Abstract
Deregulated expression of microRNAs has been associated with angiogenesis. Studying the miRNome of locally advanced breast tumors we unsuspectedly found a dramatically repression of miR-204, a small non-coding RNA with no previous involvement in tumor angiogenesis. Downregulation of miR-204 was confirmed in an independent cohort of patients and breast cancer cell lines. Gain-of-function analysis indicates that ectopic expression of miR-204 impairs cell proliferation, anchorage-independent growth, migration, invasion, and the formation of 3D capillary networks in vitro. Likewise, in vivo vascularization and angiogenesis were suppressed by miR-204 in a nu/nu mice model. Genome-wide profiling of MDA-MB-231 cells expressing miR-204 revealed changes in the expression of hundred cancer-related genes. Of these, we focused on the study of pro-angiogenic ANGPT1 and TGFβR2. Functional analysis using luciferase reporter and rescue assays confirmed that ANGPT1 and TGFβR2 are novel effectors downstream of miR-204. Accordingly, an inverse correlation between miR-204 and ANGPT1/TGFβR2 expression was found in breast tumors. Knockdown of TGFβR2, but not ANGPT1, impairs cell proliferation and migration whereas inhibition of both genes inhibits angiogenesis. Taken altogether, our findings reveal a novel role for miR-204/ANGPT1/TGFβR2 axis in tumor angiogenesis. We propose that therapeutic manipulation of miR-204 levels may represent a promising approach in breast cancer.
Over the last decade, microRNAs (miRNAs) have emerged as a prominent class of novel negative regulators of gene expression1. These non-coding single-stranded RNAs are evolutionary conserved, and function as guide molecules in posttranscriptional gene silencing by partially complementing with the 3′ untranslated region (UTR) of the target, resulting in mRNA degradation or translational repression2. Remarkably, aberrant expression of miRNAs may contribute to development of diverse neoplasia and in some cases correlates with clinical-pathological features of tumors representing potential prognostic markers and novel therapeutic targets in cancer3. One mechanism by which miRNAs may induce tumorigenesis is by affecting the production of angiogenic factors and neovascularization processes. Angiogenesis is a complex mechanism of neovascular formation from pre-existing blood vessels. Pathological formation of new blood vessels confers advantages to tumor growth and metastasis establishment; therefore molecular mechanisms of angiogenesis are major issues in the understanding of cancer progression, and constitute an important therapeutic target in cancer4. Recent investigations into molecular mechanisms of tumor angiogenesis have led to the identification of novel angiogenic targets, which have been translated into the development of promising anti-vascular agents5. In neoplasms, tumor-derived factors promote angiogenesis through the activation of multiple cellular processes such as cell division, growth, migration and invasion6. Key pro-angiogenic factors that enhance endothelial cell migration and capillary-like structure formation include the hypoxia-inducible factor 1-alpha (HIF1α), the vascular endothelial growth factor A (VEGFA), the transforming growth factor beta-1 (TGFβ1), the angiopoietins (ANGPT1 and ANGPT2), the plasminogen (PLG), and endostatins. Importantly, several genes involved in angiogenesis-dependent growth of tumors are regulated by miRNAs7,8,9,10. Therefore, deciphering the miRNAs network responsible for the modulation of angiogenesis might lead to discovery of novel therapeutic approaches for cancer. Nonetheless, the potential biological role of most miRNAs in angiogenesis regulation of breast cancer is poorly understood. Here, we analyzed the microRNome of primary breast tumors and found that miR-204 was suppressed in clinical specimens. Furthermore, we provide experimental data indicating that miR-204 inhibits diverse hallmarks of breast cancer, in particular angiogenesis through the targeting of key pro-angiogenic genes.
Results
Global miRNAs profiling of locally invasive breast tumors
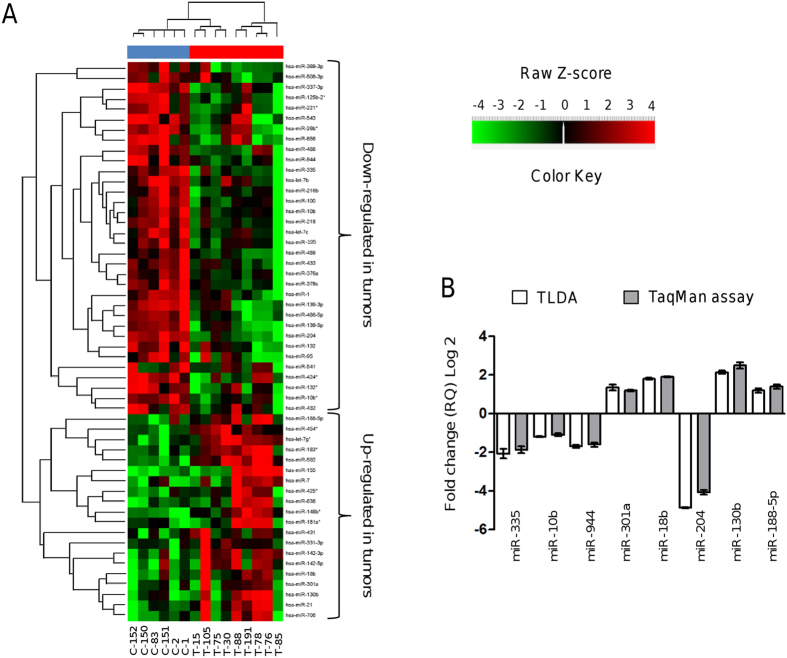
In order to identify miRNAs differentially expressed between primary breast tumors and normal mammary tissues, we profiled 667 mature miRNAs using stem-loop reverse transcription-quantitative PCR (RT-qPCR) in TaqMan low-density arrays (TLDAs). Tumors were collected from nine patients with locally invasive ductal breast carcinomas (discovery cohort). An overview of the clinical and pathological features of tumors and patients included in this study is given in Table 1. After comparative 2-ΔΔCt analyses, we identified a total of 54 miRNAs significantly deregulated in clinical specimens (|log2(T/N)| > 1.0; p < 0.05). Of these, 34 miRNAs were downregulated and 20 were upregulated (Table 2). This panel of miRNAs was able to separate the tumors group from the normal samples in the 2-way unsupervised hierarchical cluster shown in Fig. 1A indicating that differences among samples were not due to heterogeneity or the presence of different cell types in biopsies. A group of 23 miRNAs (four upregulated and 19 downregulated) were homogeneously expressed across the set of tumors (see Supplementary Table 1). These include miRNAs with known roles in breast cancer development such as miR-7, miR-10b, miR-21, miR-100, miR-155, miR-195, miR-221, and miR-218. In addition, we found that a large number of predicted target genes belong to key cellular pathways that might modulate the hallmarks of cancer, including MAP kinases, focal adhesion, WNT, TGF-β, and ERBB signaling (see Supplementary Table 2). A number of 14 miRNAs were located in chromosomal regions frequently deleted or amplified in breast cancer. In order to corroborate the differential expression of miRNAs identified by TLDAs, eight deregulated miRNAs were analyzed by RT-qPCR in biological replicates. Results showed that expression levels of miR-335, miR-10b, miR-944, miR-301a, miR-18b, miR-204, miR-130b, and miR-188-5p were similar in both assays (Fig. 1B). To further validate the arrays results, we analyzed a larger and independent dataset of miRNAs expression in 776 breast tumors and matched adjacent tissues (validation cohort) obtained from The Cancer Genome Atlas (TCGA). Results showed that of the 54 miRNAs that we previously found with differential expression in the discovery cohort 33 were reported in TCGA sets. Of these, 29 miRNAs exhibits expression levels very similar to those found in our miRNome analysis, whereas only four miRNAs showed discordant expression between the discovery and validation cohort (p < 0.0001; see Supplementary Fig. 1).
Table 1. Clinical and pathological features of breast tumors analyzed by stem-loop reverse transcription-quantitative PCR in TaqMan low-density arrays.
| Patient | Age (years) | Clinical stage | Tumor grade | Classification | Tumor size (mm) | Histological subtype | HER2 | ER | PR |
|---|---|---|---|---|---|---|---|---|---|
| 15 | 45 | II | 2 | HER2 | 35 | Infiltrating ductal carcinoma | + | − | + |
| 30 | 41 | I | 2 | Luminal B | 20 | In situ ductal carcinoma | + | + | + |
| 75 | 50 | IIB | 2 | HER2 | 25 | Infiltrating ductal carcinoma | + | − | − |
| 76 | 49 | IIB | 3 | HER2 | 20 | Infiltrating ductal carcinoma | + | − | − |
| 78 | 81 | IIIB | 3 | HER2 | 47 | Infiltrating ductal carcinoma | + | − | − |
| 85 | 47 | IIA | 3 | HER2 | 27 | Infiltrating ductal carcinoma | + | − | + |
| 88 | 63 | I | ND | Luminal A | 15 | Infiltrating ductal carcinoma | + | + | − |
| 105 | 39 | IIIC | 2 | HER2 | 23 | Infiltrating ductal carcinoma | + | − | + |
| 191 | 55 | IIB | ND | HER2 | 39 | Infiltrating ductal carcinoma | + | − | − |
HER2, Human epidermal receptor 2. ER, Estrogen receptor. PR, Progesterone receptor. ND, Not determined.
Table 2. MicroRNAs modulated in locally invasive breast tumors.
| Fold change (log2 RQ) | RQ-value | p-value | Locus | |
|---|---|---|---|---|
| Down-regulated miRNAs | ||||
| hsa-miR-204 | −4.87 | 0.0342 | 0.003 | 9q21.12 |
| hsa-miR-139-3p | −3.86 | 0.0691 | 0.005 | 11q13.4 |
| hsa-miR-125b-2* | −3.54 | 0.0859 | 0.038 | 21q21.1 |
| hsa-miR-486-5p | −3.53 | 0.0866 | 0.001 | 8p11.21 |
| hsa-miR-541 | −3.25 | 0.1054 | 0.032 | 14q32.31 |
| hsa-miR-216b | −3.04 | 0.1213 | 0.018 | 2p16.1 |
| hsa-miR-139-5p | −2.76 | 0.1481 | 0.001 | 11q13.4 |
| hsa-miR-489 | −2.60 | 0.1650 | 0.042 | 7q21.3 |
| hsa-miR-1 | −2.08 | 0.2373 | 0.021 | 20q13.33 |
| hsa-miR-335 | −2.04 | 0.2428 | 0.051 | 7q32.2 |
| hsa-miR-376c | −1.93 | 0.2623 | 0.009 | 14q32.31 |
| hsa-miR-132* | −1.89 | 0.2697 | 0.021 | 17p13.3 |
| hsa-miR-132 | −1.75 | 0.2973 | 0.010 | 17p13.3 |
| hsa-miR-369-3p | −1.73 | 0.3009 | 0.006 | 14q32.31 |
| hsa-miR-944 | −1.72 | 0.3028 | 0.025 | 3q28 |
| hsa-miR-488 | −1.66 | 0.3157 | 0.051 | 1q25.2 |
| hsa-miR-433 | −1.62 | 0.3247 | 0.006 | 14q32.2 |
| hsa-miR-424* | −1.60 | 0.3296 | 0.001 | Xq26.3 |
| hsa-miR-508-3p | −1.49 | 0.3566 | 0.030 | Xq27.3 |
| hsa-miR-218 | −1.49 | 0.3568 | 0.003 | 4p15.31 |
| hsa-miR-337-3p | −1.47 | 0.3612 | 0.044 | 14q32.2 |
| hsa-miR-10b* | −1.45 | 0.3648 | 0.001 | 2q31.1 |
| hsa-let7c | −1.42 | 0.3739 | 0.040 | 21q21.1 |
| hsa-let7b | −1.34 | 0.3962 | 0.038 | 22q13.31 |
| hsa-miR-376a | −1.32 | 0.4011 | 0.021 | 14q32.31 |
| hsa-miR-26b* | −1.29 | 0.4093 | 0.006 | 2q35 |
| hsa-miR-656 | −1.26 | 0.4161 | 0.018 | 14q32.31 |
| hsa-miR-95 | −1.26 | 0.4165 | 0.038 | 4p16.1 |
| hsa-miR-221* | −1.24 | 0.4243 | 0.016 | Xp11.3 |
| hsa-miR-543 | −1.20 | 0.4361 | 0.014 | 14q32.31 |
| hsa-miR-10b | −1.18 | 0.4420 | 0.015 | 2q31.1 |
| hsa-miR-100 | −1.18 | 0.4427 | 0.037 | 11q24.1 |
| hsa-miR-432 | −1.05 | 0.4816 | 0.035 | 14q32.2 |
| hsa-miR-195 | −1.03 | 0.4898 | 0.048 | 17p13.1 |
| Up-regulated miRNAs | ||||
| has-miR-155 | 11.91 | 3849.69 | 0.019 | 21q21.3 |
| hsa-miR-592 | 4.43 | 21.4937 | 0.003 | 7q31.33 |
| hsa-miR-638 | 3.14 | 8.8009 | 0.035 | 19p13.2 |
| hsa-miR-183* | 3.08 | 8.4533 | 0.005 | 7q32.2 |
| hsa-miR-21 | 2.73 | 6.6543 | 0.029 | 17q23.1 |
| hsa-miR-130b | 2.37 | 5.1594 | 0.044 | 22q11.21 |
| hsa-miR-142-3p | 2.26 | 4.7981 | 0.019 | 17q22 |
| hsa-miR-708 | 2.22 | 4.6601 | 0.049 | 11q14.1 |
| hsa-miR-181a* | 2.16 | 4.4810 | 0.044 | 1q32.1 |
| hsa-let7g* | 2.10 | 4.2809 | 0.004 | 3p21.1 |
| hsa-miR-148b* | 2.10 | 4.2749 | 0.010 | 12q13.13 |
| hsa-miR-18b | 1.80 | 3.4850 | 0.035 | Xq26.2 |
| hsa-miR-454* | 1.79 | 3.4608 | 0.007 | 17q23.2 |
| hsa-miR-7 | 1.71 | 3.2647 | 0.031 | 9q21.32 |
| hsa-miR-142-5p | 1.68 | 3.2130 | 0.039 | 17q22 |
| hsa-miR-431 | 1.60 | 3.0374 | 0.031 | 14q32.2 |
| hsa-miR-301a | 1.38 | 2.6071 | 0.024 | 17q22 |
| hsa-miR-188-5p | 1.32 | 2.5022 | 0.026 | Xp11.23 |
| hsa-miR-331-3p | 1.19 | 2.2822 | 0.003 | 12q22 |
| hsa-miR-425* | 1.18 | 2.2718 | 0.041 | 3p21.31 |
Figure 1. Expression profiling of microRNAs in locally invasive breast tumors and normal tissues.
(A) Unsupervised hierarchical clustering analysis displaying the differential expression of microRNAs in breast tumors relative to normal tissues from TLDA. The heat map (Spearman correlation, Euclidean distance) represents a cluster analysis of the logarithm of transformed ΔΔCt values of the differentially expressed microRNAs. Color key: upregulated microRNAs (red); downregulated microRNAs (green). Blue upper bar, normal tissues; red upper bar, tumor tissues. (B) Validation of expression levels of eight microRNAs using Taqman qRT-PCR assay (grey) in comparison with data obtained from TLDA (white). Data were expressed as mean ± S.D.
MicroRNA-204 is suppressed in breast tumors and cancer cell lines
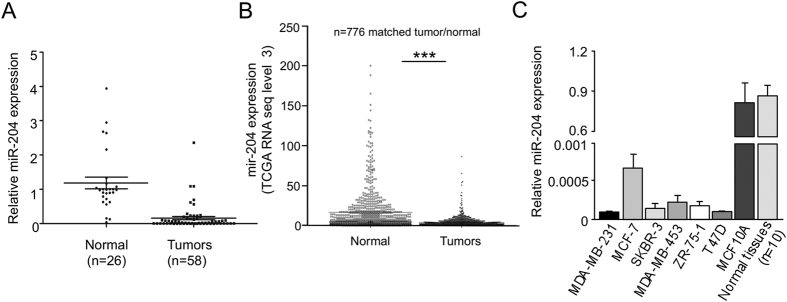
The miRNAs profiling of locally invasive breast tumors allowed us to evidence that, in particular, miR-204 was suppressed. To study the biological relevance of miR-204, we measured its expression by RT-qPCR in clinical specimens obtained from an independent cohort of 58 breast cancer patients. Clinical features of breast tumors including hormonal receptors status, tumor size, histology, clinical stage, and tumor grade are summarized in Table 3. Results indicated that miR-204 expression was significantly low (p < 0.05) in breast tumors in comparison with normal adjacent tissues (Fig. 2A). No association between miR-204 levels and the status of estrogen, progesterone and HER2/neu receptors was found. We further performed a validation analysis using 776 breast tumors and matched normal adjacent samples from the TGCA datasets. Results confirmed that miR-204 was significantly (p < 0.0001) suppressed in breast tumors in comparison to normal tissues in this large cohort of patients (Fig. 2B). Moreover, we found that miR-204 was severely downregulated in MCF-7, MDA-MB-231, MDA-MB-45, ZR-45, and T47-D breast cancer cell lines when compared with non-tumorigenic MCF-10A cells and normal tissues (Fig. 2C).
Table 3. Clinical features of breast tumors analyzed for miR-204 expression by RT-qPCR.
| Characteristics | Patients (n = 58) |
|---|---|
| Classification | |
| HER2 positive | 5 |
| Luminal A | 25 |
| Luminal B | 7 |
| Triple negative | 17 |
| Unknown | 4 |
| Histology | |
| In situ ductal carcinoma | 17 |
| Infiltrating ductal carcinoma | 20 |
| Infiltrating lobular carcinoma | 8 |
| Invasive mucinous carcinoma | 3 |
| Mixed carcinoma (ductal and lobular) | 6 |
| Others | 4 |
| Clinical stage | |
| I | 8 |
| II | 27 |
| III | 19 |
| IV | 4 |
Figure 2. MiR-204 is suppressed in breast tumors and cancer cell lines.
(A) qRT-PCR assays for miR-204 relative expression in non-tumoral adjacent breast tissues and breast tumors. (B) MiR-204 expression in 776 breast tumors and matched adjacent normal samples from TCGA datasets (***p < 0.0001). (C) qRT-PCR assays for miR-204 expression in breast cancer cell lines. Non-tumorigenic MCF-10A cell line and normal adjacent tissues pool (n = 10) were used as controls. Data were normalized with the endogenous small-nucleolar RNU44. Bars represent the mean of three independent experiments ± S.D.
MicroRNA-204 inhibits cell proliferation and anchorage-independent growth
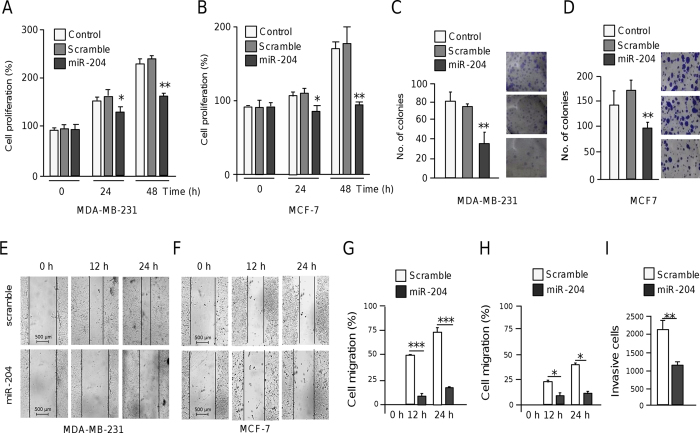
We next wondered whether ectopic expression of miR-204 could have tumor suppressor effects in vitro. Results of MTT assays showed that the growth of both MDA-MB-231 and MCF-7 cells transfected with miR-204 was significantly (p < 0.05) decreased in comparison with non-transfected and scramble transfected control cells (Fig. 3A,B). Moreover, colony formation assays indicated that anchorage-independent growth was attenuated (p < 0.05) by miR-204 in both cell lines relative to controls (Fig. 3C,D).
Figure 3. MiR-204 represses cell proliferation, growth and migration.
(A,B) MTT assays of MDA-MB-231 (A) and MCF-7 cells (B) transfected with pre-miR-204 (30 nM) or pre-miR-negative control (scramble) at 48 h and 24 h post-transfection. (C,D) Graphical representation of colony-forming assays for MDA-MB-231 (C) and MCF-7 cells (D) transfected with pre-miR-204 or scramble. (E,F) Scratch/wound-healing assays of MDA-MB-231 (E) and MCF-7 (F) cell monolayers treated with pre-miR-204 (30 nM), or scramble for 12 h and 24 h. (G,H) Transwell cell migrating assays of MDA-MB-231 (G) and MCF-7 (H) cells treated as above. (I) Matrigel invasion assays of MDA-MB-231 cells transfected with pre-miR-204 (30 nM). Results shown are the mean of three independent experiments +/− SD. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to controls. Bars represent the mean of three independent experiments ± S.D.
MicroRNA-204 impairs cell migration and invasion
To evaluate the contribution of miR-204 in cell migration and invasion we restored its expression in triple negative MDA-MB-231 (highly metastatic) and estrogen responsive MCF-7 (poorly invasive) breast cancer cell lines and then performed scratch/wound-healing and transwell assays. Results showed that monolayer restoration at 24 h was significantly (p < 0.05) delayed by 82% and 72% in MDA-MB-231 and MCF-7 cells respectively in comparison with non-transfected control cells (Fig. 3E,F). Furthermore, we found that the number of migratory cells at 24 h was reduced (p < 0.05) in miR-204-expressing MDA-MB-231 cells in comparison with control (Fig. 3G,H). Congruently, restoration of miR-204 levels also attenuated the ability of MDA-MB-231 cells to invade matrigel chambers (Fig. 3I).
MicroRNA-204 impairs angiogenesis in vitro
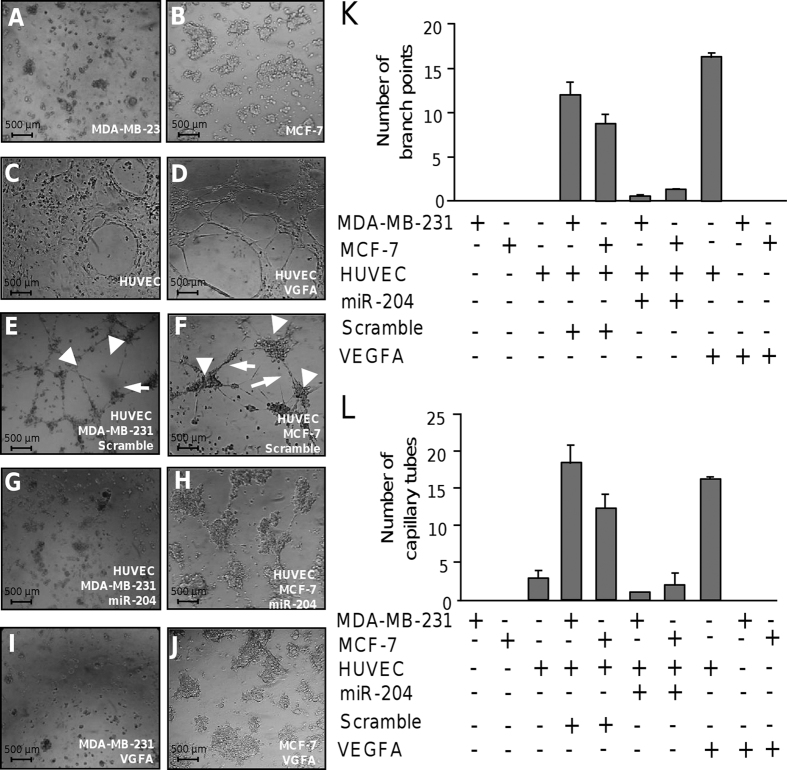
Tumor progression requires a sustained angiogenesis. As the role of miR-204 in angiogenesis is largely unknown in breast cancer, we investigated its contribution in this cancer hallmark. We carried out tube formation assays using human umbilical vein endothelial cells (HUVEC), which is one of the simple but well-established in vitro angiogenesis assays based on the ability of endothelial cells to form three-dimensional (3D) capillary-like tubular structures. Co-cultures of HUVEC with MDA-MB-231 or MCF-7 breast cancer cells in different conditions were performed. As expected MDA-MB-231, MCF-7 and HUVEC cells alone did not form tubules-like structures (Fig. 4A–C). In contrast, a strong angiogenic effect was observed in HUVEC cells treated with recombinant VEGFA used as positive control (Fig. 4D). Typical HUVEC tubular networks on the matrigel were observed at 24 h. Co-incubation of HUVEC with MDA-MB-231 or MCF-7 cells transfected with scramble control also resulted in an angiogenic behavior as the number of endothelial cells branch points and capillary tubes were increased in comparison with monoculture controls (Fig. 4E,F,K,L). Interestingly, co-culture of HUVEC with MDA-MB-231 or MCF-7 cells transfected with miR-204 resulted in a marked inhibition of endothelial cell tubules and branch point formation, and the tubular networks were disrupted at 24 h (Fig. 4G,H,K,L). Taken altogether these data indicate that miR-204 inhibits the angiogenesis-induced by breast cancer cells in endothelial cells.
Figure 4. MiR-204 inhibits angiogenesis.
Capillary tubes development was evaluated in vitro using co-cultures of HUVEC and breast cancer cells. (A) MDA-MB-231 and (B) MCF-7 cells plated onto matrigel with serum free medium. (C) HUVEC plated into matrigel with endothelial complete medium. (D) Monoculture of HUVEC in presence of VEGFA (10 ng/ml). (E) Co-culture of HUVEC with MDA-MB-231 or (D) MCF-7 cells transfected with scramble and plated into matrigel with serum free medium for 24 h. (G) Co-culture of HUVEC with MDA-MB-231 or (H) MCF-7 cells transfected with miR-204 (30 nM) and plated into matrigel with serum free medium for 24 h. (I) Mono-cultures of MDA-MB-231 or (J) MCF-7 cells in presence of VEGFA (10 ng/ml). Graphical representation of quantification of branch points (K) and capillary tubes (L) number after 24 h of co-cultures. Data were obtained by two different observers. Results shown are the mean of three independent experiments ± S.D. Arrowheads indicate branch points. Arrows denote capillary-like tubes structures.
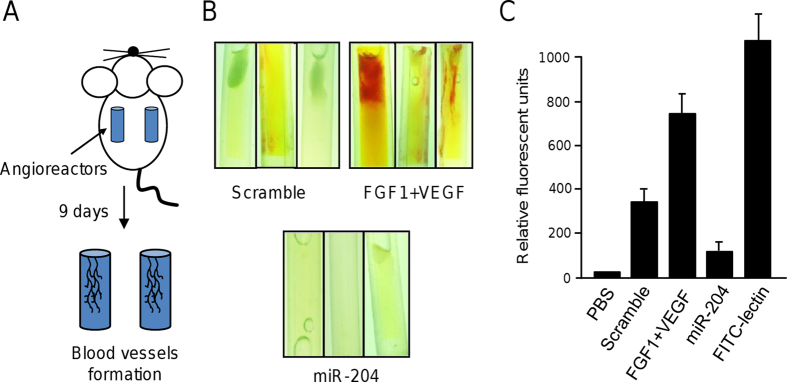
MicroRNA-204 inhibits vascularization and angiogenesis in vivo
Then we asked if miR-204 have an effect in blood vessels formation in a nude mice model. A direct in vivo angiogenesis assay (DIVAA) was setup as describes in Methods. In this assay, the cellular in vivo vascularization was evaluated using transplanted angioreactors, which provides a system to determine an effective response to angiogenic modulating factors. Angioreactors containing basement membrane extract (BME) mixed with VEGF/FGF1 (positive control), scramble transfected cells (negative control), or miR-204 transfected cells (test) were implanted in nude mice. After nine days of implantation angioreactors were removed and vascularization analyzed (Fig. 5A). Data showed a significant blood vessels infiltration in angioreactors containing pro-angiogenic factors VEGF and FGF1. In contrast, angioreactors mixed with MDA-MB-231 cells transfected with scramble showed a significant very low infiltration (Fig. 5B upper panel). Remarkably, blood vessels infiltration was severely impaired in angioreactors containing miR-204 transfected breast cancer cells (Fig. 5B bottom panel) in comparison to controls, indicating that miR-204 significantly inhibits angiogenesis in vivo.
Figure 5. MiR-204 inhibits blood vessels formation in vivo.
(A) Directed in vivo angiogenesis assay. (B) Representative images of angioreactors extirpated from nude mice 9 days after implantation. Inductor of angiogenesis VEGF and FGF1 or MDA-MB-231 cells transfected with miR-204 or scramble control were placed into angioreactors as described in Methods section. (C) Fluorometric quantification of blood vessels formation using FITC-lectin detection assay. Experiments were performed by duplicate tree times.
MicroRNA-204 modulates genes involved in cell proliferation, migration and angiogenesis
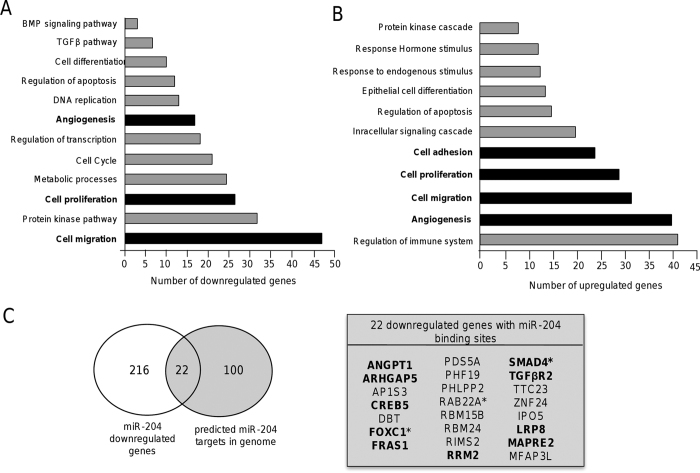
In order to identify potential target genes of miR-204 associated to the inhibition of the cancer hallmarks described above, we carried out a transcriptional profiling of MDA-MB-231 cells that ectopically express miR-204 (see Supplementary Fig. 2) using DNA microarrays. Data from two biological replicates were analyzed, normalized, and raw p-values adjusted. Only the genes with a significant fold change (FC > 1.5; p < 0.05) were included in this analysis. Genome-wide analysis showed that 549 genes (311 upregulated and 238 downregulated) were significantly modulated (see Supplementary Table 3). To validate DNA microarrays data, the mRNA expression of ten selected genes was analyzed by RT-qPCR using specific oligonucleotides (see Supplementary Table 4). Triplicates were performed for each sample and for each gene. In all cases, the mRNA expression values obtained by RT-qPCR were similar to those found by DNA microarrays analysis (see Supplementary Fig. 3). Classification of genes based on Gene ontology categories indicated that a number of genes were involved in cellular processes and pathways frequently deregulated in human cancers (Fig. 6A,B). Of the 238 downregulated genes a subset of 22 genes contains potential miR-204 binding sites at their 3′UTR as predicted by TargetScan and Pic-Tar programs (Fig. 6C), and three genes (FOXC1, RAB22A, SMAD4) were previously reported as truly miR-204 targets. Interestingly, these repressed genes are involved in functions related to cell proliferation, migration, apoptosis and angiogenesis (see Supplementary Table 5). In agreement with the suppressive role of miR-204 in cell proliferation and angiogenesis, the pro-angiogenic angiopoietin-1 (ANGPT1) and the transforming growth factor β receptor type 2 (TGFβR2) genes were downregulated in MDA-MB-231 cells with restored expression of miR-204. In particular, these genes play key roles in cell migration, invasion and in the activation of angiogenesis program of tumor cells.
Figure 6. Classification of genes modulated by miR-204.
The pathways and biological processes analysis using gene ontology terms is shown. (A) Downregulated and, (B) upregulated genes. Cellular processes of particular interest in the present study are denoted in bold. (C) Venn diagram comparing the number of genes modulated by miR-204 overexpression (white circle) and predicted as miR-204 targets (grey circle) by TargetScan and Pic-tar programs. Circles intersection indicates the 22 genes identified in both analyses. Genes related to cell proliferation, migration and angiogenesis are in bold. Previously validated targets of miR-204 are indicated with asterisks.
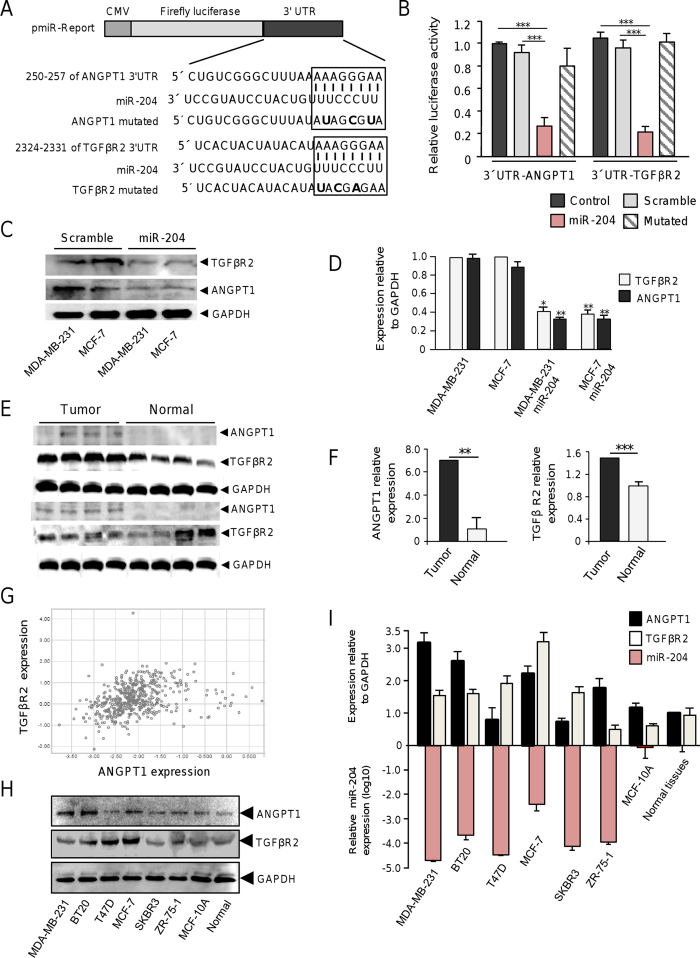
MicroRNA-204 targets the angiogenesis-related ANGPT1 and TGFβR2 genes
Increased expression of ANGPT1 and TGFβR2 promotes cell proliferation and angiogenesis in diverse types of tumors11,12, thus its reliable to propose that miR-204 may negatively regulate these cellular processes through direct targeting of both genes. To corroborate whether miR-204 can exerts posttranscriptional repression of ANGPT1 and TGFβR2, we used luciferase reporter gene assays. Nucleotides sequences corresponding to 3′UTR of either ANGPT1 or TGFβR2 genes were cloned downstream of the luciferase coding region of pmiR report vector (Fig. 6A). In addition, three point mutations in the predicted miR-204 binding site of the 3′UTR of ANGPT1 or TGFβR2 genes were included in the analysis. Recombinant pmiR-LUC-ANGPT1 and pmiR-LUC-TGFβR2 plasmids were transfected into MDA-MB-231 cells and luciferase activity was analyzed after 24 h. Data showed that ectopic expression of miR-204 and co-transfection of either pmiR-LUC-ANGPT1 or pmiR-LUC-TGFβR2 constructs resulted in a significantly reduction of the relative luciferase activity in comparison with controls (Fig. 7B). When mutated sequences of the 3′UTR of ANGPT1 and TGFβR2 genes were assayed, no significant changes in luciferase activity were found indicating that miR-204 binding was specific. Furthermore, Western blot assays corroborated that miR-204 restoration resulted in a significant decrease of the endogenous ANGPT1 and TGFβR2 proteins in both MDA-MB-231 and MCF-7 cells (Fig. 6C,D). In order to extend our observations, then we evaluated if miR-204 suppression in clinical breast tumors correlates with the increased expression of ANGPT1 and TGFβR2 proteins. Our results indicated that the mean expression of ANGPT1and TGFβR2 was increased (60%) in miR-204-deficient breast tumors in comparison with normal tissues (Fig. 6E,F). To further corroborate these findings, an integration-based approach was applied to compare the ANGPT1 and TGFβR2 expression in a larger cohort of breast cancer patients (n = 522) using datasets from TCGA. Results indicate that both ANGPT1 and TGFβR2 were downregulated in the half of samples (Fig. 6G). Furthermore, the expression of ANGPT1 and TGFβR2 was evaluated by Western blot in the six breast cancer cell lines which exhibits reduced miR-204 levels. We found higher ANGPT1 expression in MDA-MB-231, BT20, MCF-7, and ZR-75-1 cell lines, but not inT47D and SKBR3 cells, in comparison to non-tumorigenic MCF-10A and normal adjacent tissues (Fig. 6H). In a similar way high expression levels of TGFβR2 were observed in five breast cancer cell lines, except for ZR-75-1 cells, relative to MCF-10A and normal tissues. These data suggested the existence of an inverse correlation between the expression of mR-204 and ANGPT1 and TGFβR2 proteins in the majority of breast cancer cell lines tested.
Figure 7. MiR-204 targets ANGPT1 and TGFβR2.
(A) Schematic representation of p-miR report constructs containing the 3′UTR of ANGPT1 and TGFβR2 genes cloned downstream of luciferase gene. Seed sequences are indicated in boxes. Point mutations in the miR-204 binding sites of 3′UTR of ANGPT1 or TGFβR2 genes are denoted in bold. (B) Luciferase assays in MDA-MB-231 cells transfected with miR-204 and the constructs described in A. Cells transfected with p-miR report plasmid alone or not transfected were used as controls. Data represent the mean ± S.D. of three independent experiments. (C) Western blot of MDA-MB-231 and MCF-7 cells, non-transfected and transfected with pre-miR-204, using ANGPT1 (1:1000) and TGFβR2 (1:1000) primary antibodies. GAPDH antibodies were used as control. (D) Densitometric analysis of bands in panel C. Data were normalized using GAPDH expression. Images are representative of three independent experiments. (E) Western blots of two representative sets of breast tumors and non-tumoral tissues using ANGPT1 (1:1000) and TGFβR2 (1:1000) antibodies. GAPDH antibodies were used as control. Data were normalized using GAPDH expression. (F) Densitometric analysis of immunodetected bands in panel E. (G) Comparison of ANGPT1 and TGFβR2 expression in breast cancer using dataset from TCGA. (H) Western blot assays for ANGPT1 and TGFβR2 in breast cancer cell lines and normal tissues. (I) Densitometric analysis of immunodetected bands in H (upper bars), and miR-204 expression levels in the same breast cancer cell lines (bottom bars). Protein expression data were normalized using GAPDH. Data for miR-204 expression were normalized using RNU44. Bars are representative of three independent experiments ± S.D.
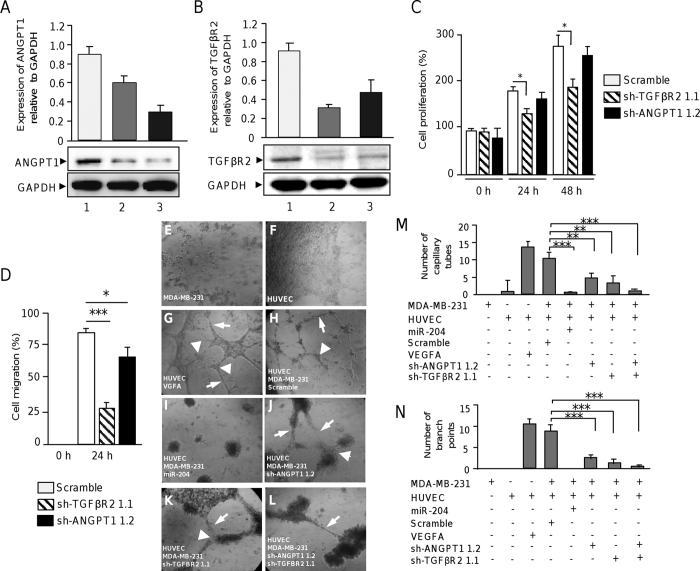
TGFβR2 gene silencing, but not ANGPT1, impairs cell proliferation and migration
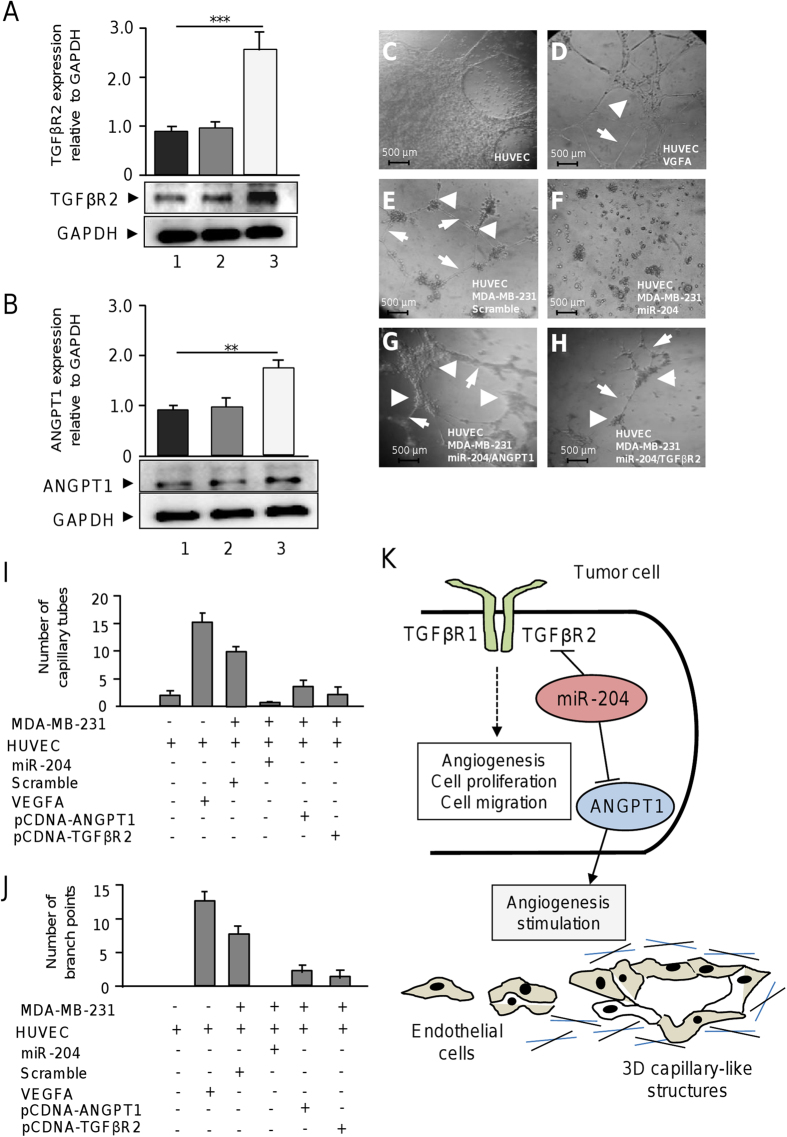
As we previously evidenced that miR-204 directly binds to 3′UTR of the pro-angiogenic ANGPT1 and TGFβR2 genes and inhibits its expression, we sought to determine if targeted inhibition of ANGPT1 and TGFβR2 could affect cell proliferation and migration. Therefore, we proceeded to knock-down its expression by RNA interference using two specific shRNAs targeting both the ANGPT1 and TGFβR2 genes (see Supplementary Table 6). The designed shRNAs dubbed as sh-ANGPT1 1.1, sh-ANGPT1 1.2, sh-TGFβR2 1.1 and sh-TGFβR2 1.2 were cloned in pSilencer vector. The constructs were individually introduced into MDA-MB-231 cells and protein expression was analyzed by Western blot 48 h after transfection. Results showed that the four short-hairpin sequences effectively down-regulate both the ANGPT1 and TGFβR2 expression (Fig. 8A,B). Densitometric analysis of immunodetected bands showed that gene silencing induced by sh-ANGPT1 1.2 and sh-TGFβR2 1.1 sequences was more effective since they suppressed ANGPT1 and TGFβR2 expression by 58% and 62%, respectively, thus they were selected for further experiments. GADPH used as a control, did not showed significantly expression changes between treatments. The effects of ANGPT1 and TGFβR2 silencing in cell proliferation were evaluated in MDA-MB-231 cells. Results of MTT assays showed that cell proliferation was significantly (p < 0.05) decreased in TGFβR2 -deficient cells at 24 h and 48 h in comparison with control (Fig. 8C). In contrast, we did not observed significant differences in cell proliferation in ANGPT1-deficient cells (Fig. 8C). Then, we sought to evaluate the effects of ANGPT1 and TGFβR2 knock-down in cell migration using scratch/wound healing assays. Our data indicate that cell monolayers restoration was significantly delayed at 24 h in both ANGPT1 (p < 0.05) and TGFβR2 (p < 0.001)-deficient cells relative to control (Fig. 8D) being more evident the inhibitory effect in the TGFβR2 silenced cells.
Figure 8. Effects of ANGPT1 and TGFβR2 targeted silencing in cell proliferation, migration and angiogenesis.
(A) Western blot analysis for ANGPT1 expression in MDA-MB-231 cells transfected with scramble (lane 1), sh-ANGPT1 1.1 (lane 2) or sh-ANGPT1 1.2 (lane 3) constructs using ANGPT1 (1:1000) antibodies. Upper panel, densitometric analysis of immunodetected bands. (B) Western blot analysis for TGFβR2 in MDA-MB-231 cells transfected with scramble (lane 1), sh-TGFβR2 1.1 (lane 2) or sh-TGFβR2 1.2 (lane 3) constructs using TGFβR2 (1:1000) antibodies. Upper panel, densitometric analysis of immunodetected bands. Data were normalized using GAPDH expression. Images are representative of three independent experiments. (C) MTT assays and (D) Scratch/wound-healing assays of MDA-MB-231 cells transfected with scramble, sh-ANGPT1 1.1 or sh-TGFβR2 1.2 plasmids. (E–N) Capillary tubes development using co-cultures of HUVEC and breast cancer cells. (E) MDA-MB-231 and (F) HUVEC cells plated into matrigel with endothelial complete medium. (G) Monoculture of HUVEC in presence of VEGFA (10 ng/ml). (H) Co-culture of HUVEC with MDA-MB-231 transfected with scramble or with (I) miR-204 (30 nM). (J) Co-culture of HUVEC with MDA-MB-231 transfected cells with sh-ANGPT1 1.2, (K) sh-TGFβR2 1.1 or (L) both sh-ANGPT1 1.2 and sh-TGFβR2 1.1 plasmids. Graphical representation of (M) branch points, and (N) capillary tubes number after 24 h of co-cultures. Arrowheads indicate branch points. Arrows denote capillary-like tubes structures. Results shown are the mean of three independent experiments +/− SD. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to controls. Bars represent the mean of three independent experiments ± S.D.
Knockdown of ANGPT1 and TGFβR2 suppresses angiogenesis
In order to define if knockdown of ANGPT1 and TGFβR2 impairs angiogenesis we carried out tube formation assays as described above. As previously observed, co-incubation of HUVEC with MDA-MB-231 cells transfected with miR-204 resulted in a dramatical reduction of the number of endothelial cells branch points and capillary tubes in comparison with controls (Fig. 8E–G). Interestingly, co-culture of HUVEC with ANGPT1-deficient cells alters the typical morphology and development of endothelial cell tubules. Moreover, the number of capillary-like structures decreased up to 70%, whereas the number of branch points diminished up to 80% in comparison to HUVEC cells treated with VGFA used as positive control (Fig. 8F,G). Moreover, typical tubular networks on the matrigel were disrupted at 24 h. The inhibition of TGFβR2 produced a similar effect in angiogenesis, as we found that the formation of tubules was also compromised (Fig. 8E–G). An additive effect was observed when ANGPT1 and TGFβR2 genes were knock-down, as the number of capillary-like structures and the branch points diminished up to 95% in comparison to control. These changes were not due to alterations in cell viability of MDA-MB-231 with reduced expression of ANGPT1 and TGFβR2.
Rescue of ANGPT1 and TGFβR2 in miR-204 expressing cells partially restore angiogenesis
To obtain additional insights confirming the role of miR-204/ANGPT1/TGFβR2 axis in angiogenesis we performed rescue assays. The complete open reading frame of ANGPT1 and TGFβR2 genes were amplified and cloned into the pcDNA3 expression vector to overexpress the proteins in breast cancer cells. MDA-MB-231 cells were cotransfected with miR-204 and then with pcDNA3-ANGPT1 or pcDNA3-TGFβR2 constructs and in vitro angiogenesis assays were performed. Western blot assays confirmed that transfection of pcDNA3-ANGPT1 or pcDNA3-TGFβR2 result in significant increased levels of ANGPT1 and TGFβR2 in MDA-MB-231 cells expressing miR-204 (Fig. 9A,B). After that tube formation assays were performed as described above. As previously observed HUVEC control cells in the presence of VGFA form tubules-like structures which were abolished after 24 h coculture with MDA-MB-231 cells expressing miR-204 (Fig. 9D,F). Interestingly, co-incubation of HUVEC with MDA-MB-231 cells transfected with miR-204 and then rescued with pcDNA3-ANGPT1 resulted in an moderate increase of angiogenic behavior as the number of endothelial cells branch points and capillary tubes were significantly increased in comparison with controls represented by cells transfected with miR-204 alone (Fig. 9G,I,J). Likewise, transfection of pcDNA3-TGFβR2 in MDA-MB-231 cells expressing miR-204 resulted in an moderate increase in the number of endothelial cells branch points and capillary tubes in comparison with control (Fig. 9H,I, J). Taken altogether these data indicate that ANGPT1 and TGFβR2 are effectors of miR-204 and that its forced expression partially restores angiogenesis in breast cancer cells.
Figure 9. Rescue of ANGPT1 and TGFβR2 in miR-204 expressing cells partially restore angiogenesis.
(A) Western blot analysis for TGFβR2 expression in (1) MDA-MB-231 cells treated with transfection agent, (2) cells transfected with empty vector and, (3) cells transfected with pCDNA3-TGFβR2 using anti-TGFβR2 (1:1000) antibodies. Upper panel, densitometric analysis of immunodetected bands. (B) Western blot analysis for ANGPT1 in (1) MDA-MB-231 cells treated with transfection agent, (2) cells transfected with empty vector and, (3) cells transfected with pCDNA3-ANGPT1 using anti-ANGPT1 (1:1000) antibodies. Upper panel, densitometric analysis of immunodetected bands. Data were normalized using GAPDH expression. (C) monoculture of HUVEC cells. (D) HUVEC cells treated with VEGFA (10 ng/ml). (E) co-culture of HUVEC and MDA-MB-231 cells transfected with scramble. (F) co-culture of HUVEC and MDA-MB-231 cells transfected with miR-204 precursor. (G) co-culture of HUVEC with MDA-MB-231 cells co-transfected with miR-204 and pcDNA3-ANGPT1 plasmid. (H) co-culture of HUVEC with MDA-MB-231 cells co-transfected with miR-204 and pcDNA3-TGFβR2 construct. Arrowheads indicate branch points. Arrows denote capillary-like tubes structures. (I) Graphical representation of the quantification of capillary tubes, and (J) branch points number after 24 h of co-cultures. Data were obtained by two different observers. Results shown are the mean of three independent experiments. Bars represent the mean of three independent experiments ± S.D. **p < 0.01, ***p < 0.001. (C–H) 3D capillary-like formation evaluated in vitro using co-cultures of HUVEC and breast cancer cells. (K) Working model of the miR-204 functions in angiogenesis of endothelial cells through the simultaneous targeting of ANGPT1 and TGFβR2 genes in breast cancer cells.
Discussion
In this study, we analyzed the miRNome of a set of ductal breast tumors, and identified a signature of 54 miRNAs differentially expressed between tumors and normal tissues. Importantly, we validated the expression of 29 miRNAs in 776 breast tumors and matched adjacent tissues obtained from TCGA. In particular, we found that miR-204, a miRNA that is frequently repressed in human malignancies, was consistently downregulated in all the clinical specimens analyzed here. Importantly, other downregulated miRNAs with key roles in tumorigenesis such as miR-216b, which targets K-RAS oncogene in nasopharyngeal carcinoma and colorectal cells, were identified13,14. MiR-376c and miR-369-3p, which target the Insulin-like Growth Factor 1 Receptor (IGF1R) in melanoma15, and c-MYC in osteosarcoma, respectively, were also identified. Additionally, we detected upregulated miRNAs targeting important tumor suppressor genes in diverse neoplasia, such as miR-638 (BRCA1), miR-130b (TP53), miR-130b (CSF1), miR-142-3p (IL1A), and miR-301a (BIM, PTEN) (Table 2). However, the role of most of these miRNAs in breast cancer remains to be elucidated. Here, we focused in the study of miR-204, as its function has not been completely addressed in breast cancer. Previous reports indicate indicated that miR-204 is downregulated in diverse malignancies16,17,18,19,20,21,22,23,24,25,26 where it is associated with a poor prognostic and a more aggressive phenotype. A potential role for miR-204 in neovascularization processes no related to cancer has been reported. For instance, miR-204 modulates vascular remodeling in human pulmonary hypertension27,28, and loss of miR-204 is associated to corneal neovascularization in mice29.
Several miRNAs have been implicated in angiogenesis in diverse types of cancers30,31. Nonetheless; the potential biological role of most miRNAs in the angiogenesis regulation of breast cancer is poorly understood. To further define the functions of miR-204 in breast cancer we restored its expression by RNA mimics in MDA-MB-231 and MCF-7 breast cancer cells and analyzed its effects by diverse cellular approaches. The expression of miR-204 resulted in a reduction in cell proliferation, migration and invasion indicating that it functions as a tumor suppressor. Transcriptome analyses showed that a number of genes involved in cell proliferation, migration and angiogenesis, including CREB5, ARHGAP5, FOXC1 MAPRE2, RAB22A and SMAD4, were suppressed by miR-204. Particularly, we found the downregulation of the ANGPT1 and TGβR2 genes which are involved in cell proliferation, migration and angiogenesis in cancer cells11,12. Congruently, our data indicated that miR-204 was able to suppress angiogenesis in vitro through the direct targeting of ANGPT1 and TGβR2, indicating an important role in the neovascularization process. Recent studies showed that both ANGPT1 and TGFβR2 could be regulated by microRNAs32, however no previous involvement of miR-204 in angiogenesis in breast cancer has been described. ANGPT1 is a secreted glycoprotein, which binds to Tie2/Tie1 receptors expressed in vascular endothelium, and exerts downstream cellular effects required for the organization and maturation of newly formed vessels33 ANGPT1 also inhibits apoptosis, stimulates migration, and cell proliferation34,35,36. On the other hand, TGFβR2 is the ligand-binding receptor for all members of TGFβ family, and previous studies in mouse models have reported that loss of TGFβR2 expression in mammary fibroblasts is linked to tumor initiation and metastasis and to cell proliferation, and angiogenesis37. TGFβ pathway plays intricate roles in tumorigenesis behaving as a tumor suppressor at early stages of carcinogenesis as well as a tumor promoter at late stages38,39. The TGFβ pathway promotes tumor progression by inducing tumor growth, epithelial mesenchymal transition, invasion, and metastasis and plays important roles in physiological and pathological angiogenesis40. Notably, the TGFβ pathway regulates a number of miRNAs such as miR-29a which promotes angiogenesis in endothelium41. Taken altogether our results suggested that miR-204 plays a role in angiogenesis through the negative regulation of ANGPT1 and TGFβR2. Congruently, we found an inverse correlation between the expression of mR-204 and ANGPT1 and TGFβR2 in breast tumors and cancer cell lines. Intriguingly, in some cases, the expression levels of ANGPT1 and TGFBR2 did not correlated with miR-204 levels as is common when compare tissues and cell lines because the heterogeneity of cancer cells. The genetic background of breast cancer cell lines confers differential changes in the expression impacting biological processes resulting from mutational, transcriptional and epigenetic changes. We observed similar findings, particularly for breast cancer cell lines that express different levels of miR-204 and ANGPT21/TGBR2 which sometimes did not correlate because the heterogeneous genetic background of the cells. It is important to note that target gene levels could be finely coregulated by others microRNAs or additional genetic or epigenetic mechanisms which may differ between the breast cancer cells lines studied here which explains, in part, the discordance between miR-204 and ANGPT21/TGBR2 levels observed in a minority of cell lines analyzed here. We found higher ANGPT1 expression in MDA-MB-231, BT20, MCF-7, and ZR-75-1 cell lines, but not inT47D and SKBR3 cells, in comparison to non-tumorigenic MCF-10A and normal adjacent tissues (Fig. 7H). In a similar way high expression levels of TGFβR2 were observed in five breast cancer cell lines, except for ZR-75-1 cells, relative to MCF-10A and normal tissues. These observations are strengthened by the results of the integration-based approach for ANGPT1 and TGFβR2 expression in a larger cohort of breast cancer patients (n = 522) using datasets from TCGA which indicate that both proteins were downregulated in the half of samples (Fig. 7G). It will be expected that the targets levels should be inversely proportional to miR-204 expression, however we don’t found this behavior in less of half of cell lines analyzed, this could be due also to different regulation mechanisms of TGFβR2 and ANGPT1 expression level, which did not involve miRNAs. The mechanism behind the discordant expression between miR-204 and these two targets in specific cell lines remains to be defined.
Finally, here we provide evidences based on the functional analysis, including knockdown and recue assays, which highlight the role of miR-204/ANGPT1/TGFβR2 axis in angiogenesis. TGFβR2 gene silencing, but not ANGPT1, was able to impair cell proliferation and migration, whereas inhibition of both genes alters angiogenesis. In addition, rescue of ANGPT1 and TGFβR2 expression in MDA-MD-231 cells expressing miR-204 resulted in a partially restoration of angiogenesis in vitro. Thus it is reliable to propose that miR-204 may exert an anti-oncogenic activity in breast cancer cells by two pivotal mechanisms depicted in the working model: i) suppression of TGFβ pathway leading to cell proliferation, migration and angiogenesis repression, and ii) repression of ANGPT1 resulting in angiogenesis blockage (Fig. 9K). Although speculative and considering the role of miR-204 in angiogenesis, we propose that the implementation of microRNA mimics approaches may represent a potential tool for RNA-based breast cancer therapy.
Methods
Cell lines
Human breast carcinoma MDA-MB-231, MCF-7, MDA-MB-453, ZR-75 and T457-D cell lines were obtained from the American Type Culture Collection, and routinely grown in Dulbecco’s modification of Eagle’s minimal medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (50 unit/ml; Invitrogen, Carlsbad, CA, USA).
Animals
Female athymic nu/nu mice at 6–8 weeks of age were used in the experiments. All animals were maintained on 12 h light/12 h dark. Food and water were available ad libitum before to angioreactors implantation Mice were anesthetized with 100 mg/ml Ketamine HCL and 20 mg/ml Xylazine.
Tissues collection
None of the patients recruited in this study received any antineoplastic therapy prior to surgery. After tumor resection clinical specimens were embedded in Tissue-Tek and snap frozen in liquid nitrogen at −80 °C. Pathologist confirmed the existence of at least 80% tumor cells in clinical specimens. All experimental protocols with human tissues were approved by the ethics committee of Institute of Breast Diseases-FUCAM.
Ethics statements
The Institute of Breast Diseases-FUCAM, Mexico provided the locally invasive breast tumors and normal tissues collection. We confirm that all experiments were performed in accordance with relevant guidelines and regulations of FUCAM ethics committee. A signed informed consent was obtained from each participant or her representative prior to release for research use. For animal studies, all experimental protocols were carried out in accordance with the approved guidelines at CINVESTAV-IPN.
RNA isolation
Tissues were lysed using a Tissue Ruptor (Qiagen Inc., Valencia, CA), and RNA was extracted using 1 ml Trizol (Invitrogen, Carlsbad, CA) per 50–100 mg tissue as described by the manufacturer. RNA integrity was assessed using capillary electrophoresis system Agilent 2100 Bioanalyzer. Samples with a RNA integrity number >6 were processed.
MicroRNAs profiling
Expression analysis of 667 miRNAs in nine ductal breast tumors and normal adjacent tissues was performed by reverse transcription and quantitative real-time polymerase chain reaction (RT-qPCR) using the Megaplex TaqMan Low-Density Arrays (TLDAs) v2.0 system (Applied Biosystems. Foster City, CA) as described by the manufacturer. Briefly, 70 ng total RNA were retro-transcribed using stem-loop primers. In order to detect low abundant miRNAs, a pre-amplification step was included. The pre-amplified product was loaded into the TLDA and amplification signal detection was performed in the 7900 FAST real time thermal cycler (ABI). Tests were normalized using RNU48 and RNU44 as controls.
Validation of microRNAs profiling data
The Cancer Genome Atlas (TCGA) was used to obtain the datasets of miRNA expression from 776 matched tumors and adjacent normal tissues available from TCGA data matrix (http://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). This dataset was compared with the miRNAs expression profile obtained here in the discovery cohort using TLDAs.
Statistical analysis of microRNAs expression
MiRNAs levels were measured by RT-qPCR in TLDAs using the comparative Ct (2−ΔΔCt) method. All analyses were done using R (HTqPCR, gplots-bioconductor). The Ct raw data was determined using an automatic baseline and a threshold of 0.2. A fold change (log2 RQ) value >1.0 was used to define the differentially expressed miRNAs. An adjusted t-test was used to evaluate the significant differences in Ct values between tumoral and non-tumoral tissues. To identify sub-groups defined by miRNAs expression profiles, an unsupervised clustering analysis using Spearman correlation and average linkage was used.
Reverse transcription and real-time polymerase chain reaction
Quantitative RT-PCR analysis of individual microRNAs was performed using MiRNA Assays (Applied Biosystems, Foster City, CA). 100 ng total RNA were reverse transcribed using a looped-RT specific primer, 0.15 μl dNTPs (100 mM), 1.0 μl reverse transcriptase MultiScribe (50 U/μl), 1.5 μl 10X buffer, 0.19 μl RNase inhibitor (20 U/μl) and 4.16 μl RNase-free water. Then, diluted retrotranscription reaction (1:15) was mixed with 10 μl master mix TaqMan (Universal PCR Master Mix, No AmpErase UNG, 2X), 7.67 μl RNase free water, and 1.0 μl PCR probe. PCR reaction was performed in a GeneAmp System 9700 (Applied Biosystems) as follows: 95 °C for 10 min, and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Tests were normalized using RNU44 and RNU48 as control.
Prediction of miRNAs targets
MiRNAs target genes were predicted using TargetScan and PicTar software. Only gene targets predicted by the two algorithms were included in further analysis.
MicroRNA-204 transfection
MiRNA-204 precursor (pre-miR-204, Life Technologies) and pre-miR-negative control scramble (AM17110, Life Technologies) were transfected at 30 nM in MDA-MB-231 and MCF-7 cells using siPORT amine transfection agent (Ambion). Briefly, pre-miR-204 was individually added to wells containing 1 × 107 cells cultured in DMEM for 48 h. Expression of miR-204 was evaluated by RT-qPCR at 48 h postransfection.
Clonogenic assays
MCF-7 and MDA-MB-231 cells were transfected with pre-miR-204 (30 nM) and scramble (30 nM). Forty-eight hours after transfection, cells were trypsinized, manually counted and seeded in six-well plates (1000 cells per well) to form colonies in 1–3 weeks. A colony was defined to consist of at least 50 cells. After 1–3 weeks, the colonies were counted and experimental data were analyzed. At least three independent experiments were performed for each cell line and data were expressed as mean ±S.D. p < 0.05 was considered as statistically significant.
Cell migration and invasion assays
Briefly, cells treated with the pre-miR-204 (30 nM), or scramble sequence (30 nM) were seeded in triplicate in a six-well plate and grown to 80% confluence. Twenty-four hours postransfection, a vertical wound was traced in the cell monolayer. After 12 and 24 h, cells were fixed with 4% paraformaldehyde and the scratched area was quantified. In transwell assays, chambers (Corning) with 6.5-mm diameter and 8-μm pore size polycarbonate membrane were used. MDA-MB-231 and MCF-7 cells (1 × 105) transfected with pre-miR-204, or scramble were transferred to 0.5 ml serum-free medium and placed in the upper chamber, whereas the lower chamber was loaded with 0.8 ml medium containing 10% fetal bovine serum. The total number of cells that migrated into the lower chamber was counted after 24 h incubation at 37 °C. Cell invasiveness was evaluated using transwell chambers coated with a layer of extracellular matrix (BD Biosciences). MDA-MB-231 cells were treated with pre-miR-204 (30 nM) and 24 h postransfection, the invasive cells were fixed with 100% methanol, stained with 1% toluidine blue (Sigma) and quantified by manual counting in randomly selected areas. Experiments were performed three times by triplicate and results were expressed as mean ± S.D. p < 0.05 was considered as statistically significant.
Angiogenesis assays in vitro
Tubules formation assays based on the ability of human umbilical vein endothelial cells (HUVEC) to form three-dimensional capillary-like tubular structures in vitro were performed using the Gibco Angiogenesis Starter Kit (A1460901), which contains media and reagents optimized for culturing HUVEC on Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix to model the formation of endothelial cell tube networks. Procedures were as follows: Wells of a 24-well plate were coated with 50 μL of geltrex matrix and incubated at 37 °C for 30 min. Then, MDA-MB-231 cells (1 × 104cells/well) transfected with pre-miR-204 (30 nM) or scramble (30 nM) negative control were added and cultured in complete DMEM medium. After confluence, medium was removed and HUVEC (1 × 104 cells) were co-cultured with MDA-MB-231 cells in DMEM without complement. Cultures of HUVEC alone or treated with VEGFA were used as negative and positive controls, respectively. After 24 h of co-culture, the formed tubules were observed under an inverted microscope (Iroscope SI-PH) and imaged. Branch points and tubular structures were individually counted by two observers. MDA-MB-231 cells (1 × 104 cells/well) silenced in ANGPT1 or TGFβR2 genes by RNA interference were also evaluated. Experiments were performed three times by triplicate and results were expressed as mean ± S.D. p < 0.05 was considered as statistically significant.
Directed in vivo angiogenesis assay
Angiogenesis in vivo was evaluated using the DIVAA kit (Trevigen) with some modifications. Briefly, angioreactors were filed with 50,000 pre-miR-204 transfected or scramble control MDA-MB-231 cells embedded in 20 μl of basement membrane extract (BME). Angioreactors were incubated at 37 °C for 1 h. For positive controls, angioreactors were filled with BME supplemented with VEGF (18 ng/ml) plus FGF1 (60 ng/ml). Two angioreactors were implanted in each immunocompromised nude mice subcutaneously in the dorsal region, (2 mice for pre-miR-204, 2 mice for scramble and 2 mice for VEGF/FGF1). The angioreactors were removed after 9 days after implantation, angioreactors were photographed. Presence of blood vessels quantified using FITC-Lectin detection, fluorescence was determined in a plate reader as mean relative fluorescence units.
DNA microarrays analysis
Global gene expression analysis was done for MDA-MB-231 cells transfected with pre-miR-204 (30 nM) or scramble (30 nM) using the NimbleGen array (Roche). RNA samples were used to synthesize double-stranded labeled cDNA using SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and NimbleGen One-Color DNA Labeling Kit. Samples were hybridized in NimbleGen array 12 × 135 K (12 × 135,000 features). After hybridization and washing, the processed slides were scanned using a NimbleGen MS200 Microarray Scanner. Raw data were extracted as pair files by NimbleScan software (version 2.5), background corrected and data were normalized. The probe level files and gene summary files were produced and imported into ANAIS software (Analysis of NimbleGen Arrays Interface) for further analysis. DNA microarrays data for 10 selected genes was validated by RT-qPCR using specific oligonucleotides.
Gene Ontology (GO) analysis
In order to determine possible pathways affected by the modulated genes by miR-204 an enrichment an analysis was performed using Gene Ontology (GO) program DAVID 6.7. Gene Ontology (GO) is an international standardized classification system for gene function, which supplies a set of controlled vocabulary to comprehensively describe the property of genes and gene products. We analyzed the 3 classifications from GO: cellular component, molecular function and biological process using the default parameters.
Luciferase reporter gene assays
The 3′UTR of ANGPT1 and TGFβR2 genes was amplified using specific primers and cloned downstream of luciferase gene into p-miR-report vector (Ambion). All constructs were verified through automatic sequencing. The recombinant pmiR-LUC-ANGPT1 and pmiR-LUC-TGFβR2 plasmids were transfected into MDA-MB-231 cells using lipofectamine 2000 (Invitrogen). At 3 h postransfection, the medium was replaced with complete fresh medium. 24 h after transfection, pre-miR-204 (30 nM) or pre-miR-negative control (scramble) were co-transfected with lipofectamine RNAi max (Invitrogen). Then 24 h after transfection, firefly and renilla luciferase activities were both measured by the Dual-Glo luciferase Assay (Promega) using a Fluoroskan Ascent™ Microplate Fluorometer (Thermo Scientific). Firefly luciferase activities were normalized with Renilla reniformis luciferase activities. P values for differences between control and pre-miR-204 transfected cells were determined by two-tailed Student’s t test.
Western blots
Proteins were separated on 10% polyacrylamide gels and transferred to 0.2 μm nitrocellulose membrane (Bio-Rad) in transfer buffer (25 mM Tris, 192 mM glycine and 10% methanol). Membranes were dried and blocked for 60 min at room temperature with TBST-1X (137 mM NaCl, 20 mM Tris, 0.1% Tween-20, at pH 7.6) containing 5% BSA (Sigma-Aldrich) and incubated overnight at 4 °C with rabbit anti-GAPDH (1:2000 Cell Signaling), rabbit anti-ANGPT1 (1:2000 Abcam), rabbit anti-CREB5 (1:1000 Abcam), and rabbit anti-TGFβR2 (1:1000 Abcam) antibodies. Membranes were washed three times in TBST-1X and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2,500, Zymed). Signal was detected and developed using ECL Western blot detection reagent (Amersham).
RNA interference assays
Four oligonucleotides (21–23 nt) encoding short hairpin RNAs (shRNAs) targeting the ANGPT1 and TGFβR2 genes were designed. To minimize the possibility of shRNAs off targeting effects, a nucleotide BLAST search was performed. The oligonucleotides codifying for specific shRNAs were cloned into pSilencer 5.1 U6 retro plasmid (Ambion) and sequences were confirmed by automatic sequencing. The resulting recombinant plasmids were dubbed as sh-ANGPT1 1.1, sh-ANGPT1 1.2, sh-TGFβR2 1.1 and sh-TGFβR2 1.2. The effectiveness of the constructs in gene silencing was tested by transient transfection of MDA-MB-231 cells (2 × 105) using lipofectamine 2000 reagent (Invitrogen, 12566014) followed by ANGPT1 and TGFβR2 detection using Western blotting at 48 h post-transfection.
ANGPT1 and TGFβR2 expression and rescue assays
The ANGPT1 (NM_001146.4) open reading frame (ORF) was amplified from human cDNA of MDA-MB-231 cells and by polymerase chain reaction (PCR) using the primers: 5′-CCCAAGCTTATGACAGTTTTCCTTTCCTTTGCT-3′ (forward) and 5′-CCGCTCGAGTGTGAACTCAAACGGCTCCA-3′ (reverse). The TGFβR2 (NM_001024847.2) ORF was amplified from human cDNA of MDA-MB-231 cells by PCR using the primers: 5′-CCCAAGCTT ATGGGTCGGGGGCTGCTCAGGG-3′ (forward) and 5′-CCGCTCGAGTTTGGTAGTGTTTAGGGAGCCG TCT-3′ (reverse). The ANGPT1 and TGFβR2 genes were orientally cloned into HindIII and XhoI restriction sites of pCDNA3 mammalian expression vector (Thermo Fisher Scientific) to generate pCDNA3-ANGPT1 and pCDNA3-TGFβR2 plasmids. All constructs were verified through automatic sequencing. 5 μg each of constructions were individually transfected into MDA-MB-231 cells using Lipofectamine 2000 (Invitrogen). After transfection Western blotting were performed to determine the overexpression of ANGPT1 and TGFβR2 proteins. For rescue assays, MDA-MB-231 cells (250,000) were transfected with miR-204 precursor or pre-miR-negative control (scramble). 24 h after miRNA transfection MDA-MB-231 cells were individually cotransfected with pCDNA3-ANGPT1 or pCDNA3-TGFβR2 constructions and after 48 h transfected cells (10,000) were prepared for angiogenesis assays as describe before. Briefly, cells were seed in BME coated well in 96 well plate in medium without supplement, and 24 h after HUVEC cells (10,000) were co-cultured with MDA-MB-231 cells as described above. Tube structures formation were analyzed and quantified 24 h after the co-cultures.
Statistical analysis
Experiments were performed three times by triplicate and results were represented as mean ±S.D. One-way analysis of variance (ANOVA) followed by Tukey’s test were used to compare the differences between means. A p < 0.05 was considered as statistically significant.
Additional Information
How to cite this article: Flores-Pérez, A. et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Sci. Rep. 6, 34504; doi: 10.1038/srep34504 (2016).
Supplementary Material
Acknowledgments
We acknowledge to CONACyT- fondo investigacion cientifica basica and SS/IMSS/ISSSTE for support in this investigation (grants 233370 and 222335). We also acknowledge to Instituto de Ciencia y Tecnología del Distrito Federal, Mexico (ICyTDF/328/2011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Ali Flores-Pérez was supported by ICyTDF-UACM (SRI/PB/64/2011), Estado de Mexico, COMECYT (14BTD0106) and CONACYT (222335) fellowships.
Footnotes
Author Contributions A.F.-P., L.A.M., C.P.-F., M.A.F.-S., J.A.G.-B., C.P.-P. and M.L.S., performed all the experiments. S.R.-C., V.B.-P. and M.S.M., provided the biopsies and clinical data. A.H.-M., assayed the microRNAs profiling. M.L.S. provided materials for in vivo assays. E.R.-G., H.A.-d.l.V. and C.P.-P provided other materials. C.L.-C., L.A.M., A.F.-P. and E.A.O., designed all the experiments. C.L.-C., L.A.M. and A.F.-P wrote the manuscript.
References
- Croce C. M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 10, 704–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. & Gregory R. I. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 22, 321–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E. MicroRNAs and cancer: Role in tumourigenesis, patient classification and therapy. Clin Transl Oncol. 9, 155–160 (2007). [DOI] [PubMed] [Google Scholar]
- Spannuth W. A., Sood A. K. & Coleman R. L. Angiogenesis as a strategic target for ovarian cancer therapy. Nat Clin Pract Oncol. 5, 194–204 (2008). [DOI] [PubMed] [Google Scholar]
- Albini A., Tosetti F., Li V. W., Noonan D. M. & Li W. W. Cancer prevention by targeting angiogenesis. Nat Rev Clin Oncol. 9, 498–509 (2012). [DOI] [PubMed] [Google Scholar]
- Ferrara N. & Kerbel R. S. Angiogenesis as a therapeutic target. Nature 438, 967–974 (2005). [DOI] [PubMed] [Google Scholar]
- Dews M. et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 38, 1060–1065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Deng Z., Wang C. H. & Yang B. B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 104, 20350–20355 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Z. et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS One. 6, e19139 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Gallagher R., Ufret-Vincenty R., Li X., Olson E. N. & Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA. 108, 8287–8292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunckhorst M. K., Xu Y., Lu R. & Yu Q. Angiopoietins promote ovarian cancer progression by establishing a procancer microenvironment. Am J Pathol. 184, 2285–2296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. et al. Prostate angiogenesis in development and inflammation. Prostate. 74, 346–358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Zhang W. & Huang J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 588, 1562–1570 (2014). [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Lee Y. H. & Bae Y. S. MiR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting α subunit of protein kinase CKII in human colorectal cancer cells. Biochem Biophys Res Commun. 429, 173–179 (2012). [DOI] [PubMed] [Google Scholar]
- Zehavi L. et al. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 11, 44–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Li W., Jin X., Zhang Q., Zhang G. & Deng X. Decreased expression of miR-204 is associated with poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 7, 3287–3292 (2014). [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li L., Su J. & Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS One. 9, e101457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bao W. et al. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 9, 155–164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. miR-204 mediated loss of myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol Cancer. 12, 105–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam J. S. et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 7, e52397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova O. et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 21, 532–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang X. & Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 14, 290–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z. et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 73, 990–999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacconi A. et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 3, e423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T. K. et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 130, 1036–1045 (2012). [DOI] [PubMed] [Google Scholar]
- Courboulin A. et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 208, 535–548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potus F., Graydon C., Provencher S. & Bonnet S. Vascular remodeling process in pulmonary arterial hypertension, with focus on miR-204 and miR-126. Pulm Circ. 4, 175–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather J. N. et al. Angiopoietin-1 Is regulated by miR-204 and contributes to corneal neovascularization in KLEIP-deficient mice. Invest Ophthalmol Vis Sci. 55, 4295–4303 (2014). [DOI] [PubMed] [Google Scholar]
- Plummer P. N. et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 73, 341–352 (2013). [DOI] [PubMed] [Google Scholar]
- Suárez Y. & Sessa W. MicroRNAs as novel regulators of angiogenesis. Circ Res. 104, 442–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N. et al. Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-b pathway and reveal microRNA regulation of TGFBR2. Silence. 14, 23–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle N. P., Saharinen P. & Alitalo K. Signaling and functions of angiopoietin-1 in vascular protection. Circ Res. 98, 1014–1023 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H. J., So J. N., Lee S. J., Kim I. & Koh G. Y. Angiopoietin-1 is an apoptosis survival factor for endothelial cells. FEBS Lett. 448, 249–253 (1999). [DOI] [PubMed] [Google Scholar]
- Fujikawa K., Scherpenseel I., Jain S. K., Presman E., Christensen R. A. & Varticovski L. Role of PI 3-kinase in angiopoietin-1-mediated migration and attachment-dependent survival of endothelial cells. Exp Cell Res. 253, 663–672 (1999). [DOI] [PubMed] [Google Scholar]
- Xiao-Hong L., Chen-Guang B., Yang Y., De-Jun G. & Sheng-Dong H. Angiopoietin-1 targeted RNA interference suppresses angiogenesis and tumor growth of esophageal cancer. World J Gastroenterol. 14, 1575–1581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S., Aca r. A., Magnusson Y., Gregersson P., Ryde’n L. & Landberg G. TGF-beta receptor type-2 expression in cancer-associated fibroblasts regulates breast cancer cell growth and survival and is a prognostic marker in pre-menopausal breast cancer. Oncogene 34, 27–38 (2015). [DOI] [PubMed] [Google Scholar]
- Scollen S. et al. TGF-β signalling pathway and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 20, 1112–1119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzinska-Ustymowicz K. & Kemona A. Transforming growth factor beta can be a parameter of aggressiveness of pT1 colorectal cancer. World J Gastroenterol. 11, 1193–1195 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Li G., Wu J., Wang L. Uhler M. & Simeone D. PKA modulates TGF-beta signaling through a direct Interaction with Smad4. J Biol Chem. 288, 8737–8749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Non-Smad pathways in TGF-β signaling. Cell Research. 19, 128–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang Y., Wang Y., Ma Y., Lan Y. & Yang X. Transforming growth factor β-regulated microRNA-29a promotes angiogenesis through targeting the phosphatase and tensin homolog in endothelium. J Biol Chem. 288, 10418–10426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.