Abstract
The study of Ebola virus (EBOV) pathogenesis in vivo has been limited to nonhuman primate models or use of an adapted virus to cause disease in rodent models. Herein we describe wild-type EBOV (Makona variant) infection of mice engrafted with human hematopoietic CD34+ stem cells (Hu-NSG™-SGM3 mice; hereafter referred to as SGM3 HuMice). SGM3 HuMice support increased development of myeloid immune cells, which are primary EBOV targets. In SGM3 HuMice, EBOV replicated to high levels, and disease was observed following either intraperitoneal or intramuscular inoculation. Despite the high levels of viral antigen and inflammatory cell infiltration in the liver, the characteristic histopathology of Ebola virus disease was not observed, and this absence of severe immunopathology may have contributed to the recovery and survival of some of the animals. Future investigations into the underlying mechanisms of the atypical disease presentation in SGM3 HuMice will provide additional insights into the immunopathogenesis of severe EBOV disease.
Keywords: Ebola, virus, hemorrhagic fever, humanized NSG-SGM3 mice, myeloid cell, immune response, immunopathology, flow cytometry
Ebola virus (EBOV; species Zaire ebolavirus; family Filoviridae) causes EBOV disease (EVD) in humans, sometimes involving a severe hemorrhagic syndrome. The 2013–2016 EBOV (Makona variant) epidemic in West Africa has caused thousands of cases and deaths [1]. The human immune system's contribution to EVD remains unclear and is based on investigations of scarce samples from EVD outbreaks and on in vitro infection of cell culture–derived monocytes, macrophages, and dendritic cells (DC) [2–5]. Nonhuman primates (NHPs) are an important pathogenesis model, but present several limitations, including sensitivity to disease; difficulties in using large numbers of animals for serial euthanasia experiments; and important differences in the innate and adaptive immune responses compared to humans [6–8]. Rodent models are even less desirable than NHPs for pathogenesis studies, as the rodent immune response and target cell populations differ greatly from human ones [9, 10]. Furthermore, rodent disease models require use of serially adapted virus, which may not accurately represent mechanisms of wild-type virus [11].
Immunodeficient mice reconstituted with a human immune system (HuMice) are unique in vivo models useful for studying interactions between pathogens and human immune system components. Commonly, HuMice are derived by engrafting human hematopoietic stem cells (HSC) into a NOD.Cg-Prkdcscid Il2rgtm1Wil (NOD-scid Il2rγnull or NSG) mouse background [12, 13]. The NOD background has diminished DC and macrophage function, and the scid mutation prevents development of B and T cells [14]. A targeted mutation of the IL-2 receptor common gamma chain (IL2rgnull) blocks high-affinity signaling by several cytokine receptors (IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21), and prevents the development of functional murine natural killer (NK) cells, allowing increased engraftment of functional human immune cells. However, following engraftment with human stem cells, the human myeloid compartment fails to fully reconstitute due to a lack of human-specific cytokine signaling.
A newly characterized HuMouse model has increased development of myeloid cells, which are the target cells for filovirus infection [15–18]. Hu-NSG™-SGM3 mice (henceforth referred to as SGM3 HuMice) are derived from NSG mice that express human stem cell factor, granulocyte/monocyte colony-stimulating factor, and IL-3 transgenes to facilitate myeloid cell homing and development. SGM3 mice are then engrafted with human CD34+ stem cells after conditioning with a sub-lethal dose of irradiation. SGM3 HuMice support human HSC engraftment, leading to the development of high levels of human myeloid cells, T cells, and B cells [19, 20].
To date, wild-type EBOV infection has been characterized in 2 NSG-derived humanized mouse models: NSG mice expressing the human HLA-A2.1 major histocompatibility complex (MHC) class I molecule (Hu-NSG-A2) engrafted with HSC, and NSG mice engrafted with human fetal liver, thymus, and HSC (Hu-NSG-BLT) [21, 22]. Given the importance of the immune response in EBOV pathogenesis, herein we longitudinally characterize infection, human immune cell responses, and pathologic changes in SGM3 HuMice following intraperitoneal or intramuscular inoculation with EBOV-Makona.
MATERIALS AND METHODS
Biosafety
All work with infectious virus or infected animals was conducted in a biosafety level 4 (BSL-4) laboratory at Rocky Mountain Laboratories (Hamilton, Montana) and adhered to established BSL-4 standard operating procedures approved by the Institutional Biosafety Committee.
Ethics Statement
All animal experiments were approved by the Institutional Animal Care and Use Committee of Rocky Mountain Laboratories and were performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) by certified staff in an AAALAC-approved facility.
Humanized Mice
Two cohorts of SGM3 HuMice (NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg[CMV-IL3,CSF2,KITLG]1Eav/MloySzJ; stock no. 013062) engrafted with CD34+ human cells (stock no. 701362) derived from 2 separate donors were obtained from Jackson Laboratories (Sacramento, CA). Data are from 62 animals: 36 for serial sampling studies and 26 for survival studies. All animals were housed in a climate-controlled laboratory with a 12-hour day/12-hour night cycle. All animals were given sterile water, bedding, and food and were group housed in an isolator-caging system (Innovive, San Diego, California). HuMice were provided commercially available mouse chow, DietGel Boost (ClearH2O), and water ad libitum. Animals were humanely euthanized with isoflurane vapors at the indicated time points or when clinical illness scores based on weight loss, piloerection, neurological signs, changes in mentation, ataxia, dehydration, or dyspnea indicated that the animal was in distress or in the terminal stages of disease.
Virus Inoculation
SGM3 HuMice (20–21 weeks after engraftment) were sham injected with sterile Dulbecco's modified Eagle's medium (DMEM) or inoculated with 10 or 103 focus-forming units (FFU) of EBOV Makona C07 [23, 24] (Ebola virus/H.sapiens-tc/GIN/2014/Makona; first passage after isolation) diluted in DMEM; inoculation was done either intraperitoneally or intramuscularly in the hind limbs.
Quantitative Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
RNA was extracted from blood and homogenized tissue samples, using the QIAamp Viral RNA Mini Kit or RNeasy Mini Kit, respectively (Qiagen, Carlsbad, California) following the manufacturer's instructions and according to established protocol [25]. RNA was quantitated using a 1-step real-time RT-PCR targeting the nucleoprotein gene with a Rotor-Gene probe kit (Qiagen) according to manufacturer's instructions (primer and probe sequences are available on request). In each run, standard dilutions of RNA extracted from a titrated virus stock were analyzed in parallel to calculate FFU equivalents in the samples.
Flow Cytometry
Intracardiac blood samples were collected in ethylenediaminetetraacetic acid (EDTA)–coated tubes. Red blood cells were lysed with RBC lysis buffer (BioLegend, San Diego); remaining cells were washed in Dulbecco's phosphate-buffered saline (PBS) and enumerated. Spleens were weighed, macerated through a 70-µM filter, and washed in R10 medium with benzonase (Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 1% penicillin-streptomycin, β-mercaptoethanol, and 25 U/mL benzonase). Red blood cells in the spleen samples were lysed with ACK lysis buffer (Gibco, Grand Island, New York), and the remaining intact cells were washed in R10 medium and enumerated. Approximately 1 × 106–2 × 106 cells were stained in fluorescence-activated cell-sorting (FACS) buffer (PBS, 2% FBS, and 2 mM EDTA) for 15 minutes at 4°C after blocking Fc receptors with Fc Block (BD Biosciences, San Jose, California). Stains included CD3-V450, CD8-APC-eFluor780, CD14-PE/Cy7, CD33-PE, CD45-V500 or eFluor450 mouse CD45-PE/Cy7, CD56-AF700, CD69-PerCP/Cy5.5, CD80-PE, CD11c-APC, CD123-BV510, HLA-DR-A700, and lineage-FITC. All flow cytometry samples were fixed with 2% paraformaldehyde and evaluated on a FACSCanto II (BD Biosciences) cytometer in the BSL-4 laboratory. Data were acquired using FACSDiva software (BD Biosciences) and analyzed using FlowJo, version 10 (Tree Star).
Histologic and Immunohistochemistry (IHC) Analyses
Tissues were fixed in 2 changes of 10% neutral buffered formalin for at least 7 days [25]. Tissues were placed in cassettes and processed with Sakura VIP-5 Tissue Tek (Sakura Finetek, Torrance, California) on a 12-hour automated schedule, using a graded series of ethanol, xylene, and ParaPlast Extra. Embedded tissues were sectioned into 5 µm and dried overnight at 42°C before staining. Specific anti-EBOV immunoreactivity was detected using polyclonal rabbit serum against EBOV VP40 diluted 1:2000 as described elsewhere [26]. The secondary antibody was a Biogenex biotinylated anti-rabbit link (Fremont, California). The tissues were then processed for IHC analysis, using the BenchMark Ultra stainer with a RedMap (Ventana Medical Systems, Tuscon, Arizona) kit.
Statistics
Flow cytometric data were analyzed using a 1-way analysis of variance with a Tukey post hoc test.
RESULTS
EBOV-Makona Infection of SGM3 HuMice
To test the susceptibility of SGM3 HuMice to infection with wild-type (WT) EBOV, we intraperitoneally inoculated SGM3 HuMice generated from 2 separate donors (DR1 and DR2) with 103 FFU of EBOV-Makona. Inoculating mice with mouse-adapted EBOV-Mayinga (MA-EBOV) intraperitoneally causes disease, while peripheral inoculation does not cause disease and even protects BALB/c mice against subsequent intraperitoneal challenge with homologous virus [11]. Thus, we also tested peripheral inoculation (intramuscularly) of WT-EBOV in SGM3 HuMice. Animals inoculated intraperitoneally or intramuscularly with 103 FFU of WT-EBOV exhibited weight loss (>10%) at approximately 5 days after inoculation and 7 days after inoculation, respectively (Figure 1A and 1B). Intraperitoneal inoculation resulted in severe disease requiring euthanasia due to weight loss in 3 of 6 animals (50.0%; 1 from DR1 and 2 from DR2). Intramuscular inoculation resulted in severe disease in 4 of 6 animals (66.7%, 2 from each donor; Figure 1C). Overall clinical outcomes in animals from each donor cohort were consistent for both inoculation routes. Additional groups of HuMice from DR1 were inoculated with 10 FFU either intraperitoneally or intramuscularly. Two of 3 animals inoculated intramuscularly required euthanasia, but intraperitoneally inoculated animals showed no signs of disease (Figure 1A–1C).
Figure 1.
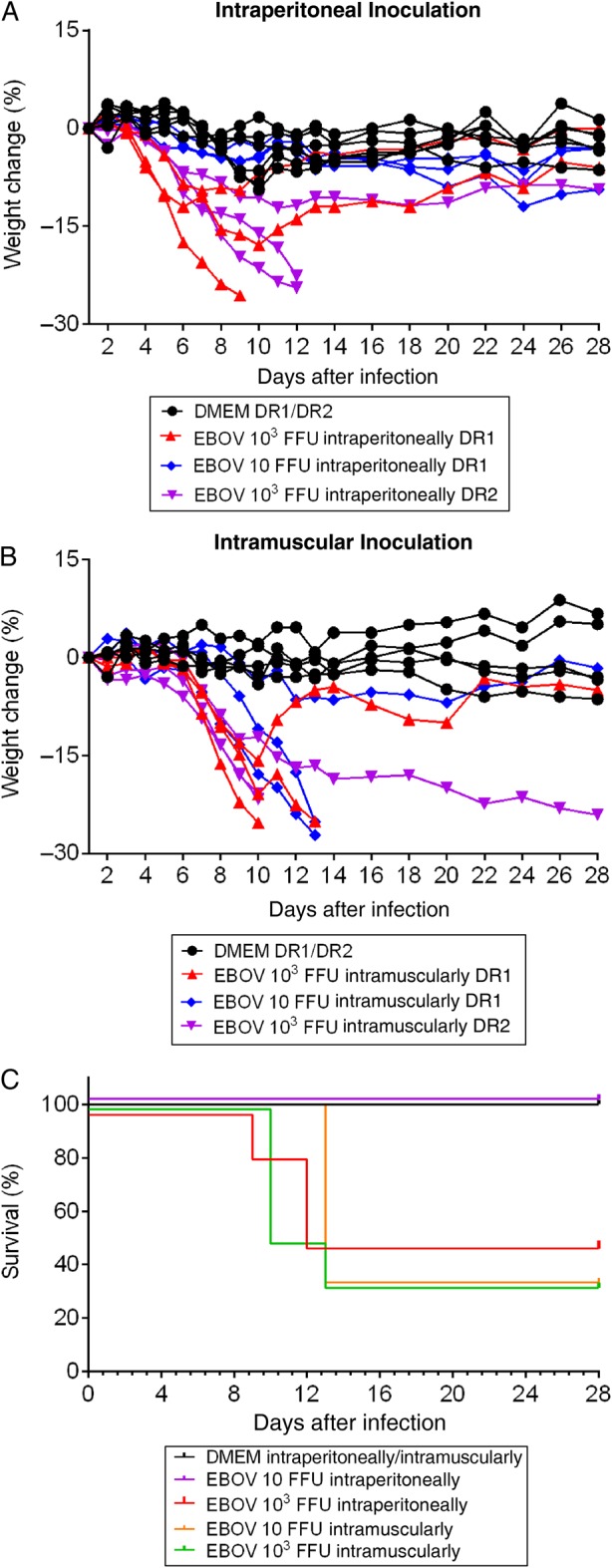
Weight loss and survival of SGM3 HuMice inoculated with Ebola virus (EBOV) Makona. A and B, Weight change in humanized mice with CD34+ cells from donor 1 (DR1) and donor 2 (DR2) inoculated intraperitoneally (A) or intramuscularly (B) with Dulbecco's modified Eagle's medium (DMEM; mock) or EBOV Makona at indicated doses. Data are presented on indicated days after infection. C, Survival in HuMice inoculated intraperitoneally or intramuscularly with DMEM (DR1 and DR2; n = 8) or with 103 focus-forming units (FFU) intraperitoneally (DR1 and DR2; n = 6), 103 FFU intramuscularly (DR1 and DR2; n = 6), 10 FFU intraperitoneally (DR1; n = 3), or 10 FFU intramuscularly (DR1; n = 3) of EBOV Makona.
High-Level EBOV Replication in SGM3 HuMice
Viral RNA levels were determined in the animals investigated above; animals were designated as terminal animals (based on end point criteria) or as survivors (study completion, 28 days after inoculation). Additionally, viral RNA was assessed in serially euthanized animals from the 2 donor cohorts previously described in survival studies. Groups of 3 animals that were mock inoculated or inoculated intramuscularly were serially euthanized on 1, 3, 5, and 8 days after inoculation; EBOV intraperitoneally inoculated animals were euthanized 1 and 3 days after inoculation only. Animals from DR1 inoculated intraperitoneally or intramuscularly had samples obtained 1 and 3 days after inoculation (103 FFU only) and at terminal or survival time points (103 FFU and 10 FFU). Based on the clinical course observed in DR1 studies, in addition to terminal and survival time points, serial studies on animals from DR2 focused on later time points (5 and 8 days after inoculation) and were limited to high-dose EBOV (103 FFU) administered intramuscularly.
Blood, spleen, and liver samples were collected for qRT-PCR analysis to determine viral RNA loads in these animals (Figure 2A–2C). At 3 days after inoculation, EBOV RNA levels in blood were higher in intraperitoneally inoculated DR1 HuMice than in intramuscularly inoculated mice from the same donor. However, blood RNA levels were comparable in all terminal animals, regardless of route, and slightly higher median levels were found in intramuscularly inoculated HuMice. Similar to blood, higher levels of viral RNA were seen in spleens and livers of intraperitoneally inoculated DR1 animals at early time points, but terminal DR1 and DR2 samples were comparable. The highest levels of viral RNA were detected in DR2 samples 8 days after inoculation, only slightly higher than in terminal animals. Surviving animals (DR1 and DR2) had spleen and liver RNA levels equal to or higher than those of terminal HuMice. Animals inoculated with 10 FFU only underwent sampling at terminal or survival time points and had viral RNA levels comparable to those in animals inoculated with 103 FFU. Viral RNA load was not associated with outcome in this model; 28 days after inoculation, survivors inoculated either intramuscularly or intraperitoneally had viral RNA levels equally high as those of animals that were euthanized earlier in the course of infection due to severe disease.
Figure 2.
Ebola virus (EBOV) RNA loads in the blood and tissues of SGM3 HuMice. RNA quantitation in blood (A), spleen (B), and liver (C) samples collected at the indicated time points from HuMice inoculated with the indicated doses of EBOV Makona. Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) was specific for the EBOV nucleoprotein (NP) gene, and focus-forming unit (FFU) equivalents were based on standards generated from stock virus. Samples collected 1 and 3 days after infection are from DR1; samples collected 5 and 8 days after infection are from intramuscularly infected DR2 animals only; no intraperitoneally infected animals were analyzed at these time points. Terminal and survivor samples are from both donors and are from animals in Figure 1 that were euthanized because of disease (terminal) or at 28 days after infection (survivor).
EBOV-Associated Histopathologic and Immunohistochemistry Analyses in SGM3 HuMice
Hematoxylin and eosin (HE) staining and IHC analyses were performed on serially euthanized intramuscularly inoculated animals (103 FFU; DR1 and DR2) to assess pathologic changes and antigen distribution. The uninfected control HuMice had mild-to-moderate portal and random hepatitis characterized by infiltration of the portal tract by small-to-moderate numbers of human macrophages, lymphocytes, plasma cells, and occasional multinucleated giant cells. Many of the macrophages and multinucleated cells contained a brown granular pigment within the cytoplasm. These inflammatory cells also formed small foci scattered randomly throughout the hepatic parenchyma. Rarely, the inflammation was more granulomatous, forming foci with a center of necrotic hepatocytes admixed with small numbers of neutrophils and a border of macrophages, lymphocytes, plasma cells, and some multinucleated giant cells.
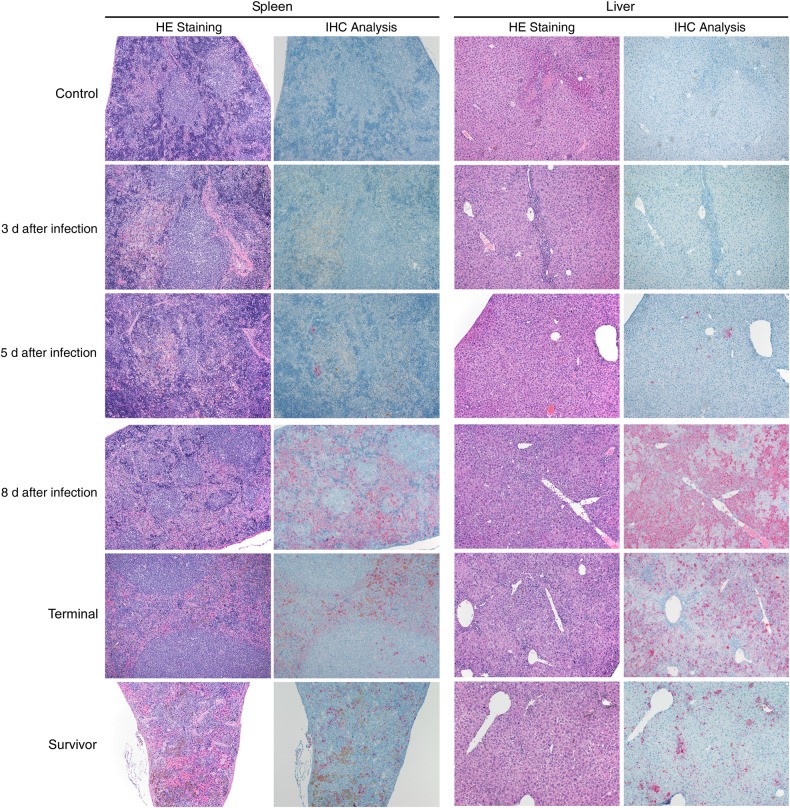
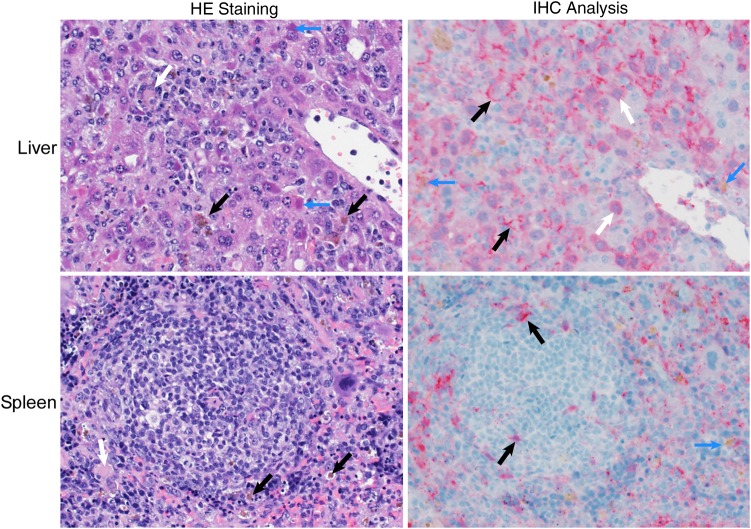
Infected HuMice demonstrated similar background hepatic lesions; however, the number of inflammatory cells within the portal tracts and parenchyma increased from 5 days after inoculation until 8 days after inoculation (Figure 3). In addition, by 8 days after inoculation, the majority of hepatocytes were swollen by small, discreet vacuoles (vacuolar degeneration, lipid type), and many hepatocytes contained eosinophilic intracytoplasmic inclusion bodies (Figures 3 and 4). HuMice in the terminal group were essentially indistinguishable from HuMice euthanized 8 days after inoculation. IHC analysis showed viral antigen in small numbers of macrophages and hepatocytes at 5 days after inoculation. By 8 days after inoculation, most hepatocytes and numerous macrophages and endothelial cells were positive for EBOV antigen. HuMice in the terminal group and the group euthanized 8 days after inoculation displayed a similar distribution of viral antigen, whereas survivors had much lower distribution of EBOV antigen within the liver.
Figure 3.
Histopathologic changes and antigen staining of spleen and liver serial samples in SGM3 HuMice inoculated with Ebola virus (EBOV). Original magnification ×100 of spleen and liver samples stained with hematoxylin-eosin (HE) or an EBOV-specific anti-VP40 antibody to assess pathologic changes and antigen distribution in animals inoculated intramuscularly (103 focus-forming units). Control animals had a baseline low level of macrophage infiltration (top row). By 5 days after infection, antigen was apparent in both spleen and liver, and by 8 days after infection, extensive macrophage infiltration appears in both the liver and spleen of infected animals. Antigen was present in the red pulp of the spleen and in most hepatocytes of the liver. Terminal samples were similar to samples collected 8 days after infection. In survivors (28 days after infection; intramuscularly or intraperitoneally inoculated), antigen was less abundant and infiltrating macrophages less apparent in both tissue types (bottom row). Abbreviation: IHC, immunohistochemistry analysis.
Figure 4.
High magnification of histopathologic changes and antigen staining in the livers of SGM3 HuMice inoculated with Ebola virus (EBOV). Tissues are stained as described in Figure 3; panels are representative of SGM3 HuMouse samples obtained 8 days after infection, and images are 400× original magnification. The hepatic architecture was disrupted by infiltrating macrophages, many of which contained a brown granular pigment (by hematoxylin-eosin [HE] staining; black arrows), and by lymphocytes and multinucleate giant cells (by HE staining; white arrows). Numerous hepatocytes contained eosinophilic intracytoplasmic inclusions (by HE staining; blue arrows). Viral antigen was detected in hepatocytes (by immunohistochemistry [IHC] analysis; white arrows) and macrophages (by IHC analysis; blue arrows) and was lining the space of Disse (by IHC analysis; black arrows). In the spleen, the red pulp contained numerous macrophages, many with a brown granular pigment (by HE staining; black arrows). Viral antigen was primarily found within macrophages (by IHC analysis; blue arrow) and extracellularly in the red pulp. Small numbers of cells morphologically consistent with dendritic cells (by IHC analysis; black arrows) were positive for viral antigen within the lymphoid follicular center.
The red pulp of the spleen in control HuMice contained small-to-moderate numbers of macrophages and occasional multinucleated giant macrophages containing the previously described pigment (Figure 3). The animals euthanized 1, 3, and 5 days after inoculation had similar histologic lesions, but by 8 days after inoculation, both macrophages and brown pigment increased within the red pulp (Figures 3 and 4). The IHC results were similar to those seen in the liver, with small amounts of viral antigen observed 5 days after inoculation in macrophages in the red pulp and in cells morphologically consistent with DC and macrophages within the white pulp. By 8 days after inoculation, viral antigen was present in most of the red pulp. The histologic and IHC findings of the terminal group were similar to those of the group euthanized 8 days after inoculation, whereas the survivor group had fewer macrophages within the red pulp and less viral antigen. The colon, brain, and ovaries were examined histologically, but IHC was not performed; no notable findings were observed in these tissues in control or infected animals at any time points (data not shown).
Levels of Human Myeloid and Lymphoid Cells in HuMice
Cells of the myeloid lineage, including macrophages and myeloid DC (mDC), are early targets of EBOV infection and replication and likely play key roles in viral dissemination [16, 17, 27]. Normal mice are refractory to WT-EBOV-induced disease, but HuMice developed disease. Thus, we analyzed several cell populations in naive SGM3 HuMice to investigate the availability of putative human target cells, particularly monocytes, macrophages, and DC.
First, we determined the percentages of human- and mouse-derived leukocytes in the spleen, using species-specific CD45 markers (Figure 5A). Approximately 9% of total splenocytes were positive for murine CD45 in naive HuMice, whereas 67% were positive for human leukocytes (hCD45+), indicating a low level of residual murine cells, which were presumably myeloid cells as NSG mice are genetically devoid of lymphocytes (Figure 5A). We then assessed additional human immune cell populations in the blood and spleens of naive animals (Figure 5B and 5C, respectively). In the blood, approximately 63% of hCD45+ cells were T cells (CD3+) and 20% expressed CD8. In the spleen, approximately 36% of hCD45+ cells were T cells (CD3+) and 27% expressed CD8. The majority of the remaining T cells were presumably CD4+ T cells. In blood and spleen hCD45+ populations, monocytes (CD14+) were present (blood, 1.4%; spleen, 0.4%); mDC (Lin−/HLA-DRhi/CD11c+) were present (blood, 0.3%; spleen, 0.4%); and NK cells (CD3−/CD56+) were present at low levels (blood, 1.9%; spleen, 2.6%) and appeared abnormal. Plasmacytoid DC (pDC; Lin−/HLA-DRhi/CD123+) were not detected.
Figure 5.
Flow cytometric assessment of the human immune system development in naive SGM3 HuMice. A, Splenocytes from naive animals (20 weeks after engraftment) stained with mouse- or human-specific CD45 to determine reconstitution and residual mouse leukocyte levels. B and C, Blood cell (B) and splenocyte (C) frequencies in naive animals were determined following staining with the indicated immune cell markers. Panels are representative of 3 animals. Abbreviations: HLA-DR, human leukocyte antigen–D related, major histocompatibility complex class II antigen; mDC, myeloid dendritic cells; NK, natural killer; pDC, plasmacytoid dendritic cells.
Characterization of Myeloid and Lymphoid Cell Responses in EBOV-Infected HuMice
We performed multicolor flow cytometry on blood and splenic lymphocytes from intramuscularly inoculated HuMice (103 FFU) to assess the immune response to EBOV infection. In the blood, leukocyte (hCD45+) frequencies were initially higher in infected animals than in uninfected controls, but these levels declined significantly by 8 days after inoculation (Figure 6A). In infected animals, a significant decrease in monocytes (CD14+) and NK cells (CD56+) was observed 5 and 8 days after inoculation (Figure 6B and 6C) as compared to earlier time points. This corresponded with a small increase in total T-cell frequency over time, which was highest 8 days after inoculation (Figure 6D). Additionally, in infected animals, both the frequency and geometric mean fluorescence intensity of CD69 expression, a marker of T-cell activation, on both CD8+ and CD8− (presumably CD4+) T cells were significantly higher 8 days after inoculation than at earlier time points (Figure 6E–6H). The highest level of CD80 expression on mDC was at 8 days after inoculation, but this difference was not significant (Figure 6I).
Figure 6.
Flow cytometric analysis of blood leukocytes of SGM3 HuMice. Red blood cells were lysed from whole blood, and remaining cells were stained with multicolor panels of antibodies specific for myeloid and lymphoid cell types and for overall leukocytes (CD45). Data for each panel are from 3 mock-inoculated HuMice (closed circles) and 3 HuMice intramuscularly inoculated with Ebola virus (103 focus-forming units; open circles) from each time point (in days). *P < .5, **P < .1, and ***P < .01, by 1-way analysis of variance, with a Tukey post hoc test. Abbreviations: gMFI, geometric mean fluorescence intensity; mDC, myeloid dendritic cells; NK, natural killer; ns, nonsignificant.
In the spleen, the percentage of hCD45+ cells in infected animals significantly increased by 8 days after inoculation (Figure 7A). Mean myeloid cell (CD33+) frequencies in infected animals increased at 3 days after inoculation, but significant decreases were seen 5 and 8 days after inoculation in both infected and control animals (Figure 7B). In infected animals, we observed a similar trend, with NK cell (CD56+) mean frequency increasing 3 and 5 days after inoculation and significantly decreasing 8 days after inoculation (Figure 7C). Monocyte (CD14+) numbers in infected animals were above control levels 1 and 3 days after inoculation but returned to control levels by 5 and 8 days after inoculation (Figure 7D). Unlike in the blood, T-cell frequencies in infected animals decreased 8 days after inoculation, compared with frequencies observed 1, 3, and 5 days after inoculation, yet these cells were activated, as seen by expression of CD69 by both CD8+ and CD8− T cells (Figure 7E–7I).
Figure 7.
Flow cytometric analysis of splenocytes of SGM3 HuMice. Single-cell suspensions from spleen samples were lysed, and splenocytes were stained with multicolor panels of antibodies specific for myeloid and lymphoid cell types and overall leukocytes (CD45). Data for each panel are from 3 mock-inoculated HuMice (closed circles) and 3 HuMice intramuscularly inoculated with Ebola virus (103 focus-forming units; open circles) from each time point (in days). *P < .5, **P < .1, and ***P < .01, by 1-way analysis of variance, with a Tukey post hoc test. Abbreviations: gMFI, geometric mean fluorescence intensity; mDC, myeloid dendritic cells; ns, nonsignificant.
DISCUSSION
Herein we describe EBOV infection in NSG-SGM3 HuMice engrafted with human HSC. Inoculating these mice intraperitoneally or intramuscularly with WT-EBOV-Makona resulted in high levels of virus replication in the blood and tissues. Consistent with previous reports suggesting that immune cells are initial targets of EBOV infection and dissemination, early targets of infection in SGM3 HuMice were macrophages in the liver and macrophages and DC in the spleen, which is primarily comprised of human-derived immune cells in these animals. Early EBOV replication occurred in the human immune cells in SGM3 HuMice, and, over the course of infection, both human and murine cells became infected by EBOV; IHC analysis revealed viral antigen both in immune and parenchymal cells in the liver and spleen.
As in previous HuMouse models of EVD, severe disease was observed in SGM3 HuMice infected with WT-EBOV. However, it remains unclear what aspects of humanization in current HuMouse models contribute to clinical disease. WT-EBOV infection of Hu-NSG-A2 (which express the HLA-A2.1 transgene) and NSG-BLT (which are transplanted with human bone marrow, liver, and thymus) HuMice caused severe weight loss, variable time to death, and pathological changes consistent with EVD [21, 22]. However, despite extensive virus replication and up to approximately 67% lethality in intramuscularly inoculated SGM3 HuMice, we did not observe the hallmarks of pathology associated with severe EVD in humans and NHP and rodent models, such as severe inflammation, hepatocyte necrosis, and lymphocyte apoptosis [11, 15, 28].
Engraftment levels in HuMice have been proposed to contribute to EVD progression and severity [21]. Leukocyte levels in SGM3 HuMice are >20% higher than in previously described models, suggesting that engraftment level may not positively correlate with disease severity in all HuMouse models and that characteristics of disease are associated with complex factors like relative immune cell populations and function. In SGM3 HuMice, human T cells are educated on murine MHC present on thymic epithelial cells and only incidentally interact with human MHC-peptide complexes presented by human antigen-resenting cells (on MHC II or cross-presented on MHC I). Furthermore, MHC I–expressing murine parenchymal cells, educated on mouse-encoded MHC, likely lack compatible costimulatory signals for initiating human T-cell responses. Immunopathology seen in the previously described HuMice strains may differ from what we observed, owing to the expression of the human MHC I transgene (in Hu-NSG-A2) or the presence of human thymic tissue (in NSG-BLT), which would allow cognate T-cell functionality. Increased disease severity and hallmarks of EVD pathology seen in NSG-BLT and Hu-NSG-A2 models, compared with what we observed in the SGM3 HuMice, are likely influenced by immune responses elicited in the more abundant residual murine innate cells (DC and macrophages).
The lack of immunopathology in SGM3 HuMice may also be a result of deficient proinflammatory cytokine responses. The extensive myeloid cell recruitment to the livers of infected SGM3 HuMice suggests that some immune signals were produced, likely originating from infected human resident tissue macrophages or DC that secreted chemokines to recruit additional myeloid cells. However, this signaling to recruit myeloid cells appears to occur without associated immunopathogenesis. It is possible that, although the human immune cells respond to infection, the inflammatory mediators cannot interact with the murine parenchymal cells, diminishing the characteristic inflammation associated with severe EVD. Human and murine immune responses differ significantly, and cross-species activation of human cytokines in murine cells is often weak and not physiological [29]. Thus, in the presence of functional human cell inflammation, murine cells may be unable to respond to interferon and other immune responses initiated by the human immune cells.
The contributors to progressive weight loss and lack of clinical resolution seen in terminal animals remain unclear. T cells were well reconstituted in SGM3 HuMice, but NK cell populations were poorly developed, and pDC were absent. Susceptibility to infection and high viral replication in SGM3 HuMice may be due to NK cell deficiency or pDC absence. NK cells are important in the early immune responses to viral infection, and pDC are the primary producers of type I interferon (IFN) and important in modulating the adaptive immune response [30]. Type I IFN protects against EVD in mice; mice inoculated subcutaneously with MA-EBOV produce high levels of IFN-α/β and do not develop disease, while blocking type I IFN in these animals results in disease [31, 32]. Adaptive immunity is also necessary for protection from EVD in mice. In scid mice, WT-EBOV causes delayed disease when administered intraperitoneally or subcutaneously, and MA-EBOV is lethal 10–15 days after inoculation when injected subcutaneously, similar to our observations with SGM3 HuMice [32]. Thus, a lack of interaction between T cells and APCs and deficient IFN signaling necessary for effective T-cell responses may also contribute to EBOV susceptibility in the SGM3 HuMice. Although defects in both the innate and adaptive responses in SGM3 HuMice likely contribute to disease susceptibility, they may also be responsible for the absence of characteristic EVD immunopathogenesis and should be investigated in more detail in subsequent studies. Furthermore, EVD is not exclusively an immunopathology-based disease. Severe disease is also a result of EBOV infection of liver parenchymal, endothelial, and andrenocortical cells, which contributes to membrane leakage, corticosteroid insufficiency, and hypotension, coupled with viral-mediated coagulation abnormalities [33–35]. Viral-mediated pathology must also be considered in future investigations of disease observed in terminal SGM3 HuMice.
A disconnect between the reconstituted human immune system and native murine parenchymal cells provides a model system that can be leveraged for elucidating how immunological events promote or inhibit severe disease. The mechanism underlying the atypical presentation of EBOV-caused disease in SGM3 HuMice, specifically the lack of immunopathology, is unclear and should be the focus of follow-up investigations. Humanized mouse models are still new in EBOV studies. Depending on background strain and type of engraftment, HuMouse models can have several limitations, like absence or paucity of particular immune cell populations, lack of appropriate T-cell education and cytokine support, and deficiencies in the adaptive immune response, including antibody production and class switching. While HuMice offer an important opportunity to investigate viral interaction and infection of human immune cells in vivo, more work must be done to determine how best to address pathogenesis questions in the unique and complex immune milieu that these animals provide.
Notes
Acknowledgments. We thank Anita Mora, for preparing the figures; Tatyana Klimova, for editing the manuscript; and Tina Thomas, Rebecca Rosenke, and Dan Long, for preparing and staining tissues for histopathologic studies.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the National Institute of Allergy and Infectious Diseases.
Financial support. This work was supported by the Centers for Disease Control and Prevention (CDC; emerging infectious disease research core funds); the CDC Research Participation Program (to J. R. S., administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the CDC); the National Institutes of Health (NIH) Loan Repayment Program (award to J. R. S.); and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Ebola situation report, 2016. http://apps.who.int/ebola/current-situation/ebola-situation-report-17-february-2016 Accessed 18 February 2016.
- 2.Ströher U, West E, Bugany H, Klenk HD, Schnittler HJ, Feldmann H. Infection and activation of monocytes by Marburg and Ebola viruses. J Virol 2001; 75:11025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldmann H, Bugany H, Mahner F, Klenk HD, Drenckhahn D, Schnittler HJ. Filovirus-induced endothelial leakage triggered by infected monocytes/macrophages. J Virol 1996; 70:2208–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosio CM, Aman MJ, Grogan C et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis 2003; 188:1630–8. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson KL, Rollin PE. Cytokine and chemokine expression in humans infected with Sudan Ebola virus. J Infect Dis 2007; 196 (suppl):S357–63. [DOI] [PubMed] [Google Scholar]
- 6.Jesudason S, Collins MG, Rogers NM, Kireta S, Coates PTH. Non-human primate dendritic cells. J Leukoc Biol 2012; 91:217–28. [DOI] [PubMed] [Google Scholar]
- 7.Magalhaes I, Vudattu NK, Ahmed RK et al. High content cellular immune profiling reveals differences between rhesus monkeys and men. Immunology 2010; 131:128–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barreiro LB, Marioni JC, Blekhman R, Stephens M, Gilad Y. Functional comparison of innate immune signaling pathways in primates. PLoS Genet 2010; 6:e1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterston RH, Lindblad-Toh K, Birney E et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002; 420:520–62. [DOI] [PubMed] [Google Scholar]
- 10.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J Immunol 2004; 172:2731–8. [DOI] [PubMed] [Google Scholar]
- 11.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 1998; 178:651–61. [DOI] [PubMed] [Google Scholar]
- 12.Shultz LD, Lyons BL, Burzenski LM et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 2005; 174:6477–89. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa F, Yasukawa M, Lyons B et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chain null mice. System 2005; 106:1565–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shultz LD, Schweitzer PA, Christianson SW et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol 1995; 154:180–91. [PubMed] [Google Scholar]
- 15.Baskerville A, Bowen ETW, Platt GS, McArdell LB, Simpson DIH. The pathology of experimental Ebola virus infection in monkeys. J Pathol 1978; 125:131–8. [DOI] [PubMed] [Google Scholar]
- 16.Ryabchikova EI, Kolesnikova LV, Netesov SV. Animal pathology of filoviral infections. Curr Top Microbiol Immunol 1999. 235:145–73. [DOI] [PubMed] [Google Scholar]
- 17.Ryabchikova EI, Kolesnikova LV, Luchko SV. An analysis of features of pathogenesis in two animal models of Ebola virus infection. J Infect Dis 1999; 179(suppl):S199–202. [DOI] [PubMed] [Google Scholar]
- 18.Geisbert TW, Hensley LE, Larsen T et al. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 2003; 163:2347–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wunderlich M, Chou F-S, Link KA et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 2010; 24:1785–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2R{gamma}null humanized mice. Blood 2011; 117:3076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lüdtke A, Oestereich L, Ruibal P et al. Ebola virus disease in mice transplanted with human hematopoietic stem cells. J Virol 2015; 89:4700–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird BH, Spengler JR, Chakrabarti AK et al. Humanized mouse model of Ebola virus disease mimics immune responses in human disease. J Infect Dis 2016; 213:703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baize S, Pannetier D, Oestereich L et al. Emergence of Zaire Ebola virus disease in Guinea—preliminary report. N Engl J Med 2014; 371:1418–25. [DOI] [PubMed] [Google Scholar]
- 24.Hoenen T, Groseth A, Feldmann F et al. Complete genome sequences of three Ebola virus isolates from the 2014 outbreak in west Africa. Genome Announc 2014; 2:e01331–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haddock E, Feldmann F, Feldmann H. Effective chemical inactivation of Ebola virus. Emerg Infect Dis 2016; 22:1292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groseth A, Marzi A, Hoenen T et al. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog 2012; 8:e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnittler HJ, Feldmann H. Molecular pathogenesis of filovirus infections: role of macrophages and endothelial cells. Curr Top Microbiol Immunol 1999; 235:175–204. [DOI] [PubMed] [Google Scholar]
- 28.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol 2015; 235:153–74. [DOI] [PubMed] [Google Scholar]
- 29.Harari D, Abramovich R, Zozulya A et al. Bridging the species divide: transgenic mice humanized for type-I interferon response. PLoS One 2014; 9:e84259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gary-Gouy H, Lebon P, Dalloul AH. Type I interferon production by plasmacytoid dendritic cells and monocytes is triggered by viruses, but the level of production is controlled by distinct cytokines. J Interferon Cytokine Res 2002; 22:653–9. [DOI] [PubMed] [Google Scholar]
- 31.Mahanty S, Gupta M, Paragas J, Bray M, Ahmed R, Rollin PE. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 2003; 312:415–24. [DOI] [PubMed] [Google Scholar]
- 32.Bray M. The role of the type I interferon response in the resistance of mice to filovirus infection. J Gen Virol 2001; 82(Pt 6):1365–73. [DOI] [PubMed] [Google Scholar]
- 33.Bray M, Hatfill S, Hensley L, Huggins JW. Haematological, biochemical and coagulation changes in mice, guinea-pigs and monkeys infected with a mouse-adapted variant of Ebola Zaire virus. J Comp Pathol 2001; 125:243–53. [DOI] [PubMed] [Google Scholar]
- 34.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McElroy AK, Erickson BR, Flietstra TD et al. Ebola hemorrhagic fever: novel biomarker correlates of clinical outcome. J Infect Dis 2014; 210:558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]