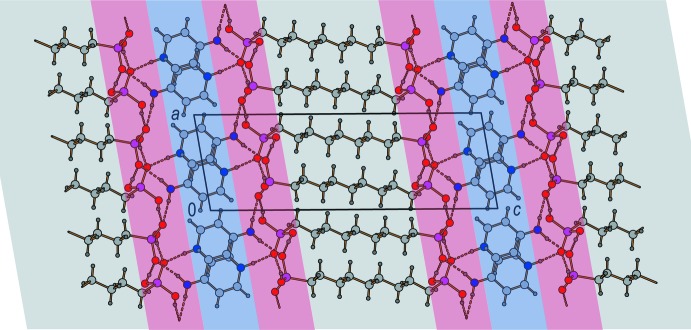
The structure of the title molecular salt, [C5H7N2][(HO)2OP(CH2)9PO2(OH)], shows a three-dimensional network with hydrogen bonding, π–π stacking, and van der Waals forces-dominated layered regions.
Keywords: crystal structure, 4-aminopyridinium, bis(phosphonate), hydrogen bonding
Abstract
The asymmetric unit of the title molecular salt, [C5H7N2 +][(HO)2OP(CH2)9PO2(OH)−], consists of one 4-aminopyridinium cation and one hydrogen (9-phosphonononyl)phosphonate anion, both in general positions. As expected, the 4-aminopyridinium moieties are protonated exclusively at their endocyclic nitrogen atom due to a mesomeric stabilization by the imine form which would not be given in the corresponding double-protonated dicationic species. In the crystal, the phosphonyl (–PO3H2) and hydrogen phosphonate (–PO3H) groups of the anions form two-dimensional O—H⋯O hydrogen-bonded networks in the ab plane built from 24-membered hydrogen-bonded ring motifs with the graph-set descriptor R 6 6(24). These networks are pairwise linked by the anions’ alkylene chains. The 4-aminopyridinium cations are stacked in parallel displaced face-to-face arrangements and connect neighboring anionic substructures via medium–strong charge-supported N—H⋯O hydrogen bonds along the c axis. The resulting three-dimensional hydrogen-bonded network shows clearly separated hydrophilic and hydrophobic structural domains.
Chemical context
Salts of organophosphonic acids with organic cations, e.g. with protonated primary (Mahmoudkhani & Langer, 2002b
▸), secondary (Wheatley et al., 2001 ▸) and tertiary amines (Kan & Ma, 2011 ▸) are of growing interest in supramolecular chemistry and crystal engineering. Besides their interesting topologies and structural diversity, they seem to be feasible model compounds for metal phosphonates as they exhibit similar structural characteristics but are less difficult to crystallize. Mostly, these organic solids establish extended hydrogen-bonded networks which are characterized by a rich diversity of strong charge-supported hydrogen bonds (Aakeröy & Seddon, 1993 ▸) and can either be one-, two- or three-dimensional. This contribution forms part of our research on the principles of the arrangement of alkane-α,ω-diphosphonic acids (van Megen et al., 2015 ▸) and their organic aminium salts (van Megen et al., 2016 ▸). Moreover, aminopyridines and the related protonated cations are of crucial interest in the field of biochemistry (Muñoz-Caro & Niño, 2002 ▸; Bolliger et al., 2011 ▸) and are also used as counter-cations to stabilize complex salts (Reiss & Leske, 2014a
▸,b
▸), in crystal engineering (Sertucha et al., 1998 ▸; Surbella III et al., 2016 ▸) as well as in polymer chemistry (Deng et al., 2015 ▸).
Structural commentary
The asymmetric unit of the title compound, [C5H7N2
+][(HO)2OP(CH2)9PO2(OH)−], consists of one 4-aminopyridinium cation and one hydrogen (9-phosphonononyl)phosphonate anion, both in general positions (Fig. 1 ▸). Generally, the first protonation of the 4-aminopyridine can take place at the exo- as well as at the endocyclic nitrogen atom. In the literature, all monoprotonated 4-aminopyridines characterized to date are protonated at the endocyclic nitrogen atom. Geometric parameters derived from the single-crystal diffraction experiment for the title compound show a short exocyclic N—C bond length [1.324 (2) Å] and slightly longer C—C and C—N bond lengths of the six-membered ring [1.350 (3)–1.425 (2) Å]. The bonding properties of this cation are best described by a pair of mesomeric structures: the enamine and the imine form (Scheme 2), which have been discussed in detail before (Koleva et al., 2008 ▸).
Figure 1.
The asymmetric unit of the title compound plus symmetry-related hydrogen-bonded atoms [displacement ellipsoids are drawn at the 50% probability level; hydrogen atoms are drawn as spheres with arbitrary radii; symmetry codes: (i) 1 + x, −1 + y, 1 + z; (ii) x, −1 + y, 1 + z; (iii) 1 − x, 2 − y, 1 − z; (iv) 1 − x, 1 − y, 1 − z; (v) −x, 1 − y, 1 − z; (vi) −1 + x, 1 + y, −1 + z, (vii) x, 1 + y, −1 + z].
For the designation of the title compound, the systematic name of the amino form is used throughout this article. The bond lengths and angles of the anion are unexceptional and lie within the expected ranges. The alkylene chain of the anion shows nearly antiperiplanar conformations. In detail, the P—OH distances of the phosphonate moieties have values between 1.5535 (13) and 1.5786 (14) Å, longer than the P=O distances [1.5045 (13)–1.5149 (12) Å].
Supramolecular features
Within the crystal of the title compound, the phosphonyl and hydrogen phosphonate groups of the anions form two-dimensional O—H⋯O hydrogen-bonded networks which propagate in the ab plane. These networks contain 24-membered rings classified as a third level graph set  (24) (Etter et al., 1990 ▸; Fig. 2 ▸; Table 1 ▸). 24-Membered hydrogen-bonded rings have been well known for decades (e.g. Mootz & Poll, 1984 ▸). In particular, the
(24) (Etter et al., 1990 ▸; Fig. 2 ▸; Table 1 ▸). 24-Membered hydrogen-bonded rings have been well known for decades (e.g. Mootz & Poll, 1984 ▸). In particular, the  (24) motif is very common (e.g. Gomathi & Muthiah, 2011 ▸; Maspoch et al., 2007 ▸). Along the c-axis direction, these networks are pairwise linked by the anions’ alkylene chains to form a three-dimensional anionic substructure. The 4-aminopyridinium cations show π–π stacking interactions. The rings are oriented in parallel displaced face-to-face arrangements (Grimme, 2008 ▸; Fig. 3 ▸). The geometry of these π–π interactions is reflected by distances of 3.25 and 3.32 Å between neighbouring pyridinium rings and centroid offsets of 2.37 and 2.42 Å. These findings are comparable to those found for other compounds containing pyridyl moieties (Janiak, 2000 ▸). Anions and cations are connected by medium–strong, charge-supported N—H⋯O hydrogen bonds (Steiner, 2002 ▸; Table 2 ▸) along the c axis. For these connections, each nitrogen-bound hydrogen atom forms one unbifurcated hydrogen bond (Fig. 1 ▸). The resulting three-dimensional hydrogen-bonded network clearly shows separated hydrophilic and hydrophobic regions (Fig. 3 ▸).
(24) motif is very common (e.g. Gomathi & Muthiah, 2011 ▸; Maspoch et al., 2007 ▸). Along the c-axis direction, these networks are pairwise linked by the anions’ alkylene chains to form a three-dimensional anionic substructure. The 4-aminopyridinium cations show π–π stacking interactions. The rings are oriented in parallel displaced face-to-face arrangements (Grimme, 2008 ▸; Fig. 3 ▸). The geometry of these π–π interactions is reflected by distances of 3.25 and 3.32 Å between neighbouring pyridinium rings and centroid offsets of 2.37 and 2.42 Å. These findings are comparable to those found for other compounds containing pyridyl moieties (Janiak, 2000 ▸). Anions and cations are connected by medium–strong, charge-supported N—H⋯O hydrogen bonds (Steiner, 2002 ▸; Table 2 ▸) along the c axis. For these connections, each nitrogen-bound hydrogen atom forms one unbifurcated hydrogen bond (Fig. 1 ▸). The resulting three-dimensional hydrogen-bonded network clearly shows separated hydrophilic and hydrophobic regions (Fig. 3 ▸).
Figure 2.
Two-dimensional hydrogen-bonded networks composed of phosphonyl and hydrogen phosphonate groups. The graph set  (24) is indicated by blue bonds.
(24) is indicated by blue bonds.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O6i | 0.78 (3) | 1.85 (3) | 2.6171 (18) | 166 (3) |
| O5—H5⋯O1ii | 0.88 (3) | 1.64 (3) | 2.5059 (18) | 168 (3) |
| O4—H4⋯O2iii | 0.91 (3) | 1.59 (3) | 2.4977 (17) | 178 (3) |
| N1—H1⋯O6 | 0.96 (2) | 1.74 (3) | 2.696 (2) | 173 (2) |
| N2—H22⋯O2iv | 0.90 (3) | 1.92 (3) | 2.806 (2) | 170 (2) |
| N2—H21⋯O1v | 0.88 (3) | 2.14 (3) | 2.965 (2) | 156 (3) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Figure 3.
View along [010] of the title structure, showing the hydrogen bonding (red), π–π stacking (blue), and van der Waals forces (grey) dominated layered regions within the three-dimensional network.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C5H7N2 +·C9H21O6P2 − |
| M r | 382.32 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 123 |
| a, b, c (Å) | 6.7275 (4), 6.8963 (4), 20.0643 (10) |
| α, β, γ (°) | 97.956 (4), 98.767 (4), 94.309 (5) |
| V (Å3) | 906.73 (9) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.27 |
| Crystal size (mm) | 0.33 × 0.07 × 0.03 |
| Data collection | |
| Diffractometer | Stoe IPDS |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8855, 4131, 3674 |
| R int | 0.029 |
| (sin θ/λ)max (Å−1) | 0.650 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.038, 0.079, 1.02 |
| No. of reflections | 4131 |
| No. of parameters | 241 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.50, −0.36 |
Related structures
For related phosphonate and bis(phosphonate) salts, see: Ferguson et al. (1998 ▸); Fu et al. (2004 ▸); Fuller & Heimer (1995 ▸); Glidewell et al. (2000 ▸); Kan & Ma (2011 ▸); Mahmoudkhani & Langer (2002a ▸,b ▸,c ▸); van Megen et al. (2016 ▸); Plabst et al. (2009 ▸); Wheatley et al. (2001 ▸). For related 4-aminopyridinium salts, see: Sertucha et al. (1998 ▸); Reiss & Leske (2014a ▸,b ▸); Surbella III et al. (2016 ▸).
Synthesis and crystallization
Equimolar quantities (0.5 mmol) of 4-aminopyridine (47.1 mg) and nonane-1,9-diphosphonic acid (144.1 mg) were dissolved in methanol, separately. The solutions were mixed and stored in an open petri dish. Within several days, colorless platelet-shaped crystals of the title compound were obtained by slow evaporation of the solvent. 4-Aminopyridine was purchased from commercial sources and nonane-1,9-diphosphonic acid was synthesized according to the literature (Schwarzenbach & Zurc, 1950 ▸; Moedritzer & Irani, 1961 ▸; Griffith et al., 1998 ▸). Elemental analysis: C14H28N2O6P2 (382.3): calculated C 44.0, H 7.4, N 7.3; found C 43.6, H 7.9, N 7.1. M. p.: 157 °C. The IR and Raman spectra of the title compound are shown in Fig. 4 ▸.
Figure 4.
The IR (blue) and Raman (red) spectra of the title compound.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms bound to either nitrogen or oxygen atoms were identified in difference syntheses and refined without any geometric constraints or restraints with individual U iso(H) values. Carbon-bound hydrogen atoms were included using a riding model (AFIX23 option of the SHELX program for the methylene groups and AFIX43 option for the methine groups).
Supplementary Material
Crystal structure: contains datablock(s) I, publication_text. DOI: 10.1107/S2056989016014298/hb7610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016014298/hb7610Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016014298/hb7610Isup3.cml
CCDC reference: 1503436
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank E. Hammes and P. Roloff for technical support.
supplementary crystallographic information
Crystal data
| C5H7N2+·C9H21O6P2− | Z = 2 |
| Mr = 382.32 | F(000) = 408 |
| Triclinic, P1 | Dx = 1.400 Mg m−3 |
| a = 6.7275 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 6.8963 (4) Å | Cell parameters from 6853 reflections |
| c = 20.0643 (10) Å | θ = 3.0–35.3° |
| α = 97.956 (4)° | µ = 0.27 mm−1 |
| β = 98.767 (4)° | T = 123 K |
| γ = 94.309 (5)° | Platelet, colourless |
| V = 906.73 (9) Å3 | 0.33 × 0.07 × 0.03 mm |
Data collection
| Stoe IPDS diffractometer | Rint = 0.029 |
| Radiation source: sealed tube | θmax = 27.5°, θmin = 3.0° |
| ω scans | h = −8→8 |
| 8855 measured reflections | k = −8→8 |
| 4131 independent reflections | l = −26→26 |
| 3674 reflections with I > 2σ(I) |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.038 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.079 | w = 1/[σ2(Fo2) + (0.011P)2 + 1.110P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.001 |
| 4131 reflections | Δρmax = 0.50 e Å−3 |
| 241 parameters | Δρmin = −0.36 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| P1 | 0.29655 (6) | 1.15924 (6) | 0.22320 (2) | 0.01579 (10) | |
| O1 | 0.14667 (19) | 1.0094 (2) | 0.17549 (6) | 0.0230 (3) | |
| N1 | 0.5995 (3) | 0.2836 (2) | 0.92948 (8) | 0.0257 (3) | |
| H1 | 0.553 (4) | 0.306 (4) | 0.8838 (13) | 0.036 (6)* | |
| C1 | 0.2470 (3) | 1.1616 (3) | 0.30883 (8) | 0.0179 (3) | |
| H1A | 0.3161 | 1.2802 | 0.3371 | 0.021* | |
| H1B | 0.1032 | 1.1663 | 0.3086 | 0.021* | |
| P2 | 0.24757 (6) | 0.29013 (6) | 0.77082 (2) | 0.01589 (10) | |
| O2 | 0.51802 (18) | 1.13618 (18) | 0.22159 (6) | 0.0199 (3) | |
| N2 | 0.7740 (3) | 0.1858 (3) | 1.12583 (8) | 0.0252 (3) | |
| H21 | 0.901 (4) | 0.169 (4) | 1.1403 (14) | 0.049 (8)* | |
| H22 | 0.681 (4) | 0.172 (4) | 1.1527 (12) | 0.035 (6)* | |
| C2 | 0.3143 (3) | 0.9827 (3) | 0.34082 (8) | 0.0191 (3) | |
| H2A | 0.2686 | 0.8640 | 0.3084 | 0.023* | |
| H2B | 0.4608 | 0.9929 | 0.3504 | 0.023* | |
| O3 | 0.2486 (2) | 1.3660 (2) | 0.20293 (7) | 0.0239 (3) | |
| C3 | 0.2306 (3) | 0.9668 (3) | 0.40681 (8) | 0.0191 (3) | |
| H3A | 0.2699 | 1.0891 | 0.4379 | 0.023* | |
| H3B | 0.0842 | 0.9503 | 0.3964 | 0.023* | |
| H3 | 0.343 (5) | 1.442 (5) | 0.2068 (17) | 0.070 (11)* | |
| O4 | 0.24107 (19) | 0.08343 (18) | 0.72796 (6) | 0.0188 (2) | |
| C4 | 0.3028 (3) | 0.7975 (3) | 0.44244 (8) | 0.0195 (3) | |
| H4A | 0.4490 | 0.8157 | 0.4542 | 0.023* | |
| H4B | 0.2664 | 0.6752 | 0.4112 | 0.023* | |
| H4 | 0.330 (5) | 0.006 (5) | 0.7471 (16) | 0.066 (9)* | |
| O5 | 0.1083 (2) | 0.27587 (19) | 0.82576 (6) | 0.0216 (3) | |
| C5 | 0.2125 (3) | 0.7822 (3) | 0.50710 (9) | 0.0195 (3) | |
| H5A | 0.2483 | 0.9055 | 0.5379 | 0.023* | |
| H5B | 0.0664 | 0.7648 | 0.4950 | 0.023* | |
| H5 | 0.017 (5) | 0.174 (5) | 0.8191 (16) | 0.064 (9)* | |
| O6 | 0.45679 (19) | 0.37295 (19) | 0.80557 (6) | 0.0220 (3) | |
| C6 | 0.2806 (3) | 0.6151 (3) | 0.54491 (8) | 0.0188 (3) | |
| H6A | 0.4264 | 0.6325 | 0.5579 | 0.023* | |
| H6B | 0.2450 | 0.4910 | 0.5146 | 0.023* | |
| C7 | 0.1838 (3) | 0.6074 (3) | 0.60867 (8) | 0.0179 (3) | |
| H7A | 0.0383 | 0.5858 | 0.5951 | 0.022* | |
| H7B | 0.2146 | 0.7341 | 0.6377 | 0.022* | |
| C8 | 0.2526 (3) | 0.4476 (3) | 0.65035 (8) | 0.0180 (3) | |
| H8A | 0.3972 | 0.4716 | 0.6661 | 0.022* | |
| H8B | 0.2261 | 0.3205 | 0.6214 | 0.022* | |
| C9 | 0.1432 (3) | 0.4431 (3) | 0.71211 (8) | 0.0175 (3) | |
| H9A | 0.0022 | 0.3966 | 0.6959 | 0.021* | |
| H9B | 0.1488 | 0.5762 | 0.7360 | 0.021* | |
| C10 | 0.7973 (3) | 0.2769 (3) | 0.95325 (10) | 0.0283 (4) | |
| H10 | 0.8919 | 0.2928 | 0.9247 | 0.034* | |
| C11 | 0.8604 (3) | 0.2470 (3) | 1.01867 (9) | 0.0272 (4) | |
| H11 | 0.9972 | 0.2420 | 1.0341 | 0.033* | |
| C12 | 0.7189 (3) | 0.2236 (3) | 1.06310 (9) | 0.0199 (3) | |
| C13 | 0.5127 (3) | 0.2384 (3) | 1.03659 (9) | 0.0214 (4) | |
| H13 | 0.4144 | 0.2288 | 1.0642 | 0.026* | |
| C14 | 0.4601 (3) | 0.2665 (3) | 0.97075 (10) | 0.0242 (4) | |
| H14 | 0.3249 | 0.2741 | 0.9537 | 0.029* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| P1 | 0.01293 (19) | 0.0198 (2) | 0.0158 (2) | 0.00022 (16) | 0.00233 (15) | 0.00733 (16) |
| O1 | 0.0224 (6) | 0.0286 (7) | 0.0167 (6) | −0.0056 (5) | 0.0023 (5) | 0.0042 (5) |
| N1 | 0.0360 (9) | 0.0236 (8) | 0.0159 (7) | 0.0009 (7) | −0.0008 (6) | 0.0036 (6) |
| C1 | 0.0179 (8) | 0.0202 (8) | 0.0161 (8) | −0.0003 (6) | 0.0038 (6) | 0.0048 (6) |
| P2 | 0.0175 (2) | 0.0167 (2) | 0.01360 (19) | −0.00040 (16) | 0.00111 (15) | 0.00535 (15) |
| O2 | 0.0163 (6) | 0.0239 (6) | 0.0228 (6) | 0.0042 (5) | 0.0057 (5) | 0.0111 (5) |
| N2 | 0.0187 (8) | 0.0392 (10) | 0.0194 (7) | 0.0036 (7) | 0.0035 (6) | 0.0097 (7) |
| C2 | 0.0185 (8) | 0.0237 (9) | 0.0162 (8) | 0.0015 (7) | 0.0030 (6) | 0.0072 (6) |
| O3 | 0.0158 (6) | 0.0267 (7) | 0.0328 (7) | 0.0026 (5) | 0.0047 (5) | 0.0162 (6) |
| C3 | 0.0195 (8) | 0.0229 (9) | 0.0163 (8) | 0.0003 (7) | 0.0047 (6) | 0.0066 (6) |
| O4 | 0.0190 (6) | 0.0194 (6) | 0.0178 (6) | 0.0029 (5) | 0.0014 (5) | 0.0036 (5) |
| C4 | 0.0201 (8) | 0.0239 (9) | 0.0161 (8) | 0.0009 (7) | 0.0038 (6) | 0.0079 (6) |
| O5 | 0.0280 (7) | 0.0206 (6) | 0.0170 (6) | −0.0021 (5) | 0.0070 (5) | 0.0041 (5) |
| C5 | 0.0209 (8) | 0.0217 (8) | 0.0169 (8) | 0.0002 (7) | 0.0039 (6) | 0.0063 (6) |
| O6 | 0.0216 (6) | 0.0228 (6) | 0.0202 (6) | −0.0045 (5) | −0.0032 (5) | 0.0091 (5) |
| C6 | 0.0193 (8) | 0.0221 (8) | 0.0161 (8) | 0.0008 (7) | 0.0041 (6) | 0.0056 (6) |
| C7 | 0.0195 (8) | 0.0199 (8) | 0.0153 (7) | 0.0014 (6) | 0.0029 (6) | 0.0060 (6) |
| C8 | 0.0191 (8) | 0.0200 (8) | 0.0157 (7) | 0.0019 (6) | 0.0027 (6) | 0.0058 (6) |
| C9 | 0.0178 (8) | 0.0198 (8) | 0.0157 (7) | 0.0020 (6) | 0.0027 (6) | 0.0054 (6) |
| C10 | 0.0305 (10) | 0.0338 (11) | 0.0206 (9) | 0.0008 (8) | 0.0077 (7) | 0.0020 (8) |
| C11 | 0.0206 (9) | 0.0390 (11) | 0.0215 (9) | 0.0018 (8) | 0.0038 (7) | 0.0032 (8) |
| C12 | 0.0212 (8) | 0.0192 (8) | 0.0184 (8) | 0.0008 (7) | 0.0025 (6) | 0.0014 (6) |
| C13 | 0.0206 (8) | 0.0207 (8) | 0.0232 (9) | 0.0010 (7) | 0.0039 (7) | 0.0047 (7) |
| C14 | 0.0249 (9) | 0.0206 (9) | 0.0254 (9) | 0.0022 (7) | −0.0020 (7) | 0.0046 (7) |
Geometric parameters (Å, º)
| P1—O1 | 1.5088 (13) | C4—H4A | 0.9700 |
| P1—O2 | 1.5149 (12) | C4—H4B | 0.9700 |
| P1—O3 | 1.5786 (14) | O5—H5 | 0.88 (3) |
| P1—C1 | 1.7974 (17) | C5—C6 | 1.526 (2) |
| N1—C10 | 1.350 (3) | C5—H5A | 0.9700 |
| N1—C14 | 1.352 (3) | C5—H5B | 0.9700 |
| N1—H1 | 0.96 (2) | C6—C7 | 1.527 (2) |
| C1—C2 | 1.534 (2) | C6—H6A | 0.9700 |
| C1—H1A | 0.9700 | C6—H6B | 0.9700 |
| C1—H1B | 0.9700 | C7—C8 | 1.530 (2) |
| P2—O6 | 1.5045 (13) | C7—H7A | 0.9700 |
| P2—O4 | 1.5535 (13) | C7—H7B | 0.9700 |
| P2—O5 | 1.5601 (13) | C8—C9 | 1.537 (2) |
| P2—C9 | 1.7880 (17) | C8—H8A | 0.9700 |
| N2—C12 | 1.324 (2) | C8—H8B | 0.9700 |
| N2—H21 | 0.88 (3) | C9—H9A | 0.9700 |
| N2—H22 | 0.90 (3) | C9—H9B | 0.9700 |
| C2—C3 | 1.530 (2) | C10—C11 | 1.365 (3) |
| C2—H2A | 0.9700 | C10—H10 | 0.9300 |
| C2—H2B | 0.9700 | C11—C12 | 1.415 (3) |
| O3—H3 | 0.78 (3) | C11—H11 | 0.9300 |
| C3—C4 | 1.523 (2) | C12—C13 | 1.425 (2) |
| C3—H3A | 0.9700 | C13—C14 | 1.359 (3) |
| C3—H3B | 0.9700 | C13—H13 | 0.9300 |
| O4—H4 | 0.91 (3) | C14—H14 | 0.9300 |
| C4—C5 | 1.527 (2) | ||
| O1—P1—O2 | 116.41 (8) | C6—C5—C4 | 114.77 (15) |
| O1—P1—O3 | 105.96 (8) | C6—C5—H5A | 108.6 |
| O2—P1—O3 | 108.76 (7) | C4—C5—H5A | 108.6 |
| O1—P1—C1 | 109.09 (8) | C6—C5—H5B | 108.6 |
| O2—P1—C1 | 109.62 (7) | C4—C5—H5B | 108.6 |
| O3—P1—C1 | 106.51 (8) | H5A—C5—H5B | 107.6 |
| C10—N1—C14 | 120.49 (16) | C5—C6—C7 | 112.06 (14) |
| C10—N1—H1 | 121.8 (15) | C5—C6—H6A | 109.2 |
| C14—N1—H1 | 117.6 (15) | C7—C6—H6A | 109.2 |
| C2—C1—P1 | 113.66 (12) | C5—C6—H6B | 109.2 |
| C2—C1—H1A | 108.8 | C7—C6—H6B | 109.2 |
| P1—C1—H1A | 108.8 | H6A—C6—H6B | 107.9 |
| C2—C1—H1B | 108.8 | C6—C7—C8 | 114.51 (14) |
| P1—C1—H1B | 108.8 | C6—C7—H7A | 108.6 |
| H1A—C1—H1B | 107.7 | C8—C7—H7A | 108.6 |
| O6—P2—O4 | 113.40 (7) | C6—C7—H7B | 108.6 |
| O6—P2—O5 | 109.15 (7) | C8—C7—H7B | 108.6 |
| O4—P2—O5 | 108.70 (7) | H7A—C7—H7B | 107.6 |
| O6—P2—C9 | 111.12 (8) | C7—C8—C9 | 111.67 (14) |
| O4—P2—C9 | 105.43 (8) | C7—C8—H8A | 109.3 |
| O5—P2—C9 | 108.89 (8) | C9—C8—H8A | 109.3 |
| C12—N2—H21 | 119.9 (18) | C7—C8—H8B | 109.3 |
| C12—N2—H22 | 119.6 (16) | C9—C8—H8B | 109.3 |
| H21—N2—H22 | 120 (2) | H8A—C8—H8B | 107.9 |
| C3—C2—C1 | 112.06 (14) | C8—C9—P2 | 113.70 (12) |
| C3—C2—H2A | 109.2 | C8—C9—H9A | 108.8 |
| C1—C2—H2A | 109.2 | P2—C9—H9A | 108.8 |
| C3—C2—H2B | 109.2 | C8—C9—H9B | 108.8 |
| C1—C2—H2B | 109.2 | P2—C9—H9B | 108.8 |
| H2A—C2—H2B | 107.9 | H9A—C9—H9B | 107.7 |
| P1—O3—H3 | 115 (2) | N1—C10—C11 | 120.95 (18) |
| C4—C3—C2 | 113.89 (15) | N1—C10—H10 | 119.5 |
| C4—C3—H3A | 108.8 | C11—C10—H10 | 119.5 |
| C2—C3—H3A | 108.8 | C10—C11—C12 | 120.38 (18) |
| C4—C3—H3B | 108.8 | C10—C11—H11 | 119.8 |
| C2—C3—H3B | 108.8 | C12—C11—H11 | 119.8 |
| H3A—C3—H3B | 107.7 | N2—C12—C11 | 121.92 (17) |
| P2—O4—H4 | 113 (2) | N2—C12—C13 | 121.32 (17) |
| C3—C4—C5 | 112.68 (15) | C11—C12—C13 | 116.75 (16) |
| C3—C4—H4A | 109.1 | C14—C13—C12 | 119.77 (17) |
| C5—C4—H4A | 109.1 | C14—C13—H13 | 120.1 |
| C3—C4—H4B | 109.1 | C12—C13—H13 | 120.1 |
| C5—C4—H4B | 109.1 | N1—C14—C13 | 121.60 (18) |
| H4A—C4—H4B | 107.8 | N1—C14—H14 | 119.2 |
| P2—O5—H5 | 117 (2) | C13—C14—H14 | 119.2 |
| O1—P1—C1—C2 | 74.30 (14) | O6—P2—C9—C8 | 66.99 (14) |
| O2—P1—C1—C2 | −54.24 (14) | O4—P2—C9—C8 | −56.25 (14) |
| O3—P1—C1—C2 | −171.74 (12) | O5—P2—C9—C8 | −172.75 (12) |
| P1—C1—C2—C3 | −167.86 (12) | C14—N1—C10—C11 | 1.9 (3) |
| C1—C2—C3—C4 | −176.88 (14) | N1—C10—C11—C12 | −0.4 (3) |
| C2—C3—C4—C5 | −178.52 (15) | C10—C11—C12—N2 | 176.98 (19) |
| C3—C4—C5—C6 | −179.83 (15) | C10—C11—C12—C13 | −1.7 (3) |
| C4—C5—C6—C7 | −179.62 (14) | N2—C12—C13—C14 | −176.38 (18) |
| C5—C6—C7—C8 | −177.72 (15) | C11—C12—C13—C14 | 2.3 (3) |
| C6—C7—C8—C9 | −177.78 (14) | C10—N1—C14—C13 | −1.3 (3) |
| C7—C8—C9—P2 | −169.70 (12) | C12—C13—C14—N1 | −0.9 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O6i | 0.78 (3) | 1.85 (3) | 2.6171 (18) | 166 (3) |
| O5—H5···O1ii | 0.88 (3) | 1.64 (3) | 2.5059 (18) | 168 (3) |
| O4—H4···O2iii | 0.91 (3) | 1.59 (3) | 2.4977 (17) | 178 (3) |
| N1—H1···O6 | 0.96 (2) | 1.74 (3) | 2.696 (2) | 173 (2) |
| N2—H22···O2iv | 0.90 (3) | 1.92 (3) | 2.806 (2) | 170 (2) |
| N2—H21···O1v | 0.88 (3) | 2.14 (3) | 2.965 (2) | 156 (3) |
Symmetry codes: (i) −x+1, −y+2, −z+1; (ii) −x, −y+1, −z+1; (iii) −x+1, −y+1, −z+1; (iv) x, y−1, z+1; (v) x+1, y−1, z+1.
References
- Aakeröy, C. B. & Seddon, K. R. (1993). Chem. Soc. Rev. 22, 397–407.
- Bolliger, J. L., Oberholzer, M. & Frech, C. M. (2011). Adv. Synth. Catal. 353, 945–954.
- Brandenburg, K. (2015). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Deng, Y., Helms, B. A. & Rolandi, M. (2015). J. Polym. Sci. Part A Polym. Chem. 53, 211–214.
- Etter, M. C., MacDonald, J. C. & Bernstein, J. (1990). Acta Cryst. B46, 256–262. [DOI] [PubMed]
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998). Acta Cryst. B54, 129–138.
- Fu, R.-B., Wu, X.-T., Hu, S.-M., Du, W.-X. & Zhang, J.-J. (2004). Chin. J. Struct. Chem. 23, 855–861.
- Fuller, J. & Heimer, N. E. (1995). J. Chem. Crystallogr. 25, 129–136.
- Glidewell, C., Ferguson, G. & Lough, A. J. (2000). Acta Cryst. C56, 855–858. [DOI] [PubMed]
- Gomathi, S. & Muthiah, P. T. (2011). Acta Cryst. E67, o2762. [DOI] [PMC free article] [PubMed]
- Griffith, J. A., McCauley, D. J., Barrans, R. E. & Herlinger, A. W. (1998). Synth. Commun. 28, 4317–4323.
- Grimme, S. (2008). Angew. Chem. Int. Ed. 47, 3430–3434. [DOI] [PubMed]
- Janiak, C. (2000). J. Chem. Soc. Dalton Trans. pp. 3885–3896.
- Kan, W.-Q. & Ma, J.-F. (2011). Z. Kristallogr. New Cryst. Struct. 226, 73–74.
- Koleva, B. B., Kolev, T., Seidel, R. W., Tsanev, T., Mayer-Frigge, H., Spiteller, M. & Sheldrick, W. S. (2008). Spectrochim. Acta part A, 71, 695–702. [DOI] [PubMed]
- Mahmoudkhani, A. H. & Langer, V. (2002a). Cryst. Growth Des. 2, 21–25.
- Mahmoudkhani, A. H. & Langer, V. (2002b). J. Mol. Struct. 609, 97–108.
- Mahmoudkhani, A. H. & Langer, V. (2002c). Phosphorus, Sulfur Silicon Relat. Elem. 177, 2941–2951.
- Maspoch, D., Domingo, N., Roques, N., Wurst, K., Tejada, J., Rovira, C., Ruiz-Molina, D. & Veciana, J. (2007). Chem. Eur. J. 13, 8153–8163. [DOI] [PubMed]
- Megen, M. van, Frank, W. & Reiss, G. J. (2015). Z. Kristallogr. 230, 485–494.
- Megen, M. van, Frank, W. & Reiss, G. J. (2016). CrystEngComm, 18, 3574–3584.
- Moedritzer, K. & Irani, R. (1961). J. Inorg. Nucl. Chem. 22, 297–304.
- Mootz, D. & Poll, W. (1984). Z. Naturforsch. Teil B, 39, 290–297.
- Muñoz-Caro, C. & Niño, A. (2002). Biophys. Chem. 96, 1–14. [DOI] [PubMed]
- Plabst, M., Stock, N. & Bein, T. (2009). Cryst. Growth Des. 9, 5049–5060.
- Reiss, G. J. & Leske, P. B. (2014a). Z. Kristallogr. New Cryst. Struct. 229, 239–240.
- Reiss, G. J. & Leske, P. B. (2014b). Z. Kristallogr. New Cryst. Struct. 229, 452–454.
- Schwarzenbach, G. & Zurc, J. (1950). Monatsh. Chem. 81, 202–212.
- Sertucha, J., Luque, A., Lloret, F. & Román, P. (1998). Polyhedron, 17, 3875–3880.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Steiner, T. (2002). Angew. Chem. Int. Ed. 41, 48–76.
- Stoe & Cie (2002). X-AREA. Stoe & Cie, Darmstadt, Germany.
- Surbella, R. G. III, Andrews, M. B. & Cahill, C. L. (2016). J. Solid State Chem. 236, 257–271.
- Wheatley, P. S., Lough, A. J., Ferguson, G., Burchell, C. J. & Glidewell, C. (2001). Acta Cryst. B57, 95–102. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, publication_text. DOI: 10.1107/S2056989016014298/hb7610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016014298/hb7610Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016014298/hb7610Isup3.cml
CCDC reference: 1503436
Additional supporting information: crystallographic information; 3D view; checkCIF report