the UCLA Animal Research Committee protocol #2004-159-23. Cell lines were cultured in DMEM media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA) and 2nM L-glutamine (Invitrogen). YUMM1.1 and YUMM1.7 cell lines were obtained from induced tumors in conditional mouse models of melanoma based on melanocyte specific BRAFV600Eactivating mutation and PTEN loss (BRAFV600E/PTEN-/-). YUMM2.1 was obtained from BRAFV600E/PTEN-/- mice crossed with mice bearing a Ctnnb1loxex3 allele (19), which targets exon 3, resulting in removal of the GSK3b kinase sites in β-catenin that are needed for ubiquitin-mediated destruction. However, analysis of the YUMM2.1 cell line showed that it had not recombined the β-catenin site (see below). YUMM cell lines were tested and authenticated by PCR and exome sequencing. Recombinant murine interferon gamma (IFNγ) was obtained from Peprotech (Rocky Hill, NJ). Tumors were followed by caliper measurement three times per week and tumor volume was calculated using the following formula: tumor volume= ((width)2 × length)/2. Mean and standard deviation of the tumor volumes per group was calculated.
Antitumor studies in mouse models
To establish subcutaneous (s.c.) tumors, 3×105 MC38, 1×106 YUMM2.1 or 1×106 YUMM1.1 cells per mouse were injected into the flanks of C57BL/6 mice. When tumor diameter reached 4 to 5 mm, four doses of 300 μg of anti–PD-1 (Cat.No.BE0146, clone RMP1-14), anti–PD-L1 (Cat.No.BE0101, clone 10F.9G2) or isotype control antibody (Cat.No.BE0090, clone LTF-2), all from BioXCell (West Lebanon, NH), were injected intraperitoneally (i.p.) every 3 days. For T-cell subset depletion studies, 250 μg of anti-CD8 (Cat.No.BE0117, clone YTS 169.4), 250 μg of anti-CD4 (Cat.No.BE0003-2, clone OKT-4), both from BioXCell, or the combination were administered every 2 days starting the day before anti–PD-1 was initiated and through the duration of the experiment. For CD103 depletion, 200 μg of CD103 (Cat.No.BE0026, clone M290) from BioXCell was administered starting the day before anti–PD-1 treatment was initiated and administered i.p. every 2 days until the end of the experiment.
Whole exome sequencing: mutation calling and copy number analysis
Sequencing of the MC38, YUMM2.1, YUMM1.7, and YUMM1.1 cell lines was performed to a mean depth of 55X, with >90% of targeted bases covered by more than 15 reads in all samples. Exonic mutations were annotated by the Ensembl Variant Effect Predictor (EVEP). MC38 was compared to tail DNA from a C57BL6 parental mouse, whereas the YUMM2.1 and YUMM1.1 were compared to tail DNA from a B6.Cg-Braftm1MmcmPtentm1HwuTg(Tyr-cre/ERT2)13Bos/BosJ mouse. Exon capture and library preparation were performed at the UCLA Clinical Microarray Core using the NimbleGenSeqCap EZ Mouse Exome Design kit (Roche NimbleGen, Madison, WI) targeting 54.3 megabases of genome. 2×100bp paired-end sequencing was carried out on the HiSeq 2000 platform (Illumina, San Diego, CA) and sequences were aligned to the UCSC mm10 reference (Burrows-Wheeler Aligner BWA-mem algorithm v0.7.9). Preprocessing followed the Genome Analysis Toolkit (GATK) Best Practices Workflow v3 (20), including duplicate removal (Picard), indel realignment and base quality score recalibration. Somatic mutations were called with methods modified from (21) using Varscan2 (22), and the GATK-HaplotypeCaller. Mutations were annotated by EVEP release 80 (23) and filtered to remove those with a known database single nucleotide polymorphism (dbSNP) reference SNP cluster identification to exclude residual strain-related differences due to imperfect backcross-dilution. Depth ratio for copy number variation was produced by Sequenza (24), with the ratio.priority option engaged.
RNA sequencing and enrichment analysis
RNA sequencing was performed using the Illumina HiSeq 2500 platform on 100-bp paired-end libraries prepared using the IlluminaTruSeq RNA sample preparation kit. Reads were mapped using TopHat2 v2.0.9 (25) and aligned to the Musmusculus genome NCBI build 37.2. Reads were quantified and normalized using Cufflinks v2.2.1 (26) and CuffNorm to generate normalized expression tables by library size using the geometric normalization method. Resulting fragments per kilobase of exon per million fragments mapped expression values were log2 transformed with an offset of 1. To identify pathways enriched in the YUMM2.1 cell line, Gene Set Enrichment Analysis (GSEA) was performed using the pre-ranked option. Genes were ranked by log2 fold changes between YUMM2.1 and YUMM1.1 cell lines. Enrichment was assessed across the curated Molecular Signatures Database C5 GO biological process gene sets (27). RNA sequencing data has been deposited in GEO repository under the accession number GSE84264.
Flow cytometry analysis
MC38, YUMM2.1, and YUMM1.1 tumors and spleens were harvested from mice at pre-defined time points. Tumors were digested with collagenase D (Roche), and stained with antibodies to CD3 BV605, Ly6C FITC, PD-L1/CD274 PE, CD8a BV421, CD45RA/B220, CD11b BV785, CD11c PECy7, CD103 PerCP Cyanine 5.5, MHC Class II (I-A/I-E) FITC (Biolegend, San Diego, CA), Ly6G (Gr-1) PerCP Cyanine 5.5, F4/80 Pacific blue/eFluor450, CD25 APC, CD4 FITC (eBioscience, San Diego, CA). Intracellular staining of Foxp3 PE (eBioscience) was done according to manufacturer's recommendations. Cells were analyzed with a LSR-II or FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), followed by Flow-Jo software (Tree-Star, Ashland, OR) analysis (28).
Western blotting and immunofluorescence staining
Western blotting was performed using standard methods on lysates from cultured murine melanoma cell lines using primary antibodies to β-catenin, GAPDH and histone H3, and secondary anti-rabbit IgG HRP-linked antibody, all from Cell Signaling (Danvers, MA), and Pdcd-1L1 (H-130) and gp100 (H-300) from Santa Cruz Biotechnology (Dallas, TX). Nuclear and cytoplasmic extraction reagents were obtained from Thermo Scientific (Waltham, MA). Proteins were visualized using ImageQuant 4000 scanner. Immunofluorescence staining was performed on tumor sections of frozen OCT blocks (Sakura Finetek, Torrance, CA) using primary antibodies to β-catenin (Cell Signaling) and CD8a (BD Biosciences) followed by normal donkey serum and rat IgG(H+L) FITC-conjugated secondary antibody (Jackson Immunoresearch Laboratories, West Groove, PA) (29).
Topflash analysis
Topflash vectors were obtained from Addgene (M51 Super 8x FOPFlash/TOPFlash mutant, Cat.No.12457; M50 Super 8x TOPFlash, Cat.No.12456). YUMM1.7 and YUMM2.1 cells (± 10 μM tamoxifen) were plated to achieve 70% confluency in 6-well plates. Cells were co-transfected with pTK-RLuc (green Renilla luciferase) along with either Topflash or Fopflash vectors. After 48 hours, cells were harvested and luciferase activity was measured using Dual-Luciferase Reporter Assay System (Cat.No.E1910) from Promega (Madison, WI), where firefly luciferase signal was normalized to its corresponding Renilla luciferase signal. Topflash/fopflash signal was determined from each treatment and graphed using Graphpad/Prism.
β-catenin down-regulation
β-catenin small hairpin RNA (shRNA) lentiviral vector (Cat.No.29210-V) and the negative control shRNA lentiviral vector (Cat.No.108080) were purchased from Santa Cruz Biotechnology. YUMM2.1 and YUMM1.1 cells were transduced at a multiplicity of infection (MOI) of 1-10 in media containing 5 μg/ml polybrene and then selected in complete DMEM with 2.5 μg/ml of puromycin for 3 weeks.
Statistical analysis
Data were analyzed with GraphPad Prism (version 5) software (GraphPad Software, La Jolla, CA). Descriptive statistics such as number of observations, mean values, and SD were reported and presented graphically for quantitative measurements. Normality assumption was checked for outcomes before statistical testing. For measurements such as tumor volume or percentage of tumor infiltrating lymphocytes (TILs), pairwise comparisons between treatment groups were performed by unpaired t tests. All hypothesis testing was two-sided, and a significance threshold of 0.05 for P value was used.
Results
In vivo syngeneic animal models with differential responses to PD-1 pathway blockade
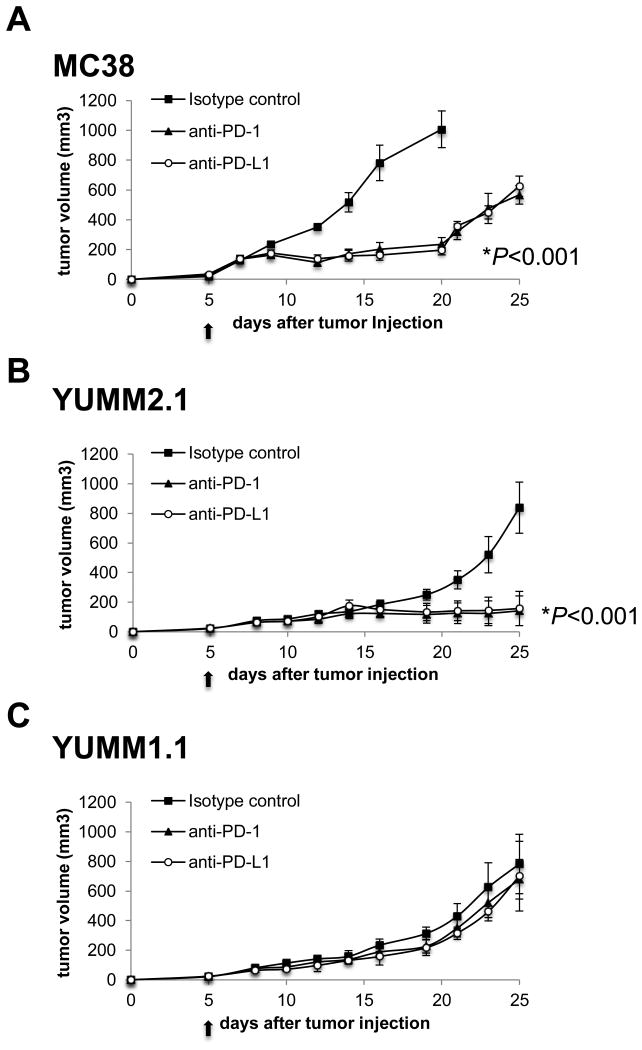
In order to have animal models that consistently respond to anti–PD-1 therapy, we tested four melanoma models, three derived from BRAFV600E/PTEN-/- genetically engineered mice (Fig. S1A) and B16, and compared them to MC38, a cell line that has been previously shown to respond well to PD-1 blockade therapy (30, 31). In three replicate studies we observed antitumor activity of anti–PD-1 or anti–PD-L1 antibody therapy against MC38 (Fig. 1A) and YUMM2.1 (Fig. 1B), but no antitumor activity against YUMM1.1 (Fig. 1C), YUMM1.7 or B16 (Fig. S1B). Of note, these responses to anti-PD1 antibody are incomplete and both MC38 and YUMM2.1 tumors start regrowing around day 35-40 after tumor injection. We decided to focus our further mechanistic studies in MC38 for a tumor that is known to respond to anti–PD-1, and studied the differential responses in YUMM1.1 and YUMM2.1.
Fig. 1. Enhanced in vivo antitumor activity with anti–PD-1 or anti–PD-L1 in MC38 and YUMM2.1 tumor models compared to YUMM1.1.
Tumor growth curves of MC38 (A), YUMM2.1 (B), and YUMM1.1 (C) with 4 mice in each group (mean ± SD) after anti–PD-1, anti–PD-L1 or isotype control. The arrow indicates the day when treatment with anti–PD-1, anti–PD-L1 or isotype control was started. *P < 0.001 by unpaired t test on day 20, anti–PD-1 versus isotype control, anti–PD-L1 versus isotype control in MC38, anti–PD-1 versus isotype control, anti–PD-L1 versus isotype control in YUMM2.1 tumors.
Similar PD-L1 expression induced in MC38, YUMM2.1, and YUMM1.1 by IFNγ
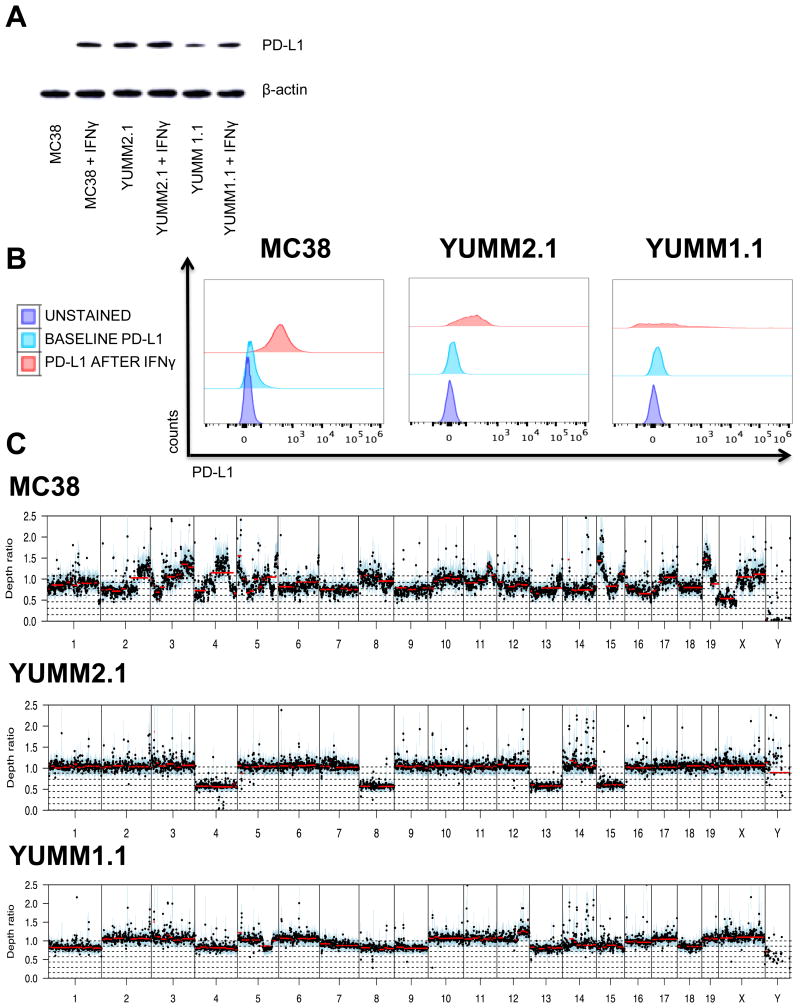
In order to investigate the mechanism of response to anti–PD-1 therapy, we first focused on induced PD-L1 expression in these three cell lines. Total cellular PD-L1 increased upon exposure to IFNγ in the three cell lines, with a higher magnitude of increase in MC38 cells than in YUMM2.1 and YUMM1.1 cells (Fig. 2A). Surface expression of PD-L1 was low at baseline, and increased upon exposure to IFNγ in the three cell lines, though less evident in the morphologically more heterogeneous YUMM1.1 cell line (Fig. 2B).
Fig. 2. Interferon gamma (IFNγ) modulates PD-L1 expression in MC38, YUMM2.1 and YUMM1.1.
(A) Western blot analysis of PD-L1. MC38, YUMM2.1 and YUMM1.1 cells were cultured with or without IFNγ for 24 hours. (B) Expression of PD-L1 by flow cytometry on MC38, YUMM2.1 and YUMM1.1 cells at baseline and after 24 hours of stimulation with IFNγ. (C) Chromosomal copy number variation in MC38, YUMM2.1 and YUMM1.1 cell lines. Y-axis represents Log2 depth ratio vs matched normal.
Increased mutational load in MC38 compared to YUMM1.1 and YUMM2.1
Next we determined whether mutational load is a contributor to the observed differential response to anti–PD-1 therapy. MC38, which was established from a mouse exposed to the carcinogen dimethylhydralazine (32), has a higher mutational load (2,778 mutations), compared to the much lower mutational rates in YUMM1.1 and YUMM2.1 (128 and 68 non-synonymous variants, respectively) (Fig. S1C). Despite independent derivation, 26 variants are shared by YUMM1.1 and YUMM2.1, which likely represent SNPs not found in the sequenced strain-matched control or in the National Center for Biotechnology Information database of genetic variation. Copy number variation analysis revealed substantial differences in chromosomal alteration patterns between the three cell lines (Fig. 2C). However, most are shallow amplifications or deletions (log2 ratio between 0.5 and 1.5).
CD8 and CD4 T cells important in response to PD-1 blockade in MC38 and YUMM2.1
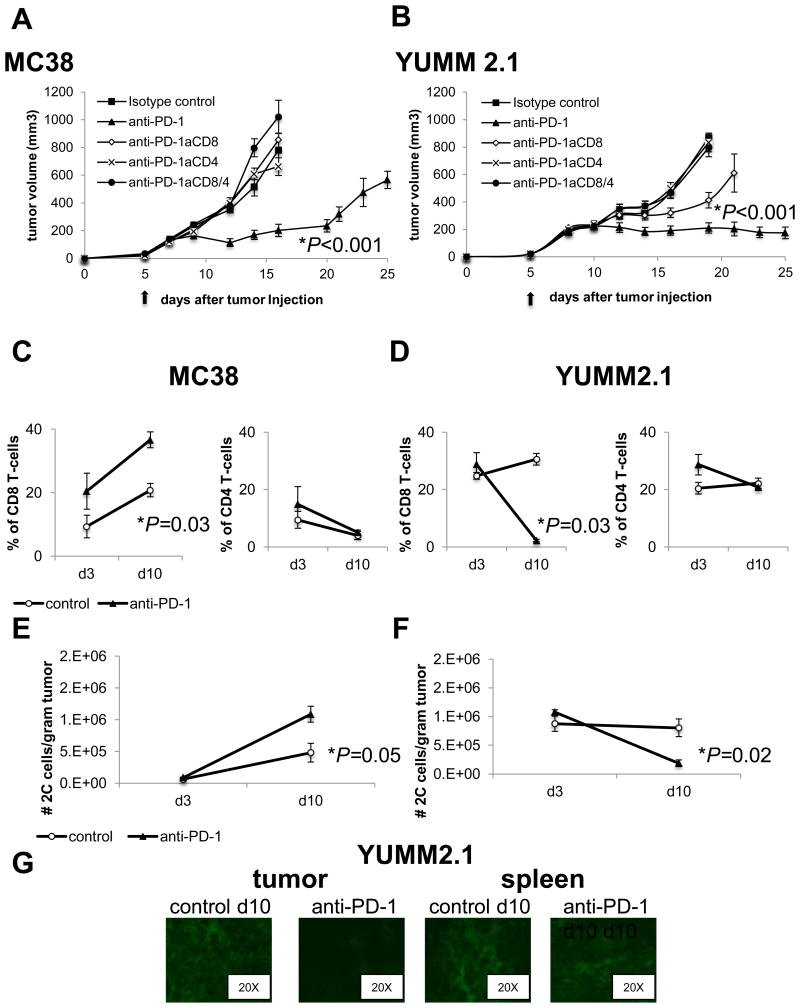
To elucidate the role of CD8 and CD4 T cells in anti–PD-1 activity, both cell subtypes were depleted in C57BL/6 mice bearing MC38 or YUMM2.1 tumors. Antibody-mediated depletion was confirmed in YUMM2.1 tumors and spleens (Fig. S2A and S2B). In the absence of CD8 cells, CD4 cells or both, antitumor response diminished in both MC38 and YUMM2.1 models (Fig. 3A and 3B). Of note, CD8 cell depletion (anti–PD-1aCD8) in the YUMM2.1 tumor model only partially abrogated the response to anti–PD-1 therapy, whereas CD4 cell depletion, or CD4 plus CD8 depletion, completely abrogated this response (Fig. 3B).
Fig. 3. Both CD8 and CD4 cells mediate response to PD-1 blockade in MC38 and YUMM2.1.
Tumor growth curves of MC38 (A) and YUMM2.1 (B) after anti–PD-1 and either anti-CD8 (anti–PD-1aCD8), anti-CD4 (anti–PD-1aCD4), anti-CD8 + anti-CD4 (anti–PD-1aCD8/4) or isotype control; 4 mice in each group, mean ± SD). (*P < 0.001 isotype control, anti–PD-1aCD8, anti–PD-1aCD4, anti–PD-1aCD8/4 versus anti–PD-1 in MC38, P < 0.001 isotype control, anti–PD-1aCD4, anti–PD-1aCD8/4 versus anti–PD-1 in YUMM2.1, unpaired t test, n=4), *P = 0.003 anti–PD-1aCD8 versus anti–PD-1, unpaired t test, n = 4), The arrow indicates the day treatment with anti–PD-1 or isotype control was started. This experiment was performed in triplicate. On day 3 (d3) and 10 (d10) after treatment with anti–PD-1 or isotype control was started, MC38 and YUMM2.1 tumors were isolated and stained with fluorescent-labelled antibodies, analysed by FACS (C and D) Percentage of CD3+CD8+ (CD8 T cells) and CD3+CD4+ (CD4 T cells) in MC38 (C) and YUMM2.1. (D) tumors are shown (mean ± SD). *P = 0.03 anti–PD-1 d10 versus control d10 in MC38; P = 0.03 anti–PD-1 d10 versus control d10 in YUMM2.1 (unpaired t test, n = 4). Results were consistent in 6 replicate experiments. (E and F) Statistical analysis of the 2C total number of CD8 T cells per gram of tumor in MC38 (E) and (F) YUMM2.1 tumors. *P = 0.05 anti PD-1 d10 versus control d10 in MC38, P = 0.02 anti–PD-1 d10 versus control d10 in YUMM2.1, unpaired t test, n = 8). (G) Representative immunofluorescence of CD8 T cells stained in YUMM2.1 tumors and spleens d10 after treatment with anti–PD-1 or isotype control was started.
Increased TILs in MC38, but decreased in YUMM2.1, upon PD-1 blockade
Three and ten days after starting treatment with anti–PD-1 or isotype control, tumors and spleens were harvested and stained for CD3, CD4, and CD8 (Fig. S2C and S2D). CD8 T-cell infiltration increased in MC38 tumors (calculated as percentage of all cells in the tumor) on day 3 and day 10 of treatment with anti–PD-1 when compared to isotype control (Fig. 3C), whereas CD8 T cells in the corresponding spleens of MC38 tumor-bearing mice remained unchanged (Fig. S2E). No significant difference in the percentage of CD4 T cells was observed in MC38 tumors (Fig. 3C) and spleens (Fig. S2F). However, CD8 T cell infiltration into YUMM2.1 tumors was significantly decreased on day 10 of anti–PD-1 therapy when compared to isotype control. This decrease in CD8 T cells was not present on day 3 (anti–PD-1 d3) compared to isotype control group (Fig. 3D). CD8 T cells did not decrease in the corresponding spleens of any of the conditions in the YUMM2.1 model (Fig. S2E). The percentage of CD4 T cells in the YUMM2.1 tumors or spleens was not significantly different across different time points or between anti–PD-1 and isotype control tumors (Fig. 3D). The YUMM1.1 tumor model did not show any CD8 T-cell variation in either tumors or spleens comparing anti–PD-1 and isotype control treated conditions (Fig. S2G). When we calculated the absolute number of CD8 T cells per gram of tumor pooled from two separate experiments, it confirmed the significant increase in CD8 T cells in the MC38 tumors (Fig. 3E), and the significant decrease in CD8 T cells in the YUMM2.1 tumors on day 10 of anti–PD-1 treatment (Fig. 3F). Immunofluorescence staining of tumors and spleens from mice in the YUMM2.1 group collected after anti–PD-1 therapy or isotype control also demonstrated a remarkable decrease in intratumoral CD8 T cells on day 10 and no change in spleen (Fig. 3G).
Wnt/β-catenin uninvolved in YUMM2.1 CD8 T-cell decrease or response to anti–PD-1
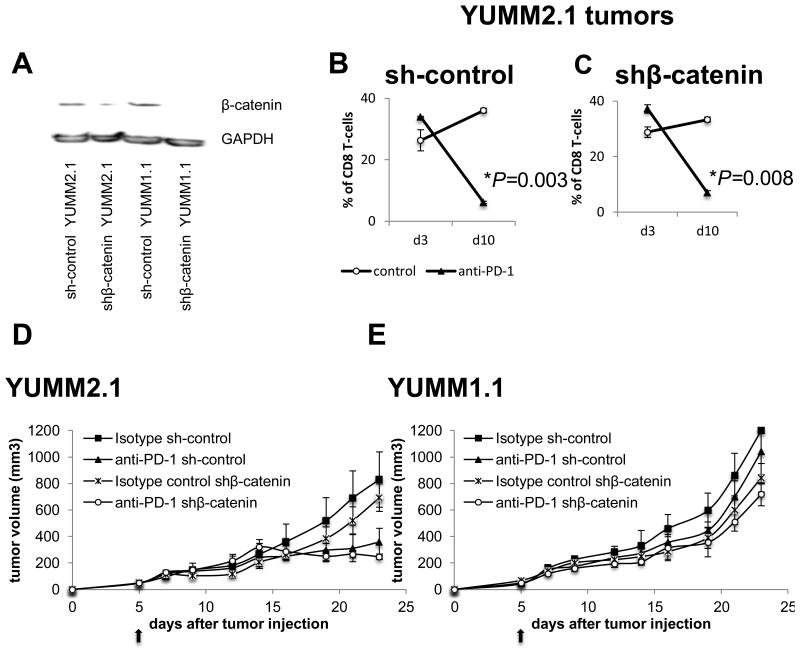
YUMM2.1 cell line was derived from a mouse with the same genetic background as YUMM1.1 but containing an additional transgenic allele that, when recombined by tamoxifen induction, produces a stabilized β-catenin, which leads to increased metastatic potential of the tumors (33). However, whole exome sequencing and PCR showed that β-catenin was unrecombined in the YUMM2.1 cell line, and the recombination could be induced by tamoxifen (4HT) (Fig. S3A and S3B). Nevertheless, we observed that YUMM2.1 cells do have more β-catenin protein expression with increased activity tested in vitro (Fig. S3C) and in macro-dissected tumor sections when implanted in mice (Fig. S3D). Active Wnt/β-catenin was linked to T-cell exclusion in tumors (34). To test if β-catenin had a role in the immunogenicity of YUMM2.1 and the loss of CD8 infiltrates on day 10 after anti–PD-1 therapy, β-catenin in both YUMM2.1 and YUMM1.1 cell lines was knocked down and confirmed at the protein level (Fig. 4A). Knockdown of β-catenin in YUMM2.1 did not change the significant decrease of CD8 T cells on day 10 with anti–PD-1 treatment when compared to the respective isotype treated controls (Fig. 4B and 4C). Silencing β-catenin did not change the antitumor response in the YUMM2.1 model (Fig. 4D), nor did it change in the non-responsive YUMM1.1 model (Fig. 4E).
Fig. 4. Wnt/β-catenin pathway is not involved in CD8 T-cell decrease or anti–PD-1 antitumor response in YUMM2.1 tumor model.
(A) Western blot analysis of β-catenin in YUMM2.1 cells transduced with shRNA without β-catenin (sh YUMM2.1) or with shβ-catenin (shβ-catenin YUMM2.1) and YUMM1.1 cells transduced with shRNA without β-catenin (sh YUMM1.1) or with shβ-catenin (shβ-catenin YUMM1.1). (B) Quantification of CD3+CD8+ (CD8 T cells). Tumor cells harvested on day 3 and 10 after anti–PD-1 or isotype control were counted and analyzed by flow cytometry for CD3/CD8 staining; three mice in each group (mean ± SD). *P = 0.003 anti–PD-1 d10 versus control d10 in sh-control YUMM2.1 tumors. (C) shβ-catenin YUMM2.1 tumors. *P = 0.008 anti–PD-1 d10 versus control d10 in shβ-catenin YUMM2.1 tumors, unpaired t test, n = 4. (D) In vivo sh and shβ-catenin YUMM2.1 and (E) sh and shβ-catenin YUMM1.1 tumor growth curves with three to four mice in each group (mean ± SD) after anti–PD-1 or isotype control.
Requirement of costimulation with PD-1 blockade in YUMM2.1
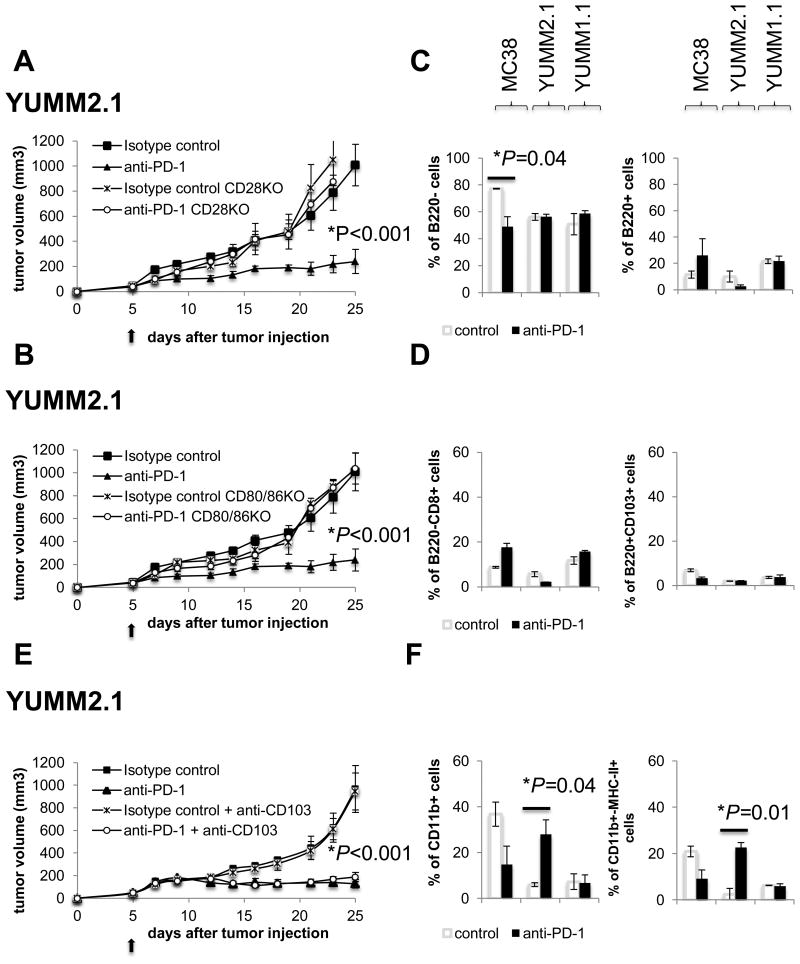
The evidence that both CD4 and CD8 cells are required for response to PD-1 blockade in the MC38 and YUMM2.1 models suggest that T-cell priming and CD4 helper function may be needed to induce the cytotoxic response to the tumors, which was further studied. The antitumor activity of PD-1 blockade against YUMM2.1 was completely abolished in CD28 knockout (KO) (Fig. 5A) and CD80/CD86 double KO mice (Fig. 5B), clearly demonstrating that costimulation is a requirement for the efficacy of anti–PD-1 blockade in this model.
Fig. 5. Increased antigen-presenting dendritic cells (DCs) in anti–PD-1 treated YUMM2.1 tumors.
(A) Tumor growth curves of CD28KO or C57BL/6 mice bearing YUMM2.1 treated with anti–PD-1 or isotype control. (B) Tumor growth curves of CD80/86KO or C57BL/6 mice bearing YUMM2.1 treated with anti–PD-1 or isotype control. Four mice in each group (mean ± SD). The arrow indicates the day treatment with anti–PD-1 or isotype control was initiated. (C) On day 10 after starting treatment, MC38, YUMM2.1 and YUMM1.1 tumors were isolated and stained with fluorescent-labeled antibodies and analyzed by FACS, with 3 mice in each group (mean ± SD). B220- and B220+ cells presented as percentage of CD11c+ cells. *P = 0.04 anti–PD-1 versus isotype control, CD11c+B220- cells in MC38 tumors, unpaired t test, n = 3. (D) B220-CD8+ and B220-CD103+ presented as percentage of CD11c+ cells. (E) In vivo YUMM2.1 growth curve after anti–PD-1 ± anti-CD103 or isotype control ± anti-CD103, 4 mice in each group (mean ± SD). The arrow indicates the day anti–PD-1 or isotype control treatment was started. (F) CD11b+ and CD11b+MHC-IIhigh DCs presented as percentage of CD11c+ cells. *P = 0.04 anti–PD-1 versus control, P = 0.01 anti–PD-1 versus control in YUMM2.1 tumors, unpaired t test, n = 3.
Increased antigen-presenting dendritic cells (DCs) in anti–PD-1–treated YUMM2.1 tumors
The next step was to identify the cells involved in antigen presentation and costimulation. We phenotyped the different subtypes of DCs by staining for CD11c+B220- (conventional) and CD11c+B220+ (plasmacytoid) subsets. Conventional DCs and can be further subdivided into CD11c+B220-CD8+ DCs, which are CD103+ in peripheral tissues and have been reported to mediate antigen cross-presentation to CD8 T cells (35), and CD11c+CD11b+MHC-IIhigh DCs, which are considered to be dedicated APCs that present peptides on MHC-II molecules to CD4 T cells (36) (gating strategy in Fig. S4A and S4B). The percentage of CD11c+B220– cells was significantly decreased in MC38 tumors of mice treated with anti–PD-1 compared to isotype control, with no significant change in YUMM2.1 or YUMM1.1 tumors (Fig. 5C). The percentage of intratumoral CD11c+B220-CD8+ and CD11c+B220-CD103+ DCs in MC38, YUMM2.1 or YUMM1.1 was not significantly different across time points or with PD-1 blockade therapy. A very small percentage of CD11c+B220-CD8+ cells in YUMM2.1 tumors were present (Fig. 5D). Growth of tumors in mice that were CD103-depleted was analogous to non-depleted mice, with or without the addition of anti–PD-1 (Fig. 5E). Of note, anti–PD-1 treated YUMM2.1 tumors exhibited a significant increase in CD11c+CD11b+ and CD11c+CD11b+MHC-IIhigh DCs compared to isotype control treated tumors (Fig. 5F). This finding was not present in MC38 tumors.
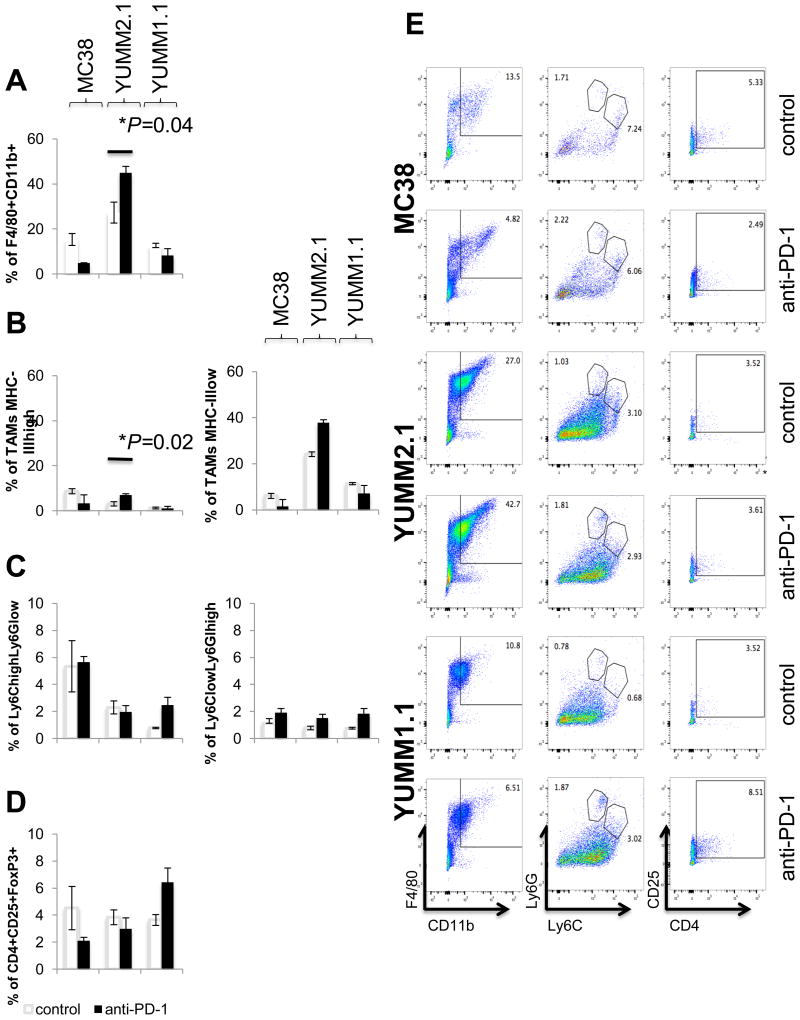
Increased tumor-associated macrophages (TAMs) in YUMM2.1 tumors treated with anti–PD-1
Another immune cell subtype potentially implicated in T-cell priming are TAMs. CD11b+F4/80+ TAMs were gated after the exclusion of dead cells (Fig. S4C). The total percentage of TAMs decreased (not statistically significant) in MC38 tumors treated with anti–PD-1 (Fig. 6A). In contrast, TAMs significantly increased in YUMM2.1 tumors on day 10 after anti–PD-1 treatment was started. Immune suppressive TAMs (CD11b+F4/80+MHC-IIlow, M2 TAMs) were more frequent in YUMM2.1 tumors with or without anti–PD-1 therapy, with an increase in the percentage of both CD11b+F4/80+MHC-IIhigh TAMs (M1 TAMs) and M2 TAMs upon PD-1 blockade (Fig. 6B). These observations were not present in YUMM1.1 tumors, where TAMs remained mostly unchanged (Fig. 6B). Taken together, TAMs may play a different role in YUMM2.1 tumors compared to MC38, although both tumor models respond to anti-PD1 blockade.
Fig. 6. Modulation of the tumor microenvironment by anti–PD-1 in MC38, YUMM2.1 and YUMM1.1.
On day 10 after anti–PD-1 or isotype control, MC38, YUMM2.1 and YUMM1.1 tumors were isolated and stained with fluorescent-labeled antibodies and analyzed by FACS, with 3 mice in each group (mean ± SD). (A) Analysis of TAMs (CD11b+F4/80+). (B) TAMs MHC-IIhigh (M1 TAMs, CD11b+F4/80+MHC-IIhigh) and TAMs MHC-IIlow (M2 TAMs, CD11b+F4/80+MHC-IIlow). *P = 0.04 anti–PD-1 d10 versus control d10 TAMs; P = 0.02 anti–PD-1 d10 versus control d10 TAMs MHC-IIhigh in YUMM2.1 tumors, unpaired t test, n = 3. (C) MO-MDSC (CD11b+Ly6ChighLy6Glow) and PMN-MDSC (CD11b+Ly6ClowLy6Ghigh) presented as percentage of CD11b+ cells. (D) Analysis of Tregs (CD4+CD25+FOXp3+). (E) Representative FACS plots in tumors.
No change in MDSCs or regulatory T cells (Tregs) with PD-1 blockade therapy
To evaluate the effect of anti–PD-1 on other cellular components of the tumor microenvironment, we harvested tumors 10 days after anti–PD-1 treatment was started and analyzed the two main subsets of MDSCs (myeloid-derived suppressor cells): monocytic MDSCs (MO-MDSC, CD11b+Ly6ChighLy6Glow) and polymorphonuclear MDSCs (PMN-MDSC, CD11b+Ly6ClowLy6Ghigh, Fig. S4D). Anti–PD-1 did not change the percentage of MO-MDSCs or PMN-MDSC in any tumors compared to isotype control (Fig. 6C). Another immune suppressive cell population, Tregs (Fig. S4E, Tregs, CD4+CD25+FOXp3+), showed a non-statistically significant trends towards a decrease in MC38 and YUMM2.1 tumors with anti–PD-1 and an increase in YUMM1.1 (Fig. 6D). Representative flow charts of TAMs, MDSCs, and Tregs are shown in Fig. 6E.
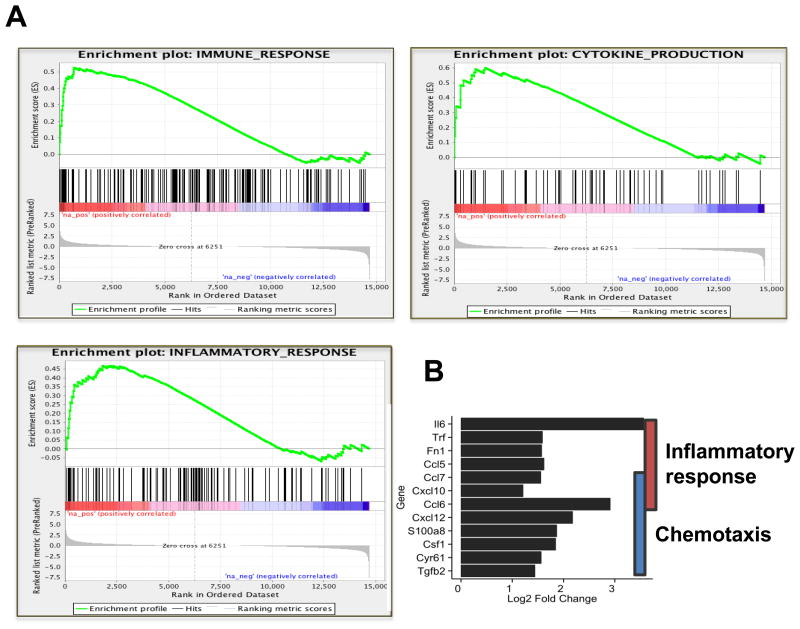
A more inflammatory gene signature profile in YUMM2.1 compared to YUMM 1.1
RNA was extracted from cultured YUMM1.1 and YUMM2.1 and subjected to RNA sequencing. GSEA and pathway analyses indicated that immune response, cytokine production and inflammatory-related genes were strongly represented in YUMM2.1 compared to YUMM =1.1 cells (Fig. 7A). Corresponding normalized enrichment scores (NES), P values and false discovery rates (FDR) of the GSEA plots are included (Fig. 7B). Analysis of genes that code for secreted proteins with a log2 -fold higher than 1 in YUMM2.1 compared to YUMM1.1 cells revealed an increase in inflammatory and chemotaxis-related genes (Fig. S4F).
Fig. 7. YUMM2.1 is more inherently immune permissive than YUMM1.1.
(A) GSEA curves for YUMM2.1 versus YUMM1.1 enriched pathways involved in immune response, cytokine production and inflammatory response. (B) Corresponding normalized enrichment scores (NES), P values and false discovery rates (FDR) of the GSEA plots.
Discussion
Immunological checkpoint blockade with anti–PD-1 or anti–PD-L1 antibodies reverses cancer immunosuppression and promotes antitumor immune responses in several cancer types. Long-term responses with minimal side effects have been reported in patients with melanoma, lung, liver, kidney, bladder, mismatch repair–deficient colon cancers and hematological malignancies, among others (1-4, 31). Why these agents exhibit antitumor responses in certain histologies and only in a percentage of patients with the same type of tumor remains unknown. Here we studied tumor models that respond differently to anti–PD-1 treatment and tested the reasons for anti–PD-1 activity in MC38 and YUMM2.1 tumors.
Upregulation of PD-L1 and its ligation to PD-1 on activated T cells is a well-described mechanism by which cancer tissues limit the host immune response, termed adaptive immune resistance (37). High baseline PD-L1–expressing tumor cells have been positively correlated with response to PD-1 blockade in patient samples (5, 6). However, PD-L1 was markedly increased upon IFNγ exposure in the three murine cell lines studied, which does not provide an explanation for the different responses to anti–PD-1.
Mutational load has been associated with a higher clinical benefit to immunotherapy (38-40). A greatly increased number of somatic mutations were observed in MC38 compared to YUMM2.1 and YUMM1.1, accompanied by high copy number variation, consistent with its origin as a carcinogen-induced cell line. The high mutational load could be at least partially responsible for the effectiveness of anti-PD1 therapy in MC38 tumors. However, both YUMM2.1 and YUMM1.1 displayed a very low number of new somatic mutations, consistent with tumors arising from genetically engineered mice driven by a strong driver oncogene and avoidance of senescence.
T-cell response has been widely accepted to be crucial for effective anti–PD-1/PD-L1 antitumor activity (41). We confirmed the essential roles of both the CD8 and CD4 T cells in anti–PD-1 effect in both MC38 and YUMM2.1 tumor models. Depletion of CD8 cells completely abrogated the antitumor effect of PD-1 blockade in the MC38 model but only had a partial effect in the YUMM2.1 model, whereas CD4 depletion completely reversed the antitumor effect in both models. Considering that anti–PD-1 also controls key T-cell inhibitory interactions between PD-L1 on APCs and PD-1 on T cells (17, 42) and that PD-1 limits CD4 T-cell clonal expansion in response to an immunogenic stimulus (43), it is not surprising that CD4 T cells are required for anti–PD-1/PD-L1 tumor response. However, another group has reported opposite observations, with increased antitumor effect seen with CD4 cell depletion combined with PD-1/PD-L1 blockade (44). Of note, none of the tumor models evaluated by this group was responsive to anti–PD-1/PD-L1 itself. The authors suggested that CD4 cell depletion effect was partially attributed to a removal of CD4 positive immunosuppressive Tregs. However, in another report (31), Tregs increased after very early analysis (48 and 72 hours) following treatment with anti–PD-1 in MC38, whereas in our tumor models, Tregs did not change with anti–PD-1 when analyzed at 10 days after starting therapy.
Next we characterized anti–PD-1 modulation of the cellular components in the tumor microenvironment. CD8 T cells were expected to increase in both anti–PD-1 responsive tumors. This was true for MC38, but in YUMM2.1 CD8 T cells decreased over time with anti–PD-1 therapy, implying that CD8 T cells may have an early role in this antitumor response. Therefore, the early activation of CD8 T cells could take place during antigen presentation to naïve T cells, where PD-1/PD-L1 costimulation has been shown to lead to T-cell receptor (TCR) down-modulation (16, 17, 42). Dendritic cells have been reported to hyperactivate CD8 T cells in the absence of PD-1/PD-L1 costimulation, which was accompanied by a higher TCR surface level and an increase in IFNγ (17). Depending on where PD-1/PD-L1 blockade takes place, T-cell activity may vary. It is unknown if the location of PD-1/PD-L1 interaction and its consecutive blockade is tumor-dependent in a short term implanted tumor model. Functional studies to determine T-cell activity shortly after anti–PD-1 is administered and further characterization of the specific CD8 T-cell phenotype could provide some explanation on how CD8 T cells exhibit their effect in this tumor model. The role of natural killer (NK) cells in this setting is unknown and technically challenging because of their low frequency in the tumor microenvironment, but certainly interesting to explore. Differences in PD-1 expression on the CD8 T cells could also be informative to address PD-1 responsiveness in the YUMM2.1 tumor model, as shown by others (31).
The correlation between tumor-intrinsic stabilized β-catenin and both T-cell exclusion and anti–PD-L1 resistance in genetically engineered mice with BRAFV600E/PTEN-/-/β-catenin stabilized tumors (34) led us to investigate the effect of β-catenin down-regulation in T-cell modulation and anti–PD-1 antitumor response. Although our analysis indicated that YUMM2.1 did not have recombined β-catenin allele that would render β-catenin more stable, it does have more β-catenin expression and activity compared to the other YUMM cell lines. We observed that T cells were reduced over time (but never upfront excluded) with anti–PD-1 therapy, and this phenomenon was independent from the β-catenin status. PD-1 blockade antitumor effect was not altered in the presence of a down-regulated Wnt/β-catenin pathway.
Looking further into the importance of costimulatory interactions during antigen presentation to naïve T cells, we demonstrated that the absence of CD28 or CD80/86 prevented the anti–PD-1 effects in YUMM2.1 tumors. This observation does not necessarily imply that the PD-1/PD-L1 inhibitory effects only take place at the APC–T-cell synapse, but suggest that PD-L1–expressing APCs are positively enhanced upon PD-1 blockade. Indeed, the priming of CD4 and CD8 T cells is more effective in the absence of PD-1/PD-L1 signaling (45), and down-modulation of PD-L1 in DCs results in increased costimulatory molecule CD80 expression and a distinct cytokine profile (46). The same group observed strong tumor growth control when using PD-L1–silenced DCs in a mouse model of lymphoma, although with no increased cure rates, possibly due to PD-L1–expressing tumor cells that might counteract CD8 T-cell activity (47).
Analysis of the different DC subsets in YUMM2.1 tumors revealed an increase in CD11c+CD11b+MHC-IIhigh DCs upon PD-1 blockade, which was not present in the other tumor models analyzed. Cross-priming of tumor antigens by BATF3-dependent DCs is crucial to the efficacy of anti–PD-1 antibodies (48). Taken together, these data imply that priming via CD4 T cells has a more important role in the antitumor efficacy of PD-1 blockade in the YUMM2.1 model.
When looking into the ability of the models to evoke an inflammatory reaction required for immune cell recruitment and DC-T-cell costimulation, YUMM2.1 exhibited an “inflammatory profile” consistent with an endogenous upregulation of immune, cytokine producing, and inflammatory response-related genes. The YUMM2.1 model could therefore intrinsically harbor inflammatory mediators necessary to couple innate recognition to T-cell mediated immunity by DCs in vivo, which is also supported by the increase in chemotactic factors such as Cxcl10, Ccl6, or Cxcl12. This observation is consistent with other reports, where chemokine-trafficking of immune cells into tumors was observed in human melanoma cell lines (49) or in mice receiving adoptive cell therapy and anti–PD-1 blockade (50).
In conclusion, T-cell priming supports anti–PD-1 antitumor responses mediated by CD4 and CD8 T cells, critically requiring costimulation in vivo.
Supplementary Material
Acknowledgments
Funding: This study was funded in part by the NIH grants P01CA168585 (to A.R. and T.G.G.), R35 CA197633, the Ressler Family Fund, the Dr. Robert Vigen Memorial Fund, the Grimaldi Family Fund, the Samuels Family Fund, the Ruby Family Fund, the Alexandra Cooper Memorial Fund, and the Garcia-Corsini Family Fund (to A.R.). B.H. was supported in part by the Rio Hortega Scholarship (08/142) from the Hospital 12 de Octubre, Madrid, Spain. G.P. was supported in part by the Division of Medical Oncology and Immunotherapy (University Hospital of Siena). J.M.Z is a member of the UCLA Medical Scientist Training Program supported by NIH NIGMS training grant GM08042. J.T. is supported by the NIH Ruth L. Kirschstein Institutional National Research Service Award #T32-CA009120. S.H-L. was supported by a Young Investigator Award and a Career Development Award from the American Society of Clinical Oncology (ASCO), a Tower Cancer Research Foundation Grant, and a Dr. Charles Coltman Fellowship Award from the Hope Foundation.
Footnotes
Competing interests: The authors declare no conflicts of interest.
References and notes
- 1.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti–PD-1) in melanoma. N Engl J Med. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribas A, Schnachter JV, Long G, Arance A, Grob JJ, Mortier L, et al. Phase III study of prembrolizumab (MK-3475) versus ipilimumab in patients with ipilimumab-naive advanced melanoma (abstract CT101) American Association for Cancer Research (Anual Meeting) 2015 [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin Cancer Res. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blank C, Kuball J, Voelkl S, Wiendl H, Becker B, Walter B, et al. Blockade of PD-L1 (B7-H1) augments human tumor-specific T cell responses in vitro. Int J Cancer. 2006;119:317–27. doi: 10.1002/ijc.21775. [DOI] [PubMed] [Google Scholar]
- 9.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latchman YE, Liang SC, Wu Y, Chernova T, Sobel RA, Klemm M, et al. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc Natl Acad Sci U S A. 2004;101:10691–6. doi: 10.1073/pnas.0307252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Pino-Lagos K, de Vries VC, Guleria I, Sayegh MH, Noelle RJ. Programmed death 1 ligand signaling regulates the generation of adaptive Foxp3+CD4+ regulatory T cells. Proc Natl Acad Sci U S A. 2008;105:9331–6. doi: 10.1073/pnas.0710441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–30. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–8. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- 14.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 15.Carter L, Fouser LA, Jussif J, Fitz L, Deng B, Wood CR, et al. PD-1:PD-L inhibitory pathway affects both CD4(+) and CD8(+) T cells and is overcome by IL-2. Eur J Immunol. 2002;32:634–43. doi: 10.1002/1521-4141(200203)32:3<634::AID-IMMU634>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Karwacz K, Arce F, Bricogne C, Kochan G, Escors D. PD-L1 co-stimulation, ligand-induced TCR down-modulation and anti-tumor immunotherapy. Oncoimmunology. 2012;1:86–88. doi: 10.4161/onci.1.1.17824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karwacz K, Bricogne C, MacDonald D, Arce F, Bennett CL, Collins M, et al. PD-L1 co-stimulation contributes to ligand-induced T cell receptor down-modulation on CD8+ T cells. EMBO Mol Med. 2011;3:581–92. doi: 10.1002/emmm.201100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med. 2012;209:1201–17. doi: 10.1084/jem.20112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–42. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper ZA, Juneja VR, Sage PT, Frederick DT, Piris A, Mitra D, et al. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol Res. 2014;2:643–54. doi: 10.1158/2326-6066.CIR-13-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koya RC, Mok S, Comin-Anduix B, Chodon T, Radu CG, Nishimura MI, et al. Kinetic phases of distribution and tumor targeting by T cell receptor engineered lymphocytes inducing robust antitumor responses. Proc Natl Acad Sci U S A. 2010;107:14286–91. doi: 10.1073/pnas.1008300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cross RS, Malaterre J, Davenport AJ, Carpinteri S, Anderson RL, Darcy PK, et al. Therapeutic DNA vaccination against colorectal cancer by targeting the MYB oncoprotein. Clin Transl Immunology. 2015;4:e30. doi: 10.1038/cti.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngiow SF, Young A, Jacquelot N, Yamazaki T, Enot D, Zitvogel L, et al. A Threshold Level of Intratumor CD8+ T-cell PD1 Expression Dictates Therapeutic Response to Anti-PD1. Cancer Res. 2015;75:3800–11. doi: 10.1158/0008-5472.CAN-15-1082. [DOI] [PubMed] [Google Scholar]
- 32.Mule JJ, Shu S, Schwarz SL, Rosenberg SA. Adoptive immunotherapy of established pulmonary metastases with LAK cells and recombinant interleukin-2. Science. 1984;225:1487–9. doi: 10.1126/science.6332379. [DOI] [PubMed] [Google Scholar]
- 33.Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–54. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 35.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–30. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganguly D, Haak S, Sisirak V, Reizis B. The role of dendritic cells in autoimmunity. Nat Rev Immunol. 2013;13:566–77. doi: 10.1038/nri3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribas A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–96. [PubMed] [Google Scholar]
- 42.Escors D, Bricogne C, Arce F, Kochan G, Karwacz K. On the Mechanism of T cell receptor down-modulation and its physiological significance. J Biosci Med. 2011;1 [PMC free article] [PubMed] [Google Scholar]
- 43.Konkel JE, Frommer F, Leech MD, Yagita H, Waisman A, Anderton SM. PD-1 signalling in CD4(+) T cells restrains their clonal expansion to an immunogenic stimulus, but is not critically required for peptide-induced tolerance. Immunology. 2010;130:92–102. doi: 10.1111/j.1365-2567.2009.03216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueha S, Yokochi S, Ishiwata Y, Ogiwara H, Chand K, Nakajima T, et al. Robust Antitumor Effects of Combined Anti-CD4-Depleting Antibody and Anti-PD-1/PD-L1 Immune Checkpoint Antibody Treatment in Mice. Cancer Immunol Res. 2015;3:631–40. doi: 10.1158/2326-6066.CIR-14-0190. [DOI] [PubMed] [Google Scholar]
- 45.Gibson A, Ogese M, Sullivan A, Wang E, Saide K, Whitaker P, et al. Negative regulation by PD-L1 during drug-specific priming of IL-22-secreting T cells and the influence of PD-1 on effector T cell function. J Immunol. 2014;192:2611–21. doi: 10.4049/jimmunol.1302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pen JJ, Keersmaecker BD, Heirman C, Corthals J, Liechtenstein T, Escors D, et al. Interference with PD-L1/PD-1 co-stimulation during antigen presentation enhances the multifunctionality of antigen-specific T cells. Gene Ther. 2014;21:262–71. doi: 10.1038/gt.2013.80. [DOI] [PubMed] [Google Scholar]
- 47.Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: implications for tumor immunotherapy. Cancer Immunol Immunother. 2005;54:307–14. doi: 10.1007/s00262-004-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Paulete AR, Cueto FJ, Martinez-Lopez M, Labiano S, Morales-Kastresana A, Rodriguez-Ruiz ME, et al. Cancer Immunotherapy with Immunomodulatory Anti-CD137 and Anti-PD-1 Monoclonal Antibodies Requires BATF3-Dependent Dendritic Cells. Cancer Discov. 2016;6:71–9. doi: 10.1158/2159-8290.CD-15-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–85. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng W, Liu C, Xu C, Lou Y, Chen J, Yang Y, et al. PD-1 blockade enhances T-cell migration to tumors by elevating IFN-gamma inducible chemokines. Cancer Res. 2012;72:5209–18. doi: 10.1158/0008-5472.CAN-12-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.