Abstract
The linear plasmid pBSSB1 mediates the flagellar phase variation in H:z66 positive Salmonella enterica serovar Typhi (S. Typhi). The gene named stp17 (S. Typhi plasmid number 17 gene) is located on pBSSB1 and encodes the protein STP17. The expression pattern at the protein-level and function of STP17 remains unknown. In this study, the recombinant protein STP17His6 was expressed, purified and used to prepare the polyclonal anti-STP17 antibody. We detected protein-level expression of stp17 in S. Typhi and further investigated the protein expression characteristics of stp17 in different growth phases by western blot analysis. The effects of STP17 on bacterial growth and motility were analyzed. In addition, the structure of STP17 was predicted and the active site of STP17 was identified by site-directed mutagenesis. The results showed that STP17 was expressed stably in the wild type strain of S. Typhi. STP17 expression at the protein level peaks when cultures reach an OD600 value of 1.2. The growth rate and motility of the Δstp17 strain were significantly decreased compared with the wild type strain (P < 0.05) and this phenotype was restored in the stp17 complementary strain. Moreover, the growth rate and motility of the stp17 over-expression strain was greater than the wild type strain. STP17 contains nine Helix segments, six Stand segments and some Coil segments in the secondary structural level. The top-ranked 3-D structure of STP17 predicted by I-TASSER contains a putative ATPase domain and the amino acid residues of GLY16, GLY19, LYS20, ASN133, LYS157, and LYS158 may be the active site residues of STP17. Finally, STP17 was able to catalyze the ATP to ADP reaction, suggesting that STP17 may be an ATPase. To our knowledge, this is the first report describing the protein expression characteristics of STP17 in S. Typhi, showing that STP17 promotes bacterial growth and motility, which may be associated with its potential ATPase activity.
Keywords: Salmonella enterica serovar Typhi, linear plasmid, pBSSB1, stp17, growth, motility, ATPase
Introduction
Salmonella enterica serovar Typhi (S. Typhi) is an important human pathogen responsible for typhoid fever (Everest et al., 2001; Wain et al., 2015). Harm caused by S. Typhi has been greatly reduced by development and use of antibiotics. However, typhoid fever remains a common disease in tropical and subtropical regions, with many drug-resistant strains having been isolated (John et al., 2016; Yap et al., 2016). In addition, S. Typhi is an important model organism for researching the expression and regulation of prokaryotic genes (Winter et al., 2014).
Most serovars of S. enterica contain fliC and fljB genes encoding the phase-1 and phase-2 flagellar antigen respectively at different chromosomal loci, and show biphasic characteristics (Simon et al., 1980). Biphasic serovars of S. enterica alternatively express two flagellar antigens through a process called “phase variation,” which is mediated by a hin located upstream fljBA (Henderson et al., 1999). S. Typhi normally do not possess flagellar antigen phase variation because they only harbor the fliC gene (H:d) and lack the fljB gene. However, some isolates of S. Typhi, from Indonesia contain an fljB gene equivalent (fljBz66) which encode the novel flagellin named H:z66 (Guinee et al., 1981). fljBz66 is located on linear plasmid pBSSB1 which is the first non-bacteriophage-related linear plasmid found in Enterobacteriaceae and mediates the unidirectional flagellar phase variation of the S. Typhi z66-positive strain (Baker et al., 2007a,b). S. Typhi z66-positive strain was isolated mainly from Indonesia and caused the incidences of serious typhoid fever which may be due to the escaping immunologic surveillance through its unique unidirectional flagellar phase variation (Baker et al., 2008; Hatta et al., 2011).
Thirty-three putative genes are encoded on pBSSB1 and most remain to be elucidated with the exception of three genes: 030, fljBz66, and fljAz66 (Baker et al., 2007a,b). The genes fljBz66 and fljAz66 have been well studied in terms of gene expression regulation and have been found to mediate the flagellar variation in z66-positive S. Typhi (Huang et al., 2004; Xu et al., 2008, 2010; Zou et al., 2009). The seventeenth gene, here named stp17 (S. Typhi plasmid number 17 gene), is immediately adjacent to the possible replication origin of pBSSB1 and is predicted to encode a putative nucleotide binding protein STP17 (Baker et al., 2007a). In 2014, the expression characteristic of stp17 in mRNA level was demonstrated (Zhao et al., 2014). However, the function of stp17 has not previously been described. Here, we demonstrate that stp17 may promote bacterial growth and motility through the ATPase activity of STP17.
Materials and methods
Bacterial strains, plasmids, and culture conditions
A z66-positive wild-type strain, S. Typhi GIFU10007, was used in this study. Mutants and plasmids used in this work are listed in Table 1. Primer sequences are described in Table 2. Bacteria were cultured in Luria–Bertani (LB) broth at 37°C.
Table 1.
Strains and plasmids used in the study.
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| STRAINS | ||
| S. Typhi GIFU10007 | Wild-type strain, Z66+ | Laboratory collection |
| SY372λpir | E. coli host strain of suicide plasmid | Laboratory collection |
| Δstp17 | GIFU10007(Δstp17) | This work |
| Δstp17(pACYC184) | Δstp17containing pACYC184 empty vector | This work |
| Δstp17(pACYC184-stp17) | Δstp17 containing pACYC184-stp17,recombinant plasmid | This work |
| wt(pBAD) | GIFU10007 containing pBAD, empty vector | This work |
| wt(pBAD-stp17) | GIFU10007 containing pBAD-stp17,recombinant plasmid | This work |
| DH5a(pET28b-stp17) | E. coli DH5 containing pET28b-stp17,recombinant plasmid | This work |
| BL21(pET28b-stp17) | E. coli BL21 containing pET28b-stp17,recombinant plasmid | This work |
| ΔpBSSB | GIFU10007 cured pBSSB1 | This work |
| ΔpBSSB (pBAD) | ΔpBSSB containing pBAD empety vector | This work |
| ΔpBSSB (pBAD-stp17) | ΔpBSSB containing pBAD-stp17 recombinant plasmid | This work |
| PLASMIDS | ||
| pGMB151 | Suicide plasmid; sacB; Ampr | Laboratory collection |
| pGMB-stp17 | pGMB151 containing stp17 deleted homologous fragments | This work |
| pACYC184 | Complementary vector; Chlr, Tetr | Laboratory collection |
| pACYC184-stp17 | pACYC184 containing stp17 gene | This work |
| pBAD/gIII | Expression vector; Ampr | Laboratory collection |
| pBAD-stp17 | pBAD/gIII containing stp17 gene | This work |
| pET28b | Expression vector; Kanar | Laboratory collection |
| pET28b-stp17 | pET28b containing stp17 gene | This work |
Table 2.
Primers used in this study.
| Primers | Sequence(5′-) | Purpose |
|---|---|---|
| F1A(BamH I) | TTAGGATCCAGTTCCGAATCCCATAGGC | stp17 mutant construction |
| F1B | CGAATAGATAACACCTCCCTTATAGTTCCA | |
| F2A | AGGGAGGTGTTATCTATTCGGAAGGTACAGG | |
| F2B(BamH I) | CTAGGATCCAGCAGCATTATTACTATGTGC | |
| C-stp17-A(Xba I) | CGTCTAGAGGCAACTCCTTAGTTATG | pACYC184-stp17 construction |
| C-stp17-B(Hind III) | GCAAGCTTGTAAGAGTCACCGGCATT | |
| O-stp17-A(Nco I) | GTACCATGGGTATGTTAGGGGGTTTTATGAT | pBAD-stp17 construction |
| O-stp17-B(Hind III) | GCCAAGCTTTTATGCCTTCTCTTTTGCTTTC | |
| P-stp17-A(Nco I) | GACCATGGATATGTTAGGGGGTTTTATG | pET28b-stp17 construction |
| P-stp17-B(Xho I) | ATCTCGAGTGCCTTCTCTTTTGCTTT |
Underline means the sequences recognized by specific restriction endonucleases.
Construction of stp17 mutant, complementation and over-expression strains
The stp17 gene deletion mutant of S. Typhi was prepared through homologous recombination mediated by the suicide plasmid pGMB151, as previously described (Huang et al., 2004; Zhang et al., 2012a). The stp17 deletion mutant was confirmed by sequencing analysis and designated as Δstp17.
For complementary expression of stp17 in Δstp17, the CDS of stp17 was amplified with pfu DNA polymerase by PCR. The amplicon was inserted into the complementary vector pACYC184 to form the recombinant plasmid pACYC184-stp17, which was verified by sequencing analysis. Then, the strain Δstp17 was transformed by the recombinant plasmid pACYC184-stp17 and designated as the stp17 complementary strain Δstp17(pACYC184-stp17). As a control, the strain Δstp17 was also transformed with the empty vector pACYC184 and named Δstp17(pACYC184).
The stp17 ORF (a 642-bp DNA fragment) was cloned into the expression vector pBAD/gIII which can be induced by L-arabinose to generate the recombinant plasmid (pBAD-stp17). The recombinant plasmid pBAD-stp17 was confirmed by sequence analysis and subsequently transformed into the wild type strain S. Typhi GIFU10007 to generate the over-expression strain wt(pBAD-stp17). As a control, the strain S. Typhi GIFU10007 was also transformed with the empty vector pBAD/gIII and named wt(pBAD). Over-expression of stp17 in wt(pBAD-stp17) was induced by L-arabinose (0.1% wt/vol).
Expression and purification of the recombinant protein STP17His6
The entire stp17 ORF (a 642-bp DNA fragment) containing a Nco I site (5′-end) and a Xho I site (3′-end) was amplified by PCR. Then, the resulting PCR product was digested with Nco I and Xho I and cloned into plasmid pET28b which carries a N-terminal His-tag digested with the same enzymes. The resulting stp17 recombinant expression plasmid pET28b-stp17 was transformed into E. coli BL21(DE3) cells. The cell cultures were incubated at 37°C in LB medium until the OD600 reached 0.6. To induce expression of the recombinant protein, IPTG was added at a final concentration of 0.03 mM. The culture was grown for 5 h at 37°C and harvested by centrifugation (4000 r/min, 10 min, 4°C). Bacteria were re-suspended in 20 ml PBS. After bacteria were lysed with an ultrasonic cell disruptor, bacterial lysate was purified using a Ni-NTA agarose column as directed by the manufacturer (QIAGEN). Recombinant protein STP17His6 was eluted with elution buffer containing 250 mM imidazole. Purified STP17His6 from the elution buffer was concentrated in PBS using Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-10 membrane, according to the manufacturer's protocol (Millipore Corporation, Bedford, MA, USA). Purified STP17His6 was analyzed by SDS-PAGE.
Production of polyclonal antibody of STP17
To prepare the polyclonal antibody of STP17, purified STP17His6 was mixed completely with an equal volume of Freund's complete adjuvant and 1 ml of mixture which contained 50 μg STP17His6 was injected subcutaneously into each rabbit. After 1 week, purified STP17His6 was mixed completely with an equal volume of Freund's un-complete adjuvant and the mixture was used to immunize the rabbits as above every 2 weeks. The rabbits were immunized for total five times. Finally, the antiserum was obtained from the carotid artery, and was purified through salting out extraction with ammonium sulfate to prepare the rabbit polyclonal antibody of STP17.
Verifying STP17 expression in S. Typhi GIFU10007 by western blot analysis
S. Typhi wild type and mutant Δstp17 strains were cultured overnight with shaking (250 rpm) at 37°C. Bacteria were normalized to OD600 0.6. Proteins were separated by 15% SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was blocked with 5% dried skim milk. Rabbit anti-STP17 antiserum as the primary antibody was used at a dilution of 1:500 and incubated with the membrane for 2 h at room temperature. HRP-conjugated goat anti-rabbit-IgG antibody (Sigma-Aldrich, St. Louis, Missouri, USA) was used at a dilution of 1:10,000 as the secondary antibody. Horseradish peroxidase-antibody conjugates were detected by chemiluminesence.
Investigation of STP17 expression in S. Typhi GIFU10007 by western blot analysis
The expression characteristics of STP17 protein in S. Typhi GIFU10007 under different growth phases were investigated by western blot. S. Typhi cells were collected when their OD600 values were 0.2, 0.5, 0.8, 1.2, and 1.6, respectively. Western blotting experiments in triplicate were performed as previously described.
Plasmid curing and complementation of stp17 into pBSSB1-dificient derivatives of S. Typhi
The plasmid curing method was referred to the reference (El-Mansi et al., 2000) with some following modifications. Sodium dodecyl sulfate (SDS) was added to LB media and the final concentration of SDS was adjusted to 1%. The wild-type strain S. Typhi GIFU10007 was inoculated into the above SDS containing LB and cultures were incubated overnight with shaking at 43°C. Subsequently, the cultures were inoculated into normal LB and incubated overnight with shaking at 43°C. The cultures were inoculated into SDS containing LB again and cultured overnight at 43°C with shaking. Then, the cultures were placed on to SS agar plates to select the possible pBSSB1-cured derivatives of S. Typhi through the amplification of the gene fljBz66 located on pBSSB1 by PCR. Finally, the possible pBSSB1-cured derivatives of S. Typhi were verified by Southern-blot as previous described (Zhang et al., 2012b) and designated as ΔpBSSB1. Then, the strain ΔpBSSB1 was transformed by the recombinant plasmid pACYC184-stp17 and designated as the stp17 complementary ΔpBSSB1 strain ΔpBSSB1(pACYC184-stp17). As a control, the strain ΔpBSSB1 was also transformed with the empty vector pACYC184 and named ΔpBSSB1 (pACYC184).
Structure prediction of STP17 and mutagenesis of stp17 gene
Based on the amino acid sequence of protein STP17 which were retrieved from GenBank (accession no. AM419040.1; Baker et al., 2007a), its secondary structure and three-dimensional structure were predicted by I-TASSER online server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) as previous described (Cai et al., 2013). In addition, the conserved ATPase domain of STP17 was predicted, and the possible ATP binding site residues of STP17 was analyzed through I-TASSER online server as well. According to the predicted possible ATP binding site residues of STP17, the site-directed mutations were introduced into pACYC184-stp17 using standard PCR mutagenesis techniques, and mutations were confirmed by DNA sequencing. The mutants generated were the following: Δstp17:: pACYC184-stp17G16A, Δstp17:: pACYC184-stp17G17A, Δstp17:: pACYC184-stp17G19A, Δstp17:: pACYC184-stp17K20A, Δstp17:: pACYC184-stp17N133A, Δstp17:: pACYC184-stp17R134A, Δstp17:: pACYC184-stp17K157A, Δstp17:: pACYC184-stp17K158A.
Growth curve assay
Bacteria were grown in LB medium at 37°C. A single colony of bacteria from a LB agar plate was inoculated into 2 ml of LB broth and incubated at 37°C with shaking (250 rpm) overnight. Then, the culture was diluted 1/100 in fresh LB broth (containing L-arabinose (0.1% wt/vol) for the culturing of strains harboring pBAD). Cell growth was monitored at OD600 every hour using a BioPhotometer (Eppendorf, Hamburg, Germany). The experiments were repeated at least three times.
Motility assay
For detecting the motility of bacteria, different strains were incubated until their OD600 reached 0.4. From each culture, 2 μl was inoculated onto a 0.3% semisolid LB agar plate (containing L-arabinose 0.1% wt/vol) for the strains harboring pBAD). The plates were incubated at 37°C for 10 h and motility was assessed qualitatively by examining the diameter of circular swimming, which was formed by growing motile bacterial cells. Meanwhile, the flagella gene fljBz66 and fliC expression of different strains were examined by qRT-PCR.
STP17 ATPase activity assay
The potential ATPase activity of STP17 was determined by the ATP assay kit (Beyotime, China) according to the manufacturer's recommended protocol. Briefly, STP17His6 protein was added to the ATP solution provided in the assay kit at a concentration of 10 μM and then incubated at 25°C for 10-, 30-, and 50 min, respectively. The ATP concentration of the above reaction mixtures was measured by a F-4500 fluorescent spectrophotometer (Hitachi, Japan). As a control, BSA protein which lacks ATPase activity was mixed with the ATP solution and analyzed on the spectrophotometer. Because ATPase can catalyze ATP into ADP and free phosphate, the reduction of ATP concentration in the reaction mixture indicates the presence of ATPase activity, which should be dependent on the added protein being studied (STP17His6). These experiments were performed at least three times.
Statistical analysis
Data were analyzed using Prism 5 software. The Student's t-test was used to determine significant differences between results. Significance was defined as P < 0.05.
Results
The gene stp17 is expressed and translated into STP17 protein in S. Typhi
The gene stp17 is located on a linear plasmid named pBSSB1 in S. Typhi and is predicted to encode a putative nucleotide binding protein, STP17. In this study, expression of stp17 in E. coli BL21(DE3) was generated with the expression vector pET28b. Results showed that the purified recombinant protein STP17His6 was successfully obtained (Figure 1A). To verify whether the stp17 gene is expressed and translated into STP17 protein in S. Typhi, western blotting was performed to identify STP17 levels in the S. Typhi wild type strain and the mutant strain Δstp17 (Figure 1B). The results showed that the gene stp17 can be translated into STP17 protein in S. Typhi.
Figure 1.
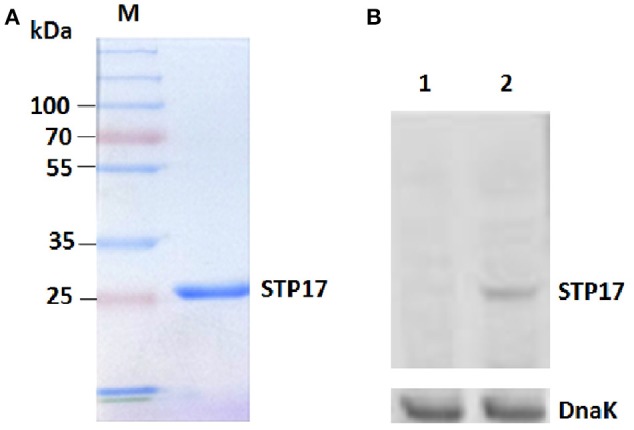
Identification of stp17 expression in S. Typhi. (A) SDS-PAGE analysis of STP17His6 protein purification; (B) Western blot analysis using the purified anti-STP17His6 polyclonal antibody. Lane 1, Δstp17; Lane 2, S. Typhi GIFU 10007. DnaK: the loading control marker.
Protein expression characteristics of STP17 in different growth phases
We investigated the expression characteristics of STP17 in different growth phases by western blot analysis. Five OD600 values (0.2, 0.5, 0.8, 1.2, and 1.6) corresponding to early log phase, mid-log phase, late log phase, early stationary phase and late stationary phase, respectively, were monitored. As shown in Figure 2, stp17 can be expressed stably in normal growth conditions with expression levels increasing with OD600 values throughout log phase. The expression level of STP17 reaches a peak value at an OD600 value of 1.2. Then, expression levels decrease with OD600 values at stationary phase.
Figure 2.
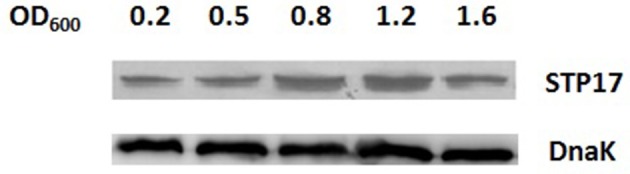
Western blot analysis of STP17 protein levels in S. Typhi in different growth phases. OD600 values (0.2, 0.5, 0.8, 1.2, and 1.6) corresponding to early log phase, mid-log phase, late log phase, early stationary phase and late stationary phase, respectively. DnaK: the loading control marker.
The gene stp17 affects the growth of S. Typhi in a pBSSB1 independent manner
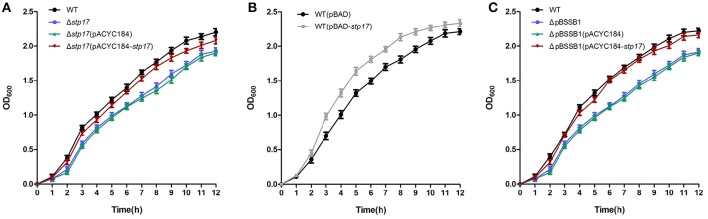
To investigate the role of the stp17 gene in S. Typhi, the stp17 mutant was constructed by homologous recombination mediated by the suicide plasmid. We measured growth of the wild-type and Δstp17 strain and found that growth of stp17 mutant strain was significantly slower compared to wild type (P < 0.05; Figure 3A). In addition, the growth of complementary strain Δstp17(pACYC184-stp17) restored to the wild type level (Figure 3A). At the same time, we constructed the S. Typhi stp17 over-expression strain wt(pBAD-stp17) and found that cell growth upon STP17 over-expression was significantly increased compared with the wild type strain (P < 0.05; Figure 3B). As shown in Figure 3C, the growth of pBSSB1-deficient strain ΔpBSSB1 was obviously decreased compared with the wild type strain because stp17 was absent due to the loss of pBSSB1. However, the growth rate of stp17 complementary ΔpBSSB1 strain ΔpBSSB1(pACYC184-stp17) was similar to the wild type strain. In addition, we compared the stability of pBSSB1 in wild type and Δstp17, and found no obvious difference presenting after deletion of stp17 (data not shown). These results suggest that stp17 has an effect on S. Typhi cell growth in a pBSSB1 independent manner.
Figure 3.
Effect of stp17 on the growth rate of S. Typhi. (A) Growth curve of wild type strain, stp17 deletion mutant and stp17 complementary strain. (B) growth curve of over-expression strain. (C) growth curve of wild type strain, pBSSB1-cured derivative and pBSSB1 complementary strain.
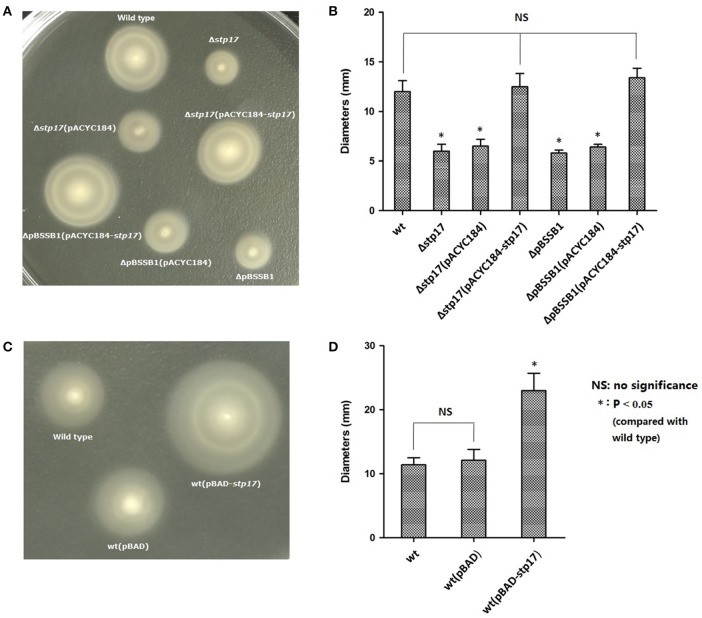
The gene stp17 enhances the motility of S. Typhi in a pBSSB1 independent manner
As shown in Figure 4A, the motility of Δstp17 was greatly decreased compared with the wild type strain, and the motility of pBSSB1-deficient strain ΔpBSSB1 was obviously decreased as well because stp17 was absent due to the loss of pBSSB1. However, bacterial motility was restored obviously in the complementary strain Δstp17(pACYC184-stp17) and ΔpBSSB1(pACYC184-stp17). Moreover, the motility of stp17 over-expression strain wt(pBAD-stp17) was much greater compared with the control strain wt(pBAD) (Figure 4C). The ring diameters for various strains are shown in Figures 4B,D. Moreover, the expression of flagellar gene fljBz66 and fljC show no obvious difference when stp17 was deleted (data not shown). This result suggests that the differences of bacterial motility may be not due to the different expression of flagellar gene. The data demonstrate that stp17 affects the motility of S. Typhi in a pBSSB1 independent manner.
Figure 4.
Motility ring and diameter of S. Typhi. (A,B) Effect of stp17 and pBSSB1 on the motility of S. Typhi. (C,D) Effect of stp17 over-expression on the motility of S. Typhi.
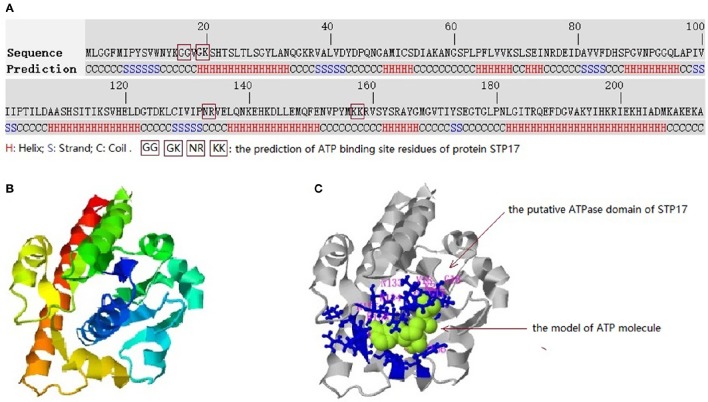
Structure prediction of STP17
The structure of STP17 was predicted by I-TASSER online server, as explained in Materials and Methods. As shown in Figure 5A, the 213 amino acids of STP17 contains nine Helix segments, six Stand segments and some Coil segments in the secondary structural level. The top-ranked 3-D structure of STP17 predicted by I-TASSER was shown in Figure 5B, and it contains a putative ATPase domain like a pocket (Figure 5C), with the top-ranked predicted residues GLY16, GLY17, GLY19, LYS20, ASN133, ARG134, LYS157, and LYS158 as ATPase active site residues (Figure 5A).
Figure 5.
Predicted 3-D structures and conserved domains of the protein STP17. (A) The amino acid sequence of STP17 and the predicted secondary structure. (B) The top-ranked 3-D structure of STP17 predicted by I-TASSER. (C) The putative ATPase domain, the ligand molecule of STP17 is indicated by green and the top-ranked predicted active residues GLY16, GLY17, GLY19, LYS20, ASN133, ARG134, LYS157, and LYS158 is shown by blue.
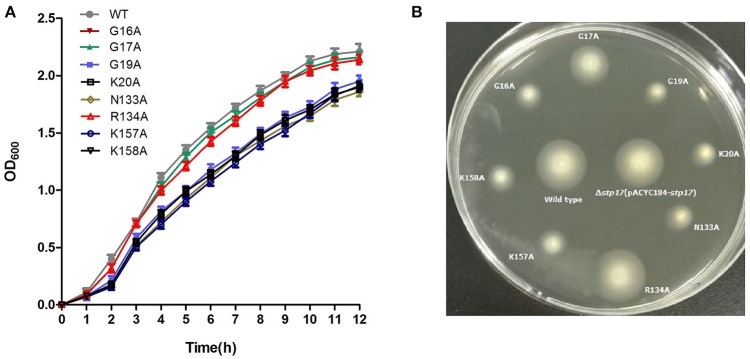
Identification of the active site residues of STP17
To define the active site of STP17 activity, eight mutants of site-directed mutagenesis were generated, and their growth and motility were compared to the wild type strain. As shown in Figure 6, when the residues of GLY16, GLY17, GLY19, LYS20, ASN133, ARG134, LYS157, and LYS158 mentioned above were mutated, the growth and motility of these mutant strains Δstp17::pACYC184-stp17G16A, Δstp17::pACYC184-stp17G19A,_Δstp17::pACYC184- stp17K20A,_ Δstp17::pACYC184- stp17N133A,_Δstp17::pACYC184- stp17K157A, and Δstp17::pACYC184-stp17K158A were significantly decreased compared with that of wild type strain, while there was no obviously change in growth and motility of mutant strains Δstp17::pACYC184-stp17G17A and Δstp17::pACYC184-stp17R134A. In addition, the stability of wild type strain and above mutants shows no significant difference (data not shown). All these results suggest that the amino acid residues of GLY16, GLY19, LYS20, ASN133, LYS157, and LYS158 may be the active site residues of STP17.
Figure 6.
Identification of the active site residues of STP17. (A) Growth cure of site-directed mutants of STP17. (B) Motility comparison of site-directed mutants of STP17. G16A, G17A, G19A, K20A, N133A, R134A, K157A, and K158A represent eight mutants through site-directed mutagenesis.
Analysis of ATPase activity of STP17
To identify the potential ATPase activity of STP17, we investigated whether the ATP concentration in solution could be decreased in the presence of STP17. As shown in Figure 7, the concentration of ATP in solution decreased markedly after incubation with STP17 for 10 min and continued to decrease after incubation for 30- and 50-min, while the concentration of ATP in solution containing the control protein BSA which lacks the ATPase activity did not decreased. The reduction in ATP concentration indicates catalytic activity of STP17, suggesting that STP17 can catalyze ATP into ADP and free phosphate.
Figure 7.
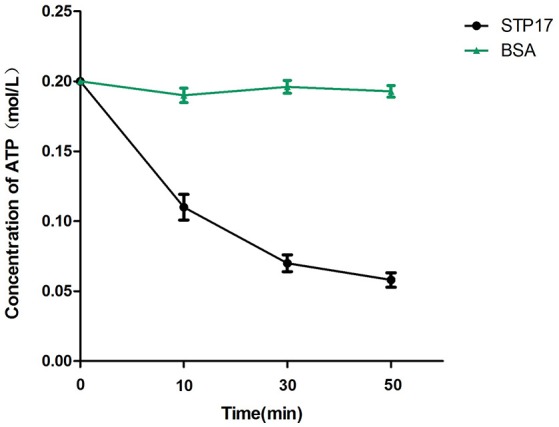
Analysis of ATP reduction by STP17. BSA (Bovine Serum Albumin), which lacks the ATPase activity, was used as a negative control.
Discussion
Plasmids are extra chromosomal, self-replicating genetic elements with additional functions, such as antibiotic resistance, that are complementary to chromosomal DNA. In many cases, bacterial plasmid DNA is circular. In 1979, the first linear plasmid in a prokaryote was found in Streptomyces rochei (Hayakawa et al., 1979). Subsequently, another kind of linear plasmid was also found in Borrelia (Plasterk et al., 1985). However, most linear plasmids have been found in about a dozen of Streptomyces, and their molecular size is between 12 and 640 kb (Hinnebusch and Tilly, 1993; Zhong et al., 2010). In 2007, Baker et al. reported an approximately 27 kb linear plasmid named pBSSB1 in the S. Typhi z66-positive strain and indicated this plasmid mediates unidirectional flagellar phase variation (Baker et al., 2007a,b). The pBSSB1 plasmid is the first non-bacteriophage-related linear plasmid found in Enterobacteriaceae (Baker et al., 2007a). It was found that the factor for inversion stimulation (Fis) encoded by the fis gene can affect the stability of pBSSB1 in S. Typhi (Zhang et al., 2012b). There are 33 putative ORFs on pBSSB1 including the operon fljBAz66, which encodes the phase-2 flagellin FljBZ66 and the repressor FljAZ66 of the phase-1 flagellin gene fliC, respectively (Huang et al., 2004; Baker et al., 2007a,b; Zou et al., 2009). There is also a notion that most of these genes encoded on pBSSB1, except the fljBAz66, are pseudogenes (Simon et al., 1980).
In this study, the gene stp17 was cloned and expressed successfully in E. coli. The purified STP17His6 protein was gained and the polyclonal anti-STP17 antibody was prepared in rabbit. Then, the anti-STP17 antibody was used to investigate whether the stp17 gene is expressed at the protein level through western blot analysis. The results showed that STP17 was expressed well in the wild type strain of S. Typhi. Gene expression of stp17 has already been verified by qRT-PCR and Northern-blot (Zhao et al., 2014). Therefore, the gene stp17 is not a pseudogene and can be expressed at both the mRNA and protein levels.
In 2014, we investigated the transcriptional expression of several genes located on pBSSB1 in different growth phases and environmental stresses and found that the expression of stp17 increases with the OD600 value in log phase and is not influenced by acidic stress, oxidative stress or osmotic stress (Zhao et al., 2014). Here, the translational expression level of stp17 was studied by western blot analysis. It was found that the expression level of STP17 increased with the OD600 value throughout log phase and reached a peak value at an OD600 of 1.2, then decreased in stationary phase. The expression profile of stp17 at the mRNA and protein level under different growth phases is very similar and shows that S. Typhi needs more STP17 when cells are in the growth period. When stp17 was deleted, growth of the Δstp17 strain was obviously slower than the wild type strain. Moreover, cell growth under stp17 over-expression was faster than the control strain. Therefore, STP17 may be involved in promoting bacterial growth.
The DNA sequence of pBSSB1 shows that the stp17 gene is supposed to encode a putative nucleotide binding protein and it is immediately adjacent to the possible replication origin of pBSSB1 (Baker et al., 2007a). In addition, stp17 is predicted to encode a protein containing a conserved domain, which is shared by the ParA protein. ParA is involved in the segregation of plasmids and bacterial chromosomal DNA (Motallebi-Veshareh et al., 1990; Volante and Alonso, 2015). Therefore, stp17 is suggested to be involved in the replication of pBSSB1. Although, it was shown that the stability is not affected by stp17 in this study, future studies are required to determine whether stp17 is involved in replication or segregation of pBSSB1.
The amino acid sequence of STP17 was analyzed in this study and an ATPase domain was predicted. The predicted activity of STP17 was also identified in this study. Moreover, ParA, which shares a conserved domain with STP17, was reported to possess the sequence of a conserved and widespread family of ATPases (Motallebi-Veshareh et al., 1990; Bignell and Thomas, 2001). ATPases is very important to bacterial motility (Bai et al., 2014; Lin et al., 2015). In bacteria, a specific protein export apparatus, which utilizes ATP and proton motive force as the energy source to transport component proteins to the distal growing end, is crucial for self-assembly of the bacterial flagellum (Erhardt et al., 2014; Minamino et al., 2014). It was reported that bacterial motility was obviously reduced due to infrequent ATP hydrolysis caused by mutation of the FliI6-FliJ complex, which is similar to the FOF1-ATPase (Imada et al., 2016; Minamino et al., 2016). It was shown that the motility of Δstp17 was significantly decreased compared to the wild type strain in this study. One explanation for this phenotype is that STP17 could utilize ATP and proton motive force as the energy source through its ATPase activity to help bacterial motility.
In summary, this study is the first to show that the stp17 gene is not a pseudogene and is expressed well at the protein level. Furthermore, our results show that stp17 plays an important role in promoting the growth and motility of bacteria.
Author contributions
Conceived and designed the experiments: HZ, XH, HX. Performed the experiments: HZ, YuZ, XX, MW, YiZ, MG, BN. Analyzed the data: HZ, YuZ, HD, SX. Wrote the manuscript: HZ, YuZ, XH.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31000076, 81371780, 81572032, 81401636, 31300122), the Natural Science Foundation for Colleges and Universities in Jiangsu Province (16KJB320006), the Startup Fund of Soochow University (SDY2015B07), and the Science and Technology Program of Suzhou (SS201638, SYS201551).
References
- Bai F., Morimoto Y. V., Yoshimura S. D., Hara N., Kami-Ike N., Namba K., et al. (2014). Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci. Rep. 4:6528. 10.1038/srep06528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Hardy J., Sanderson K. E., Quail M., Goodhead I., Kingsley R. A., et al. (2007a). A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 3:e59. 10.1371/journal.ppat.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Holt K., van de Vosse E., Roumagnac P., Whitehead S., King E., et al. (2008). High-throughput genotyping of Salmonella enterica serovar Typhi allowing geographical assignment of haplotypes and pathotypes within an urban District of Jakarta, Indonesia. J. Clin. Microbiol. 46, 1741–1746. 10.1128/JCM.02249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Holt K., Whitehead S., Goodhead I., Perkins T., Stocker B., et al. (2007b). A linear plasmid truncation induces unidirectional flagellar phase change in H:z66 positive Salmonella Typhi. Mol. Microbiol. 66, 1207–1218. 10.1111/j.1365-2958.2007.05995.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell C., Thomas C. M. (2001). The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91, 1–34. 10.1016/S0168-1656(01)00293-0 [DOI] [PubMed] [Google Scholar]
- Cai W., Wannemuehler Y., Dell'anna G., Nicholson B., Barbieri N. L., Kariyawasam S., et al. (2013). A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundanthost metabolites. PLoS Pathog. 9:e1003428. 10.1371/journal.ppat.1003428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mansi M., Anderson K. J., Inche C. A., Knowles L. K., Platt D. J. (2000). Isolation and curing of the Klebsiella pneumoniae large indigenous plasmid using sodium dodecyl sulphate. Res Microbiol. 151, 201–208. 10.1016/S0923-2508(00)00140-6 [DOI] [PubMed] [Google Scholar]
- Erhardt M., Mertens M. E., Fabiani F. D., Hughes K. T. (2014). ATPase-independent type-III protein secretion in Salmonella enterica. PLoS Genet. 10:e1004800. 10.1371/journal.pgen.1004800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everest P., Wain J., Roberts M., Rook G., Dougan G. (2001). The molecular mechanisms of severe typhoid fever. Trends Microbiol. 9, 316–320. 10.1016/S0966-842X(01)02067-4 [DOI] [PubMed] [Google Scholar]
- Guinee P. A., Jansen W. H., Maas H. M., Le Minor L., Beaud R. (1981). An unusual H antigen (Z66) in strains of Salmonella typhi. Ann. Microbiol. (Paris) 132, 331–334. [PubMed] [Google Scholar]
- Hatta M., Sultan A. R., Pastoor R., Smits H. L. (2011). New flagellin gene for Salmonella enterica serovar Typhi from the East Indonesian archipelago. Am. J. Trop. Med. Hyg. 84, 429–434. 10.4269/ajtmh.2011.10-0605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T., Otake N., Yonehara H., Tanaka T., Sakaguchi K. (1979). Isolation and characterization of plasmids from Streptomyces. J Antibiot. 32, 1348–1350. [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Owen P., Nataro J. P. (1999). Molecular switches–the ON and OFF of bacterial phase variation. Mol. Microbiol. 33, 919–932. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J., Tilly K. (1993). Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 10, 917–922. [DOI] [PubMed] [Google Scholar]
- Huang X., Phung L. V., Dejsirilert S., Tishyadhigama P., Li Y., Liu H., et al. (2004). Cloning and characterization of the gene encoding the z66 antigen of Salmonella enterica serovar Typhi. FEMS Microbiol. Lett. 234, 239–246. 10.1111/j.1574-6968.2004.tb09539.x [DOI] [PubMed] [Google Scholar]
- Imada K., Minamino T., Uchida Y., Kinoshita M., Namba K. (2016). Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator. Proc. Natl. Acad. Sci. U.S.A. 113, 3633–3638. 10.1073/pnas.1524025113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J., Van Aart C. J., Grassly N. C. (2016). The burden oftyphoidand paratyphoid in India: systematic review and meta-analysis. PLoS Negl. Trop. Dis. 10:e0004616. 10.1371/journal.pntd.0004616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Gao L., Zhao X., Liu J., Norris S. J. (2015). Mutations in the Borrelia burgdorferi flagellar type III secretion system genes fliH and fliI profoundly affect spirochete flagellar assembly, morphology, motility, structure, and cell division. MBio 6, e00579–e00515. 10.1128/mBio.00579-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Morimoto Y. V., Hara N., Aldridge P. D., Namba K. (2016). The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12:e1005495. 10.1371/journal.ppat.1005495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Morimoto Y. V., Kinoshita M., Aldridge P. D., Namba K. (2014). The bacterial flagellar protein export apparatus processively transports flagellar proteins even with extremely infrequent ATP hydrolysis. Sci. Rep. 4:7579. 10.1038/srep07579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motallebi-Veshareh M., Rouch D. A., Thomas C. M. (1990). A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 9, 1455–1463. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Simon M. I., Barbour A. G. (1985). Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature 318, 257–263. [DOI] [PubMed] [Google Scholar]
- Simon M., Zieg J., Silverman M., Mandel G., Doolittle R. (1980). Phase variation: evolution of a controlling element. Science 209, 1370–1374. [DOI] [PubMed] [Google Scholar]
- Volante A., Alonso J. C. (2015). Molecular anatomy of ParA-ParA and ParA-ParB interactions during plasmid partitioning. J. Biol. Chem. 290, 18782–18795. 10.1074/jbc.M115.649632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain J., Hendriksen R. S., Mikoleit M. L., Keddy K. H., Ochiai R. L. (2015). Typhoid fever. Lancet 385, 1136–1145. 10.1016/S0140-6736(13)62708-7 [DOI] [PubMed] [Google Scholar]
- Winter S. E., Winter M. G., Poon V., Keestra A. M., Sterzenbach T., Faber F., et al. (2014). Salmonella enterica serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog. 10:e1004207. 10.1371/journal.ppat.1004207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Zhang H., Sheng X., Xu H., Huang X. (2008). Transcriptional expression of fljB:z66, a flagellin gene located on a novel linear plasmid of Salmonella enterica serovar Typhi under environmental stresses. New Microbiol. 31, 241–247. [PubMed] [Google Scholar]
- Xu S., Zou X., Sheng X., Zhang H., Mao L., Du H., et al. (2010). Expression of fljB:z66 on a novel linear plasmid of Salmonella enterica serovar Typhi is dependent on FliA and FlhDC and regulated by OmpR. Braz. J. Microbiol. 41, 729–740. 10.1590/S1517-83822010000300025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. P., Ho W. S., Gan H. M., Chai L. C., Thong K. L. (2016). Global MLST of Salmonella Typhi revisited in post-genomic era: genetic conservation, population structure, and comparative genomics of rare sequence types. Front. Microbiol. 7:270. 10.3389/fmicb.2016.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Du H., Ji X., Ni B., Mao L., Xu S., et al. (2012a). OmpR may regulate the putative YehU/YehT two-component system in Salmonella enterica serovar Typhi under hypotonic growth condition. Curr. Microbiol. 64, 283–289. 10.1007/s00284-011-0066-3 [DOI] [PubMed] [Google Scholar]
- Zhang H., Ni B., Zhao X., Dadzie I., Du H., Wang Q., et al. (2012b). Fis is essential for the stability of linear plasmid pBSSB1 and affects the motility of Salmonella enterica serovar Typhi. PLoS ONE 7:e37462. 10.1371/journal.pone.0037462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Zhu Y., Zhang H., Du H., Ni B., Huang X. (2014). Transcriptional expression of 6 genes located on pBSSB1 of Salmonella enterica serovar Typhi in different growth phases and environmental stresses. Curr. Microbiol. 3, 252–257. 10.1007/s00284-014-0577-9 [DOI] [PubMed] [Google Scholar]
- Zhong L., Cheng Q., Tian X., Zhao L., Qin Z. (2010). Characterization of the replication, transfer, and plasmid/lytic phage cycle of the Streptomyces plasmidphage pZL12. J. Bacteriol. 192, 3747–3754. 10.1128/JB.00123-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Huang X., Xu S., Zhou L., Sheng X., Zhang H., et al. (2009). Identification of fljA located on a linear plasmid as a repressor gene of fliC in Salmonella enterica serovar Typhi. Microbiol. Immunol. 53, 191–197. 10.1111/j.1348-0421.2009.00106.x [DOI] [PubMed] [Google Scholar]