Abstract
Almost all eukaryotic proteins are subject to post-translational modifications during mitosis and cell cycle, and in particular, reversible phosphorylation being a key event. The recent use of high-throughput experimental analyses has revealed that more than 70% of all eukaryotic proteins are regulated by phosphorylation; however, the mechanism of dephosphorylation, counteracting phosphorylation, is relatively unknown. Recent discoveries have shown that many of the protein phosphatases are involved in the temporal and spatial control of mitotic events, such as mitotic entry, mitotic spindle assembly, chromosome architecture changes and cohesion, and mitotic exit. This implies that certain phosphatases are tightly regulated for timely dephosphorylation of key mitotic phosphoproteins and are essential for control of various mitotic processes. This review describes the physiological and pathological roles of mitotic phosphatases, as well as the versatile role of various protein phosphatases in several mitotic events.
Keywords: chromosome, human disease, kinase, mitosis, protein phosphatase, therapeutics
INTRODUCTION
Reversible protein phosphorylation is the primary type of post-translational modification in eukaryotic cell division and cell cycle. Dynamic changes in phosphorylation state of many cell cycle-mediated proteins regulate their molecular and cellular fates, and aberrant protein phosphorylation can contribute to many pathological conditions and diseases. In fact, more than 70% of all cellular proteins are regulated by phosphorylation (Dephoure et al., 2008; Macek et al., 2009; Olsen et al., 2006; 2010). In addition, through the human genome analyses, a recent report indicated that 518 kinases are either Tyrosine (Tyr) kinases or Serine/Threonine (Ser/Thr) kinases (Johnson and Hunter, 2005; Manning et al., 2002), whereas the total number of Tyr phosphatases and Ser/Thr phosphatases is approximately 137, much fewer than the number of aforementioned kinases (Brautigan, 2013; Tonks, 2006). There are around 107 Tyr phosphatases in the human genome and, overall, the number of tyrosine kinases is similar to those of Tyr phosphatases. However, more than 400 of serine/threonine kinases are identified in the human genome (Manning et al., 2002) and the bulk phosphorylation at serine/threonine residues is more than 98% of the total protein phosphorylation in mitosis. In contrast, the number of genes encoding Ser/Thr phosphatases is about 30 in the human genome (Shi, 2009). This numerical paradox is explained by the facts that some of the Ser/Thr phosphatases form diverse complexes with many interacting and catalytic subunits for substrate specificity; in addition, there also exist dual specificity phosphatases that can dephosphorylate both phospho-Tyr and phospho-Ser/phospho-Thr residues within one substrate. In this regard, it is becoming apparent that protein phosphatases need to be highly and properly regulated, depending on the specific substrates, particularly in terms of the balance with their kinase counterparts during mitosis. Here, we will focus on recent advances depicting the importance of phosphatase functions in the dynamic control of phosphorylation, including mitotic entry, mitotic spindle assembly, chromosome architecture changes and cohesion, and mitotic exit. We also discuss the roles and molecular mechanisms of mitotic phosphatases in human health and disease, including various cancers.
COMPOSITION OF MITOTIC PHOSPHATASES
The genomes of higher eukaryotes contain about 140 phosphatases, which are primarily classified into three groups (Table 1): the classical tyrosine phosphatases (cTPs), the dual specificity phosphatases (DSPs) in tyrosine phosphatase (TP), and the serine/threonine phosphatases (STPs) (Hunter, 1995; Shi, 2009; Virshup and Shenolikar, 2009). The cTP superfamily containing the cysteine-based classical Tyr phosphatase domain is encoded about 38 genes in the mammalian genome (Manning et al., 2002).
Table 1.
Classification of protein phosphatases
| Phosphatase superfamily | Phosphatase subfamily | Examples of member | Identified mitotic phosphatases |
|---|---|---|---|
| Classical Tyrosine phosphatase | Non-receptor phosphatase | PTP1B, TCPTP, SHP1, LYP, MEG2 | n.d. |
| Transmembrane phosphatase | PTPα, CD45, CD148, IA-2, GLEPP1 | ||
| Dual specificity phosphatase | MAP kinase phosphatase | MKP1-5, PAC1 | CDC14 CDC25 Ssu72 |
| Atypical DSPs | VHR, PIR, Laforin, VHZ, STYX | ||
| Slingshots | SSH1, SSH2, SSH3 | ||
| PRLs | PRL-1, PRL-2, PRL-3 | ||
| CDC14 proteins | CDC14A, KAP | ||
| PTENs | PTEN, TPIP | ||
| Myotubularins | MTM1, MTMR1-15 | ||
| Low molecular weight | Ssu72 | ||
| Cdc25 proteins | Cdc25A, Cdc25B, Cdc25C | ||
| Serine/Threonine phosphatase | PPP subfamily | PP1, PP2A, Calcineurin, PP5 | PP1 PP2A PP4 PP6 Fcp1 |
| PPM subfamily | PP2C | ||
| Fcp family | Fcp1 |
n.d. not yet determined
Protein tyrosine phosphorylation and dephosphorylation are the common post-translational modification that can create specific recognition motifs for cellular localization, protein stability, enzyme activity and protein interactions (Tonks, 2006). Tyrosine-specific phosphatases remove the phosphate group attached to a tyrosine residue by using the cysteinyl-phosphate enzyme intermediate. The DSP superfamily belongs to the tyrosine phosphatase (TP) superfamily and shares catalytic mechanism similarity to the TP superfamily by which a conserved cysteine residue forms a covalent intermediate with the phosphate group to be removed. DSPs can dephosphorylate the phospho-serine/phospho-threonine residues as well as the phospho-tyrosine residues, implying that they function as a possible alternative for the insufficient number of the serine/threonine phosphatases. The number of genes for cTP and DSP superfamilies is about 107 in mammalian genomes, which is similar to the gene number of tyrosine kinases. However, for cTPs and DSPs, only Cdc14, Cdc25 and Ssu72 play an essential role in mitosis (Kim et al., 2010; 2013; Queralt and Uhlmann, 2008; Trinkle-Mulcahy and Lamond, 2006).
The number of genes encoding STP proteins is about 30 in mammalian genomes (Shi, 2009). There are 10 times fewer genes encoding serine/threonine phosphatases than serine/threonine kinases. Therefore, this imbalance in number implies that STPs have extensive activity in dephosphorylation of various substrates during mitosis and, in fact, the diversity of STPs predominantly results from their association with distinct regulatory subunits to make holoenzyme complexes. STP proteins have many regulatory proteins that bind to the phosphatase catalytic subunits and determine their cellular localization, substrate specificity and phosphatase activity. In particular, PP2A forms over 100 heterotrimeric holoenzymes depending on specific regulatory proteins. Similarly, protein phosphatase 1 (PP1) forms over 400 heterodimeric holoenzymes (Bollen et al., 2010; Eichhorn et al., 2009; Guo et al., 2014; Shi, 2009). The STPs superfamily is further divided into three groups: phosphoprotein phosphatases (PPPs), metal-dependent protein phosphatases (PPMs) and aspartate-based phosphatases (Shi, 2009). The PPP group is the largest group of STPs superfamily, and many PPPs are involved in cell cycle regulation, including PP2A (Hunter, 1995; Shi, 2009; Virshup and Shenolikar, 2009). However, previous reports from various organisms have only implicated a limited number of phosphatases in the control of mitosis with PP2A, PP1, PP4 and PP6 (STP superfamily) and Cdc14, Cdc25, Fcp1 and Ssu72 (TP and DSP superfamilies) (Della Monica et al., 2015; Mehta et al., 2012; Nilsson and Hoffmann, 2000; Queralt and Uhlmann, 2008; Sullivan and Morgan, 2007; Trinkle-Mulcahy and Lamond, 2006; Zeng et al., 2010).
FUNCTION OF PHOSPHATASES DURING MITOTIC ENTRY
The requirement of specific protein kinases for accurate regulation of mitosis implies the existence of specific protein phosphatases as well (Fig. 1; Barr et al., 2004; Lindqvist et al., 2009; O’Farrell, 2001; Ruchaud et al., 2007). Among the various mitotic kinases, the cyclin-dependent protein kinase 1 (Cdk1) has a versatile role as it drives mitotic entry. Interestingly, multiple phosphatases are associated with proper regulation of Cdk1 and regulated by their intracellular localization. Activation of Cdk1 triggers the mitotic entry and this situation requires removal of Wee1-mediated inhibitory phosphorylations by dual specific phosphatases Cdc25B and Cdc25C at both centrosomes and nucleus (Bonnet et al., 2008; Hoffmann et al., 1993; Izumi and Maller, 1993; Mueller et al., 1995). During interphase, Cdc25 phosphatases are generally kept inactive by phosphorylation of Ser287 residue, which provides a docking site for 14-3-3 protein and masks the nuclear localization signal (Kumagai and Dunphy, 1999; Kumagai et al., 1998). The dephosphorylation of Ser287 by PP1 partially activates Cdc25C, which then activates a small pool of Cdk1 for the G2-M transition (Perdiguero and Nebreda, 2004). A positive feedback between Cdc25C and Cdk1 along with increased interaction of PP1 lead to full activation of both enzymes. Activated Cdk1 is generally assumed to be sufficient to drive cells into mitosis, and degra dation of cyclin B and inactivation of Cdk1 are sufficient to allow mitotic exit (Deibler and Kirschner, 2010; O’Farrell, 2001; Pomerening et al., 2003; Potapova et al., 2006).
Fig. 1.
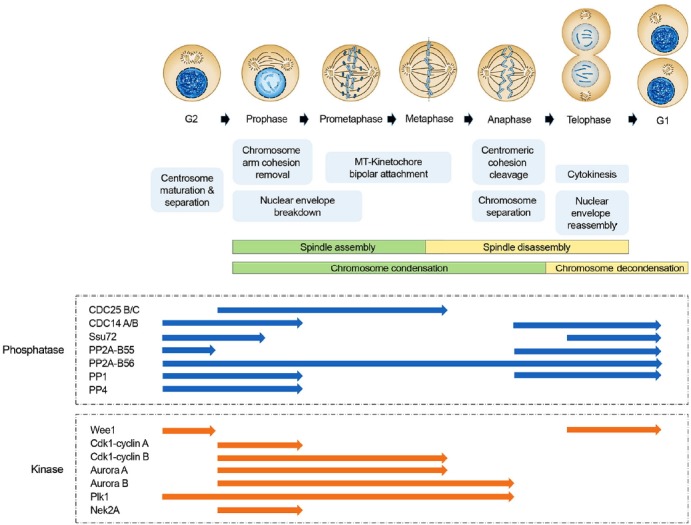
Role of protein kinases and phosphatases in mitotic events. Schematic model of the composition of centrosomes, chromosome, microtubules, and nucleus during cell cycle is shown (upper panel). Key functional events at each stage during cell cycle are also depicted (middle panel). The phases in activation status both major mitotic kinases and phosphatases during cell cycle are indicated (bottom pane). The blue and orange arrows represent functional stages in kinase and phosphatase activation, respectively.
Greatwall is a kinase that was initially characterized as a mutant displaying defective chromosome condensation in mitosis of the Drosophila melanogaster (Archambault et al., 2007; Yu et al., 2004). More recently, similar findings were identified for the human homologue microtubule-associated serine/threonine kinase-like enzyme (MASTL) (Burgess et al., 2010; Voets and Wolthuis, 2010). When cells enter mitosis, Greatwall highly increases its kinase activity and is auto-phosphorylated, in part by Cdk1. Interestingly, even in the presence of high Cdk1 activity, loss of Greatwall maintains the dephosphorylation status of several mitotic phosphoproteins, suggesting that Greatwall is an inhibitor of the protein phosphatases that antagonize Cdk1. In fact, a series of studies has revealed that Greatwall promotes the mitotic entry, not only by the activation of Cdk1 but also by inhibition of PP2A (Burgess et al., 2010; Castilho et al., 2009; Vigneron et al., 2009). Intriguingly, this inhibitory effect is not mediated by direct phosphorylation of PP2A catalytic and/or regulatory subunits, but involves small heat-stable proteins called ENSA and ARPP-19 (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Greatwall directly phosphorylates ENSA and ARPP-19 at serine 67 residue, highly conserved among the different species. The phosphorylated ENSA and ARPP-19 then specifically interact with PP2A-B55δ complexes and do not bind to other complexes of PP2A containing B′ (B56)-, B″ (PR72)-, and B′″ (PR93/PR110)-subunits (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). These interactions cause the inhibition of PP2A-B55δ activity and promote mitotic entry. Importantly, even in the presence of full Cdk1 activation, depletion of Greatwall or over-expression of PP2A-B55δ inhibits the entry into mitosis (Mochida et al., 2009). Collectively, these results suggest that several phosphatases, along with kinases, could contribute to the fine-tuning of mitotic entry.
FUNCTION OF PHOSPHATASES IN SPINDLE ASSEMBLY AND ITS CHECKPOINT
During mitosis, the centrosomes move to the bilateral poles and their microtubule-assembly is activated to promote bipolar spindle formation. Formation of the stable bipolar spindle begins with outgrowth of microtubules at centrosomes, a process that is associated with diverse phosphatases. In C. elegans, PP2A-B″ includes RSA1 and 2 and this complex is localized to centrosomes (Schlaitz et al., 2007). The complex coordinates the microtubule outgrowth for both the centrosome and kinetochore, and maintains microtubule stability. In particular, the RSA-PP2A complex regulates the microtubule depolymerizing activity of MCAK and spindle formation by TPX2 in centrosomes (Schlaitz et al., 2007). In human cells, PP1, as a physiological antagonist of Nek2 kinase, regulates the centrosome splitting (Meraldi and Nigg, 2001). In addition, the PP1-Mars complex stabilizes microtubules through the dephosphorylation of microtubule stabilizer TACC during Dro sophila embryogenesis(Tan et al., 2008). PP4 is also required for the centrosome maturation and organization of microtubules at the centro somes (Helps et al., 1998; Sumiyoshi et al., 2002). Human Cdc14A binds with interphase centrosomes and is required as a physiological regulator of centrosome duplication cycle (Mailand et al., 2002). Also, Cdc14B can interact with, bundle, and stabilize microtubules in vitro and in vivo (Cho et al., 2005)and microtubule bundles of Cdc14B are enriched with acetylated α-tubulins (Mailand et al., 2002). Finally, Cdc25B is also associated within the assembly of microtubules and centrosomal gamma-tubulin (Boutros et al., 2007). A recent report suggests that Cdc25B contributes to the natural turnover of centrosomal proteins (Mps1 and centrin2) and controls centrosome number and chromosome instability (Boutros et al., 2013).
As a component of the chromosomal passenger complex (CPC), Aurora B kinase is bound to the inner centromere during mitosis and plays an essential role in both chromosome biorientation and spindle checkpoint. The tension across centromeres and centrosomes establishes a spatial gradient of high phosphorylation activity at the inner centromere and low phosphorylation activity at the outer kinetochores. These gradients are generated by the recruitment of PP1 to the outer kinetochores. In this context, PP1 functions by dephosphorylating Aurora B substrates, including Ndc80, KNL1, and CENP-E (Kelly and Funabiki, 2009; Kotwaliwale and Biggins, 2006; Ruchaud et al., 2007), and then stabilizes kinetochore-microtubule interaction and silences the spindle checkpoint when chromosome bi-orientation is achieved (Kelly and Funabiki, 2009; Ruchaud et al., 2007). Another phosphatase PP2A-B56 also tunes the balance of phosphorylation to promote chromosome-spindle interactions (Foley et al., 2011). PP2A-B56 is enriched at centromere and kinetochore of unattached chromosomes and could regulate the level of phosphorylation of kinetochore substrates by Aurora B and Plk1 kinases (Foley et al., 2011). Indeed, BubR1, as a mitotic checkpoint protein, recruits PP2A-B56 complex to kinetochores and this complex contributes to stabilization of kinetochore-microtubule interaction by counteracting Aurora B kinase activity (Kruse et al., 2013, Suijkerbuijk et al., 2012). Thus, the functional inter play between kinases and phosphatases regulates microtubule binding to kinetochores.
The spindle assembly checkpoint (SAC) blocks the chromosome segregation until all chromosomes have attached to opposite spindle poles (London and Biggins, 2014). This checkpoint is generated by unattached kinetochores and microtubules and forms the huge inhibitor complex, called as the mitotic checkpoint complex (MCC), which is composed of an assembly of the proteins Mad2, BubR1, Bub3 and Cdc2 (Izawa and Pines, 2015). Strikingly, the recruitment of the MCC to kinetochores requires hyper-phosphorylated kinetochore proteins, and PP1 has been suggested to control of SAC inactivation through the phosphatase activity (Musacchio and Salmon, 2007; Sassoon et al., 1999). The multiple pools of kinetochore-associated PP1 may mainly counteract kinase activities and their functions will increase until the threshold levels of PP1 activity required for anaphase onset is reached. Recent study has also suggested that the spindle- and the kinetochore-associated (SKA) complex may aid the PP1 recruitment at kinetochores and oppose the SAC kinases signaling, thereby controlling proper chromosome alignment with full recruitment of PP1 (Sivakumar et al., 2016). Therefore, it is possible that the various mitotic kinases, involving Aurora B and SAC kinases, restrain the activity of such a phosphatase by phosphorylation of its regulatory or catalytic subunit. Conversely, phosphatases such as PP1 and PP2A could also inhibit the activity of mitotic kinases and kinase substrates by dephosphorylation until both enzyme types are physically separated through the dynamic stretching between the centromeres. Also, we do not exclude the possibility that dual specificity phosphatases, such as Cdc14 and Ssu72, exert pivotal effects in the spindle assembly and the SAC signaling pathways.
FUNCTION OF PHOSPHATASES IN CHROMOSOME ARCHITECTURE CHANGES AND COHESION FORMATION
Sister chromatid cohesion and separation from chromosome replication until the anaphase onset depend on the regulation of ring-like cohesin complex onto chromosomes. In vertebrate cell, most cohesin complexes dissociate from chromosome arms by a cellular mechanism called the prophase pathway, which depends on phosphorylation of SA2 cohesin subunit by Plk1 and Aurora B (Peters et al., 2008; Waizenegger et al., 2000). Interestingly, recent studies have suggested that Ssu72 phosphatase, as a cohesin binding protein, is capable of regulating cohesion between the sister chromatid arms in human cells (Fig. 2; Kim et al., 2010; 2013). Ssu72 directly interacts with cohesin complex and counteracts SA2 phosphorylations in vitro and in vivo. Loss of Ssu72 phosphatase activity by Aurora B kinase-mediated phosphorylation results in premature dissociation of chromosome arm cohesion, whereas the overexpression of Ssu72 causes resistance to arm cohesion (Kim et al., 2010; 2013). Additionally, Ssu72 is implicated in formation of gene loops by crosslinking with the promoter and the terminator (Tan-Wong et al., 2012), implying that Ssu72 phosphatase is deeply involved in the formation of chromosome architectures.
Fig. 2.
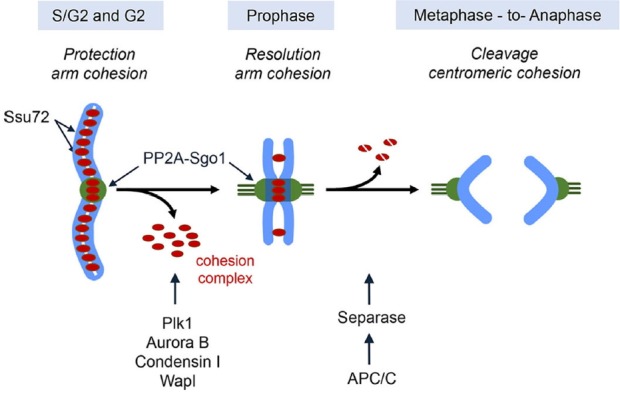
Role of phosphatases in sister chromatid cohesion and separation. Recruitment of Ssu72 onto the chromatin during the S and G2 phases serves to maintain sister chromatid arm cohesion until early prophase. During prophase, the bulk of arm cohesin removes from mitotic chromosome and this removal is controlled by Plk1, Aurora B, condensin I, and Wapl. At the same time, Sgo1-PP2A complex protects the centromeric cohesion from the phosphorylation of cohesin subunits by mitotic kinase, such as Plk1 and Aurora B. At the metaphase to anaphase transition, activated Seperase by anaphase promoting complex/cyclosome (APC/C) ensure the complete cleavage of existing cohesin complex from the chromosome.
The centromeric cohesin during early mitosis is protected by the prophase pathway. Specifically, PP2A and Shugoshin (Sgo1) complex localizes to the centromere and prevents the phosphorylations of centromeric cohesins by Plk1 and Aurora B (Fig. 2; Kitajima et al., 2006; Tang et al., 2006). Consistent with this, depletion of PP2A or Sgo1 causes precocious chromosome segregation of centromeric cohesion (Kitajima et al., 2006; Tang et al., 2006). It can be concluded that the PP2A-Sgo1 interaction is required for centromeric localization of PP2A-Sgo1 protein complex and for proper centromeric cohesion and separation. However, the exact molecular details of PP2A and Sgo1 interaction with respect to specific centromeric localizations are unclear. PP2A also contributes to proper chromosome compaction though dephosphorylation and inactivation of condensin near gene promoters during mitosis (Xing et al., 2008).
The TATA-binding protein (TBP) remains bound to DNA during mitosis and recruits PP2A. TBP-PP2A complex also interacts with condensin at the promoter, thereby allowing efficient regulation of chromosome compaction (Xing et al., 2008). This implies that PP2A complex bound to chromosome may contribute to the mitotic chromosome structure. PP2A has been implicated in the final step of cohesin removal, which involves the proteolytic cleavage of the remaining centromeric Rad21/Scc1 cohesin subunit by called Separase. The activation of Separase is triggered by APCCdc20-mediated ubiquitinational degradation of the associated inhibitor Securin/Pttg1 (Nagao and Yanagida, 2002). PP2A directly interacts and stabilizes the human Se-curin/Pttg1 through its dephosphorylation (Gil-Bernabe et al., 2006). Furthermore, PP2A associates with Separase through the B56 regulatory subunit and this complex induces the Separase autocleavage, which is necessary to activate its enzyme activity (Clift et al., 2009; Holland et al., 2007).
So far, although there has been tremendous advance in understanding of the dynamic phospho-regulations and the specific roles of various protein kinases and phosphatases in mitosis, many questions still remain to be answered.
FUNCTION OF PHOSPHATASES IN MITOTIC EXIT
The proteolytic degradation of cyclin B, by APCCdc20 activation after exact spindle attachment, and coupled with partial inactivation of Cdk1 are not sufficient to drive mitotic exit (Lopez-Aviles et al., 2009; Potapova et al., 2006). Mitotic exit is marked with reactivation of numerous protein phosphatases whose dephosphorylation activities ensure unidirectionality and irreversibility of the cell cycle (Rogers et al., 2016; Yang and Ferrell, 2013). Moreover, critical processes are tightly coordinated both spatially and temporally by the activity of protein phosphatases. A recent phosphoproteomic map of mitotic exit revealed approximately 16,000 phospho-regulation sites on over 3,300 proteins, and ∼10% of those sites were dephosphorylated during early mitotic exit (McCloy et al., 2015).
One of the well-studied phosphatases, Cdc14, although not conserved, has been shown to completely inactivate CDK1 and its substrates in yeast. Cdc14 remains sequestered in the nucleolus by forming a complex with a dephosphorylated Net1 protein known as RENT (regulator of nucleolar silencing and telophase exit) (Visintin et al., 1999). The nuclear localization of Cdc14 during the initial stages of mitotic exit is triggered by the FEAR (fourteen early anaphase release) regulatory pathway and later sustained with the MEN (mitotic exit network) regulatory pathway during late anaphase (De Wulf et al., 2009; Queralt and Uhlmann, 2008). Both of these pathways promote the release of Cdc14 through the phosphorylation of Net1 (De Wulf et al., 2009; Queralt and Uhlmann, 2008). The initial release of Cdc14 through the FEAR regulatory pathway is mediated by Cdk1 phosphorylation of Net1. In addition, working in opposition to Cdk1 is another phosphatase PP2A. As through its B55 regulatory unit, PP2A serves to prevent the release of Cdc14 just prior to anaphase (Yellman and Burke, 2006).
Although the molecular details are poorly understood, it is believed that proteins like Separase and nuclear export Zds1 and Zds2 are able to downregulate PP2A prior to mitotic exit (Calabria et al., 2012). The subsequent depletion of Cdk1, as cell cycle progresses to late anaphase, could trigger the MEN pathway, which is activated to achieve full Cdc14 phosphatase activity. Consequently, yeast Cdc14 is the primary phosphatase responsible for counterbalancing Cdk1 activity and a proper mitotic exit (Bouchoux and Uhlmann, 2011). However, in human cells, Cdc14 does not appear to play an essential role (Mocciaro and Schiebel, 2010). Ultimately, it is unclear which phosphatases control the mitotic exit in higher eukaryotic cells.
As mentioned above, the phosphatase activity of PP1 and PP2A are negatively regulated by the Cdk1-cyclin B complex. Both the high level and activity of Cdk1-cyclin B complex ensure that both the phosphatases are kept in the inactivated form. In Xenopus egg extracts, declining levels of Cdk1 promote the induction of PP1 activity through dephosphorylation of phosphorylated residue Thr309 during mitotic exit (Dohadwala et al., 1994; Wu et al., 2009). Similarly, PP2A-B55 in human and Xenopus systems has been proposed as a central phosphatase for regulating the counterbalancing Cdk1 activity during mitotic exit (Mochida et al., 2009; Schmitz et al., 2010). Additionally, it has been suggested that PP1 could contribute to the activation of PP2A-B55, which is able to promote the reactivation of PP2A-B56 subunit by counteracting the PP1-docking site in PP2A-B56 (Grallert et al., 2015). Consequently, PP1 both directly and indirectly contributes to regulation of PP2A-B55 and PP2A-B56 activities in a variety of mitotic signaling events, involving the mitotic exit.
CORRELATION BETWEEN MITOTIC PHOSPHATASES AND DISEASES
As mentioned earlier, the human genome analysis has revealed the existence of about 137 phosphatases, while there are approximate about 518 kinases. This indicates that phosphatases potentially have multi-functional roles in phospho-regulation of various cellular pathways, involving cell cycle regulation. For instance, although PP2A plays a major role in the mitotic entry, mitotic spindle assembly, chromosome cohesion, and mitotic exit through the combination with different regulatory subunits (Fig. 1), it also could regulate other cellular activities like autophagy and transcription (Janssens and Goris, 2001; Seshacharyulu et al., 2013). Furthermore, correct regulation of mitosis progression by several phosphatases is crucial for cell fate determination, tissue organization, and organ development. In effect, mutations and/or alterations in expression of phosphatases have been associated with and also been able to affect various diseases in human and mice. For instance, a recent study showed that selective inhibition of PP1 in mitosis resulted in a reduction of tumor growth via severe defects in spindle-formation and chromosome congression (Winkler et al., 2015). In addition, PP1 is often found with genetic and expression alterations in various form of cancer (Stebbing et al., 2014; Takakura et al., 2001). Furthermore, several regulatory units of PP1 holoenzyme play specific roles in a number of human diseases (Table 2). Also, inhibition of PP1 activity by HBx, as a regulatory unit, enhances viral transcription (Cougot et al., 2012). Similarly, the Tat protein of human immunodeficiency virus (HIV) could activate viral transcription through PP1 inhibition (Ammosova et al., 2011). This report also showed that disruption of the complex between HIV-1 Tat and PP1 by an 82 residue PP1 inhibitory protein (cdNIPP1) strongly repressed viral transcription and replication. Therefore, it has been suggested that the control of phosphatase activity by using regula tory units could be a valuable approach towards the development of new therapeutics. PP2A also is implicated in cancer initiation and progression. It has been reported that decreased activity of PP2A could induce cancer recurrence in many types of cancer, including colorectal and breast cancers (Table 2; Baldacchino et al., 2014; Cristobal et al., 2014). In many cancers, PP2A is inactivated via several mechanisms, including somatic mutation and phosphorylation. These genetic alterations prevent the formation of PP2A complex, resulting in suppression of PP2A enzyme activity. As such, understanding of these defects is necessary for the development of PP2A targeted therapeutics.
Table 2.
The regulatory units and associated diseases of mitotic phosphatases
| Mitotic phosphatases | Regulatory unit | Associated disease |
|---|---|---|
| PP1 | HBx | Hepatitis |
| HIV-TAT1 | AIDS | |
| PP2A | A (α,β) | Breast cancer, Colon cancer, Lung cancer, Melanoma, Glioma, Uterine cancer, Ovarian cancer, HCC, Cervical cancer |
| B55 (α,β,γ,ɛ) | Breast cancer, Prostate cancer, Myeloma, Colon cancer | |
| B56 (α,γ) | Melanoma, Lung cancer | |
| Cα | Prostate cancer, AML | |
| PP4C | R1,R2, R3 | Breast cancer, Lung cancer, Pancreatic ductal adenocarcinoma, Colorectal cancer |
| PP6 | n.d. | Melaonoma |
| Ssu72 | n.d. | NAFLD, HCC (?) |
| Cdc14 | Spo, Net1 | n.d. |
| Cdc25 | M-Cdk | Gastric cancer, Breast cancer, Pancreatic cancer, HCC, Colorectal cancer, Esophagial cell carcinoma, Ovarian cancer |
n.d., not yet determined; HCC, hepatocellular carcinoma; AML, acute myeloid leukemia; NAFLD, Non-alcoholic Fatty Liver Disease
Ssu72, a newly studied phosphatase with dual specificity, is also associated with liver homeostasis and its depletion causes the non-alcoholic fatty liver disease including steatosis, steatohepatitis, and fibrosis through the production of aberrant Rb-mediated uncontrolled chromosome polyploidy (Kim et al., 2016). According to recent several reports, Ssu72 prefers the cis configuration of serine-proline motif within its substrate by regulating Pin1, in contrast to other phosphatases (Wemer-Allen et al., 2011, Xiang et al., 2010). This evidence is likely to provide a novel hint in the design of selective Ssu72 phosphatase modulators, which are potential targets for therapeutics for liver disorders and cancer.
Thus, although linking phosphatase defects to various diseases with human genome sequence analysis and mouse models is emerging as a superior tool, attempts to study certain phosphatases such as PP1, PP2A and Ssu72 require techniques other than using generating conventional knockout mice as these tend to be embryonic lethal at a developmental stage, and do not allow viable embryos to be born for the study of various diseases (Kim et al., 2016; Larsen et al., 2003). To determine the role of mitotic phosphatase in the various human diseases, involving cancer, future studies need to study tissue-specific and cytopathological changes using conditional knockout or conditional transgenic mice at the adult stage.
CONCLUSION
Over the past few years, there have been tremendous advances in our understanding of mitotic phosphatases in control of mitotic entry, spindle assembly, chromosome cohesion, and exit. The growing evidence depicts the mitotic phosphatases being clearly involved in various human disease states, including cancer, as well as being required for strict mitotic cell cycle regulation and cellular homeostasis. Thus far, however, there is a paucity of studies on the associated pathological states and diseases with mitotic phosphatase defects, and further research is required to pinpoint the relationship between physiological mechanisms and phosphatase activity for exact understanding of the role of each phosphatase in human diseases and model mice. It also remains to be discovered how the localization of several phosphatases determines their access to substrates. In the case of PP2A, this is mediated by having various regulatory subunits, whereas in other phosphatases, in particularly for dual specificity phosphatases, specific access to substrates requires release from certain sequestration. Therefore, the identification of the physiological substrates and regulatory proteins for each phosphatase is an important task, which will likely be aided by comprehensive phospho-proteome analyses. Lastly, future studies into the control of phosphatase activity may lead to capabilities in modulating various important biological processes and in developing new therapeutics.
Acknowledgments
This work was supported by a National Research Foundation grant funded by the Korean government (MEST) (2014R1A2A1 A10050775 and 2016R1C1B2014438). The authors have no conflicting financial interests.
REFERENCES
- Archambault V., Zhao X., White-Cooper H., Carpenter A.T., Glover D.M. Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 2007;3:e200. doi: 10.1371/journal.pgen.0030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T., Yedavalli V.R., Niu X., Jerebtsova M., Van Eynde A., Beullens M., Bollen M., Jeang K.T., Nekhai S. Expression of a protein phosphatase 1 inhibitor, cdNIPP1, increases CDK9 threonine 186 phosphorylation and inhibits HIV-1 transcription. J. Biol. Chem. 2011;286:3798–3804. doi: 10.1074/jbc.M110.196493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacchino S., Saliba C., Petroni V., Fenech A.G., Borg N., Grech G. Deregulation of the phosphatase, PP2A is a common event in breast cancer, predicting sensitivity to FTY720. EPMA J. 2014;5:3. doi: 10.1186/1878-5085-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Sillje H.H., Nigg E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bollen M., Peti W., Ragusa M.J., Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Coopman P., Morris M.C. Characterization of centrosomal localization and dynamics of Cdc25C phosphatase in mitosis. Cell Cycle. 2008;7:1991–1998. doi: 10.4161/cc.7.13.6095. [DOI] [PubMed] [Google Scholar]
- Bouchoux C., Uhlmann F. A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell. 2011;147:803–814. doi: 10.1016/j.cell.2011.09.047. [DOI] [PubMed] [Google Scholar]
- Boutros R., Lobjois V., Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- Boutros R., Mondesert O., Lorenzo C., Astuti P., McArthur G., Chircop M., Ducommun B., Gabrielli B. CDC25B overexpression stabilises centrin 2 and promotes the formation of excess centriolar foci. PloS one. 2013;8:e67822. doi: 10.1371/journal.pone.0067822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigan D.L. Protein Ser/Thr phosphatases--the ugly ducklings of cell signalling. FEBS J. 2013;280:324–345. doi: 10.1111/j.1742-4658.2012.08609.x. [DOI] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbe J.C., Lorca T., Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA. 2010;107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabria I., Baro B., Rodriguez-Rodriguez J.A., Russinol N., Queralt E. Zds1 regulates PP2A(Cdc55) activity and Cdc14 activation during mitotic exit through its Zds_C motif. J. Cell Sci. 2012;125:2875–2884. doi: 10.1242/jcs.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho P.V., Williams B.C., Mochida S., Zhao Y., Goldberg M.L. The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55delta, a phosphatase directed against CDK phosphosites. Mol. Biol. Cell. 2009;20:4777–4789. doi: 10.1091/mbc.E09-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.P., Liu Y., Gomez M., Dunlap J., Tyers M., Wang Y. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol. Cell Biol. 2005;25:4541–4551. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D., Bizzari F., Marston A.L. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 2009;23:766–780. doi: 10.1101/gad.507509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot D., Allemand E., Rivière L., Benhenda S., Duroure K., Levillayer F., Muchardt C., Buendia M.A., Neuveut C. Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci. Signal. 2012;5:ra1. doi: 10.1126/scisignal.2001906. [DOI] [PubMed] [Google Scholar]
- Cristobal I., Rincon R., Manso R., Madoz-Gurpide J., Carames C., del Puerto-Nevado L., Rojo F., Garcia-Foncillas J. Hyperphosphorylation of PP2A in colorectal cancer and the potential therapeutic value showed by its forskolin-induced dephosphorylation and activation. Biochim. Biophys. Acta. 2014;1842:1823–1829. doi: 10.1016/j.bbadis.2014.06.032. [DOI] [PubMed] [Google Scholar]
- De Wulf P., Montani F., Visintin R. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 2009;21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Deibler R.W., Kirschner M.W. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol. Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Monica R., Visconti R., Cervone N., Serpico A.F., Grieco D. Fcp1 phosphatase controls Greatwall kinase to promote PP2A-B55 activation and mitotic progression. eLife. 2015;4 doi: 10.7554/eLife.10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villen J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. US A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala M., da Cruz e Silva E.F., Hall F.L., Williams R.T., Carbonaro-Hall D.A., Nairn A.C., Greengard P., Berndt N. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA. 1994;91:6408–6412. doi: 10.1073/pnas.91.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn P.J., Creyghton M.P., Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Foley E.A., Maldonado M., Kapoor T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbe J.C., Burgess A., Vigneron S., Strub J.M., Brioudes E., Van-Dorsselaer A., Castro A., Lorca T. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science. 2010;330:1673–1677. doi: 10.1126/science.1197048. [DOI] [PubMed] [Google Scholar]
- Gil-Bernabe A.M., Romero F., Limon-Mortes M.C., Tortolero M. Protein phosphatase 2A stabilizes human securin, whose phosphorylated forms are degraded via the SCF ubiquitin ligase. Mol. Cell. Biol. 2006;26:4017–4027. doi: 10.1128/MCB.01904-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A., Boke E., Hagting A., Hodgson B., Connolly Y., Griffiths J.R., Smith D.L., Pines J., Hagan I.M. A PP1-PP2A phosphatase relay controls mitotic progression. Nature. 2015;517:94–98. doi: 10.1038/nature14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Stanevich V., Wlodarchak N., Sengupta R., Jiang L., Satyshur K.A., Xing Y. Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 2014;24:190–203. doi: 10.1038/cr.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps N.R., Brewis N.D., Lineruth K., Davis T., Kaiser K., Cohen P.T. Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J. Cell Sci. 1998;111(Pt 10):1331–1340. doi: 10.1242/jcs.111.10.1331. [DOI] [PubMed] [Google Scholar]
- Hoffmann I., Clarke P.R., Marcote M.J., Karsenti E., Draetta G. Phosphorylation and activation of human cdc25-C by cdc2--cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Bottger F., Stemmann O., Taylor S.S. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J. Biol. Chem. 2007;282:24623–24632. doi: 10.1074/jbc.M702545200. [DOI] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Izawa D., Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T., Maller J.L. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol. Biol. Cell. 1993;4:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.A., Hunter T. Kinomics: methods for deciphering the kinome. Nat. Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 2009;21:51–58. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Baek K.H., Ha G.H., Lee J.C., Kim Y.N., Lee J., Park H.Y., Lee N.R., Lee H., Cho Y., et al. The hsSsu72 phosphatase is a cohesin-binding protein that regulates the resolution of sister chromatid arm cohesion. EMBO J. 2010;29:3544–3557. doi: 10.1038/emboj.2010.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Kim S.H., Park H.Y., Lee J., Yoon J.H., Choi S., Ryu S.H., Lee H., Cho H.S., Lee C.W. Functional interplay between Aurora B kinase and Ssu72 phosphatase regulates sister chromatid cohesion. Nat. Commun. 2013;4:2631. doi: 10.1038/ncomms3631. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Jeon Y., Kim H.S., Lee J.K., Lim H.J., Kang D., Cho H., Park C.K., Lee H., Lee C.W. Hepatocyte homeostasis for chromosome ploidization and liver function is regulated by Ssu72 protein phosphatase. Hepatology. 2016;63:247–259. doi: 10.1002/hep.28281. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kotwaliwale C., Biggins S. Microtubule capture: a concerted effort. Cell. 2006;127:1105–1108. doi: 10.1016/j.cell.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Yakowec P.S., Dunphy W.G. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol. Biol. Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., Nilsson J. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 2013;1:1086–1092. doi: 10.1242/jcs.122481. [DOI] [PubMed] [Google Scholar]
- Larsen M., Tremblay M.L., Yamada K.M. Phosphatases in cell-matrix adhesion and migration. Nat. Rev. Mol. Cell Biol. 2003;4:700–711. doi: 10.1038/nrm1199. [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Rodriguez-Bravo V., Medema R.H. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Biggins S. Signalling dynamics in the spindle checkpoint response. Nature reviews Mol. Cell Biol. 2014;15:736–747. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Aviles S., Kapuy O., Novak B., Uhlmann F. Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature. 2009;459:592–595. doi: 10.1038/nature07984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B., Mann M., Olsen J.V. Global and site-specific quantitative phosphoproteomics: principles and applications. Ann. Rev. Pharmacol. Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- Mailand N., Lukas C., Kaiser B.K., Jackson P.K., Bartek J., Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 2002;4:317–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- McCloy R.A., Parker B.L., Rogers S., Chaudhuri R., Gayevskiy V., Hoffman N.J., Ali N., Watkins D.N., Daly R.J., James D.E., et al. Global phosphoproteomic mapping of early mitotic exit in human cells identifies novel substrate dephosphorylation motifs. Mol. Cell. Proteomics. 2015;14:2194–2212. doi: 10.1074/mcp.M114.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta G.D., Rizvi S.M., Ghosh S.K. Cohesin: a guardian of genome integrity. Biochim. Biophys. Acta. 2012;1823:1324–1342. doi: 10.1016/j.bbamcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Nigg E.A. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- Mocciaro A., Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 2010;123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- Mochida S., Ikeo S., Gannon J., Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S.L., Skehel M., Hunt T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 2010;330:1670–1673. doi: 10.1126/science.1195689. [DOI] [PubMed] [Google Scholar]
- Mueller P.R., Coleman T.R., Dunphy W.G. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol. Biol. Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nagao K., Yanagida M. Regulating sister chromatid separation by separase phosphorylation. Dev. Cell. 2002;2:2–4. doi: 10.1016/s1534-5807(01)00112-5. [DOI] [PubMed] [Google Scholar]
- Nilsson I., Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- O’Farrell P.H. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 2001;11:512–519. doi: 10.1016/s0962-8924(01)02142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Perdiguero E., Nebreda A.R. Regulation of Cdc25C activity during the meiotic G2/M transition. Cell cycle. 2004;3:733–737. [PubMed] [Google Scholar]
- Peters J.M., Tedeschi A., Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Pomerening J.R., Sontag E.D., Ferrell J.E., Jr. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- Potapova T.A., Daum J.R., Pittman B.D., Hudson J.R., Jones T.N., Satinover D.L., Stukenberg P.T., Gorbsky G.J. The reversibility of mitotic exit in vertebrate cells. Nature. 2006;440:954–958. doi: 10.1038/nature04652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E., Uhlmann F. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 2008;20:661–668. doi: 10.1016/j.ceb.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Fey D., McCloy R.A., Parker B.L., Mitchell N.J., Payne R.J., Daly R.J., James D.E., Caldon C.E., Watkins D.N., et al. PP1 initiates the dephosphorylation of MASTL, triggering mitotic exit and bistability in human cells. J. Cell Sci. 2016;129:1340–1354. doi: 10.1242/jcs.179754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sassoon I., Severin F.F., Andrews P.D., Taba M.R., Kaplan K.B., Ashford A.J., Stark M.J., Sorger P.K., Hyman A.A. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz A.L., Srayko M., Dammermann A., Quintin S., Wielsch N., MacLeod I., de Robillard Q., Zinke A., Yates J.R., 3rd, Muller-Reichert T., et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.H., Held M., Janssens V., Hutchins J.R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A.I., Poser I., et al. Live-cell imaging RNAi screen identifies PP2AB55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshacharyulu P., Pandey P., Datta K., Batra S.K. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Letters. 2013;335:9–18. doi: 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Sivakumar S., Janczyk P.L., Qu Q., Brautigam C.A., Stukenberg P.T., Yu H., Gorbsky G.J. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife. 2016;5 doi: 10.7554/eLife.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Lit L.C., Zhang H., Darrington R.S., Melaiu O., Rudraraju B., Giamas G. The regulatory roles of phosphatases in cancer. Oncogene. 2014;33:939–953. doi: 10.1038/onc.2013.80. [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk S. J. E., Vleugel M., Teixeira A., Kops G. J. P. L. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi E., Sugimoto A., Yamamoto M. Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans . J. Cell Sci. 2002;115:1403–1410. doi: 10.1242/jcs.115.7.1403. [DOI] [PubMed] [Google Scholar]
- Takakura S., Kohno T., Manda R., Okamoto A., Tanaka T., Yokota J. Genetic alterations and expression of the protein phosphatase 1 genes in human cancers. Internat. J. Oncology. 2001;18:817–824. doi: 10.3892/ijo.18.4.817. [DOI] [PubMed] [Google Scholar]
- Tan S., Lyulcheva E., Dean J., Bennett D. Mars promotes dTACC dephosphorylation on mitotic spindles to ensure spindle stability. J. Cell Biol. 2008;182:27–33. doi: 10.1083/jcb.200712080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong S.M., Zaugg J.B., Camblong J., Xu Z., Zhang D.W., Mischo H.E., Ansari A.Z., Luscombe N.M., Steinmetz L.M., Proudfoot N.J. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tonks N.K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Lamond A.I. Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 2006;18:623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Vigneron S., Brioudes E., Burgess A., Labbe J.C., Lorca T., Castro A. Greatwall maintains mitosis through regulation of PP2A. EMBO J. 2009;28:2786–2793. doi: 10.1038/emboj.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D.M., Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Voets E., Wolthuis R.M. MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle. 2010;9:3591–3601. doi: 10.4161/cc.9.17.12832. [DOI] [PubMed] [Google Scholar]
- Waizenegger I.C., Hauf S., Meinke A., Peters J.M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Werner-Allen J.W., Lee C.J., Liu P., Nicely N.I., Wang S., Greenleaf A.L., Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C., De Munter S., Van Dessel N., Lesage B., Heroes E., Boens S., Beullens M., Van Eynde A., Bollen M. The selective inhibition of protein phosphatase-1 results in mitotic catastrophe and impaired tumor growth. J. Cell Sci. 2015;128:4526–4537. doi: 10.1242/jcs.175588. [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Guo J.Y., Tang W., Yang C.S., Freel C.D., Chen C., Nairn A.C., Kornbluth S. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 2009;11:644–651. doi: 10.1038/ncb1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K., Nagaike T., Xiang S., Kilic T., Beh M.M., Manley J.L., Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Vanderford N.L., Sarge K.D. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Ferrell J.E., Jr. The Cdk1-APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat. Cell Biol. 2013;15:519–525. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman C.M., Burke D.J. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Fleming S.L., Williams B., Williams E.V., Li Z., Somma P., Rieder C.L., Goldberg M.L. Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J. Cell Biol. 2004;164:487–492. doi: 10.1083/jcb.200310059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Bastos R.N., Barr F.A., Gruneberg U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 2010;191:1315–1332. doi: 10.1083/jcb.201008106. [DOI] [PMC free article] [PubMed] [Google Scholar]


