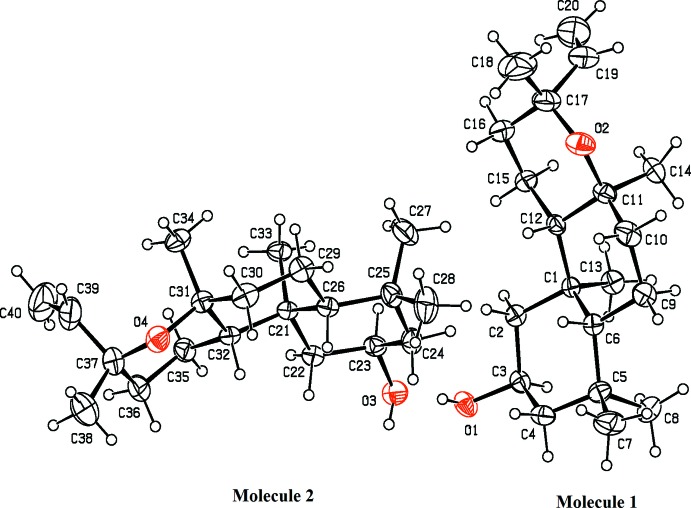
In the two independent molecules in the asymmetric unit of the title compound, the cyclohexane rings adopt a chair conformation, while the oxane rings are also puckered. In the crystal, O—H⋯ O hydrogen bonds connect adjacent molecules, forming a C(6) helical chain running along the [100] direction.
Keywords: crystal structure, cyclohexane ring, semi-empirical PM3 method, HOMO, LUMO
Abstract
The asymmetric unit of the title compound, C20H34O2, contains two crystallographically independent molecules (1 and 2) with similar conformations. In both molecules, the cyclohexane rings adopt a chair conformation, while the oxane rings are also puckered. In the crystal, O—H⋯O hydrogen bonds connect adjacent molecules, forming C(6) helical chains located around a 21 screw axis and running along the crystallographic a axis. The packing of these chains is governed only by van der Waals interactions. Semi-empirical PM3 quantum chemical calculations are in a satisfactory agreement with the structural results of the X-ray structure analysis. The absolute structure was indeterminate in the present experiment.
Chemical context
The Sideritis genus belonging to the Lamiaceae family is represented by more than 150 species in the world (Duman 2000 ▸). Sideritis species have been reported to have a broad spectrum of biological activities such as anti-inflammatory, anti-oxidant, anti-ulcerogenic, analgesic, antimicrobial, antiproliferative, anti-HIV and antifeedant activities (González-Burgos et al. 2011 ▸), and they have been consumed as teas, as flavoring agents, for therapeutic purposes, etc. In particular, Sideritis teas have been used for gastrointestinal disorders such as stomach ache and indigestion, to alleviate common colds, fever, flu and sore throats (Topçu et al. 2002 ▸). Phytochemical investigations of the species have revealed the presence of terpenes (Fraga et al. 2003 ▸), flavonoids, essential oils and other secondary metabolites (Barberan et al. 1985 ▸). As part of our studies in this area, we now describe the isolation and structure of the title compound, (I).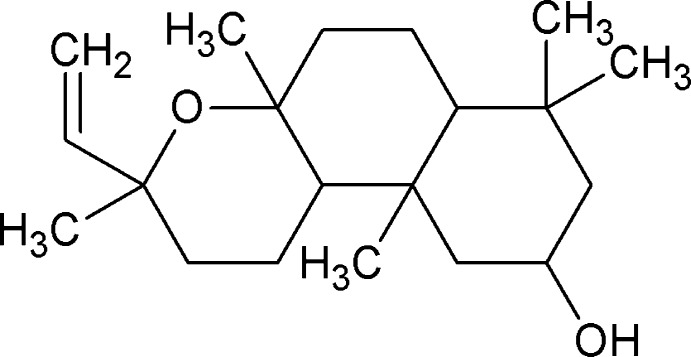
Structural commentary
In the title compound (Fig. 1 ▸), the asymmetric unit contains two crystallographically independent molecules, 1 and 2, with a similar conformations. In molecule 1, the cyclohexane ring (C1–C6) attached to the OH group and the central cyclohexane ring (C1/C6/C9–C12) each adopt a chair conformation with puckering parameters Q T = 0.536 (3) Å, θ = 0.0 (3), φ = 270 (81)° and Q T = 0.584 (3) Å, θ = 4.4 (3), φ = 59 (4)°, respectively. The oxane ring (O2/C11/C12/C15–C17) is also puckered, with puckering parameters Q T = 0.551 (3) Å, θ = 12.1 (3) and φ = 133.5 (16)°. The equivalent rings in molecule 2 (C21–C16, C21/C26/C29–C32 and O4/C31/C32/C35–C37) have as puckering parameters Q T = 0.534 (3) Å, θ = 1.9 (3), φ = 296 (11)°, Q T = 0.583 (3) Å, θ = 5.0 (3), φ = 72 (3)° and Q T = 0.554 (3) Å, θ = 11.9 (3), φ = 127.2 (15)°, respectively. Bond lengths and angles are within normal range, comparable with each other and with those reported for similar structures in the literature (e.g., Evans et al., 2011 ▸).
Figure 1.
A view of the title compound, showing the atom-numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 30% probability level. The minor component of the disorder is not shown for clarity.
Supramolecular features
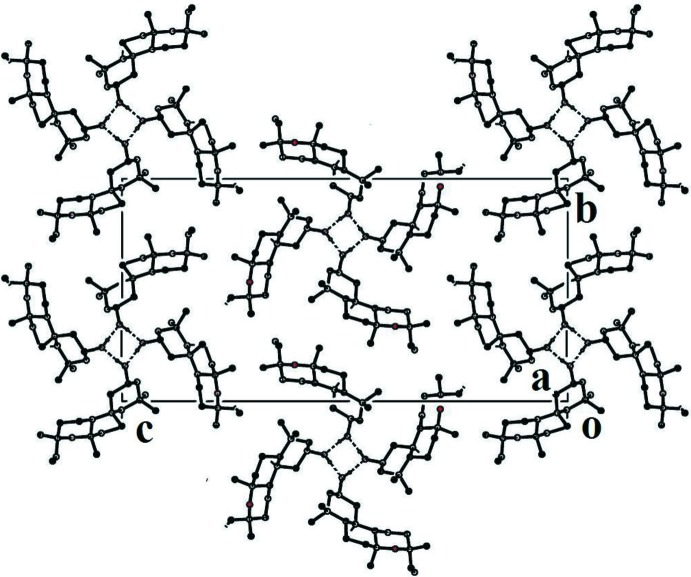
Intermolecular O—H. . . O hydrogen bonds connect adjacent molecules, forming C(6) helical chains located around a 21 screw axis running along the crystallographic a axis (Table 1 ▸ and Fig. 2 ▸). The crystal packing of these chains is governed only by van der Waals interactions. The two asymmetric molecules lead to pseudo-41 symmetry in space group P212121.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯O3 | 0.80 (4) | 1.99 (4) | 2.784 (3) | 170 (4) |
| O3—H3O⋯O1i | 0.81 (4) | 2.00 (4) | 2.804 (3) | 169 (4) |
Symmetry code: (i) 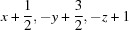 .
.
Figure 2.
A view along the a axis of the crystal packing of the title compound. H atoms not involved in hydrogen bonding (dashed lines) have been omitted for clarity.
Theoretical calculations
PM3 (parameterized model number 3) is a semi-empirical method for the quantum calculation of the molecular electronic structure in computational chemistry. It is based on the neglect of differential diatomic overlap integral approximation. The semi-empirical PM3 parameterization used in the MOPAC program is widely used to derive charges, dipole moments and bond lengths. The computed quantum chemical descriptors include bond lengths, bond angles, torsion angles, atom charges, HOMO and LUMO energy levels, dipole moment, polarizability, etc. In the present case, the geometry of the molecule of the title compound was calculated with a semi-empirical PM3 method (Stewart, 1985 ▸). A spatial view is included in the Supporting information.
The calculated net charges at atoms O1 and O2 are −0.257 and −0.309 e−, respectively. The total energy and dipole moment of the title molecule are −3514.7 eV and 1.695 Debye. The HOMO and LUMO energy levels are −10.36 and 2.71 eV, respectively.
Calculated values for the geometrical parameter are consistent with those obtained by the X-ray structure determination, within the error limits (see Table S1 in the Supporting information), with the sole exception of the angles in the methoxy groups. This may be ascribed to the steric interactions between adjacent molecules in the crystal structure.
Synthesis and crystallization
The aerial part of the plant material (5 g) was extracted with ethyl acetate (3 × 20 mL). After removal of the solvent by rotary evaporator, the extract was subjected to column chromatography (2.5 × 70 cm); sephadex LH-20 (50 g) was used as a stationary phase and methanol was used as a mobile phase with a 0.25 ml min−1 flow rate. 16 fractions, each of which was 150 mL, were collected. Similar fractions were combined according to the TLC profile. Further purification was carried out with silica gel column chromatography to isolate the title compound. Colourless prisms were recrystallized from ethanol solution.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. H atoms bound to oxygen were found from difference Fourier maps and their positional parameters were refined with U iso fixed at 1.5 times U eq(O). H atoms bound to carbon were positioned geometrically and allowed to ride on their parent atoms with U iso = 1.2U eq(C) (C—H = 0.93 Å for aromatic, 0.97 Å for methylene and 0.98 Å for methine) and with U iso = 1.5U eq(C) (C—H = 0.96 Å) for methyl H atoms. The absolute structure was indeterminate in the present experiment.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C20H34O2 |
| M r | 306.47 |
| Crystal system, space group | Orthorhombic, P212121 |
| Temperature (K) | 296 |
| a, b, c (Å) | 7.1114 (4), 16.3899 (12), 32.812 (2) |
| V (Å3) | 3824.4 (4) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.07 |
| Crystal size (mm) | 0.14 × 0.11 × 0.08 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 2003 ▸) |
| T min, T max | 0.635, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 36728, 9449, 5384 |
| R int | 0.074 |
| (sin θ/λ)max (Å−1) | 0.667 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.060, 0.130, 1.02 |
| No. of reflections | 9449 |
| No. of parameters | 413 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.17, −0.23 |
| Absolute structure | Flack (1983 ▸), 4144 Friedel pairs |
| Absolute structure parameter | 0.4 (15) |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989016013864/bg2593sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016013864/bg2593Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593sup3.pdf
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593sup4.tif
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593Isup5.cml
CCDC reference: 1501445
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are indebted to the X-ray laboratory of Sinop University Scientific and Technological Applied and Research Center, Sinop, Turkey, for use of the X-ray diffractometer.
supplementary crystallographic information
Crystal data
| C20H34O2 | F(000) = 1360 |
| Mr = 306.47 | Dx = 1.065 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 7578 reflections |
| a = 7.1114 (4) Å | θ = 2.9–25.0° |
| b = 16.3899 (12) Å | µ = 0.07 mm−1 |
| c = 32.812 (2) Å | T = 296 K |
| V = 3824.4 (4) Å3 | Prism, colourless |
| Z = 8 | 0.14 × 0.11 × 0.08 mm |
Data collection
| Bruker APEXII CCD diffractometer | 5384 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.074 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2003) | θmax = 28.3°, θmin = 2.9° |
| Tmin = 0.635, Tmax = 0.746 | h = −8→9 |
| 36728 measured reflections | k = −19→21 |
| 9449 independent reflections | l = −43→42 |
Refinement
| Refinement on F2 | H atoms treated by a mixture of independent and constrained refinement |
| Least-squares matrix: full | w = 1/[σ2(Fo2) + (0.0617P)2 + 0.0101P] where P = (Fo2 + 2Fc2)/3 |
| R[F2 > 2σ(F2)] = 0.060 | (Δ/σ)max < 0.001 |
| wR(F2) = 0.130 | Δρmax = 0.17 e Å−3 |
| S = 1.02 | Δρmin = −0.23 e Å−3 |
| 9449 reflections | Absolute structure: Flack (1983), 4144 Friedel pairs |
| 413 parameters | Absolute structure parameter: 0.4 (15) |
| 0 restraints |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > 2sigma(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.0636 (3) | 0.76038 (13) | 0.54574 (6) | 0.0525 (8) | |
| O2 | 0.4700 (3) | 0.53531 (14) | 0.71395 (6) | 0.0656 (8) | |
| C1 | 0.2450 (3) | 0.70848 (15) | 0.65200 (7) | 0.0323 (8) | |
| C2 | 0.1533 (4) | 0.69891 (16) | 0.60996 (8) | 0.0388 (9) | |
| C3 | 0.1457 (4) | 0.77677 (17) | 0.58487 (8) | 0.0400 (9) | |
| O3 | 0.3152 (3) | 0.65702 (13) | 0.50623 (6) | 0.0548 (8) | |
| C4 | 0.3381 (4) | 0.81502 (18) | 0.58077 (8) | 0.0461 (10) | |
| O4 | 0.7389 (3) | 0.33641 (12) | 0.38715 (6) | 0.0558 (7) | |
| C5 | 0.4396 (4) | 0.83089 (17) | 0.62121 (8) | 0.0440 (10) | |
| C6 | 0.4392 (3) | 0.75033 (16) | 0.64591 (8) | 0.0358 (8) | |
| C7 | 0.6443 (5) | 0.8531 (2) | 0.61049 (11) | 0.0723 (12) | |
| C8 | 0.3532 (5) | 0.90435 (19) | 0.64330 (10) | 0.0639 (11) | |
| C9 | 0.5515 (4) | 0.75220 (19) | 0.68563 (9) | 0.0510 (10) | |
| C10 | 0.5874 (4) | 0.6660 (2) | 0.70119 (9) | 0.0576 (11) | |
| C11 | 0.4068 (4) | 0.61840 (17) | 0.70812 (8) | 0.0467 (10) | |
| C12 | 0.2877 (4) | 0.62125 (16) | 0.66896 (7) | 0.0367 (8) | |
| C13 | 0.1074 (4) | 0.75550 (18) | 0.67990 (8) | 0.0460 (10) | |
| C14 | 0.3115 (5) | 0.6485 (2) | 0.74714 (8) | 0.0598 (11) | |
| C15 | 0.1228 (5) | 0.56256 (17) | 0.67286 (9) | 0.0531 (11) | |
| C16 | 0.1983 (6) | 0.47707 (19) | 0.67885 (10) | 0.0679 (14) | |
| C17 | 0.3343 (6) | 0.4701 (2) | 0.71481 (10) | 0.0713 (14) | |
| C18 | 0.4553 (8) | 0.3932 (3) | 0.71083 (15) | 0.118 (2) | |
| C19 | 0.2373 (8) | 0.4663 (2) | 0.75573 (12) | 0.0880 (16) | |
| C20 | 0.0645 (9) | 0.4471 (3) | 0.76331 (15) | 0.128 (3) | |
| C21 | 0.5044 (3) | 0.44774 (15) | 0.47669 (7) | 0.0333 (8) | |
| C22 | 0.4083 (4) | 0.53118 (15) | 0.47366 (8) | 0.0388 (9) | |
| C23 | 0.3984 (4) | 0.57841 (16) | 0.51329 (8) | 0.0424 (9) | |
| C24 | 0.5903 (4) | 0.58643 (18) | 0.53327 (9) | 0.0519 (10) | |
| C25 | 0.6949 (4) | 0.50615 (19) | 0.53944 (8) | 0.0492 (10) | |
| C26 | 0.6985 (3) | 0.46057 (16) | 0.49801 (8) | 0.0378 (9) | |
| C27 | 0.6092 (6) | 0.4573 (2) | 0.57489 (9) | 0.0747 (15) | |
| C28 | 0.8980 (5) | 0.5281 (3) | 0.55159 (12) | 0.0800 (16) | |
| C29 | 0.8158 (4) | 0.38262 (18) | 0.49711 (9) | 0.0502 (10) | |
| C30 | 0.8551 (4) | 0.35654 (18) | 0.45324 (9) | 0.0514 (10) | |
| C31 | 0.6751 (4) | 0.34233 (16) | 0.42919 (8) | 0.0419 (9) | |
| C32 | 0.5496 (4) | 0.41801 (15) | 0.43262 (8) | 0.0356 (8) | |
| C33 | 0.3720 (4) | 0.38850 (17) | 0.49895 (8) | 0.0453 (10) | |
| C34 | 0.5885 (5) | 0.26093 (16) | 0.44237 (9) | 0.0542 (10) | |
| C35 | 0.3826 (4) | 0.40988 (18) | 0.40359 (8) | 0.0478 (10) | |
| C36 | 0.4571 (5) | 0.4026 (2) | 0.36037 (9) | 0.0619 (11) | |
| C37 | 0.6014 (5) | 0.3344 (2) | 0.35483 (9) | 0.0637 (11) | |
| C38 | 0.7165 (7) | 0.3489 (3) | 0.31656 (11) | 0.0987 (18) | |
| C39 | 0.5169 (7) | 0.2505 (2) | 0.35154 (12) | 0.0843 (16) | |
| C40 | 0.3428 (8) | 0.2309 (3) | 0.34365 (15) | 0.122 (2) | |
| H1O | 0.125 (6) | 0.727 (2) | 0.5337 (12) | 0.0980* | |
| H2A | 0.22230 | 0.65800 | 0.59470 | 0.0470* | |
| H2B | 0.02610 | 0.67880 | 0.61370 | 0.0470* | |
| H3 | 0.06420 | 0.81560 | 0.59910 | 0.0480* | |
| H4A | 0.32520 | 0.86640 | 0.56640 | 0.0550* | |
| H4B | 0.41600 | 0.77950 | 0.56420 | 0.0550* | |
| H6 | 0.50890 | 0.71210 | 0.62860 | 0.0430* | |
| H7A | 0.70930 | 0.86980 | 0.63470 | 0.1080* | |
| H7B | 0.64500 | 0.89690 | 0.59110 | 0.1080* | |
| H7C | 0.70600 | 0.80640 | 0.59900 | 0.1080* | |
| H8A | 0.40310 | 0.90760 | 0.67040 | 0.0960* | |
| H8B | 0.21910 | 0.89800 | 0.64460 | 0.0960* | |
| H8C | 0.38330 | 0.95340 | 0.62870 | 0.0960* | |
| H9A | 0.67060 | 0.77950 | 0.68110 | 0.0610* | |
| H9B | 0.48240 | 0.78280 | 0.70600 | 0.0610* | |
| H10A | 0.66470 | 0.63700 | 0.68160 | 0.0690* | |
| H10B | 0.65680 | 0.66890 | 0.72660 | 0.0690* | |
| H12 | 0.36830 | 0.59640 | 0.64820 | 0.0440* | |
| H13A | 0.17640 | 0.78130 | 0.70150 | 0.0690* | |
| H13B | 0.01770 | 0.71820 | 0.69130 | 0.0690* | |
| H13C | 0.04270 | 0.79630 | 0.66420 | 0.0690* | |
| H14A | 0.18570 | 0.62730 | 0.74840 | 0.0900* | |
| H14B | 0.30740 | 0.70710 | 0.74710 | 0.0900* | |
| H14C | 0.38140 | 0.63000 | 0.77040 | 0.0900* | |
| H15A | 0.04510 | 0.57790 | 0.69590 | 0.0640* | |
| H15B | 0.04600 | 0.56470 | 0.64840 | 0.0640* | |
| H16A | 0.26260 | 0.46010 | 0.65420 | 0.0820* | |
| H16B | 0.09370 | 0.44010 | 0.68320 | 0.0820* | |
| H18A | 0.54110 | 0.39030 | 0.73340 | 0.1780* | |
| H18B | 0.52490 | 0.39530 | 0.68580 | 0.1780* | |
| H18C | 0.37570 | 0.34590 | 0.71080 | 0.1780* | |
| H19 | 0.31090 | 0.47940 | 0.77820 | 0.1050* | |
| H20A | −0.01600 | 0.43330 | 0.74210 | 0.1540* | |
| H20B | 0.02070 | 0.44690 | 0.79000 | 0.1540* | |
| H3O | 0.376 (6) | 0.680 (2) | 0.4887 (11) | 0.0840* | |
| H22A | 0.47530 | 0.56390 | 0.45380 | 0.0470* | |
| H22B | 0.28140 | 0.52330 | 0.46360 | 0.0470* | |
| H23 | 0.31650 | 0.54830 | 0.53200 | 0.0510* | |
| H24A | 0.57450 | 0.61250 | 0.55960 | 0.0620* | |
| H24B | 0.66760 | 0.62210 | 0.51660 | 0.0620* | |
| H26 | 0.76590 | 0.49780 | 0.47980 | 0.0450* | |
| H27A | 0.67140 | 0.40550 | 0.57700 | 0.1120* | |
| H27B | 0.47760 | 0.44880 | 0.56990 | 0.1120* | |
| H27C | 0.62520 | 0.48710 | 0.59980 | 0.1120* | |
| H28A | 0.96390 | 0.47950 | 0.55950 | 0.1200* | |
| H28B | 0.89580 | 0.56560 | 0.57400 | 0.1200* | |
| H28C | 0.96080 | 0.55290 | 0.52880 | 0.1200* | |
| H29A | 0.74900 | 0.33940 | 0.51120 | 0.0600* | |
| H29B | 0.93390 | 0.39180 | 0.51120 | 0.0600* | |
| H30A | 0.92890 | 0.39840 | 0.43980 | 0.0620* | |
| H30B | 0.92870 | 0.30670 | 0.45340 | 0.0620* | |
| H32 | 0.62550 | 0.46210 | 0.42090 | 0.0430* | |
| H33A | 0.44450 | 0.34590 | 0.51140 | 0.0680* | |
| H33B | 0.28550 | 0.36510 | 0.47980 | 0.0680* | |
| H33C | 0.30330 | 0.41750 | 0.51950 | 0.0680* | |
| H34A | 0.54640 | 0.26490 | 0.47010 | 0.0820* | |
| H34B | 0.48360 | 0.24830 | 0.42510 | 0.0820* | |
| H34C | 0.68110 | 0.21860 | 0.44020 | 0.0820* | |
| H35A | 0.30950 | 0.36190 | 0.41050 | 0.0570* | |
| H35B | 0.30180 | 0.45740 | 0.40590 | 0.0570* | |
| H36A | 0.51450 | 0.45400 | 0.35270 | 0.0740* | |
| H36B | 0.35230 | 0.39290 | 0.34210 | 0.0740* | |
| H38A | 0.77500 | 0.40160 | 0.31800 | 0.1480* | |
| H38B | 0.63560 | 0.34670 | 0.29320 | 0.1480* | |
| H38C | 0.81150 | 0.30760 | 0.31430 | 0.1480* | |
| H39 | 0.59900 | 0.20710 | 0.35570 | 0.1010* | |
| H40A | 0.25380 | 0.27150 | 0.33920 | 0.1460* | |
| H40B | 0.30770 | 0.17630 | 0.34240 | 0.1460* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0556 (13) | 0.0615 (14) | 0.0404 (12) | 0.0157 (11) | −0.0096 (10) | 0.0025 (10) |
| O2 | 0.0720 (15) | 0.0624 (15) | 0.0625 (14) | 0.0127 (13) | −0.0031 (12) | 0.0215 (12) |
| C1 | 0.0288 (13) | 0.0360 (15) | 0.0321 (14) | 0.0008 (11) | 0.0022 (11) | −0.0020 (11) |
| C2 | 0.0342 (15) | 0.0406 (15) | 0.0417 (15) | −0.0004 (12) | −0.0010 (12) | 0.0001 (12) |
| C3 | 0.0392 (15) | 0.0447 (16) | 0.0361 (15) | 0.0083 (13) | −0.0022 (12) | −0.0010 (12) |
| O3 | 0.0585 (14) | 0.0481 (13) | 0.0578 (13) | 0.0106 (11) | 0.0178 (11) | −0.0004 (10) |
| C4 | 0.0497 (18) | 0.0437 (17) | 0.0450 (16) | 0.0072 (14) | 0.0078 (14) | 0.0109 (13) |
| O4 | 0.0561 (13) | 0.0539 (12) | 0.0575 (13) | 0.0036 (10) | 0.0109 (11) | −0.0072 (11) |
| C5 | 0.0401 (16) | 0.0412 (17) | 0.0506 (17) | −0.0040 (13) | 0.0038 (13) | 0.0035 (13) |
| C6 | 0.0280 (13) | 0.0409 (15) | 0.0385 (14) | 0.0009 (12) | 0.0045 (12) | −0.0010 (12) |
| C7 | 0.054 (2) | 0.084 (2) | 0.079 (2) | −0.0273 (19) | 0.0005 (18) | 0.027 (2) |
| C8 | 0.079 (2) | 0.0426 (18) | 0.070 (2) | −0.0066 (17) | −0.0012 (19) | −0.0047 (16) |
| C9 | 0.0372 (16) | 0.065 (2) | 0.0509 (17) | −0.0135 (15) | −0.0063 (14) | 0.0059 (15) |
| C10 | 0.0403 (17) | 0.078 (2) | 0.0545 (18) | −0.0012 (17) | −0.0152 (14) | 0.0132 (17) |
| C11 | 0.0520 (18) | 0.0482 (17) | 0.0399 (16) | 0.0053 (15) | −0.0018 (14) | 0.0093 (13) |
| C12 | 0.0416 (16) | 0.0395 (15) | 0.0289 (13) | 0.0033 (12) | 0.0036 (12) | −0.0004 (11) |
| C13 | 0.0369 (15) | 0.0509 (18) | 0.0501 (17) | 0.0036 (14) | 0.0101 (13) | −0.0013 (14) |
| C14 | 0.076 (2) | 0.068 (2) | 0.0355 (16) | −0.0062 (18) | −0.0027 (16) | −0.0003 (15) |
| C15 | 0.066 (2) | 0.0467 (18) | 0.0465 (17) | −0.0170 (16) | −0.0032 (16) | 0.0043 (14) |
| C16 | 0.102 (3) | 0.0448 (19) | 0.057 (2) | −0.0143 (19) | 0.012 (2) | 0.0058 (15) |
| C17 | 0.103 (3) | 0.051 (2) | 0.060 (2) | 0.008 (2) | 0.005 (2) | 0.0181 (17) |
| C18 | 0.167 (5) | 0.063 (3) | 0.125 (4) | 0.036 (3) | 0.021 (4) | 0.027 (3) |
| C19 | 0.133 (4) | 0.065 (2) | 0.066 (2) | −0.013 (3) | 0.010 (3) | 0.022 (2) |
| C20 | 0.178 (6) | 0.118 (4) | 0.089 (3) | −0.063 (4) | 0.041 (4) | 0.004 (3) |
| C21 | 0.0268 (13) | 0.0368 (15) | 0.0363 (14) | −0.0016 (11) | −0.0019 (11) | 0.0078 (12) |
| C22 | 0.0325 (14) | 0.0433 (16) | 0.0407 (15) | 0.0019 (13) | −0.0002 (12) | 0.0056 (12) |
| C23 | 0.0424 (16) | 0.0436 (17) | 0.0413 (15) | 0.0005 (14) | 0.0107 (13) | 0.0045 (12) |
| C24 | 0.0532 (18) | 0.0572 (19) | 0.0453 (17) | −0.0051 (16) | 0.0053 (15) | −0.0095 (14) |
| C25 | 0.0435 (17) | 0.062 (2) | 0.0420 (17) | −0.0046 (15) | −0.0106 (15) | −0.0011 (14) |
| C26 | 0.0284 (14) | 0.0431 (16) | 0.0418 (15) | −0.0043 (12) | −0.0025 (12) | 0.0077 (12) |
| C27 | 0.096 (3) | 0.088 (3) | 0.0400 (18) | 0.006 (2) | −0.0099 (19) | 0.0103 (18) |
| C28 | 0.062 (2) | 0.094 (3) | 0.084 (3) | 0.003 (2) | −0.029 (2) | −0.026 (2) |
| C29 | 0.0309 (15) | 0.0564 (19) | 0.0633 (19) | 0.0026 (13) | −0.0155 (15) | 0.0046 (16) |
| C30 | 0.0325 (15) | 0.0507 (18) | 0.071 (2) | 0.0104 (13) | 0.0004 (15) | 0.0047 (15) |
| C31 | 0.0389 (15) | 0.0390 (16) | 0.0478 (16) | 0.0028 (13) | 0.0039 (13) | 0.0008 (13) |
| C32 | 0.0338 (14) | 0.0329 (14) | 0.0402 (14) | −0.0008 (12) | −0.0008 (12) | 0.0067 (12) |
| C33 | 0.0374 (16) | 0.0485 (17) | 0.0501 (17) | −0.0068 (13) | 0.0023 (13) | 0.0112 (14) |
| C34 | 0.0558 (18) | 0.0378 (16) | 0.069 (2) | 0.0038 (15) | −0.0042 (16) | 0.0059 (14) |
| C35 | 0.0494 (18) | 0.0487 (17) | 0.0452 (16) | 0.0060 (14) | −0.0115 (14) | 0.0012 (13) |
| C36 | 0.078 (2) | 0.064 (2) | 0.0438 (18) | 0.0015 (19) | −0.0134 (16) | −0.0016 (15) |
| C37 | 0.081 (2) | 0.063 (2) | 0.0471 (18) | −0.002 (2) | 0.0030 (18) | −0.0111 (16) |
| C38 | 0.128 (4) | 0.110 (3) | 0.058 (2) | 0.000 (3) | 0.029 (2) | −0.012 (2) |
| C39 | 0.110 (3) | 0.070 (3) | 0.073 (2) | −0.001 (2) | −0.013 (2) | −0.023 (2) |
| C40 | 0.137 (4) | 0.100 (4) | 0.129 (4) | −0.030 (3) | −0.050 (4) | −0.018 (3) |
Geometric parameters (Å, º)
| O1—C3 | 1.436 (3) | C18—H18A | 0.9600 |
| O2—C11 | 1.447 (4) | C19—H19 | 0.9300 |
| O2—C17 | 1.440 (4) | C20—H20A | 0.9300 |
| O1—H1O | 0.80 (4) | C20—H20B | 0.9300 |
| C1—C6 | 1.555 (3) | C21—C26 | 1.562 (3) |
| C1—C12 | 1.564 (4) | C21—C32 | 1.559 (3) |
| C1—C2 | 1.534 (4) | C21—C22 | 1.532 (3) |
| C1—C13 | 1.546 (4) | C21—C33 | 1.537 (4) |
| C2—C3 | 1.520 (4) | C22—C23 | 1.515 (4) |
| C3—C4 | 1.511 (4) | C23—C24 | 1.520 (4) |
| O3—C23 | 1.437 (3) | C24—C25 | 1.525 (4) |
| C4—C5 | 1.533 (4) | C25—C27 | 1.538 (4) |
| O4—C37 | 1.443 (4) | C25—C28 | 1.541 (5) |
| O4—C31 | 1.455 (3) | C25—C26 | 1.551 (4) |
| C5—C7 | 1.541 (5) | C26—C29 | 1.526 (4) |
| C5—C6 | 1.549 (4) | C29—C30 | 1.527 (4) |
| C5—C8 | 1.534 (4) | C30—C31 | 1.522 (4) |
| C6—C9 | 1.529 (4) | C31—C32 | 1.532 (4) |
| C9—C10 | 1.524 (4) | C31—C34 | 1.532 (4) |
| C10—C11 | 1.520 (4) | C32—C35 | 1.528 (4) |
| C11—C12 | 1.540 (4) | C35—C36 | 1.519 (4) |
| C11—C14 | 1.530 (4) | C36—C37 | 1.528 (5) |
| C12—C15 | 1.522 (4) | C37—C39 | 1.505 (5) |
| C15—C16 | 1.513 (4) | C37—C38 | 1.518 (5) |
| C16—C17 | 1.530 (5) | C39—C40 | 1.305 (7) |
| C17—C19 | 1.511 (6) | C22—H22A | 0.9700 |
| C17—C18 | 1.532 (6) | C22—H22B | 0.9700 |
| C19—C20 | 1.293 (8) | C23—H23 | 0.9800 |
| C2—H2A | 0.9700 | C24—H24A | 0.9700 |
| C2—H2B | 0.9700 | C24—H24B | 0.9700 |
| O3—H3O | 0.81 (4) | C26—H26 | 0.9800 |
| C3—H3 | 0.9800 | C27—H27A | 0.9600 |
| C4—H4B | 0.9700 | C27—H27B | 0.9600 |
| C4—H4A | 0.9700 | C27—H27C | 0.9600 |
| C6—H6 | 0.9800 | C28—H28A | 0.9600 |
| C7—H7B | 0.9600 | C28—H28B | 0.9600 |
| C7—H7A | 0.9600 | C28—H28C | 0.9600 |
| C7—H7C | 0.9600 | C29—H29A | 0.9700 |
| C8—H8B | 0.9600 | C29—H29B | 0.9700 |
| C8—H8A | 0.9600 | C30—H30A | 0.9700 |
| C8—H8C | 0.9600 | C30—H30B | 0.9700 |
| C9—H9B | 0.9700 | C32—H32 | 0.9800 |
| C9—H9A | 0.9700 | C33—H33A | 0.9600 |
| C10—H10B | 0.9700 | C33—H33B | 0.9600 |
| C10—H10A | 0.9700 | C33—H33C | 0.9600 |
| C12—H12 | 0.9800 | C34—H34A | 0.9600 |
| C13—H13C | 0.9600 | C34—H34B | 0.9600 |
| C13—H13A | 0.9600 | C34—H34C | 0.9600 |
| C13—H13B | 0.9600 | C35—H35A | 0.9700 |
| C14—H14C | 0.9600 | C35—H35B | 0.9700 |
| C14—H14A | 0.9600 | C36—H36A | 0.9700 |
| C14—H14B | 0.9600 | C36—H36B | 0.9700 |
| C15—H15A | 0.9700 | C38—H38A | 0.9600 |
| C15—H15B | 0.9700 | C38—H38B | 0.9600 |
| C16—H16A | 0.9700 | C38—H38C | 0.9600 |
| C16—H16B | 0.9700 | C39—H39 | 0.9300 |
| C18—H18C | 0.9600 | C40—H40A | 0.9300 |
| C18—H18B | 0.9600 | C40—H40B | 0.9300 |
| C11—O2—C17 | 119.5 (2) | H20A—C20—H20B | 120.00 |
| C3—O1—H1O | 110 (3) | C19—C20—H20A | 120.00 |
| C2—C1—C6 | 107.9 (2) | C22—C21—C26 | 107.7 (2) |
| C2—C1—C12 | 108.0 (2) | C22—C21—C32 | 108.10 (19) |
| C6—C1—C12 | 106.05 (19) | C26—C21—C32 | 105.98 (19) |
| C6—C1—C13 | 114.7 (2) | C26—C21—C33 | 114.4 (2) |
| C12—C1—C13 | 111.6 (2) | C32—C21—C33 | 111.7 (2) |
| C2—C1—C13 | 108.3 (2) | C22—C21—C33 | 108.8 (2) |
| C1—C2—C3 | 114.6 (2) | C21—C22—C23 | 114.9 (2) |
| O1—C3—C2 | 110.0 (2) | O3—C23—C22 | 109.8 (2) |
| O1—C3—C4 | 111.5 (2) | O3—C23—C24 | 111.2 (2) |
| C2—C3—C4 | 111.4 (2) | C22—C23—C24 | 111.9 (2) |
| C3—C4—C5 | 114.8 (2) | C23—C24—C25 | 114.9 (2) |
| C31—O4—C37 | 119.1 (2) | C24—C25—C27 | 110.9 (2) |
| C4—C5—C7 | 106.7 (2) | C24—C25—C28 | 106.9 (3) |
| C4—C5—C8 | 110.7 (2) | C26—C25—C27 | 114.7 (2) |
| C6—C5—C8 | 114.9 (2) | C26—C25—C28 | 108.9 (2) |
| C7—C5—C8 | 107.5 (2) | C27—C25—C28 | 107.3 (3) |
| C4—C5—C6 | 107.9 (2) | C24—C25—C26 | 107.9 (2) |
| C6—C5—C7 | 108.8 (2) | C21—C26—C25 | 116.3 (2) |
| C1—C6—C5 | 116.4 (2) | C25—C26—C29 | 115.4 (2) |
| C5—C6—C9 | 115.3 (2) | C21—C26—C29 | 111.2 (2) |
| C1—C6—C9 | 111.3 (2) | C26—C29—C30 | 110.6 (2) |
| C6—C9—C10 | 110.8 (2) | C29—C30—C31 | 112.2 (2) |
| C9—C10—C11 | 112.6 (2) | O4—C31—C32 | 107.8 (2) |
| O2—C11—C12 | 108.1 (2) | O4—C31—C34 | 109.6 (2) |
| O2—C11—C14 | 109.3 (2) | O4—C31—C30 | 103.9 (2) |
| C10—C11—C14 | 109.5 (2) | C30—C31—C34 | 109.0 (2) |
| C12—C11—C14 | 116.4 (2) | C32—C31—C34 | 116.8 (2) |
| O2—C11—C10 | 103.9 (2) | C30—C31—C32 | 109.2 (2) |
| C10—C11—C12 | 108.9 (2) | C21—C32—C31 | 116.2 (2) |
| C1—C12—C11 | 115.6 (2) | C21—C32—C35 | 116.4 (2) |
| C1—C12—C15 | 117.3 (2) | C31—C32—C35 | 109.7 (2) |
| C11—C12—C15 | 109.5 (2) | C32—C35—C36 | 108.5 (2) |
| C12—C15—C16 | 108.8 (3) | C35—C36—C37 | 113.8 (3) |
| C15—C16—C17 | 113.2 (3) | O4—C37—C38 | 103.8 (3) |
| O2—C17—C19 | 110.7 (3) | O4—C37—C39 | 110.1 (3) |
| C16—C17—C18 | 110.5 (3) | C36—C37—C38 | 110.2 (3) |
| O2—C17—C18 | 103.5 (3) | C36—C37—C39 | 114.1 (3) |
| C18—C17—C19 | 107.4 (3) | C38—C37—C39 | 107.4 (3) |
| C16—C17—C19 | 113.6 (4) | O4—C37—C36 | 110.5 (2) |
| O2—C17—C16 | 110.7 (3) | C37—C39—C40 | 128.2 (4) |
| C17—C19—C20 | 128.0 (4) | C21—C22—H22A | 109.00 |
| C1—C2—H2A | 109.00 | C21—C22—H22B | 109.00 |
| C3—C2—H2A | 109.00 | C23—C22—H22A | 108.00 |
| C3—C2—H2B | 109.00 | C23—C22—H22B | 108.00 |
| C1—C2—H2B | 108.00 | H22A—C22—H22B | 108.00 |
| H2A—C2—H2B | 108.00 | O3—C23—H23 | 108.00 |
| C2—C3—H3 | 108.00 | C22—C23—H23 | 108.00 |
| C4—C3—H3 | 108.00 | C24—C23—H23 | 108.00 |
| O1—C3—H3 | 108.00 | C23—C24—H24A | 109.00 |
| C23—O3—H3O | 108 (3) | C23—C24—H24B | 109.00 |
| C3—C4—H4B | 109.00 | C25—C24—H24A | 109.00 |
| C5—C4—H4A | 109.00 | C25—C24—H24B | 109.00 |
| H4A—C4—H4B | 108.00 | H24A—C24—H24B | 108.00 |
| C5—C4—H4B | 109.00 | C21—C26—H26 | 104.00 |
| C3—C4—H4A | 109.00 | C25—C26—H26 | 104.00 |
| C9—C6—H6 | 104.00 | C29—C26—H26 | 104.00 |
| C1—C6—H6 | 104.00 | C25—C27—H27A | 109.00 |
| C5—C6—H6 | 104.00 | C25—C27—H27B | 109.00 |
| C5—C7—H7B | 109.00 | C25—C27—H27C | 109.00 |
| C5—C7—H7A | 109.00 | H27A—C27—H27B | 109.00 |
| H7A—C7—H7C | 109.00 | H27A—C27—H27C | 110.00 |
| C5—C7—H7C | 109.00 | H27B—C27—H27C | 110.00 |
| H7A—C7—H7B | 109.00 | C25—C28—H28A | 110.00 |
| H7B—C7—H7C | 110.00 | C25—C28—H28B | 109.00 |
| C5—C8—H8B | 109.00 | C25—C28—H28C | 109.00 |
| C5—C8—H8C | 109.00 | H28A—C28—H28B | 109.00 |
| H8A—C8—H8C | 109.00 | H28A—C28—H28C | 110.00 |
| H8B—C8—H8C | 110.00 | H28B—C28—H28C | 109.00 |
| H8A—C8—H8B | 109.00 | C26—C29—H29A | 110.00 |
| C5—C8—H8A | 109.00 | C26—C29—H29B | 110.00 |
| C6—C9—H9B | 109.00 | C30—C29—H29A | 110.00 |
| C10—C9—H9B | 109.00 | C30—C29—H29B | 110.00 |
| H9A—C9—H9B | 108.00 | H29A—C29—H29B | 108.00 |
| C6—C9—H9A | 110.00 | C29—C30—H30A | 109.00 |
| C10—C9—H9A | 109.00 | C29—C30—H30B | 109.00 |
| C11—C10—H10A | 109.00 | C31—C30—H30A | 109.00 |
| C9—C10—H10A | 109.00 | C31—C30—H30B | 109.00 |
| C9—C10—H10B | 109.00 | H30A—C30—H30B | 108.00 |
| H10A—C10—H10B | 108.00 | C21—C32—H32 | 104.00 |
| C11—C10—H10B | 109.00 | C31—C32—H32 | 104.00 |
| C11—C12—H12 | 104.00 | C35—C32—H32 | 104.00 |
| C1—C12—H12 | 104.00 | C21—C33—H33A | 109.00 |
| C15—C12—H12 | 104.00 | C21—C33—H33B | 110.00 |
| C1—C13—H13A | 109.00 | C21—C33—H33C | 109.00 |
| H13A—C13—H13B | 109.00 | H33A—C33—H33B | 109.00 |
| C1—C13—H13B | 110.00 | H33A—C33—H33C | 110.00 |
| H13B—C13—H13C | 110.00 | H33B—C33—H33C | 109.00 |
| C1—C13—H13C | 109.00 | C31—C34—H34A | 110.00 |
| H13A—C13—H13C | 109.00 | C31—C34—H34B | 109.00 |
| C11—C14—H14B | 110.00 | C31—C34—H34C | 109.00 |
| C11—C14—H14A | 109.00 | H34A—C34—H34B | 109.00 |
| H14A—C14—H14C | 109.00 | H34A—C34—H34C | 109.00 |
| C11—C14—H14C | 110.00 | H34B—C34—H34C | 110.00 |
| H14A—C14—H14B | 109.00 | C32—C35—H35A | 110.00 |
| H14B—C14—H14C | 109.00 | C32—C35—H35B | 110.00 |
| C12—C15—H15B | 110.00 | C36—C35—H35A | 110.00 |
| C16—C15—H15A | 110.00 | C36—C35—H35B | 110.00 |
| H15A—C15—H15B | 108.00 | H35A—C35—H35B | 108.00 |
| C16—C15—H15B | 110.00 | C35—C36—H36A | 109.00 |
| C12—C15—H15A | 110.00 | C35—C36—H36B | 109.00 |
| C15—C16—H16A | 109.00 | C37—C36—H36A | 109.00 |
| C17—C16—H16A | 109.00 | C37—C36—H36B | 109.00 |
| C17—C16—H16B | 109.00 | H36A—C36—H36B | 108.00 |
| H16A—C16—H16B | 108.00 | C37—C38—H38A | 110.00 |
| C15—C16—H16B | 109.00 | C37—C38—H38B | 109.00 |
| C17—C18—H18C | 109.00 | C37—C38—H38C | 109.00 |
| C17—C18—H18A | 109.00 | H38A—C38—H38B | 109.00 |
| C17—C18—H18B | 109.00 | H38A—C38—H38C | 109.00 |
| H18B—C18—H18C | 109.00 | H38B—C38—H38C | 110.00 |
| H18A—C18—H18B | 110.00 | C37—C39—H39 | 116.00 |
| H18A—C18—H18C | 110.00 | C40—C39—H39 | 116.00 |
| C20—C19—H19 | 116.00 | C39—C40—H40A | 120.00 |
| C17—C19—H19 | 116.00 | C39—C40—H40B | 120.00 |
| C19—C20—H20B | 120.00 | H40A—C40—H40B | 120.00 |
| C17—O2—C11—C10 | −169.9 (2) | C15—C16—C17—C19 | 78.7 (4) |
| C17—O2—C11—C12 | −54.3 (3) | C15—C16—C17—C18 | −160.6 (3) |
| C17—O2—C11—C14 | 73.2 (3) | C15—C16—C17—O2 | −46.6 (4) |
| C11—O2—C17—C19 | −78.7 (4) | C18—C17—C19—C20 | −102.8 (5) |
| C11—O2—C17—C16 | 48.2 (4) | O2—C17—C19—C20 | 145.0 (4) |
| C11—O2—C17—C18 | 166.6 (3) | C16—C17—C19—C20 | 19.7 (5) |
| C12—C1—C2—C3 | −165.9 (2) | C32—C21—C22—C23 | −165.4 (2) |
| C13—C1—C6—C5 | −68.4 (3) | C33—C21—C26—C25 | −68.0 (3) |
| C13—C1—C2—C3 | 73.1 (3) | C33—C21—C22—C23 | 73.2 (3) |
| C6—C1—C2—C3 | −51.7 (3) | C26—C21—C22—C23 | −51.3 (3) |
| C2—C1—C12—C15 | −57.2 (3) | C22—C21—C32—C35 | −58.3 (3) |
| C13—C1—C6—C9 | 66.6 (3) | C33—C21—C26—C29 | 66.8 (3) |
| C2—C1—C12—C11 | 171.2 (2) | C22—C21—C32—C31 | 170.2 (2) |
| C12—C1—C6—C9 | −57.1 (3) | C32—C21—C26—C29 | −56.7 (3) |
| C13—C1—C12—C15 | 61.8 (3) | C33—C21—C32—C35 | 61.3 (3) |
| C6—C1—C12—C11 | 55.8 (3) | C26—C21—C32—C31 | 55.0 (3) |
| C6—C1—C12—C15 | −172.6 (2) | C26—C21—C32—C35 | −173.5 (2) |
| C13—C1—C12—C11 | −69.8 (3) | C33—C21—C32—C31 | −70.2 (3) |
| C12—C1—C6—C5 | 168.0 (2) | C32—C21—C26—C25 | 168.5 (2) |
| C2—C1—C6—C9 | −172.6 (2) | C22—C21—C26—C29 | −172.2 (2) |
| C2—C1—C6—C5 | 52.5 (3) | C22—C21—C26—C25 | 53.0 (3) |
| C1—C2—C3—C4 | 53.9 (3) | C21—C22—C23—C24 | 52.8 (3) |
| C1—C2—C3—O1 | 178.0 (2) | C21—C22—C23—O3 | 176.9 (2) |
| O1—C3—C4—C5 | −177.3 (2) | O3—C23—C24—C25 | −176.2 (2) |
| C2—C3—C4—C5 | −54.1 (3) | C22—C23—C24—C25 | −53.0 (3) |
| C3—C4—C5—C8 | −74.5 (3) | C23—C24—C25—C28 | 168.7 (3) |
| C3—C4—C5—C7 | 168.9 (2) | C23—C24—C25—C27 | −74.6 (3) |
| C3—C4—C5—C6 | 52.1 (3) | C23—C24—C25—C26 | 51.8 (3) |
| C37—O4—C31—C32 | −55.5 (3) | C27—C25—C26—C21 | 70.9 (3) |
| C37—O4—C31—C30 | −171.2 (2) | C27—C25—C26—C29 | −62.1 (3) |
| C37—O4—C31—C34 | 72.5 (3) | C24—C25—C26—C29 | 173.8 (2) |
| C31—O4—C37—C38 | 166.7 (3) | C28—C25—C26—C29 | 58.2 (3) |
| C31—O4—C37—C39 | −78.5 (3) | C28—C25—C26—C21 | −168.9 (3) |
| C31—O4—C37—C36 | 48.5 (3) | C24—C25—C26—C21 | −53.3 (3) |
| C7—C5—C6—C1 | −167.9 (2) | C25—C26—C29—C30 | −164.0 (2) |
| C7—C5—C6—C9 | 58.9 (3) | C21—C26—C29—C30 | 60.8 (3) |
| C4—C5—C6—C9 | 174.3 (2) | C26—C29—C30—C31 | −58.3 (3) |
| C8—C5—C6—C1 | 71.5 (3) | C29—C30—C31—C32 | 53.0 (3) |
| C8—C5—C6—C9 | −61.6 (3) | C29—C30—C31—C34 | −75.5 (3) |
| C4—C5—C6—C1 | −52.5 (3) | C29—C30—C31—O4 | 167.8 (2) |
| C5—C6—C9—C10 | −164.3 (2) | O4—C31—C32—C35 | 59.4 (3) |
| C1—C6—C9—C10 | 60.3 (3) | C34—C31—C32—C21 | 70.2 (3) |
| C6—C9—C10—C11 | −57.8 (3) | C30—C31—C32—C21 | −53.9 (3) |
| C9—C10—C11—C14 | −75.3 (3) | C30—C31—C32—C35 | 171.6 (2) |
| C9—C10—C11—C12 | 53.1 (3) | O4—C31—C32—C21 | −166.0 (2) |
| C9—C10—C11—O2 | 168.1 (2) | C34—C31—C32—C35 | −64.4 (3) |
| C10—C11—C12—C1 | −54.3 (3) | C31—C32—C35—C36 | −60.3 (3) |
| C10—C11—C12—C15 | 170.6 (2) | C21—C32—C35—C36 | 165.2 (2) |
| O2—C11—C12—C15 | 58.3 (3) | C32—C35—C36—C37 | 53.9 (3) |
| C14—C11—C12—C15 | −65.0 (3) | C35—C36—C37—O4 | −46.2 (4) |
| C14—C11—C12—C1 | 70.1 (3) | C35—C36—C37—C38 | −160.4 (3) |
| O2—C11—C12—C1 | −166.5 (2) | C35—C36—C37—C39 | 78.6 (3) |
| C11—C12—C15—C16 | −60.5 (3) | O4—C37—C39—C40 | 142.0 (4) |
| C1—C12—C15—C16 | 165.3 (2) | C36—C37—C39—C40 | 17.0 (6) |
| C12—C15—C16—C17 | 54.8 (4) | C38—C37—C39—C40 | −105.6 (5) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···O3 | 0.80 (4) | 1.99 (4) | 2.784 (3) | 170 (4) |
| O3—H3O···O1i | 0.81 (4) | 2.00 (4) | 2.804 (3) | 169 (4) |
Symmetry code: (i) x+1/2, −y+3/2, −z+1.
References
- Barberan, F. A. T., Nuñez, J. M. & Tomas, F. (1985). Phytochemistry, 24, 1285–1288.
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Duman, H. (2000). Sideritis L, in Flora of Turkey and East Aegean Islands (Supplement 2), Vol. 11, pp. 201–205. Edinburgh: University Press.
- Evans, G. B. & Gainsford, G. J. (2011). Acta Cryst. E67, o2870. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Fraga, B. M., Reina, M., Luis, J. G. & Rodriguez, M. L. (2003). Z. Naturforsch. Teil C, 58, 621–625. [DOI] [PubMed]
- González-Burgos, E., Carretero, M. E. & Gómez-Serranillos, M. P. (2011). J. Ethnopharmacol. 135, 209–225. [DOI] [PubMed]
- Sheldrick, G. M. (2003). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stewart, J. J. P. (1985). MOPAC. QCPE Program 445. Quantum Chemistry Program Exchange, Indiana University, Bloomington, IN 47405, USA.
- Topçu, G., Gören, A. C., Kıliç, T., Kemal Yıldız, Y. & Tümen, G. (2002). Nat. Prod. Lett. 16, 33–37. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989016013864/bg2593sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016013864/bg2593Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593sup3.pdf
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593sup4.tif
Supporting information file. DOI: 10.1107/S2056989016013864/bg2593Isup5.cml
CCDC reference: 1501445
Additional supporting information: crystallographic information; 3D view; checkCIF report