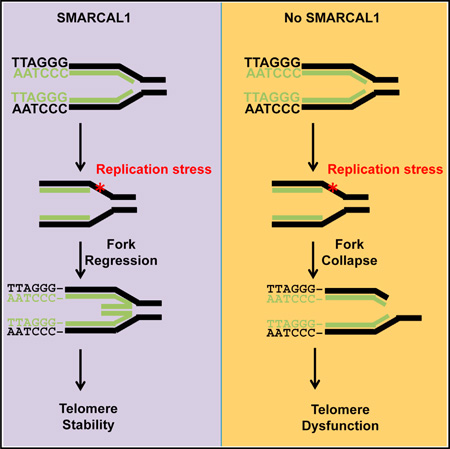
SUMMARY
Cancer cells overcome replicative senescence by exploiting mechanisms of telomere elongation, a process often accomplished by reactivation of the enzyme telomerase. However, a subset of cancer cells lack telomerase activity and rely on the alternative lengthening of telomeres (ALT) pathway, a recombination-based mechanism of telomere elongation. Although the mechanisms regulating ALT are not fully defined, chronic replication stress at telomeres might prime these fragile regions for recombination. Here, we demonstrate that the replication stress response protein SMARCAL1 is a critical regulator of ALT activity. SMARCAL1 associates with ALT telomeres to resolve replication stress and ensure telomere stability. In the absence of SMARCAL1, persistently stalled replication forks at ALT telomeres deteriorate into DNA double-strand breaks promoting the formation of chromosome fusions. Our studies not only define a role for SMARCAL1 in ALT telomere maintenance, but also demonstrate that resolution of replication stress is a crucial step in the ALT mechanism.
Graphical Abstract
INTRODUCTION
Telomeres cap the ends of chromosomes and function as a barrier shielding the human genome from nucleolytic degradation and illegitimate recombination. Telomeres are composed of double-stranded TTAGGG hexameric repeats that are organized into a lariat, or T-loop, at the end of each chromosome (Palm and de Lange, 2008). While this structure is essential to prevent chromosome ends from being recognized as DNA double strand breaks (DSBs), T-loops pose a natural impediment to DNA replication (Fouché et al., 2006; Poulet et al., 2009; Uringa et al., 2012; Vannier et al., 2013; Sarek et al., 2015). In addition, the G-rich sequence drives Hoogsteen base-pairing between guanosine nucleotides generating G-quadruplex structures that pose a threat to the processivity of the replication machinery (Sen and Gilbert, 1988; Sundquist and Klug, 1989). The repetitive nature and structural complexities of the telomeric DNA induce frequent replication fork stalling and chromosome breakage demonstrating that telomeres are common fragile sites within the genome (Verdun and Karlseder, 2006; Sfeir et al., 2009; Martínez et al., 2009). Therefore, understanding how cells have evolved to navigate the replication stress associated with telomeric DNA is essential to our understanding of genome stability.
Common fragile sites are associated with an increased rate of recombination suggesting that telomere fragility promotes recombination at telomere ends (Glover and Stein, 1987; Schwartz et al., 2005). The link between telomere fragility and recombination is most evident in cancer cells that rely on the alternative lengthening of telomeres (ALT) pathway. The ALT pathway promotes telomere elongation using homology-directed recombination between telomeric DNA sequences (Bryan et al., 1995; Dunham et al., 2000; Londoño-Vallejo et al., 2004). Although in cancer cells telomere elongation is often achieved by reactivation of the enzyme telomerase, subsets of cancer cells activate the ALT pathway for telomere maintenance (Henson and Reddel, 2010). The exact mechanisms driving activation and maintenance of the ALT pathway are not fully defined, however, telomere deprotection and changes in chromatin dynamics may represent early events in the ALT process. In ALT cells, the telomere sequence has evolved such that in addition to the canonical TTAGGG sequence these telomeres have an increase in variant hexameric repeats including TCAGGG, TTCGGG, and GTAGGG (Conomos et al., 2012; Varley et al., 2002). Variant repeats drive telomere deprotection by disrupting binding of the telomere repeat factors TRF1 and TRF2; components of the telomere capping complex, Shelterin. Loss of TRF1 increases replication fork stalling and enhances telomere fragility (Sfeir et al., 2009; Martínez et al., 2009). This fragility is exacerbated by defects in nucleosome assembly at telomeres as genetic mutations in the chromatin remodeling complex α-thalassemia/mental retardation syndrome X-linked/death-domain-associated protein (ATRX/DAXX) and histone variant H3.3 are highly correlated with ALT positive cancers (Heaphy et al., 2011; Schwartzentruber et al., 2012). In addition, loss of the histone chaperone ASF1 (anti-silencing factor 1) in mammalian cells, which promotes histone transfer during replication, leads to the induction of ALT-like phenotypes (O’Sullivan et al., 2014). Therefore, limited telomere end protection and altered chromatin dynamics stress an already fragile repetitive region during replication, enhancing replication stress and consequently promoting telomere instability. Together, these events may prime telomeres for recombination and as a result promote ALT activity.
The telomeric DNA in ALT cells is incredibly dynamic constantly undergoing rapid attrition and elongation providing one of the first indications that telomeres in ALT cells rely on recombination to promote telomere elongation (Bryan et al., 1995). This was later supported by studies demonstrating that the telomeres in ALT cells are recruited into nuclear foci that, in addition to the promyelocytic leukemia (PML) protein, contain a number of recombination and repair factors including RPA, RAD51, RAD52, BRCA1, MRE11, RAD50, and NBS1 (Yeager et al., 1999; Wu et al., 2000, 2003; Grudic et al., 2007). The formation of the ALT-associated PML bodies (or APB) led to early speculations that APB functioned as platforms for recombination. More recently, it was demonstrated that inhibition of replication, or the sensing of replication stress, could disrupt APB formation (O’Sullivan et al., 2014; Flynn et al., 2015). These findings raise the possibility that as common fragile sites, replication fork stalling at the telomere drives APB formation to either promote fork restart or salvage collapsed forks through recombination. Given the repetitive nature of telomeres, recombination can occur between nonhomologous chromosomes, sister chromatids, or extrachromosomal telomeric repeat (ECTR) DNA. ECTR DNA can exist in both linear and circular forms composed of double stranded or partially single-stranded, C-rich or G-rich telomeric sequences (Tokutake et al., 1998; Nabetani and Ishikawa, 2009). Partially single-stranded C-rich circular ECTR, or C-circles are unique to ALT cells and have been demonstrated to directly correlate with ALT activity (Lau et al., 2013; Henson et al., 2009). ECTR are likely generated as a by product of recombination, however, they may also perpetuate the ALT phenotype by functioning as a template for recombination (Henson and Reddel, 2010). Similar to the formation of APB, C-circle formation is significantly reduced following inhibition of replication, or the sensing of replication stress (O’Sullivan et al., 2014; Flynn et al., 2015). Taken together, these cellular phenotypes highlight the contribution of replication stress to telomere recombination, and ultimately, provocation of the ALT pathway.
Given the fragility of telomeric DNA, understanding how the telomere responds to replication stress will undoubtedly further our understanding of both telomere maintenance and the progression toward cancer. Recently, the annealing helicase SMARCAL1 was identified as one of the most abundant proteins bound to persistently stalled, or collapsed, replication forks (Sirbu et al., 2013; Dungrawala et al., 2015). SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like 1), also known as the HepA-related protein (HARP), is an ATP-dependent DNA annealing helicase that remodels chromatin surrounding stalled replication forks to promote replication restart (Yusufzai and Kadonaga, 2008; Bansbach et al., 2009; Bétous et al., 2012; Yuan et al., 2009). Therefore, we speculated that SMARCAL1 might function to remodel stalled replication forks at telomeric DNA and bolster replication through a particularly fragile region. Here, we demonstrate that SMARCAL1 is enriched at telomeric DNA in cells that rely on the ALT pathway for telomere maintenance indicating that ALT telomeres are prone to chronic replication stress. In the absence of SMARCAL1, persistently stalled replication forks at ALT telomeres form DNA double-strand breaks, induce RAD51-dependent telomere clustering, promote chromosomal fusions, and drive genome instability. Our studies demonstrate a function for SMARCAL1 in the resolution of replication stress at telomeric DNA and also define SMARCAL1 as a critical regulator of the ALT pathway. Defining the molecular mechanisms regulating maintenance of the ALT pathway is critical to both our understanding of telomere biology and also the progression toward cancer.
RESULTS AND DISCUSSION
SMARCAL1 Is Significantly Enriched at ALT Telomeres
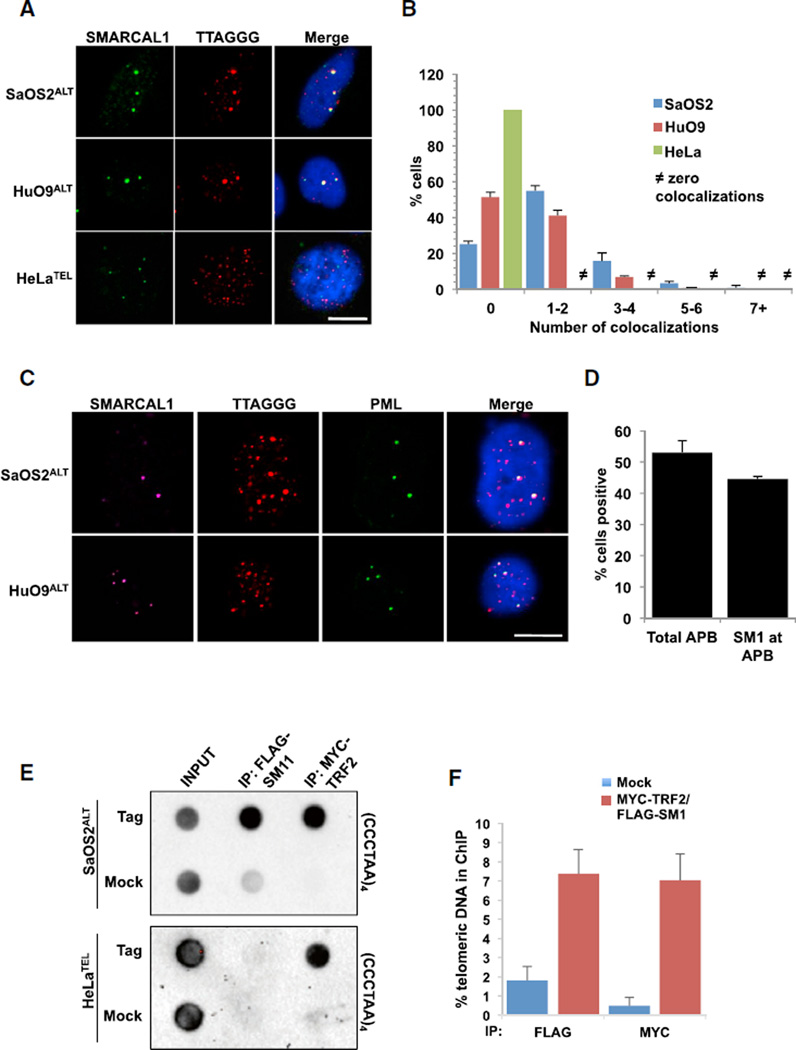
SMARCAL1 was identified as a chromatin remodeling enzyme that functions to restart stalled replication forks by catalyzing branch migration and fork regression (Bétous et al., 2012; Bansbach et al., 2009; Yuan et al., 2009). Given the prevalence of replication stress at telomeres, we asked whether SMARCAL1 functioned to alleviate replication stress and promote telomere maintenance. To do this, we initially asked whether SMARCAL1 associated with telomeric DNA in unperturbed mammalian cells. Using a combined immunofluorescence and in situ hybridization (IF-FISH) approach, we detected SMARCAL1 protein at telomeres in ALT positive SaOS2ALT and HuO9ALT cells (Figures 1A and 1B), however, we did not detect SMARCAL1 at telomeres in telomerase-positive HeLaTEL or untransformed RPEUNT cells (Figures 1A, 1B, and S1A). The association of SMARCAL1 with telomeres in ALT positive cells was robust with ~60% of cells demonstrating at least one SMARCAL1-telomere colocalization event (Figure 1B). Remarkably, this association of SMARCAL1 with telomeres was restricted exclusively to APB as we were unable to detect SMARCAL1 at telomeres that had not been recruited to PML bodies (Figures 1C and 1D). The association of SMARCAL1 with ALT telomeres was not simply attributed to highly repetitive regions as we could not detect SMARCAL1 at centromeric DNA by IF-FISH (Figure S1B). To further validate the specificity of SMARCAL1 binding at telomeric DNA, we performed chromatin immunoprecipitation assays. Similar to the results from our IF-FISH experiments, SMARCAL1 was significantly enriched at the telomeric DNA in SaOS2ALT cells but not in HeLaTEL cells (Figures 1E and 1F). Moreover, the binding of SMARCAL1 at ALT telomeres was significantly enriched over SMARCAL1 binding to the Alu repeats further highlighting the specificity of SMARCAL1 binding at ALT telomeres (Figures S1C and S1D).
Figure 1. SMARCAL1 Accumulates at ALT Telomeres.
(A) Combined immunofluorescence and DNA fluorescence in situ hybridization (IF-FISH) analyses of SMARCAL1 and telomeres in SaOS2, HuO9, and HeLa cells. Representative images are shown. Scale bar, 10 µm.
(B) The percentage of cells positive for 0, 1–2, 3–4, 5–6, and 7 or greater incidences of SMARCAL1 colocalizing with telomeres in SaOS2, HuO9, and HeLa cells are graphed as the mean of three independent experiments (n = 3). Scale bar, 10 µm.
(C) IF-FISH analyses of SMARCAL1, PML, and telomeres in SaOS2 and HuO9 cells. Scale bar, 10 µm.
(D) Quantification of experiments performed in (C). Percentage of cells positive for APB and SMARCAL1 colocalizing with APB are graphed as the mean ± SD (n = 2).
(E) ChIP for telomeric DNA associated with SMARCAL1 and TRF2 in SaOS2 and HeLa cells co-expressing FLAG-SMARCAL1 and MYCTRF2.
(F) Quantification of dot blots performed in (E). Graph represents average percent of telomeric DNA recovered in two independent experiments ± SD. See also Figure S1.
To further understand the association of SMARCAL1 with ALT telomeres, we asked whether we could promote SMARCAL1 relocalization to non-ALT telomeres by inducing replication stress. To do this, we took advantage of a HeLaTEL cell line derivative that maintains long telomeres, HeLaTEL1.2.11 and consequently, is vulnerable to replication stress. In fact, even in undamaged HeLaTEL1.2.11 cells, we could detect rare instances of colocalization between SMARCAL1 and telomeric DNA by IF-FISH. To determine whether these infrequent colocalization events are truly representative of sites of replication stress, we asked whether we could increase the frequency of these events by inducing replication stress specifically at telomere ends. Considering the role of TRF1 in telomere replication, we asked whether loss of TRF1 would induce frequent fork stalling at telomeres and promote the accumulation of SMARCAL1 at telomeric DNA in HeLaTEL1.2.11 cells. Therefore, we depleted TRF1 from HeLaTEL1.2.11 cells and either left them untreated or exacerbated replication stress with aphidicolin and analyzed the association of SMARCAL1 with telomeric DNA by IF-FISH. In the absence of TRF1, cells treated with aphidicolin demonstrated an increase in the phosphorylated form of RPA pS4/S8 at telomere ends, and these foci colocalized with SMARCAL1 suggesting the formation of irreversibly stalled replication forks (Figures S1E–S1G) (Niu et al., 1997; Vassin et al., 2004; Maréchal and Zou, 2015). These results demonstrate that SMARCAL1 does in fact associate with non-ALT telomeres, but this association is highly dependent on replication stress (Poole et al., 2015). Moreover, the abundance of SMARCAL1 at telomeres in ALT cells in the absence of exogenous replication stress suggests that ALT telomeres undergo chronic replication stress highlighting an unexplored function for SMARCAL1 in maintenance of the ALT pathway.
Loss of SMARCAL1 Promotes Telomere Clustering in ALT
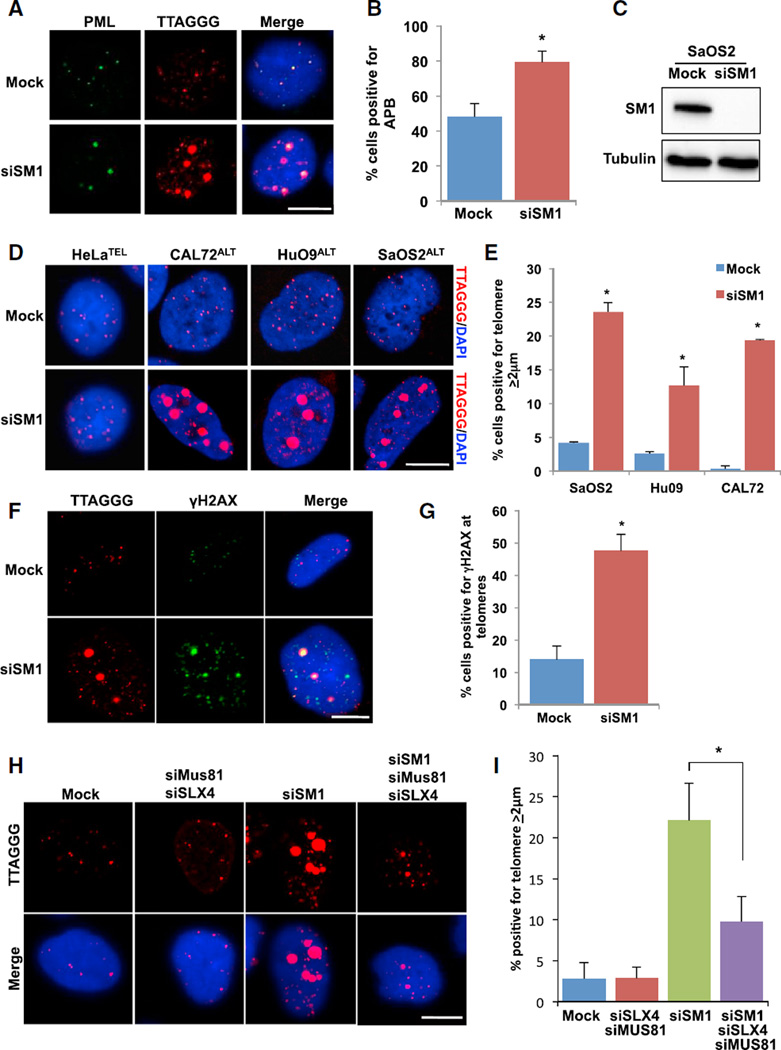
The association of SMARCAL1 with ALT telomeres led us to hypothesize that SMARCAL1 localizes to telomeres in ALT to resolve persistently stalled replication forks and promote telomere stability. Thus, we predicted that loss of SMARCAL1 in ALT would lead to defects in replication fork restart, promote the accumulation of collapsed replication forks, and consequently increase APB formation. Consistent with this reasoning, loss of SMARCAL1 led to a significant increase in the percentage of cells positive for APB (Figures 2A–2C) highlighting the contribution of replication stress to APB formation. SMARCAL1 loss did not lead to changes in the distribution of cells throughout the cell cycle, ruling out the possibility that the accumulation of APB was a result of cell-cycle arrest (Figure S2A). In addition to the increase in APB, we observed a ~17-fold increase in mean telomere foci size in a subset of cells depleted for SMARCAL1, as compared to the mean telomere foci size in control cells (Figure S2B). The percentage of cells with large telomere foci increased by ~4-fold following SMARCAL1 depletion in the ALT positive cells SaOS2ALT, CAL72ALT, and HuO9ALT (Figures 2D and S2C). In contrast, these large telomeric foci were entirely absent in HeLaTEL cells further supporting a unique role for SMARCAL1 at ALT telomeres (Figure 2D). Furthermore, we also observed an increase in the large telomere foci size using an alternative small interfering RNA (siRNA) targeting the 3′ UTR of SMARCAL1. Importantly, we could rescue the large telomere phenotype in these cells with exogenously expressed Flag-SMARCAL1 (Figures S2D and S2E).
Figure 2. SMARCAL1 Inhibits the Formation of Double-Stranded DNA Breaks at Telomeres.
(A) SaOS2 cells were mock-treated or treated with SMARCAL1 siRNA for 72 hr, and PML and telomere foci were analyzed by IF-FISH. Scale bar, 10 µm.
(B) The percentage of cells positive for APB (colocalization of PML and telomere) was graphed as the mean ± SD (n = 3). *p < 0.005.
(C) Western blot of SMARCAL1 in SaOS2 cells either mock-treated or treated with SMARCAL1 siRNA for 72 hr. Tubulin is used as a loading control.
(D) Representative images of DNA FISH analyses of telomere foci in HeLa, CAL72, HuO9, and SaOS2 cells either mock-treated or treated with SMARCAL1 siRNA for 72 hr. Scale bar, 10 µm.
(E) Quantification of experiments performed in (D). Graph represents percentage of cells positive for telomere ≥2 µm was graphed as the mean ± SD (n = 3 SaOS2, n = 2 HuO9, and CAL72). *p < 0.0001 (SaOs2), *p < 0.05 (HuO9), *p < 0.001 (CAL72).
(F) SaOS2 cells were mock-treated or treated with SMARCAL1 siRNA for 72 hr, and γH2AX and telomere foci were analyzed by IF-FISH. Scale bar, 10 µm.
(G) Quantification of experiments performed in (F). Graph represents percentage of cells positive for colocalization of γH2AX and telomere foci was graphed as the mean ± SD (n = 3). *p < 0.005.
(H) SaOS2 cells were either mock-treated, co-treated with Mus81 siRNA and SLX4 siRNA, treated with SMARCAL1 siRNA alone, or treated with SMARCAL1 siRNA, Mus81 siRNA, and SLX4 siRNA for 72 hr. Telomere foci were analyzed by DNA FISH. Scale bar, 10 µm.
(I) Quantification of experiments performed in (H). Graph represents average percentage of cells positive for telomere foci ≥2 µm ± SD (n = 5 Mock, n = 2 siSLX4/siMus81, n = 5 siSM1, n = 5 siSM1/siSLX4/siMus81). *p < 0.05. See also Figure S2.
Large telomere foci size has been linked to the aggregation of telomeric DNA following the formation of DNA DSBs (Cho et al., 2014). Therefore, we considered the possibility that the larger telomeric foci following SMARCAL1 knockdown may reflect sites of persistent stalled replication forks that breakdown into DNA DSBs. The percentage of cells demonstrating the large telomere phenotype steadily accumulated over time, reaching a maximum of ~60% cells containing larger telomeric foci after 5 days (Figures S2F and S2G). These large foci were specific to telomeric DNA as we did not observe an increase in centromeric DNA foci size in the absence of SMARCAL1 (Figure S2H). Finally, the large telomeric foci demonstrated a significant increase in colocalization with γH2AX reinforcing the speculation that these telomeres represent sites of DSBs (Figures 2F and 2G).
Stalled replication forks represent a roadblock to cellular proliferation. Therefore, persistently stalled forks are subject to cleavage by the SLX-MUS endonuclease complex (Fekairi et al., 2009; Hanada et al., 2007; Wyatt et al., 2013). Once cleaved, these replication forks are repaired through homologous recombination (Muñoz et al., 2009; Petermann et al., 2010). The SLX4 and MUS81 nuclease have been shown to localize to APB in ALT cells and regulate ALT activity (Zeng et al., 2009; Sarkar et al., 2015). Therefore, we asked whether the SLX-MUS endonuclease complex was responsible for the generation of the large telomere foci formed in ALT cells following SMARCAL1 knockdown. In fact, in SMARCAL1-deficient ALT cells also containing siRNA for MUS81 and SLX4, we observed a significant decrease in the percentage of cells containing the large telomeric foci (Figures 2H, 2I, S2I, and S2J). Previous studies have demonstrated that SMARCAL1 prevents the formation of MUS81-dependent DNA DSBs (Bétous et al., 2012). Therefore, our data suggest that loss of SMARCAL1 in ALT cells leads to irreversibly stalled replication forks at telomeric DNA that are recognized and cleaved by the SLX-MUS endonuclease complex driving DSB formation.
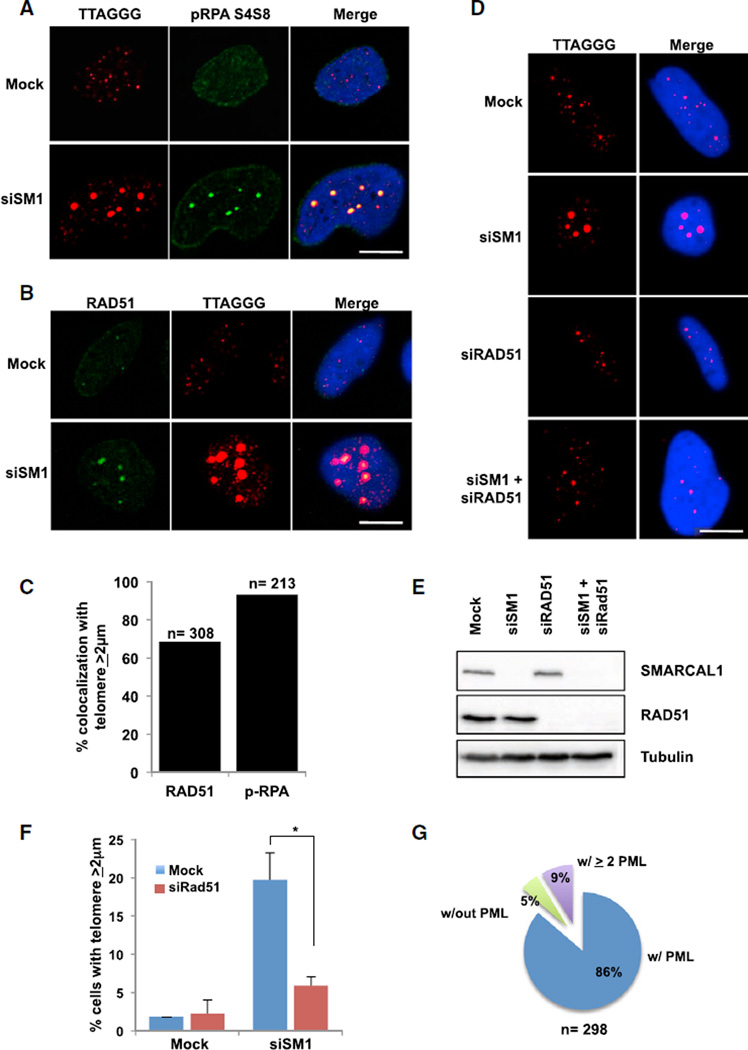
The generation of DSBs at ALT telomeres promotes RAD51-dependent telomeric clustering and consequently, primes telomeres for homology-directed repair (Cho et al., 2014). Thus, we asked whether the DSBs at telomeric DNA in ALT cells following loss of SMARCAL1 also promote telomere clustering. In fact, following SMARCAL1 knockdown, we observed a significant increase in the association of pRPA S4/S8 with large telomere foci suggesting that these telomere ends contain irreversibly stalled replication forks that have collapsed into DSBs (Figures 3A and 3C) (Niu et al., 1997; Vassin et al., 2004; Maréchal and Zou, 2015). Concomitant with RPA-coated telomeric DNA was the accumulation of the recombination protein RAD51 (Figures 3B and 3C). Notably, formation of the large telomere foci was dependent on RAD51 as SMARCAL1-deficient cells treated with RAD51 siRNA showed a significant reduction in large telomere foci (Figures 3D–3F). These foci were largely devoid of 53BP1 although in some instances we could detect 53BP1 on the periphery of the foci highlighting the increase in DNA DSBs and damage signaling at these telomere ends (Figure S3A). Similar to previous reports, these telomere clusters colocalized with PML, and a single large telomere often contained more than a single PML foci reinforcing the model that PML functions to promote telomere-telomere interactions (Figures 3G and S3B) (Cho et al., 2014; Draskovic et al., 2009). Taken together, our data suggest that loss of SMARCAL1 induces DSBs at ALT telomeres and triggers RAD51-dependent telomere clustering.
Figure 3. Loss of SMARCAL1 Promotes RAD51-Dependent Telomere Clustering.
(A) SaOS2 cells were either mock-treated or treated with SMARCAL1 siRNA for 72 hr, pRPA S4/S8 and telomere foci were analyzed by IF-FISH. Scale bar, 10 µm.
(B) SaOS2 cells were either mock-treated or treated with SMARCAL1 siRNA for 72 hr, and RAD51 foci were analyzed by IF-FISH. Scale bar, 10 µm.
(C) Quantification of experiments performed in (A) and (B). Graph represents the average percentage of cells positive for colocalization of RAD51 (left bar) and pRPA S4/S8 (right bar) with telomere foci ≥2 µm.
(D) SaOS2 cells were either mock-treated, treated with SMARCAL1 siRNA alone, RAD51 siRNA alone, or co-treated with SMARCAL1 siRNA and RAD51 siRNA for 72 hr. Telomere foci were analyzed by DNA FISH. Scale bar, 10 µm.
(E) Western blot of SMARCAL1 and RAD51 in SaOS2 cells either mock-treated, treated with SMARCAL1 siRNA alone, RAD51 siRNA alone, or co-treated with SMARCAL1 siRNA and RAD51 siRNA after 72 hr. Tubulin was used as a loading control.
(F) Quantification of experiments performed in (D). Graph represents average percentage of cells with telomere foci ≥2 µm ± SD (n = 3). *p < 0.05.
(G) SaOS2 cells were either mock-treated or treated with SMARCAL1 siRNA for 72 hr, and PML and telomere foci were analyzed by IF-FISH. Graph represents telomere foci ≥ 2 µm colocalized with zero, one, or two or greater PML foci. See also Figure S3.
SMARCAL1 Regulates ALT Activity
To further define the role of SMARCAL1 in ALT telomere maintenance, we asked whether DSBs and subsequent telomere clustering induced by SMARCAL1 loss would lead to significant changes in overall telomere heterogeneity. Therefore, we performed telomere restriction fragment (TRF) analysis on genomic DNA isolated from SMARCAL1-deficient cells. Loss of SMARCAL1 led to an increase in smaller telomeric DNA fragments that migrate below the bulk telomere signal as well as an increase in larger telomeric DNA fragments migrating above the bulk telomere signal (Figure 4A). These findings suggest that loss of SMARCAL1 leads to gross changes in telomere heterogeneity and supports a role for SMARCAL1 in maintaining ALT telomere stability. While the increase in smaller telomeric DNA fragments could be explained by the increase in DNA DSBs following SMARCAL1 knockdown, we speculated that the larger telomeric DNA fragments could represent unresolved recombination intermediates.
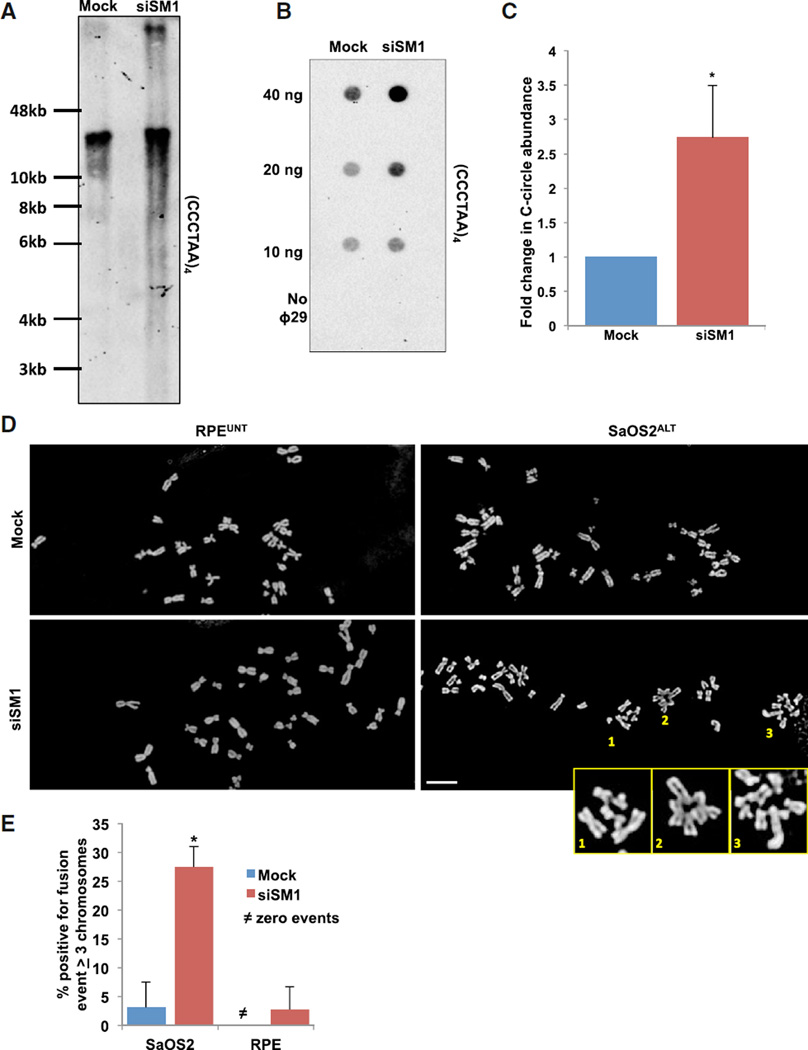
Figure 4. Loss of SMARCAL1 Increases ALT Activity and Induces Structural Chromosome Abnormalities.
(A) SaOS2 cells were either mock-treated or treated with SMARCAL1 siRNA for 12 days before isolation of genomic DNA. The isolated DNA was then digested and telomere restriction fragments were analyzed by Southern blot using telomere-specific probes.
(B) SaOS2 cells were either mock-treated or treated with SMARCAL1 siRNA for 72 hr. C-circle amplification products were loaded onto membranes by dot blot and analyzed by Southern blot using a telomere-specific probe.
(C) The levels of C-circles were graphed as the mean ± SD (n = 3). *p < 0.05.
(D) Representative metaphase spread of mock-treated (top) or SMARCAL1 siRNA-treated (bottom) RPE (left) and SaOS2 (right) cells after 72 hr. Enlarged images represent incidence of chromatid-type fusions. Scale bar, 10 µm.
(E) Quantification of experiments performed in (D). Graph represents percentage of metaphase spreads positive for fusion events containing ≥ 3 chromosomes and was graphed as mean ± SD (n = 2, ≥ 16 spreads per experiment). See also Figure S4.
The sheer abundance of telomeric DNA throughout the genome allows recombination during ALT to occur between homologous and/or nonhomologous chromosomes, chromosome fragments, and/or ECTR DNA. The C-rich circular ECTR DNA species, or C-circles, are unique to ALT-positive cells and have been shown to directly correlate with ALT activity. C-circles are thought to arise as by-products of telomeric recombination and can be readily detected by Southern blot following rolling-circle amplification. If the DSBs formed after loss of SMARCAL1 promote recombination, we predicted that SMARCAL1 knockdown in ALT cells would lead to an increase in C-circle formation. As predicted, following SMARCAL1 knockdown, we demonstrate a 3-fold increase in C-circle abundance in ALT cells (Figures 4B and 4C). However, in stark contrast, SMARCAL1 knockdown did not induce C-circle formation in HeLaTEL cells demonstrating that loss of SMARCAL1 can increase, but cannot induce, ALT activity (Figures S4A–S4C). To determine whether the increase in ALT activity and large telomere DNA fragments are indicative of unresolved recombination intermediates, we analyzed metaphase chromosome spreads from SMARCAL1-deficient SaOS2ALT and RPEUNT cells. Remarkably, in the absence of SMARCAL1 in SaOS2ALT cells we observed metaphase spreads containing chromatid-type fusions (Figures 4D and 4E). However, these structural chromosome abnormalities were absent in RPEUNT cells following SMARCAL1 knockdown (Figures 4D and 4E). Conceivably, in the absence of SMARCAL1, DNA DSBs formed by persistently stalled replication forks at ALT telomeres are forced to undergo recombination leading to an increase in ALT phenotypes. The increase in ALT telomeres primed for recombination overwhelms the repair machinery leading to defects in the resolution of recombination intermediates and emergence of structural chromosome abnormalities.
In conclusion, we have demonstrated that the annealing helicase SMARCAL1 is a critical regulator of replication stress at telomeric DNA. The enrichment of SMARCAL1 at telomeres in ALT cells demonstrates that ALT telomeres experience chronic replication stress and highlight SMARCAL1 as a critical regulator of the ALT pathway. SMARCAL1 associates with telomeric DNA in ALT cells and functions to mitigate replication stress and regulate ALT activity. Together, our data support a model in which SMARCAL1 binds ALT telomeres to resolve replication stress and facilitate telomere elongation. However, in the absence of SMARCAL1 stalled replication forks fail to restart and consequently, become substrates for cleavage by the SLX-MUS endonuclease complex. The formation of telomeric DNA DSBs drives telomere clustering and facilitates recombination to salvage collapsed replication forks and maintain telomere stability. Nevertheless, the accumulation of telomeric DNA DSBs following SMARCAL1 depletion saturates the capabilities of the repair machinery leading to the formation of unresolved recombination intermediates and genome instability. The enrichment of SMARCAL1 at ALT telomeres highlights both the prevalence of replication stress at ALT telomeres and also how resolution of this replication stress is critical for the maintenance of ALT activity. The ALT pathway is active in ~10% of all cancers; thus, further defining the mechanisms regulating ALT activity could provide an opportunity for targeted therapeutic development.
EXPERIMENTAL PROCEDURES
Cell Culture
SaOS2 cells were grown in McCoy’s 5A, 15% FBS, and 1% penicillin/streptomycin. HeLa and HeLa 1.2.11 cells were cultured in DMEM, 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. RPE and Cal72 cells were grown in DMEM F12, 10% FBS, 1% and penicillin/streptomycin. SJSA1 and HuO9 cells were grown in RPMI 1640, 5% FBS, 1% sodium pyruvate, and 1% penicillin/streptomycin.
siRNAs, Probes, Antibodies, and Plasmids
All siRNA transfections were performed using Invitrogen RNAi MAX according to manufacturer’s instructions. Plasmids were transfected using Fugene transfection reagent. Additional information described in the Supplemental Experimental Procedures.
Combined Immunofluorescence FISH
Cells were rinsed with PBS, treated with cytobuffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [pH 7], 0.1% Triton X-100), and fixed in 4% paraformaldehyde. Cells were then permeabilized in 0.5% NP40/PBS and blocked in PBG (0.5% BSA, 0.2% fish gelatin, PBS). Cells were then incubated with indicated antibodies diluted in PBG. Cells were washed with PBS and incubated with secondary antibody diluted in PBG. The cells were washed with PBS and fixed in 4% paraformaldehyde. This was followed by digestion with RNaseA 200 mg/ml. Cells were then dehydrated in a series of ethanol washes 70%, 85%, 100%, and the coverslips were dried. Ten nanomolar PNA-TAMRA-(CCCTAA) probe in hybridization buffer (50% formamide, 2× SSC, 2 mg/ml BSA, 10% dextran sulfate) was added to coverslips and DNA was and then placed in a humidified chamber overnight. The coverslips were washed in 2× SSC +50% formamide, 2× SSC alone, and finally in 2× SSC containing DAPI. The coverslips were mounted on glass slides with Vectashield and analyzed using a Zeiss LSM-710 confocal microscope.
Chromatin Immunoprecipitation Assay
Cells were fixed with 1% formaldehyde, quenched with 0.125 M glycine, and pelleted in PBS. Pellets were lysed in buffer A (5 mM PIPES, 85 mM KCl, 0.5% NP40). Nuclear pellets were isolated by centrifugation and lysed in buffer B (50 mM Tris [pH 8], 10 mM EDTA [pH 8], 0.2% SDS) and sonicated. The chromatin was normalized and incubated with 2 µg of the indicated antibodies overnight. Chromatin-antibody conjugates were precipitated with magnetic beads and washed with Dilution IP buffer (16.7 mM Tris [pH 8], 1.2 mM EDTA, 167 mM NaCl, 0.1% SDS, 1.1% Triton X-100), TSE (20 mM Tris [pH 8], 2 mM EDTA [pH 8], 500 mM NaCl, 1% Triton X-100, and 0.1% SDS), LiCl buffer (100 mM Tris [pH 8], 500 mM LiCl, 1% deoxycholic acid, and 1% NP40), and TE (10 mM Tris [pH 8] and 1 mM EDTA [pH 8]). Beads were eluted in 50 mM NaHCO3, 140 mM NaCl, and 1% SDS, decrosslinked, and bound DNA was analyzed by dot blot using a telomere-specific probe.
Terminal Restriction Fragment Analysis
Genomic DNA was purified using the QIAGEN DNA Blood Mini Kit according to the manufacturer’s instructions. DNA was digested with AluI and MboI restriction enzymes and then electrophoresed on 0.7% agarose gel in 0.5× TBE buffer. After electrophoresis, the DNA was transferred to a Hybond XL membrane capillary action, and telomeric restriction fragments were detected by Southern blot using a DIG-labeled probe (CCCTAA)4.
C-circle Assay
Genomic DNA was purified and digested with AluI and MboI restriction enzymes. The digested DNA was again purified, and the DNA was quantified by spectrophotometer. The DNA (40 ng) was diluted in 10 µl 1 × Φ29 Buffer containing BSA (0.2 mg/ml), 0.1% Tween, 0.2 mM each dATP, dGTP, dTTP, and incubated in the presence or absence of 7.5 U ΦDNA polymerase at 30°C for 8 hr, followed by 65°C for 20 min. C-circle amplification products were detected by dot blot using a DIG-labeled probe (CCCTAA)4.
Metaphase Spreads
Cells were incubated in nocodazole for 2 hr, collected by trypsinization, and then incubated in 75 mM KCl at 37°C for 20 min. Cells were fixed in ice cold fixative (3:1 methanol/acetic acid) before dropping on glass slides. Slides were incubated with Giemsa for 20 min and analyzed with a Nikon Eclipse Ti at 63×.
Supplementary Material
In Brief.
The alternative lengthening of telomeres pathway accounts for cellular immortality in 10% of all cancers; however, the mechanisms regulating ALT activity have not been fully elucidated. Cox et al. demonstrate that the replication stress response protein SMARCAL1 is significantly enriched at ALT telomeres to resolve replication stress and promote ALT activity.
Highlights.
SMARCAL1 associates with telomeric DNA after replication stress
SMARCAL1 is significantly enriched at ALT telomeres
Depletion of SMARCAL1 causes replication fork collapse and ALT telomere instability
Chronic replication stress prevails at ALT telomeres
Acknowledgments
We thank Dr. S.B. Cantor and members of the R.L.F. lab for helpful discussions. We would also like to acknowledge the BUMC Cellular Imaging Core facility and personnel, including Technical Director, Dr. Michael T. Kirber. R.L.F. is supported by the NIH Pathway to Independence Award (CA166729), the Karin Grunebaum Cancer Research Foundation, and a Peter Paul Professorship. A.M. is supported by funding from the Cancer Research Society/Sociétéde recherche sur le cancer.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.01.011.
AUTHOR CONTRIBUTIONS
K.E.C. conducted experiments, analyzed the data, and wrote the manuscript. R.L.F. designed experiments, analyzed the data, and wrote the manuscript. A.M. conducted preliminary experiments and reviewed the manuscript.
REFERENCES
- Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos D, Stutz MD, Hills M, Neumann AA, Bryan TM, Reddel RR, Pickett HA. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012;199:893–906. doi: 10.1083/jcb.201207189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londoño-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. USA. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D. The replication checkpoint prevents two types of Fork collapse without regulating replisome stability. Mol. Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suvá ML, Benes CH, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouché N, Cesare AJ, Willcox S, Ozgür S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am. J. Hum. Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- Grudic A, Jul-Larsen A, Haring SJ, Wold MS, Lønning PE, Bjerkvig R, Bøe SO. Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res. 2007;35:7267–7278. doi: 10.1093/nar/gkm738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Reddel RR. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- Lau LMS, Dagg RA, Henson JD, Au AY, Royds JA, Reddel RR. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 2013;41:e34. doi: 10.1093/nar/gks781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 2009;29:703–713. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Erdjument-Bromage H, Pan ZQ, Lee SH, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LA, Zhao R, Glick GG, Lovejoy CA, Eischen CM, Cortez D. SMARCAL1 maintains telomere integrity during DNA replication. Proc. Natl. Acad. Sci. USA. 2015;112:14864–14869. doi: 10.1073/pnas.1510750112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar J, Wan B, Yin J, Vallabhaneni H, Horvath K, Kulikowicz T, Bohr VA, Zhang Y, Lei M, Liu Y. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic Acids Res. 2015;43:5912–5923. doi: 10.1093/nar/gkv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, Chen BP, Chen DJ, Agami R, Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, et al. Corrigendum: driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;484:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, Kavanaugh GM, Chen Y, Tabb DL, Cortez D. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013;288:31458–31467. doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Tokutake Y, Matsumoto T, Watanabe T, Maeda S, Tahara H, Sakamoto S, Niida H, Sugimoto M, Ide T, Furuichi Y. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun. 1998;247:765–772. doi: 10.1006/bbrc.1998.8876. [DOI] [PubMed] [Google Scholar]
- Uringa E-J, Lisaingo K, Pickett HA, Brind’Amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol. Biol. Cell. 2012;23:2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J-B, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- Varley H, Pickett HA, Foxon JL, Reddel RR, Royle NJ. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 2002;30:301–305. doi: 10.1038/ng834. [DOI] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- Wu G, Jiang X, Lee W. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- Wyatt HDM, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body advances. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.