Abstract
Despite arduous efforts and recent technological developments structural investigation of integral membrane proteins remains a challenge. The primary deterrents include difficulties with their expression, low inherent solubility and various problems associated with existing membrane mimicking systems. A relatively new class of membrane mimetics, nanodiscs, has been developed as a promising alternative. Although nanodiscs have been proven successful for several biophysical applications, they yet remain to become the system of preferred choice for structure determination. We have hereby made nanodiscs more suitable for solution NMR applications by reducing the diameter of the self-assembly complex to its potential limit. We achieved a noticeable improvement in the quality of NMR spectra obtained for the transmembrane and cytoplasmic domains of integrin αIIb incorporated into these smaller discs rendering them susceptible for a thorough structural investigation. We also present an on-column method for a rapid, efficient, single step preparation of protein incorporated nanodiscs at high concentrations. These discs have been fully characterized by transmission electron microscopy, dynamic light scattering and differential scanning calorimetry.
Keywords: nanodiscs, lipid bilayer, integral membrane protein, bacteriorhodopsin, integrin
Introduction
Integral membrane proteins (IMPs) comprise one third of the genomic content in eukaryotes (1) and are targets for more than sixty percent of the drug leads (2). While significantly important, they are less understood and structurally characterized relative to their soluble counter parts. Notorious for the difficulties associated with over expression, purification, and inherent insolubility, they remain challenging subjects for high resolution structure determination both by X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy. To facilitate studies of membrane proteins using solution NMR, several different membrane mimicking systems including organic solvent mixtures, amphipols, micelles and bicelles have been employed (3). Though useful they come with certain caveats like surface curvature artifacts, narrow range of stable temperatures, limited diversity of detergent or lipid types, and poor control over sample homogeneity (4). A relatively new class of soluble membrane mimetic called nanodiscs, with controlled diameter and thickness of a lipid bilayer membrane, has been introduced for studying membrane proteins. Since its development, this system has been used for the biophysical characterization of several receptors, enzymes, channels and transporters (5).
Nanodiscs, originally developed by Sligar and colleagues in 2002, are discoidal nanolipoproteins, having a nanoscale phospholipid bilayer held together by an amphipathic membrane scaffold protein (MSP), which forms the outer protein belt enclosing the lipid molecules and protecting its hydrophobic aliphatic chains from polar environment (6–8). MSP is derived from apolipoprotein A-I and contains several alpha helical repeats. The size, defined as the diameter of nanodisc, is varied by using MSPs that differ in their number of helical repeats (9). The lipid content, which defines the thickness of the bilayer, is varied by the use of multifarious lipids like DMPC, POPC, DPPC, E. coli lipid extracts, etc. (10). Due to their soluble nature, ease in concentration, monodispersity, temperature stability, and compatibility with cell free protein expression (11), nanodiscs present a competitive edge over other systems available for studying membrane proteins (5). Apart from reconstitution of several proteins, including rhodopsins, SecYEG, tar, cytochrome P450, β2 adrenergic receptor and epidermal growth factor receptor (10), nanodiscs have been uniquely used for studying cell signaling (12) and nanomechanical detection of cholera toxin (13). Using solution NMR (14,15), nanodiscs have been compared to micelles in their ability to provide a proper environment for studying membrane protein interactions. Additionally, larger nanodiscs “macrodiscs” have been shown viable as an alignment media for measurement of residual dipolar couplings (16). However, up till now the application of nanodisc for high resolution structural determination by solution NMR was deterred by its large size. Up till now the application of nanodiscs for high resolution structural determination by solution NMR was deterred by its large size. While in preparation of the revised manuscript we came across a compelling paper by the Wagner group exemplifying for the first time the feasibility of the method for structure determination in smaller nanodiscs (17).
Investigating membrane proteins by solution NMR requires a model membrane system small enough to achieve a comparatively short rotational correlation time. In addition, the method requires that the model system stays homogenous under micro molar sample concentrations. In order to achieve shorter rotational correlation time and render nanodiscs more suitable for NMR applications, we reduced the overall diameter of the entire complex by reducing the number of helical repeats in MSP. These smaller discs, termed as minimal nanodiscs, clearly show a significant improvement in the NMR spectrum of the incorporated protein - integrin αIIb transmembrane and cytoplasmic domains when compared to the existing nanodiscs. Reconstituting membrane proteins within nanodisc using the original protocol (18) yields low concentration samples which require additional purification steps for separating empty discs from protein incorporated ones. Hence we developed a simpler, single step on-column method for preparing nanodiscs by concentrating proteins on the resin. On-column methods have long been in use for the incorporation of membrane proteins into lipid vesicles and detergent micelles. (19,20). In addition to a three times reduction in the preparation time we have achieved a concurrent separation of empty and protein incorporated discs obtaining concentrated homogenous preparations. Since discs are formed bound to the resin, we explored the two possible ways of preparing discs on-column: (i) discs are formed bound to the resin through MSP, and (ii) discs are formed bound to the resin through the target protein.
Materials and Methods
The buffers listed hereunder are the same as used in the original protocol for nanodiscs preparation (18): binding buffer: 20 mM NaH2PO4*H2O, 1 mM PMSF, pH 7.4; wash buffer-1: 40 mM Tris/HCl, 0.3 M NaCl, 1% Triton X-100, pH 8.0; wash buffer-2: 40 mM Tris/HCl, 0.3 M NaCl, 50 mM Na-cholate, 20 mM imidazole, pH 8.0; wash buffer-3: 40 mM Tris/HCl, 0.3 M NaCl, 50 mM imidazole, pH 8.0; elution buffer: dialysis buffer + 0.4 M imidazole; dialysis buffer: 20 mM Tris/HCl, 0.1 M NaCl,0.5 mM EDTA, pH 7.4. Additional buffers are detailed within corresponding sections.
Cloning and Expression
From the several available MSP constructs, we chose MSP1E3D1 and MSP1D1, pET28a vectors with N-terminal His-tag and kanamycin resistance, generously provided by Dr. Nathan Alder. MSP1D1 vector was used as template to generate the several deletion mutants, D7, D6 and D5. Mutants were created by successively deleting single helices from the template till the MSP was reduced to five helices using the site directed mutagenesis kit (Agilent, USA). The construct for integrin αIIb (A2b) fused to maltose binding protein (MBP) with ampicillin resistance was gifted by Dr. Jun Qin. Integrin αIIb consisting of a single trans-membrane and the C-terminal cytoplasmic domains (TMCD), 54 amino acids in total, are inserted into a modified pMAL-C2 (NEB, USA) vector containing a 6X-His tag and TEV cutting site in the order of MBP-HIS-TEV-αIIb (21). All constructs were transformed into E. coli BL21 (DE3) cells and induced with 1mM IPTG in LB for four hours. 15N αIIb was prepared by induction with 1mM IPTG in minimal media for six hours.
Purification
We found that 0.5 L of frozen cells gave us an average of 12 to 13 mg of MSP1E3D1. Two liters of culture was lysed and split into different parts giving us 1.65, 3.3, 6.6, 12 and 24 mg protein. 1ml of Ni-NTA (QIAGEN, USA) resin was centrifuged and mixed with each of the above protein concentrations. This gave us five samples labeled as NDMT1, NDMT2, NDMT3, NDMT4, NDMT5 having respectively 1.65, 3.3, 6.6, 12 and 24 mg of protein bound per ml of Ni-NTA resin. In conclusion, we have 5 samples doubling in the concentration of protein bound per ml of resin. This resin was washed according to Sligar’s protocol with wash buffers 1, 2 and 3, and additionally with five column volumes of three buffers: (1) 40mM Tris, 200mM NaCl, 50mM Imidazole, pH 8, (2) 40mM Tris, 200mM NaCl, pH 8, and (3) dialysis buffer. MSP mutants D5, D6 and D7 were purified from 0.5 L culture mixed with 1 ml of Ni-NTA resin. Each mutant preparation was washed with the above five buffers and later used for the preparation of empty nanodiscs. Bacteriorhodopsin in purple membrane was generously provided by Dr. Robert Birge and was purified using the protocol described elsewhere (22). An average of 10 – 11 mg of Integrin αIIb was obtained from 1 liter of culture, which was lysed and mixed overnight with amylose resin (NEB, USA) in a buffer containing 20mM Tris, 0.2M NaCl, 1mM EDTA, pH7.4 and complete protease inhibitor tablet (Roche, USA). This integrin bound resin was washed with 10 column volumes of the same buffer plus additional 5 column volumes of dialysis buffer before being subjected to nanodisc preparation. MSP proteins (MSP1E3D1, and D7) used for the formation of integrin containing nanodiscs were purified using the standard solution protocol (18).
On-column formation of Nanodiscs
Powdered version of DMPC (dimyristoyl-sn-glycero-3-phosphocholine) (Avanti Polar, USA) lipid was used for preparing the several nanodiscs. Nanodiscs were prepared using the “General Method for On-column Nanodisc Formation” given hereunder. Empty discs containing MSP1E3D1, was prepared at a protein to lipid ratio of 1:160; MSP1D1, at a protein to lipid ratio of 1:80 (18); and D5, D6, D7, at an optimized (data not shown) protein to lipid ratio of 1:30. Bacteriorhodopsin (bR) in purple membrane was mixed overnight with 4% Triton X-100. Discs were prepared by mixing bacteriorhodopsin with MSP (bound at 6.6 mg/ml of Ni-NTA resin) and solubilized DMPC at a MSP1E3D1:bR:DMPC molar ratio of 2:3:160 (23). Integrin αIIb (A2b) containing nanodisc was prepared by mixing MSP with the resin bound αIIb at a MSP1E3D1:A2b:DMPC molar ratio of 1:0.5:160 and D7:A2b:DMPC molar ratio of 1:0.5:20. Once nanodiscs were formed free lipids and unbound proteins were washed out with 2 column volumes of dialysis buffer. Empty and bacteriorhodopsin containing discs were then eluted with 4 column volumes of elution buffer while αIIb containing discs were eluted with 4 column volumes of 20 mM maltose, 20mM Tris, 0.2M NaCl, 1mM EDTA, pH 7.4. All the eluates were loaded onto 16/60 Superdex 200 column pre-equilibrated with dialysis buffer from which the relevant nanodisc fractions were pooled and used for further analysis. Nanodisc containing 15N labeled αIIb was eluted using 10mM KPi, 10mM KCl, 0.05mM EDTA, pH 6.5 buffer containing TEV protease and concentrated for NMR experiments.
General Method for On-column Nanodisc Formation
All experiments were done at room temperature. Powdered version of DMPC was solubilized in dialysis buffer containing twice the concentration of sodium cholate in a glass vial by sonication. Solubilized lipids were added at an appropriate ratio to the resin containing bound MSP1E3D1 or target protein or both in a short flex column using a Hamilton syringe (Hamilton, USA). The volume of the reaction mixture was adjusted with dialysis buffer to get a crucial final cholate concentration between 12–15 mM. The column was capped and kept on a nutator mixer for one hour. To this mixture wet BioBeads SM-2 (Sigma, USA) were proportionally added at a ratio of 0.6 gm/ml of the reaction volume. Prior to adding the beads they were preconditioned as recommended by the manufacturer and finally washed with dialysis buffer. This mixture was left horizontally shaking, suspending the BioBeads in solution for 3 hours after which the column was washed with appropriate buffer and eluted. Ni-NTA resin was salvaged from BioBeads by using a fine mesh sieve thereby making it available for regeneration.
Nuclear Magnetic Resonance (NMR)
HSQC experiments were run on Varian 800 MHz spectrometer (Agilent, USA) equipped with the cryoprobe using TROSY pulse sequence (24) at 40°C for 16 hours (256 scans in t2 and 128 increments in t1) in the above mentioned buffer of pH 6.5. Line-width analysis of the peaks obtained from the spectra was performed using the CCPN Analysis software (25). T1, T2 relaxation experiments were carried out using the INVREC and CPMGT2 pulse sequences at 40°C.
Transmission Electron Microscopy (TEM) and Imaging
NDMT samples were adjusted to an absorbance A280 value of around 0.46. Each sample was diluted 80 times with Milli Q water (Millipore, USA) and prepared on a glow discharged copper grid (mesh 400) covered with carbon film. Grids were negatively stained with aqueous 1% uranyl acetate and air dried after wicking away excess stain. Electron micrographs were taken with AMT XR-40 camera slide mounted on a Tecnai Biotwin G2 Spirit Transmission Electron Microscope (FEI, USA). Images were taken at a direct magnification of 180000x. NDbR and NDA2b were adjusted to an absorbance A280 value of 1 and 0.5 respectively and diluted 40 times with Milli Q water (Millipore, USA). ImageJ (NIH, USA) software was used for measuring the diameters of the two extreme NDMT samples NDMT1 and NDMT5. Electron micrograph images were reverse contrasted and converted to a binary format. The diameter was automatically counted using the Analyze Particles macro, after setting the scale to the calibrated value on the micrograph. The table obtained was exported and plotted using Origin. A total of 646 and 443 nanodiscs were counted for NDMT1 and NDMT5 respectively.
Dynamic Light Scattering (DLS)
All experiments were done at 25°C with Zetasizer NanoS (Malvern Instruments, USA) detecting backscatter at 173°. Six scans were each taken for sixty seconds and averaged to get the mean diameter. The particle size distribution by volume data was exported, fitted and overlaid in Origin.
Differential Scanning Calorimetry (DSC)
The most monodisperse sample, NDMT5, at a concentration of 0.95 mg/ml was used for the experiment. This sample was referenced against the buffer obtained as a filtrate from the concentration step. Prior to the run, both were filtered with 0.22 μm and degassed using vacuum. Thermograms were obtained by scanning from 10 °C to 50 °C at 35 °C/hour at a constant pressure of 3 atm (26). In total, 3 sets of heating, cooling cycles were done using NanoDSC (TA Instruments, USA). The data was corrected for baseline with a thermogram obtained from buffer scans and fitted with residuals of 1.6 using the two state scaled model through the NanoAnalyze (TA Instruments, USA) software.
Results
1. On-Column Preparation of Empty Nanodiscs
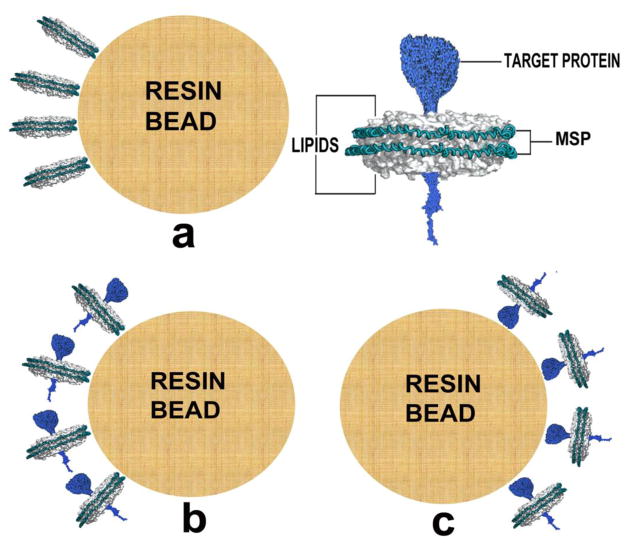
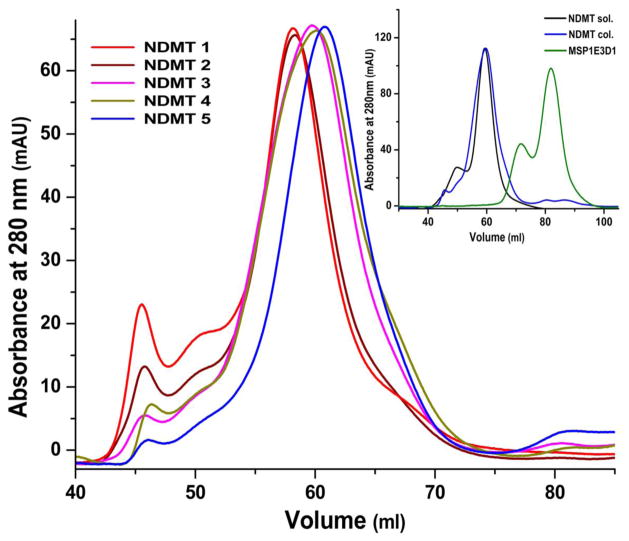
In this method nanodiscs are formed bound to the resin through their outer belt protein MSP (Figure 1a). We chose MSP1E3D1 and DMPC lipids as they have been extensively used in reconstituting several membrane proteins within nanodiscs. The histidine tagged MSP1E3D1 was used for its ability to bind Ni-NTA resin. Discs were formed at the recommended MSP1E3D1:DMPC ratio of 1:160. Five samples, NDMT1 through NDMT5, were prepared successively doubling the protein concentration giving 1.65, 3.3, 6.6, 12 and 24 mg MSP bound per ml of resin. We stopped at 24 mg/ml to avoid crossing the binding capacity of the resin. This prevents any loss of protein in the flow-through, which could perturb the final MSP to DMPC ratio. All the five concentrations of MSP was prepared from a single cell lysate and mixed with solubilized DMPC obtained from the same stock preparation. An overlay of the chromatograms obtained from each of these samples through size-exclusion chromatography (SEC) on Superdex 200 column is shown in Figure 2. The seemingly homogenous major peaks, from each of these samples, spread over a total difference of 5 ml in their elution volumes, with little to no free MSP. Interestingly, with increasing protein concentration bound per ml of resin, we saw a decrease in the amount of aggregates and a shift in the elution volumes. Nanodiscs were also prepared using the original solution method (18) as a control to compare with discs obtained from the on-column method. The inset in Figure 2 shows a perfect overlay of the chromatograms from the control and one of our preparations, NDMT4, and the free MSP protein eluting at 83 ml. The elution volume of the rightmost peak in our preparation is around 61 ml corresponding to a molecular weight of about 275 kDa. This is in good accordance with 268.6 kDa calculated from the molecular weight of two MSP1E3D1 (32.6 kDa each) and a bilayer of 300 DMPC molecules (0.67 kDa each) (10).
Figure 1.
Model diagram showing the different components of a protein incorporated nanodisc and different schemes of the on-column method: (a) Nanodiscs bound to the resin through MSP eg. NDMT; (b) Nanodisc with target protein bound to the resin through MSP eg. NDbR; (c) Nanodisc with target protein bound to the resin through the target protein eg. NDA2b.
Figure 2.
Chromatogram overlay from size exclusion chromatography (SEC) of the different NDMT samples: NDMT1 (red), NDMT2 (brown), NDMT3 (magenta), NDMT4 (khaki) and NDMT5 (blue). Inset: SEC comparing nanodiscs made in solution (NDMT sol: black) and on-column (NDMT col: blue).
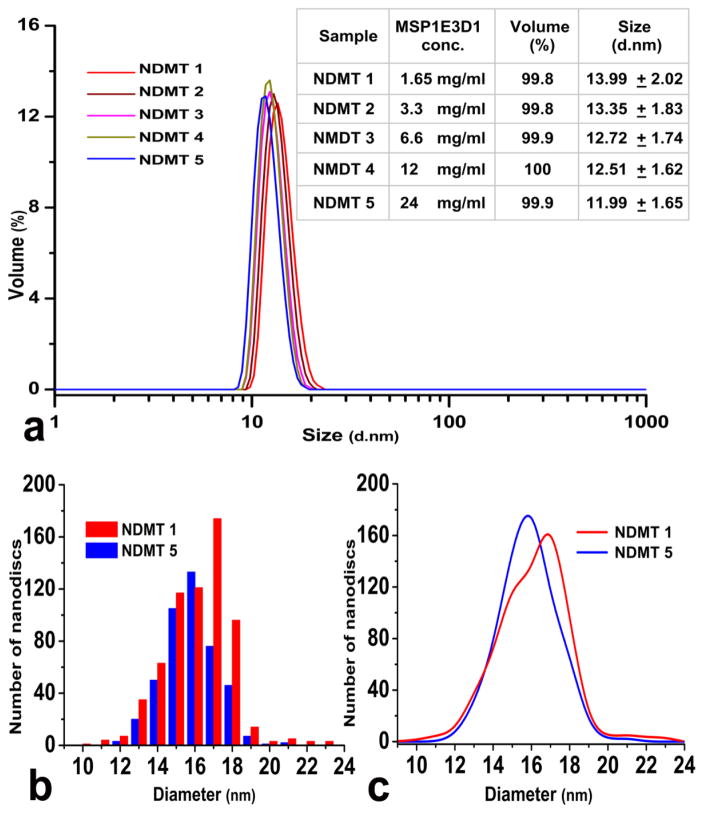
TEM and DSC were used to determine the identity of nanodiscs. Electron micrographs obtained from TEM of the five NDMT samples (Figure S1a) show discoidal nano-lipoproteins against a negative background. NDMT5 was further used to check for the characteristic transition temperatures obtained for nanodiscs through DSC. The thermogram in Figure S1b shows a broad phase transition, typical for nanodiscs, with a pre-transition temperature at 24°C and transition temperature at 27.6°C. Both temperatures corresponded well with the phase transition temperatures previously reported for DMPC nanodiscs (26). DLS was further used to confirm the purity and to quantitatively differentiate our five preparations. An overlay of the normalized particle size distributions by volume is shown in Figure 3a with impressive samples homogeneity ranging from 100% to 99.8%. The table shown as inset in Figure 3a gives the mean Stokes diameters ranging from 13.99 ± 2.02 nm for NDMT1 (1.65 mg/ml) to 11.99 ± 1.65 nm for NDMT5 (24 mg/ml). This correlates well with the originally reported diameter of 12 nm for MSP1E3D1 discs (18).
Figure 3.
(a) Overlay of the particle size distribution by volume for different nanodisc samples obtained through DLS: NDMT1 (red), NDMT2 (brown), NDMT3 (magenta), NDMT4 (khaki) and NDMT5 (blue). Inset: Table showing the mean Stokes diameters for the different NDMT samples. (b) Frequency distribution of nanodisc diameters for the two extreme samples NDMT1 (red) and NDMT5 (blue). (c) Normalized curve fitted to the same frequency distribution data.
The above results raise an interesting question as to why discs, prepared by the same method varying only in the amount of MSP bound per ml of resin, gave different mean diameters. Hence, we sought to analyze the dispersity of our samples by measuring the diameters of the individual discs over an entire electron micrograph using ImageJ software (27). 646 and 443 nanodiscs were respectively used for the two extreme samples, NDMT1 and NDMT5. Figure 3b shows the frequency distribution data, and Figure 3c shows the normalized curves fitted to this data. This clearly indicates that the NDMT5 preparation is more monodisperse than NDMT1. In electron micrographs the orientation of nanodiscs, deposition of stain around them along with the granular negative background leads to artifacts in defining the edges of the specimen at higher magnifications (28). Hence, the populations were distributed around slightly bigger diameters as seen from the x-axis. None the less, these systematic artifacts do not affect the assessment of population dispersity.
2. On-Column Incorporation of Target IMPs into Nanodiscs, Two Case Studies
The primary aim behind making nanodiscs was to incorporate target membrane proteins. This was efficiently achieved by using the on-column method. Two types of proteins are involved in the process, MSP, which forms the outer layer covering the disc; and a target protein, which is encapsulated within the disc. Thus two potential approaches could be envisioned: (i) resin immobilized MSP is mixed with the target protein forming nanodiscs bound to the resin through MSP as shown in Figure 1b, or (ii) resin immobilized target protein is mixed with MSP forming nanodiscs bound to the resin through the target protein as shown in Figure 1c. We have used bacteriorhodopsin to demonstrate the first approach and MBP-fused integrin αIIb for the second. It is worthwhile to note that the second approach has been uniquely applied before for reconstituting anthrax toxin pore into nanodiscs (29).
Bacteriorhodopsin in nanodiscs (NDbR)
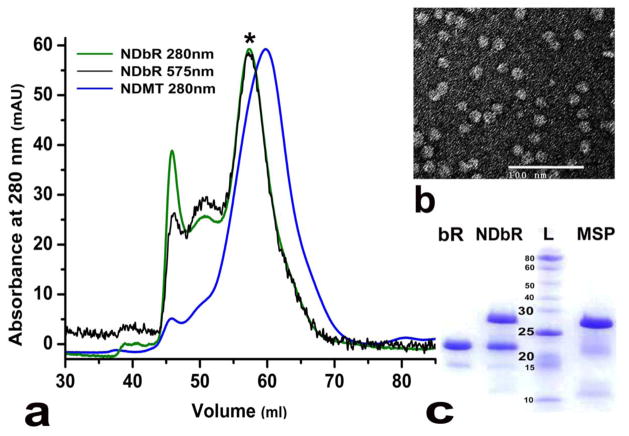
Bacteriorhodopsin (bR) in purple membrane was purified as described elsewhere (22) and solubilized overnight in 4% Triton X-100 (30). It was mixed with MSP bound to Ni-NTA resin in a previously optimized MSP1E3D1:bR:DMPC ratio of 2:3:160 (23). Figure 4a presents the overlay of the chromatograms obtained from SEC on Superdex 200 column for NDbR and NDMT monitored at 280 nm. Additionally, NDbR was monitored at 575 nm, a characteristic absorption wavelength for bacteriorhodopsin. The peak containing bacteriorhodopsin in nanodisc is shown by the asterisk sign to separate it from the other smaller peaks corresponding to higher order aggregates. The shift in elution volumes for the two chromatograms is around 2 ml. This corresponds to a difference in molecular weight of approximately 75 kDa, roughly indicating a homotrimeric arrangement of bacteriorhodopsin molecules within a single nanodisc as has been previously reported (23). Additionally, when the fractions were pooled and concentrated, the sample exhibited a light purple color with two absorbance maxima at 280 and 556 nm as compared to the empty discs with a single absorption maximum at 280 nm (data not shown). Furthermore, TEM confirmed the formation of discs as indicated by the presence of discoidal particles in the electron micrograph (Figure 4b). SDS-PAGE analysis confirmed the presence of bacteriorhodopsin within nanodiscs as seen from lane two in Figure 4c showing two bands: one, with MW of about 23 kDa, corresponding to bacteriorhodopsin shown also as a control in lane one; and the second, with a MW of about 28 kDa, corresponding to MSP1E3D1 also shown as a control in lane four. Each lane was compared against lane three representing molecular weight markers. In conclusion, we were successful in reconstituting bacteriorhodopsin within nanodiscs on Ni-NTA resin.
Figure 4.
(a) Size exclusion chromatography showing NDbR (green) and NDMT (blue) at 280nm and NDbR (black) at 575nm. (b) TEM image of NDbR negatively stained with 1% uranyl acetate. (c) SDS-PAGE analysis showing: lane 1 - bacteriorhodopsin (bR); lane 2 - nanodiscs with bacteriorhodopsin (NDbR); lane 3 - protein ladder (L); and lane 4 - and MSP1E3D1 (MSP).
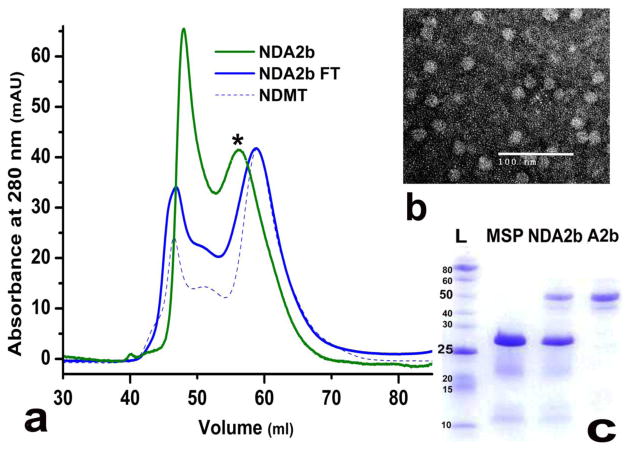
Integrin αIIb Trans-Membrane and Cytoplasmic Domains in Nanodisc (NDA2b)
We used MBP fused transmembrane and cytoplasmic domains of integrin αIIb (A2b). After lysing the cells the protein was left bound to the amylose resin to which we directly added MSP and lipids in a MSP1E3D1:A2b:DMPC ratio of 1:0.5:160. This ratio was calculated on the basis of having one αIIb molecule per disc. Two types of discs were formed during this process: empty discs and protein incorporated discs. The discs containing αIIb stayed bound to amylose resin through the MBP tag while empty discs which were unable to bind got washed out in the flow-through. Next, discs containing αIIb were eluted using maltose. The flow-through and elution fractions were run on Superdex 200 column. As a control, empty discs prepared using Sligar’s protocol was also run on the column. Figure 5a presents an overlay of the chromatograms obtained from these samples. The peak containing αIIb discs are shown by the asterisk sign to separate it from the other higher order aggregates. As expected, both the flow-through peak and the empty disc prepared as a control had the same elution volume. The peak corresponding to the αIIb disc was shifted from the flow-through by approximately 3 ml, correlating with an increase in MW of about 100 kDa. This suggests that there are roughly two αIIb (52 kDa each) molecules per nanodisc, an interesting observation consistent with the previous reports for αIIb-TMCD homo dimerization in dodecyl-phosphocoline micelles (31). Again, TEM confirmed the formation of discs as indicated by the presence of discoidal particles in the electron micrograph (Figure 5b). SDS-PAGE analysis confirmed the presence of αIIb within nanodiscs as seen from lane three in Figure 5c showing two bands: one, with MW of about 50 kDa, corresponding to MBP-integrin αIIb shown also as a control in lane four; and the second, with a MW of about 28 kDa, corresponding to MSP1E3D1, shown also as a control in lane two. Each lane was compared against lane one representing molecular weight markers. In conclusion, we successfully incorporated MBP-fused integrin αIIb-TMCD into nanodiscs on amylose resin, with the concurrent separation of empty and protein incorporated discs, in a single purification step.
Figure 5.
(a) Size exclusion chromatography showing NDA2b (green), NDA2b flow-through (solid blue) and the control NDMT (dashed blue). (b) TEM image of NDA2b negatively stained with 1% uranyl acetate. (c) SDS-PAGE analysis showing: lane 1 - protein ladder (L); lane 2 - MSP1E3D1 (MSP); lane 3 - nanodisc with MBP-integrin αIIb (NDA2b); and lane 4 - MBP-integrin αIIb (A2b).
3. Minimal Nanodiscs
Preparation
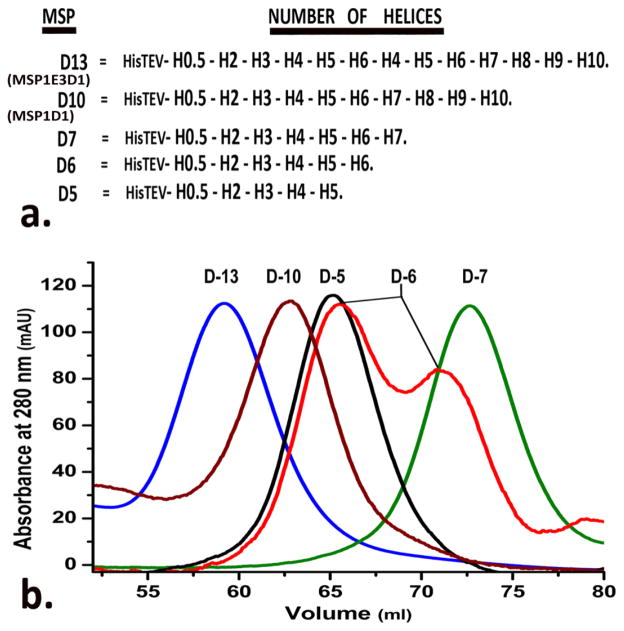
The overall diameter of nanodiscs is governed by the length of the outer coat protein, MSP. We created smaller discs by shortening the length of MSP. The popularly employed constructs of MSP, MSP1D1 and MSP1 E3D1, differ in the addition of three helical repeats in the center of the protein insinuating the possible interaction of terminal helices in the formation of nanodisc. Hence we created C-terminal deletion mutants to additionally ascertain the importance of these interactions. For the sake of simplicity, we will refer to the different sized MSPs as “D” followed by the number of helical repeats. So, the two types of nanodiscs made from the existing MSP1E3D1 and MSP1D1 are called D13 and D10 as they have thirteen and ten helical repeats respectively. We will focus on the MSP truncation mutants with five, six and seven helical repeats (Figure 6a), where major differences in the disc size were observed. Discs were prepared from each of these mutants in an optimized (data not shown) MSP to DMPC ratio of 1:30 and analyzed on Superdex 200 column. The chromatograms obtained were overlaid for comparison. Interestingly, instead of continuous disc size reduction following the truncation of MSP length, the limit was reached at D7. After this point, the discs sizes increased with any successive deletion of helices. D7 discs, with the smallest size, have the lowest elution volume followed by D6 discs with two peaks, one close to the elution volume of D7 and the other to D5 (Figure 6b). In order to confirm that the peaks fractions indeed contained nanodiscs we performed TEM. Figure S2 shows the presence of discoidal nano-lipoproteins against a negative background for each of the D5, D6 and D7 discs. The D7 discs were further characterized by DLS giving a mean diameter of 7 nm (Figure S3a); and SEC on a Superdex 75 column giving a molecular weight of 62 kDa (Figure S3b, which is in good accordance with the molecular weight of two D7 MSP (17.7 kDa each) and a bilayer of 40 DMPC molecules (0.67 kDa each).
Figure 6.
(a) Comparison of the different MSP mutants created by deletion of its terminal helices. (b) Overlay of the chromatograms obtained from size exclusion chromatography of discs prepared with different MSP mutants.
NMR Spectra
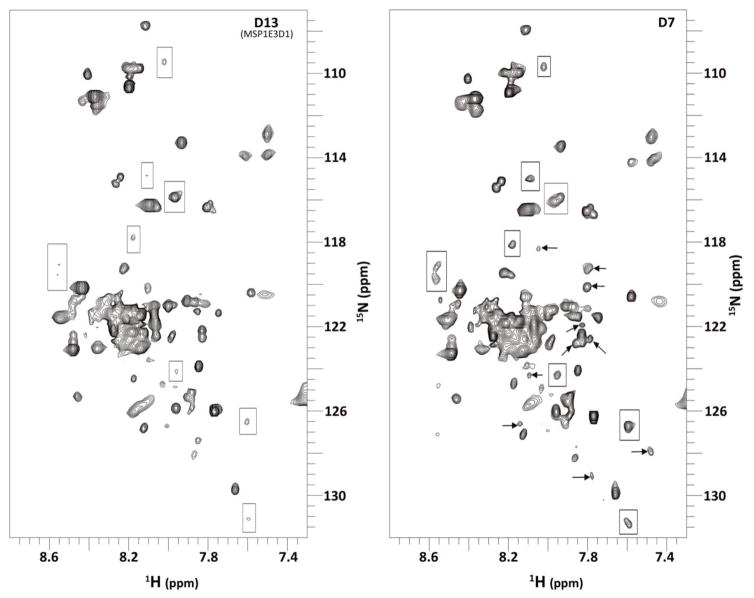
Our major goal behind developing smaller discs was to use them for studying integral membrane proteins by solution NMR. We incorporated integrin αIIb into our smallest D7 discs using the on-column method at an optimum MSP: αIIb: DMPC ratio of 1:0.5:20. In order to show that nanodiscs can be successfully used for solution NMR we obtained 15N-TROSY-HSQC spectra for integrin αIIb-TMCD incorporated into D13, D10, and D7 discs on an 800 MHz spectrometer. Although we did not see much difference between the D13 and D10 spectra (data not shown), significant improvement in spectrum quality was observed for D7 when compared to D13 discs. All amide peaks in the spectra could be divided in two major groups. Group I peaks represent the amides belonging to the cytoplasmic (~13 amino acids) and extracellular (9 amino acids) parts of the protein which, as a result of fast local motion, are equally sharp in both spectra (Figure 7). Group II peaks, corresponding to ~20 trans-membrane and ~8 membrane-proximal amino acids, are much broader, reflective of the longer overall rotational correlation time of the larger disc complex. The spectrum improvement obtained in smaller discs is associated with this group: some peaks, which are almost scarcely visible in D13 discs (shown by boxes), become much more prominent in D7 discs; while the others (shown by arrows) only appear in D7 discs bringing the overall number of peaks to an expected value of ~ 50. Interestingly, even among these broader peaks in D7 discs we observe noticeable heterogeneity in the peak widths, which is possibly an indication of some residual local motion associated with the depth of membrane insertion. We have further analyzed the line-width differences in 1H dimension for the two spectra using CCPN Analysis software (25), and confirmed the improvement as exemplified by the table presented in Figure S4 for several well resolved peaks. Though significant, the accomplishments with the smaller discs come with the caveat of having an incomplete understanding of its morphology and bilayer properties at such low lipid concentrations. We additionally measured the longitudinal and transverse relaxation rates of the acyl chains in the DMPC lipids within nanodiscs (-(CH2)n- peak at ~ 1.3 ppm). We found approximately a 1.6 times reduction in the overall rotational correlation times (τc) of the two disc complexes (T1/T2 ratio in D13 discs of ~55 is reduced to ~35 in D7 discs).
Figure 7.
15N TROSY-HSQC obtained from integrin αIIb-TMCD incorporated into larger D13 (MSP1E3D1) discs (left) and smaller D7 discs (right). Boxes represent sharper peaks while arrows represent new peaks.
Discussion
Our interest in nanodiscs as a suitable membrane mimetic for studying membrane proteins using solution NMR led us to develop a simplified method for their preparation. In this method the entire preparation occurs in a single purification step. Nanodiscs are formed bound to the resin and are eluted directly from the column. MSP’s tendency to aggregate at higher concentrations in solution (18) is circumvented in our method where high concentration of MSP is achieved due to the large binding capacity of the resin. As the result, more concentrated discs samples are prepared in lower reaction volumes. Though the binding of MSP to the resin is random, its distribution across the volume of resin is directly proportional to the concentration. Thus it would be logical to imagine that some MSP molecules are held further apart at lower concentrations than at higher ones. We suspect that this forced physical separation might lead to the formation of some discs with slightly larger diameters than regular optimal ones. NDMT1 and NDMT5 samples have respectively the lowest and highest concentration of protein bound per ml of resin. The frequency distribution data (Figure 3b) obtained for NDMT1 clearly shows the presence of a heterogeneous population distribution when compared to NDMT5. This explains why the average mean diameter obtained from the DLS experiment is higher for NDMT1 over NDMT5. Though more rigorous experimentation could explain what exactly led to this population dispersity, our experiments clearly indicate that NDMT5 (having the highest protein concentration per volume of resin) produces more homogenous and monodisperse nanodisc preparation than NDMT1. Thus, the use of higher protein concentration bound per ml of resin is preferable for obtaining a homogenous preparation of nanodiscs. DSC thermogram obtained for NDMT5 shows the characteristic broad phase transition (Figure S1b), with transition temperatures identical to the ones previously reported for nanodiscs (26). Since there is no observed difference in bilayer characteristics of nanodiscs produced by the two methods, replacement of the chloroform solubilized DMPC with its powdered form worked really well and seems like a feasible alternative. Given the stability of resin beads at different temperatures, this method can be extended to other lipids like DPPC or POPC; which in combination with different MSP constructs, will make our method applicable over the entire catalog of nanodiscs.
The major advantage of our on-column method is highlighted while incorporating proteins within nanodiscs. Not only do we achieve a rapid, single step, homogenous preparation but we do so with the concurrent separation of protein incorporated discs from the empty discs. This is beneficial as it improves the yield by reducing the number of purification steps, providing a more pure homogeneous sample, typically required for structural studies, in a shorter period of time. A quick graphical comparison of the two methods as shown in Figure S5 clearly delineates the advantages of our method. In order to test the robustness of the presented method we used different target proteins to exemplify the two strategies for incorporating membrane proteins within nanodiscs.
The first strategy entails the formation of protein incorporated discs bound to the resin through MSP. We used bacteriorhodopsin, a well characterized protein within nanodiscs, as a target protein to demonstrate the feasibility of this approach. The washing step removed free lipids and any unincorporated bacteriorhodopsin. Following elution, SEC confirmed the absence of the empty discs through the comparison of the UV absorption profiles at 280 and 575 nm.
However, it was difficult to separate the reconstituted discs from the empty ones due to their similar sizes. This drawback was overcome by our second strategy where the protein incorporated discs are formed bound to the resin through the target protein. We used MBP tagged transmembrane and cytoplasmic domains of integrin αIIb as our target protein to demonstrate this case. The amylose resin selectively binds to the MBP tagged protein. Since MSP lacks the MBP tag it cannot bind to the resin. This feature helped in the removal of unincorporated MSP and empty discs along with free lipids during the washing step. Integrin αllb containing discs could then be eluted either by using maltose or by cutting the MBP tag with TEV protease. The latter approach was used when preparing samples for solution NMR studies.
A limiting factor affecting the study of membrane proteins within any system using solution NMR lies in the effective size of the entire complex. Thus we developed smaller discs to achieve shorter rotational correlation time. Since all the deletion mutants formed nanodisc, it implies that interaction of terminal helices are not crucial for the formation of discs. MSP mutant containing seven helical repeats produced the smallest nanodisc. Any further reduction in the helical repeats led to a successive increase in the size of the nanodisc. Interestingly, D5 discs, formed with MSP containing five helical repeats, were similar in size to D10 discs, containing MSP with ten helical repeats. The larger size of D5 discs insinuates the possibility of having more than two MSP proteins per nanodisc. The above pattern of disc formation with several separate helical peptides derived from MSP possibly explains the self-assembly of 30 nm “macrodiscs” developed as an alignment media for residual dipolar coupling experiments (16). The advantage of our smallest D7 discs, with a size of 7 nm and molecular weight of 62 kDa, as compared to D13(MSP1E3D1) discs, (size of 12 nm and molecular weight of 275 kDa), is clearly evident from the presented 15N TROSY-HSQC data. The improvements in line-width are associated with the broadest peaks in the spectrum, corresponding to the amides within the lipid bilayer as confirmed by relaxation measurements showing the reduction in overall rotational correlation time. Clearly, with such relatively large system the deuteration of lipids and incorporated proteins would be required to avoid unwanted dipolar-dipolar interactions. In our view, D7 discs provide a suitable system for structural investigation of small integral membrane proteins as exemplified by their application to integrin αIIb transmembrane and cytoplasmic domains.
Conclusions
Through our study presented in this paper we have developed nanodiscs with the size optimal for structural studies using solution NMR. This has been confirmed by the significant improvement in the spectrum quality of αIIb integrin TMCD incorporated into the newly developed minimal nanodiscs. Furthermore, we have developed an on-column method for making protein incorporated nanodiscs. Through this method one could achieve a single step preparation of discs both in higher concentration and purity typically required for structural studies. The sole deterrent towards employing the on-column method used to be the uncertainty regarding the exact concentration of MSP governing the ratio of protein to lipids. In this paper we presented the experimental evidence, through the preparation of several different types of nanodiscs, that this self-assembly system is extremely robust and can very well tolerate any slight discrepancies in concentrations.
Supplementary Material
Acknowledgments
We would like to thank Drs. Nathan Alder for the MSP1E3D1 plasmid, Jun Qin for the αIIb plasmid and Robert Birge for bacteriorhodopsin in purple membrane. We would also like to acknowledge Tobias Neef and Steven Daniels for their help with DLS and TEM experiments and thank Dr. Vitaliy Gorbatyuk for critical reading of the manuscript. This work was supported in parts by AHA grant to O.V.
Abbreviations
- NMR
Nuclear Magnetic Resonance
- IMP
Integral Membrane Protein
- MSP
Membrane Scaffold Protein
- D13
Membrane Scaffold Protein E3D1 or MSP1E3D1
- NDMT
Empty Nanodiscs
- bR
Bacteriorhodopsin
- NDbR
Bacteriorhodopsin in Nanodiscs
- A2b
Integrin αIIb Trans-Membrane and Cytoplasmic Domains (TMCD)
- NDA2b
Integrin αIIb (TMCD) in Nanodiscs
- MBP
Maltose Binding Protein
- DMPC
dimyristoyl-sn-glycero-3-phosphocholine
- DLS
Dynamic Light Scattering
- DSC
Differential Scanning Calorimetry
- TEM
Transmission Electron Microscopy
- SEC
Size Exclusion Chromatography
References
- 1.Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davey J. Expert Opin Ther Targets. 2004;8:165–170. doi: 10.1517/14728222.8.2.165. [DOI] [PubMed] [Google Scholar]
- 3.Sanders CR, Oxenoid K. Biochimica et biophysica acta. 2000;1508:129–145. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 4.Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand E, Marcotte I. Biochimica et biophysica acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Borch J, Hamann T. Biological chemistry. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 6.Sligar SG. Biochem Biophys Res Commun. 2003;312:115–119. doi: 10.1016/j.bbrc.2003.09.188. [DOI] [PubMed] [Google Scholar]
- 7.Bayburt TH, Sligar SG. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayburt TH, Grinkova YV, Sligar SG. NANO Letters. 2002;2:853–856. [Google Scholar]
- 9.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 10.Bayburt TH, Sligar SG. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kopeina GS, Shulepko MA, Paramonov AS, Mineev KS, Tikhonov RV, Shingarova LN, Petrovskaya LE, Dolgikh DA, Arseniev AS, Kirpichnikov MP. Biochimica et biophysica acta. 2012;1818:349–358. doi: 10.1016/j.bbamem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Hazelbauer GL. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tark SH, Das A, Sligar S, Dravid VP. Nanotechnology. 2010;21:435502. doi: 10.1088/0957-4484/21/43/435502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu TY, Raschle T, Hiller S, Wagner G. Biochimica et biophysica acta. 2011 doi: 10.1016/j.bbamem.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raschle T, Hiller S, Yu TY, Rice AJ, Walz T, Wagner G. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ. Biochemistry. 2011;50:8983–8985. doi: 10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagn F, Etzkorn M, Raschle T, Wagner G. J Am Chem Soc. 2013 doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigaud JL, Pitard B, Levy D. Biochimica et biophysica acta. 1995;1231:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 20.Page RC, Moore JD, Nguyen HB, Sharma M, Chase R, Gao FP, Mobley CK, Sanders CR, Ma L, Sonnichsen FD, Lee S, Howell SC, Opella SJ, Cross TA. Journal of structural and functional genomics. 2006;7:51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J. Proc Natl Acad Sci U S A. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oesterhelt D, Stoeckenius W. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 23.Bayburt TH, Grinkova YV, Sligar SG. Archives of biochemistry and biophysics. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. Proc Natl Acad Sci U S A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 26.Shaw AW, McLean MA, Sligar SG. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 27.Abramoff MD, Magalhaes PJ, Ram SJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 28.Pyrz WD, Buttrey DJ. Langmuir: the ACS journal of surfaces and colloids. 2008;24:11350–11360. doi: 10.1021/la801367j. [DOI] [PubMed] [Google Scholar]
- 29.Katayama H, Wang J, Tama F, Chollet L, Gogol EP, Collier RJ, Fisher MT. Proc Natl Acad Sci U S A. 2010;107:3453–3457. doi: 10.1073/pnas.1000100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dencher NA, Heyn MP. FEBS Lett. 1978;96:322–326. doi: 10.1016/0014-5793(78)80427-x. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Babu CR, Lear JD, Wand AJ, Bennett JS, DeGrado WF. Proc Natl Acad Sci U S A. 2001;98:12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.