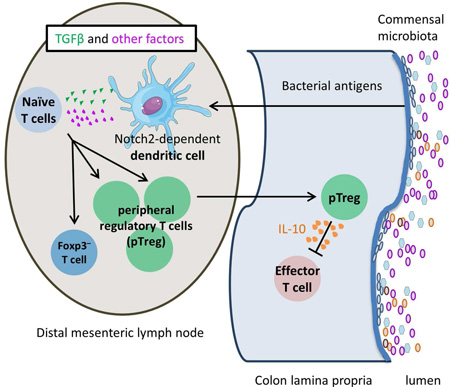
Summary
Commensal bacteria shape the colonic regulatory T (Treg) cell population required for intestinal tolerance. However, little is known about this process. Here, we use the transfer of naïve commensal-reactive transgenic T cells expressing colonic Treg TCRs to study peripheral Treg (pTreg) cell development in normal hosts. We found that T cells were activated primarily in the distal mesenteric lymph node. Treg cell induction was rapid, generating >40% Foxp3+ cells one week post-transfer. Contrary to prior reports, Foxp3+ cells underwent the most cell divisions, demonstrating that pTreg cell generation can be the dominant outcome from naïve T cell activation. Moreover, Notch2-dependent but not Batf3-dependent, dendritic cells were involved in Treg cell selection. Finally, neither deletion of the CNS1 region in Foxp3, nor blockade of TGFβ-receptor signaling, completely abrogated Foxp3 induction. Thus, these data show that pTreg cell selection to commensal bacteria is rapid, robust, and may be specified by TGFβ-independent signals.
Graphical abstract
Introduction
The trillions of commensal bacteria that live within the intestines play important roles in host health, including metabolism of food and defense against pathogens, but immune responses to commensal bacteria may be harmful and lead to inflammatory bowel disease (Elson and Cong, 2012; Jostins et al., 2012). For example, germ-free mice are highly resistant to murine models of inflammatory bowel disease, suggesting that bacteria are required to drive intestinal inflammation. In normal mice, an essential component of intestinal tolerance to gut bacteria is CD4+Foxp3+ regulatory T (Treg) cells (Belkaid and Hand, 2014; Maloy and Powrie, 2011). However, the process by which Treg cells are generated in response to commensal bacteria is unclear.
One controversy is whether the colonic Treg cell population is primarily generated in the thymus (tTreg) or in the periphery (pTreg) from naïve T cells. Some studies have favored pTreg cell differentiation. For example, putative markers of tTreg cells such as Helios and Nrp-1 are frequent on colonic Treg cells in germ free, but not conventionally housed, mice (Atarashi et al., 2011; Weiss et al., 2012). Introduction of Clostridial or other bacterial species into germ-free mice increased colonic Treg cells and lowered the frequency of Helioshi cells, consistent with pTreg cell induction (Atarashi et al., 2011; Geuking et al., 2011). Moreover, deletion of the conserved nucleotide sequence (CNS1) portion of Foxp3 resulted in a defect in pTreg cell selection, with the loss of Treg cells in the intestines and eventual colitis (Josefowicz et al., 2012; Schlenner et al., 2012; Zheng et al., 2010). Finally, our analysis using a fixed T cell receptor-β (TCRβ) model suggested that colonic Treg TCRs are unable to facilitate tTreg cell selection (Lathrop et al., 2011). However, a recent study favored tTreg cell generation to commensal bacteria also using a limited TCR repertoire approach (Cebula et al., 2013). As they observed changes in the colonic Treg TCR repertoire after antibiotics, they concluded that commensal bacteria induce the proliferation or retention of tTregs reactive to bacterial antigens. Similarly, bacteria-derived short chain fatty acids (SCFA) may act by promoting the expansion of pre-existing Treg cells in the gut (Smith et al., 2013). Thus, the origin of colonic Treg cells is unresolved.
Although TGFβ is thought to be critical for pTreg cell selection to commensal antigens, this has not been carefully studied in vivo. The importance of TGFβ was suggested by the observation that TGFβ alone is sufficient for induction of Foxp3 in vitro (Chen et al., 2003). Moreover, transgenic (Tg) expression of a dominant negative TGFβRII (dnTGFβRII) blocks both in vitro and in vivo generation of pTreg cells (Kretschmer et al., 2005), and results in the development of spontaneous colitis (Gorelik and Flavell, 2000), consistent with a defect in pTreg cell selection. As TGFβ levels are increased in the intestines relative to other tissues, it has been proposed that TGFβ is a specification factor that directs naïve T cells into the Treg cell lineage in the gut (Konkel and Chen, 2011).
Finally, the role of dendritic cell (DC) subsets in colon Treg cells is not well established. The CD103+ DC subset has been associated with the induction of Treg cells in the intestine (Coombes et al., 2007), and supported by a recent study using human Langerin-DTA BATF3−/− mice to deplete this subset in vivo (Welty et al., 2013). However, it is unclear whether the decrease in total Treg cell numbers with DC subset depletion are due to decreased responses to commensal bacteria.
To address these questions regarding pTreg cell selection to commensal bacteria, we generated two TCR Tg lines that express naturally occurring Treg TCRs (Lathrop et al., 2011). Using adoptive transfer of naïve TCR Tg cells into normal lymphoreplete hosts, we analyzed the kinetics and localization of T cell activation, proliferation, and Treg cell selection. We also examined the role of specific factors in pTreg cell generation such as the CNS1-region of Foxp3, dendritic cells, and TGFβ signaling.
Results
TCR transgenic models for studying peripheral Treg cell selection
The conflict over the source of the colonic Treg cell population may be attributed to the different indirect approaches used to address this question, including TCR repertoire analyses (Cebula et al., 2013), assessments of thymic selection (Lathrop et al., 2011), and the use of putative markers of tTreg cells (Atarashi et al., 2011). We reasoned that a direct analysis using TCR Tg T cells, an approach used previously to study tTreg cell selection (Bautista et al., 2009; Leung et al., 2009), may be useful for understanding the process of colonic Treg cell selection.
We generated TCR Tg lines expressing the microbiota-dependent colonic Treg TCRs CT2 and CT6 (Lathrop et al., 2011). tTreg cells were not detected by routine flow cytometric analysis of CT2/CT6 TCR Tg mice (Figure S1A–B and (Lathrop et al., 2011)), consistent with the lack of tTreg cell selection upon retroviral expression of these TCRs in thymocytes (Lathrop et al., 2011). We did observe Treg cells in the periphery of these mice, with increased numbers in the colon (Figure S1B), consistent with the anatomic distribution of these TCRs in the repertoire (Lathrop et al., 2011). However, the majority of T cells in the secondary lymphoid tissues of these TCR Tg mice were phenotypically naïve (CD44loCD62Lhi Foxp3−, Figure S1C) and therefore suitable for adoptive transfer experiments.
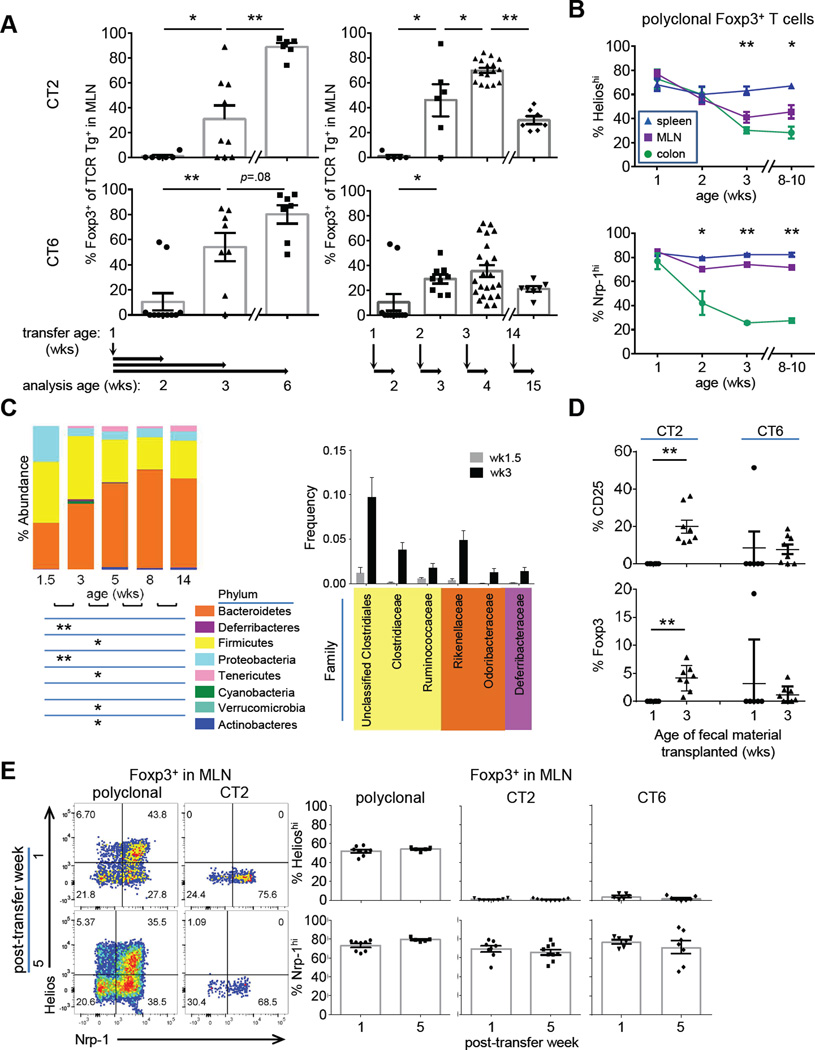
To determine when during ontogeny CT2/CT6 mediate pTreg cell selection, we injected naïve TCR Tg cells into congenically marked 1 wk old lymphoreplete hosts. It took 2 wks before we observed substantial frequencies of Foxp3+ CT2 or CT6 T cells in the mesenteric lymph nodes (MLN) (Figure 1A, left). Treg cell frequency continued to increase by 5 wks after transfer such that typically over 80% of TCR Tg cells were Foxp3+. Thus, transfer of naïve CT2/CT6 TCR Tg cells into normal mice results in robust pTreg cell selection around the age of weaning.
Figure 1. A TCR transgenic model for peripheral Treg cell selection to commensal antigens.
(A) Effect of host age on Treg cell selection. Left, sorted naive (CD44loCD62LhiCD4+) CT2/CT6 Tg cells (5×104) were injected into 1 wk old congenically marked Foxp3gfp lymphoreplete mice. The percentage of transferred cells that become Foxp3+ in the MLN was analyzed by flow cytometry at the indicated time point. Right, naïve Tg cells were injected into 1,2, 3 or 14-wk-old hosts and analyzed 1 wk later (expt=2–3, n=2+). (B) Changes in putative tTreg cell markers with age in polyclonal Treg cells. Foxp3gfp mice were analyzed by Nrp-1 or intracellular staining for Helios at different ages (expt=2–3, n=4–6). (C) Effect of host age on bacterial composition in the gut. 16s rDNA was analyzed in terminal fecal pellets from 1.5–14wk old mice. Changes in phyla (left) and selected families (right) are shown. Families shown on right are those that increase from 1.5 to 3 wk old mice by ≥1% (5–15 mice/age). BH adjusted Mann-Whitney U p-values are used. (D) Transplant of fecal material from older mice can induce CT2/CT6 Treg cell development in neonatal mice. One wk old mice were gavaged with fecal material from 1 or 3 wk old mice concurrent with i.p. injection of 2.5–5×104 each of CT2/CT6 Tg cells. TCR Tg cells were analyzed by flow cytometry for CD25 and Foxp3 at 3 days (expt=2, n=3). (E) Expression of Helios and Nrp-1 on Tg cells. Naïve Tg cells were injected into 3–4 wk old mice and analyzed for Helios and Nrp-1 expression after 1 and 5 wks. Representative FACS plots and summary graphs show Tg+ Treg cells versus endogenous polyclonal T cells from the MLN (expt=2–3, n=2–5). See also Figure S1.
There are several notable characteristics of these TCR Tg models. First, the antigen(s) that drive Treg cell selection are naturally occurring during normal gut physiology, in contrast with a different TCR Tg CBir reactive to commensal bacterial flagellin, where T cell activation required intestinal insult (Hand et al., 2012). Second, the injection of large numbers of cells decreased the efficiency of Foxp3-induction (Figure S1D), consistent with intraclonal competition for limiting antigen also seen in the thymus (Bautista et al., 2009; Leung et al., 2009). Studies of TCR Tg mice themselves could therefore affect the analysis of pTreg cell generation. Third, there are differences between CT2 and CT6 in the kinetics and efficiency of Foxp3-induction (Figure 1A), which might be predicted as they do not recognize the same epitope in vitro (Lathrop et al., 2011). While we consider CT2 and CT6 similar in that they represent two examples of colonic Treg TCRs, they should not be considered identical. Last, lymphopenia markedly alters the outcome of T cell selection. Transfer of naïve Tg cells into Rag1−/− hosts resulted in very low frequencies of Treg cells and favored effector cell differentiation (Figure S1E). Thus, the use of lymphopenic hosts to study intestinal tolerance, e.g. the classic Powrie transfer model, may not appropriately permit pTreg cell selection to establish immune homeostasis.
Age dependent effects on peripheral Treg cell selection
The lack of Foxp3+ Tg cells 1 wk after transfer of naïve cells into 1 wk old mice could be due to the age of the host. To address this, we transferred cells into mice of different ages and assessed Foxp3-induction after 1 wk. Mice greater than 2 wks old facilitated pTreg cell selection (Figure 1A, right; S1F–I), suggesting the environment of young mice is unable to support pTreg cell selection of CT2/CT6.
To determine whether the timing of Treg cell selection seen with CT2/CT6 is representative of a polyclonal population, we used Helios and Nrp-1, which have been reported to mark tTreg cells (Weiss et al., 2012; Yadav et al., 2012). In germ-free mice, most colonic Treg cells express tTreg cell markers (Atarashi et al., 2011; Geuking et al., 2011; Lathrop et al., 2011). Similarly, colonic Treg cells 1 wk after birth are mostly tTreg cells (Figure 1B). However, the ratio of tTreg/pTreg cells is reversed after 3 wks of age (Figure 1B), consistent with the CT2/CT6 data suggesting pTreg cell generation at that time (Figure 1A).
The change tTreg/pTreg proportions by these markers is associated with a major change in the composition of the gut microbiota around wk 3 of life (Pantoja-Feliciano et al., 2013), which we verified by 16S rDNA profiling (Figure 1C, S1J). To test the hypothesis that the absence of adult microbiota explains the lack of Foxp3 induction in young mice (Figure 1A), we performed fecal transplant from 1 or 3 wk old donors into 1 wk old hosts concurrent with i.p. transfer of naive CT2/CT6 cells. After 3 days, a portion of CT2/CT6 cells upregulated CD25 and Foxp3 in mice that received fecal material from 3, but not 1, wk old mice (Figure 1D), suggesting that young mice have the ability to present commensal antigens, but normally do not have the requisite microbiota for CT2/CT6. One mouse receiving 1 wk old fecal material showed substantial CD25 and Foxp3 induction in CT6, which we speculate results from stochastic colonization of young mice with bacteria recognized by CT6 (Figure 1A). Thus, data from both polyclonal and monoclonal TCR Tg studies suggest that there is a dramatic shift in the Treg cell populations in the MLN and colon around the period of weaning associated with marked changes in microbial composition coinciding with pTreg cell selection.
Colonic Treg TCR transgenic cells express low level of Helios, but not Nrp-1
The use of Helios and Nrp-1 in denoting tTreg cells has been controversial (Akimova et al., 2011; Gottschalk et al., 2012; Haribhai et al., 2011). We therefore assessed these markers on CT2/CT6 TCR Tg cells after induction of Foxp3. Whereas we observed few Helioshi cells at 1 or 5 wks after transfer, a substantial proportion of Tg Treg cells express Nrp-1 (Figure 1E). This difference is consistent with the higher frequency of Nrp-1hi vs. Helioshi subset in polyclonal MLN Treg cells (Figure 1B). Thus, our data using Tg cells suggest that Helioslo is better correlated with colonic pTreg cells than Nrp-1lo, and supports the notion that Helioslo may be a useful, albeit imperfect, surrogate for pTreg cells during homeostasis.
Initial naïve T cell activation is rapid and first seen in the distal mesenteric lymph node
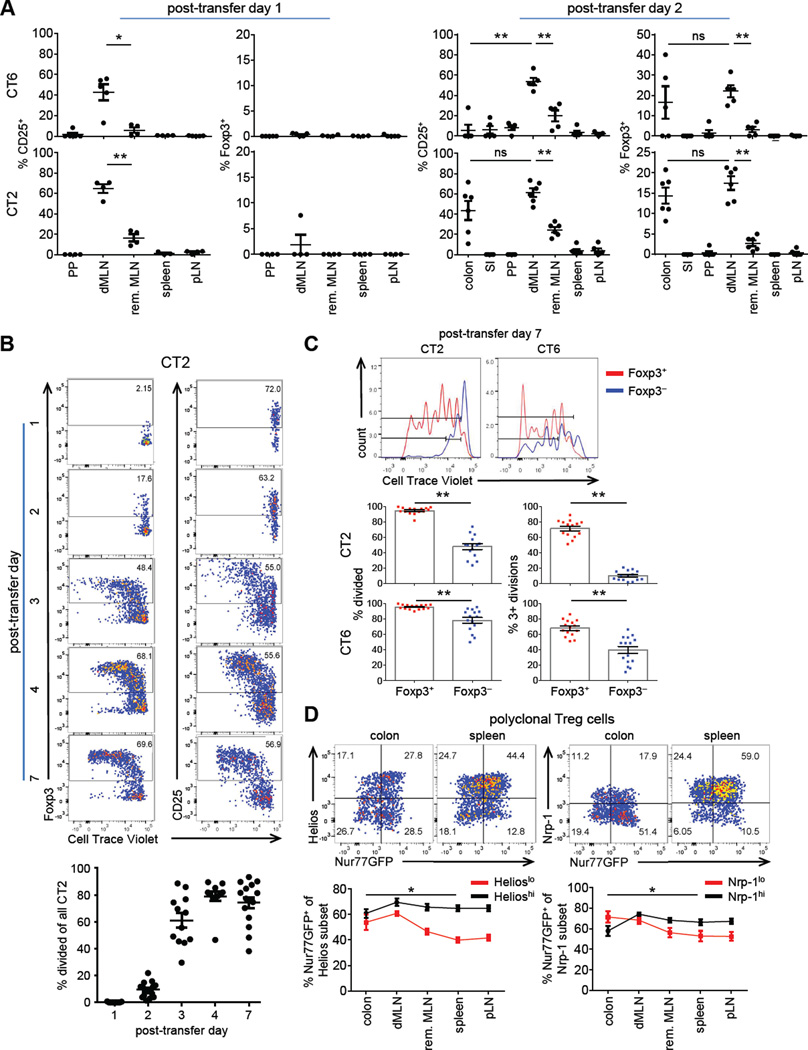
It is unknown if the site of initial activation of commensal antigen-specific T cells is the same as oral food antigens (Worbs et al., 2006). We therefore examined the kinetics of T cell activation and Foxp3 induction of transferred naïve CT2/CT6 cells in various gut associated lymphoid tissues. We first detected T cell activation as evidenced by CD25 upregulation 1 day after transfer primarily in the most distal MLN (dMLN) nearest to the cecum (Figure 2A, S2A), which drains the cecum and descending colon (Mowat and Agace, 2014). Notably, CD25+ CT2/CT6 cells were not enriched in Peyer’s patches, a site important for IgA responses to commensal bacteria (Corthesy, 2013).
Figure 2. Kinetics and biogeography of T cell activation of CT2 and CT6.
(A) Analysis of early time points after transfer of naïve CT6/CT2 T cells. Naïve Tg cells were injected into 3–4 wk old mice and analyzed after 1–2 days by flow cytometry for CD25 or Foxp3IRES-GFP (expt=2, n=2–4). Tissues: SI, small intestine; PP, Peyer’s patch; dMLN, distal MLN; rem. MLN, remaining MLN; pLN, peripheral LN (pooled axial, brachial and inguinal). (B,C) Proliferation of CT2/CT6 cells after transfer. CTV labelled naïve CT2/CT6 cells were injected into 3–4 wk old mice. Foxp3 expression and cell division were assessed by flow cytometry of dMLN at the indicated times. Representative FACS plots of CT2 (expt=2–4, n=2–5) (B), and of CT2/CT6 at day 7 after transfer (expt=4) (C) are shown. Data include points from Figure 1A. In (C), the percentage cells that have undergone any (left) or 3+ divisions (right) by CTV are shown. (D) Increased TCR activation in colonic Treg cells during homeostasis. T cells were analyzed from 6–8 wk old mice Tg for Nur77-GFP, a reporter for recent TCR activation. Nrp-1hi and Helioshi were used as markers for tTreg cells. Numbers in representative FACS plots of Foxp3+ cells indicate frequency in the quadrant. Summary data are the percentage Nur77GFP by Helios or Nrp-1 subset. Asterisk indicates significance in Nrp1lo or Helioslo colon versus spleen (expt=2, n=1–2). See also Figure S2.
By 2 days post-transfer, we observed Foxp3IRES-GFP in CT2/CT6 cells in the dMLN and colon (Figure 2A; S2B–C). However, the potential for T cell trafficking makes the primary site of pTreg cell differentiation uncertain. The relative frequency of CT2/CT6 cells amongst total CD4+ T cells was also increased in the dMLN by day 2 (Figure S2D), suggesting local retention or proliferation. Thus, T cell activation to commensal antigens and Treg cell selection is rapid and easily detected in a specific anatomic location, the distal MLN.
Colonic Treg cells appear chronically stimulated during homeostasis
Previous data examining pTreg cell generation to non-commensal antigens showed that Foxp3 induction could occur within 4 days after antigen recognition in vivo (Gottschalk et al., 2010; Kretschmer et al., 2005; Weissler et al., 2015). Notably, Foxp3+ cells were preferentially found among cells that showed fewer cell divisions in these experiments, suggesting that pTreg cell selection represented an alternative cell fate of naïve T cell activation. To address whether this happens in commensal-dependent responses, Cell Trace Violet (CTV) labelled naïve CT2 Tg cells were transferred into congenically marked 3–4 wk old mice. Tg cell proliferation in the dMLN began on day 2 along with appearance of Foxp3+ cells (Figure 2B). However, many Foxp3+ cells had not diluted CTV, suggesting that cell division is not required for upregulation of Foxp3. At later time points, a higher percentage of Foxp3+ than Foxp3− cells had divided (Figure 2C), although there were differences in proliferation between CT2 and CT6 cells reflective of the differences in Foxp3+ frequency (Figure 1A). In contrast with previous studies, our data demonstrate that pTreg cell generation can be the dominant outcome after naïve T cell activation in the periphery.
The extensive proliferation (Figure 2B,C) and ability of Tg cells to be activated at different host ages (Figure 1A) imply that intestinal Treg cells are constantly exposed to commensal antigens during immune homeostasis. To assess whether colonic Treg cells are activated in a polyclonal population, we utilized a Nur77GFP BAC-Tg line as an in vivo marker for TCR activation (Moran et al., 2011). In the colon and dMLN, ~55–60% of Helioslo Treg cells are Nur77GFPhi, whereas this is seen in only ~40% of Helioslo Treg cells in non-gut-associated tissues such as the spleen (Figure 2D). By contrast, Helioshi Treg cells show only minor changes in the frequency of Nur77GFPhi cells with location. A similar pattern is seen in Nrp-1lo and Nrp-1hi cells. Together with the TCR Tg studies (Figure 2A), these data suggest that pTreg cells are enriched in the colon due to chronic activation by commensal antigens.
Induction of IL-10 reporter occurs post Treg cell selection
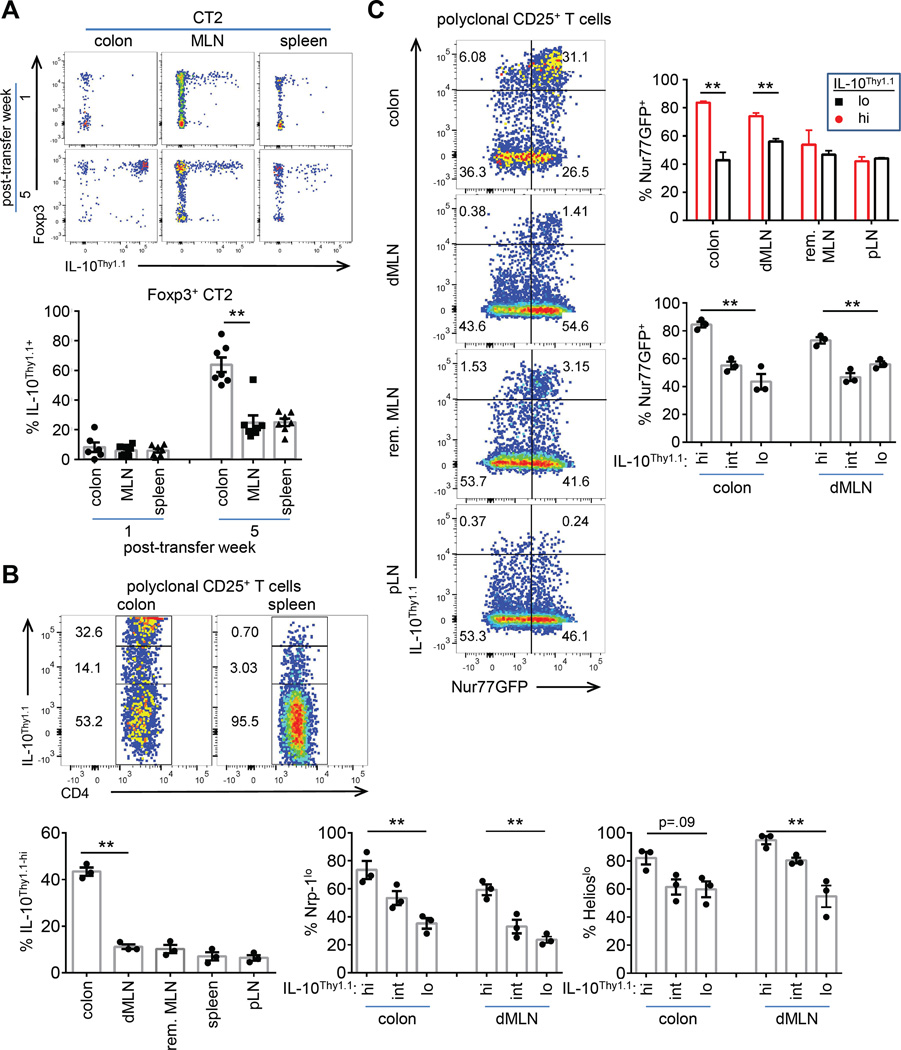
An important effector molecule produced by intestinal Treg cells is IL-10. We used the 10BiT reporter, in which Thy1.1 expression reflects current and historic IL-10 expression (Maynard et al., 2007). We observed that IL-10 reporter expression substantially lagged that of Foxp3 expression in CT2 cells (Figure 3A, S3A–C). In preliminary experiments, similar results were observed with CT6 in the MLN (not shown), but IL-10+ frequencies in the colon were difficult to assess due to lower cell recoveries (Figure S1I). These data suggest that the potential for IL-10 expression is induced post-Treg cell selection and that Foxp3− IL-10-expressing Tr1 cells are not commonly generated from naïve CT2 cells.
Figure 3. IL-10 reporter is induced in the colon post Foxp3 induction.
(A) Induction of IL-10 reporter on CT2 cells. Naïve 10BiT CT2 Tg cells were injected into 3–4 wk old mice and analyzed at the indicated time points. Representative FACS plots and summary of 10BiT in Foxp3+ cells are shown (expt=2, n=2–4). (B,C) Analysis of polyclonal CD25+ Treg cells for IL-10 reporter (B) and correlation with TCR activation (C). 2–3 month old 10BiT Nur77GFP mice were analyzed by flow cytometry (expt=2, n=1–2). Representative FACS plots and summary plots are shown (B,C). The frequency of Helioslo/Nrp-1lo by IL-10Thy1.1 expression is show in (B). The frequency of Nur77GFPhi by IL-10Thy1.1 expression in the CD25+ subset is shown (C). See also Figure S3.
At 5 wks, we observed that the frequency of IL-10+ cells amongst CT2 Treg cells were increased in the colon, and that the level of IL-10 reporter expression was also much higher (Figure 3A, S3A). Consistent with the CT2 data, we found that IL-10 reporter expressing CD4+CD25+ Treg cells in polyclonal populations is enriched in the colon (Figure 3B). Moreover, we observed that the IL-10+ population is enriched in the polyclonal pTreg cell subset using Helioslo or Nrp-1lo markers (Figure 3B), in agreement with previous data (Atarashi et al., 2011). Finally, we found that IL-10hi Treg cells in the colon are mostly Nur77GFPhi compared to cells in peripheral lymph nodes (Figure 3C). The CT2 and polyclonal data are therefore consistent with the hypothesis that many of the IL-10 producing CD4+ T cells in the colon are pTreg cells that recognize antigens presented during homeostasis.
Tuning of pTreg cell numbers to commensals
In our previous study, we showed several lines of evidence supporting the colonic commensal antigen reactivity of CT2/CT6, including in vitro reactivity to an unclassified Clostridium species for CT6 (Lathrop et al., 2011). We therefore tested whether CT2/CT6 cells could respond in vivo to a consortia of Clostridia species maintained in gnotobiotic mice (Stefka et al., 2014). However, we did not observe CD25, CD44, or Foxp3 upregulation in CT2/CT6 cells 1 wk after naïve cell transfer into either germ free or Clostridia colonized mice (4–5 mice per group, data not shown).
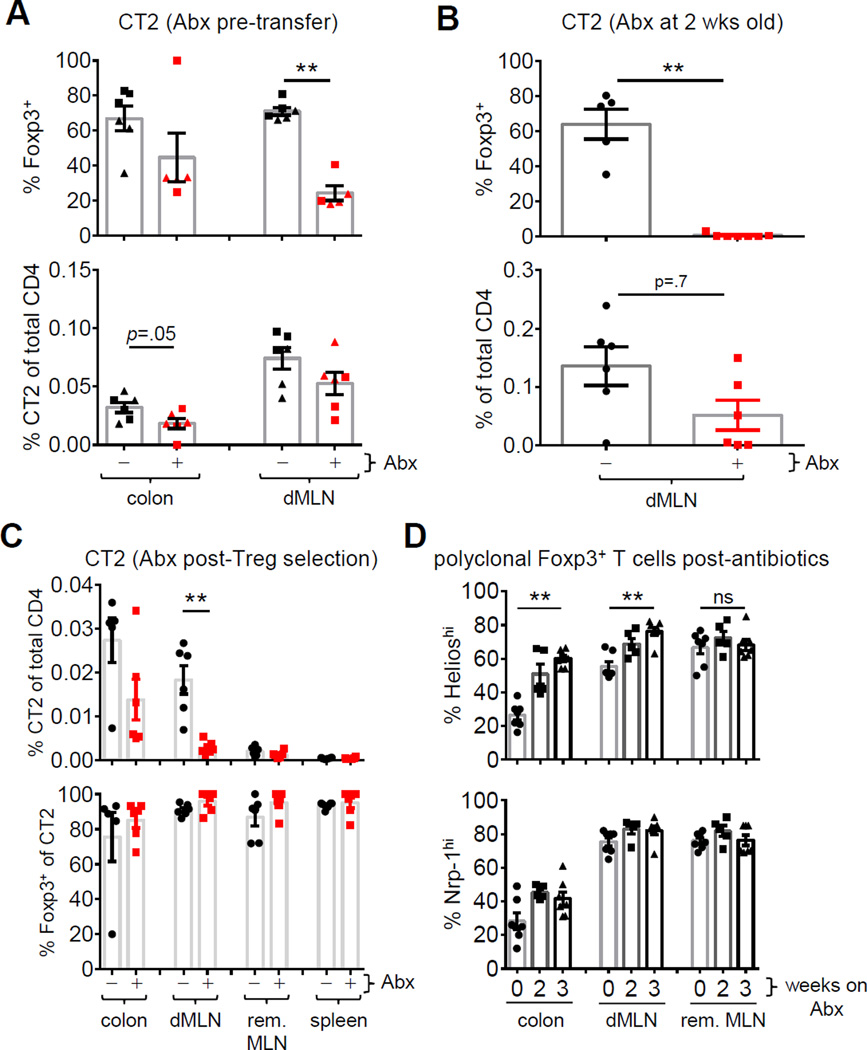
We then asked whether microbial depletion with antibiotics would affect the development of CT2/CT6. 3 wk old mice started on ad libitum vancomycin, ampicillin, metronidazole, and neomycin (VAMN) (Rakoff-Nahoum et al., 2004) showed a partial inhibition of Treg cell selection and expansion of transferred naïve CT2 cells (Figure 4A). For CT6, we found ad libitum clindamycin and streptomycin was more effective at blocking Treg conversion and proliferation (Figure S4A) than VAMN in preliminary studies (not shown), consistent with the notion that CT2 and CT6 recognize different bacteria. No individual antibiotic was responsible for the activity in preliminary experiments (not shown). The incomplete blockade of Treg induction suggests the antigens were only partially eliminated, as seen in the incomplete blockade of Th17 development with vancomycin (Ivanov et al., 2009). Thus, the activation and Treg selection of CT2/CT6 can be at least partially blocked by antibiotics.
Figure 4. Antibiotics affect pTreg cell induction of CT2 but not stability of Treg cell percentage.
(A,B) Effect of antibiotics on CT2 pTreg cell selection at weaning (A) or before (B). In (A), weaned mice were treated with VAMN in drinking water for 5 (squares) or 14 days (triangles) prior to transfer of naïve CT2 Tg cells. One wk later, CT2 cells were analyzed by flow cytometry for Foxp3 expression and the percentage of CT2 cells within the CD4 population. In (B), 2 wk old mice were gavaged with VAMN for 1 wk prior to transfer of CT2. (C) Effect of antibiotics on differentiated CT2 pTreg cells. Naïve CT2 cells were transferred into 3 wk old mice and allowed to develop into Treg cells for 2 wks. Mice were then treated with VAMN in drinking water for 3 wks and then analyzed by flow cytometry. Data shown are the percentage of CT2 cells amongst the entire CD4 population and the frequency of Foxp3+ cells within CT2 cells (expt=2, n=2+). (D) Effect of antibiotics on polyclonal Treg cells. Foxp3gfp mice were treated with VAMN in drinking water for 2–3 wks before assessing the expression of Nrp-1 and Helios in Foxp3+ cells (expt=3, n=2–3). See also Figure S4.
Antibiotic gavage of pre-weaning mice has been reported to more result in more complete bacterial elimination (Stefka et al., 2014). We tested this protocol on CT2 as: (1) it had higher frequencies in the colon (Figure 4A, S4A); and (2) an oral antibiotic cocktail similar to VAMN had been used at 2 wks of life (Stefka et al., 2014). We found that treatment of younger mice blocked the induction of Foxp3+ cells (Figure 4B), as well as decreased the percentage of CT2 cells within the total CD4 population, suggestive of poor T cell expansion (Figure 4B). Moreover, it drastically altered microbial composition and diversity as assessed by 16S rDNA sequencing (Figure S4B–C). Thus, these data show that it is possible with the appropriate protocol to abrogate Treg cell selection of CT2 with antibiotics.
Since antibiotics are commonly used in humans, we asked if antibiotics could affect the existing pTreg cell population. We injected naïve CT2 Tg cells into 3 wk old mice, allowed them to develop into pTreg cells, and then treated the mice with antibiotics for 3 wks. Because of the length of the experiment, the ad libitum protocol was used. Although the frequency of Foxp3+ cells amongst the TCR Tg population did not decrease (Figure 4C), the frequency of total CT2 Tg cells was reduced–indicative of decreased commensal antigen stimulation of CT2 (Figure 4C). The polyclonal T cell population also showed decreases in colonic Nrp-1lo and Helioslo pTreg cell fraction after antibiotics consistent with the overall decrease in CT2 cells (Figure 4D). Thus, these data suggest that the gut Treg cell population is maintained during homeostasis by continuous commensal bacterial antigen stimulation.
pTreg cell selection in older mice is associated with lower Foxp3 and CD25 expression
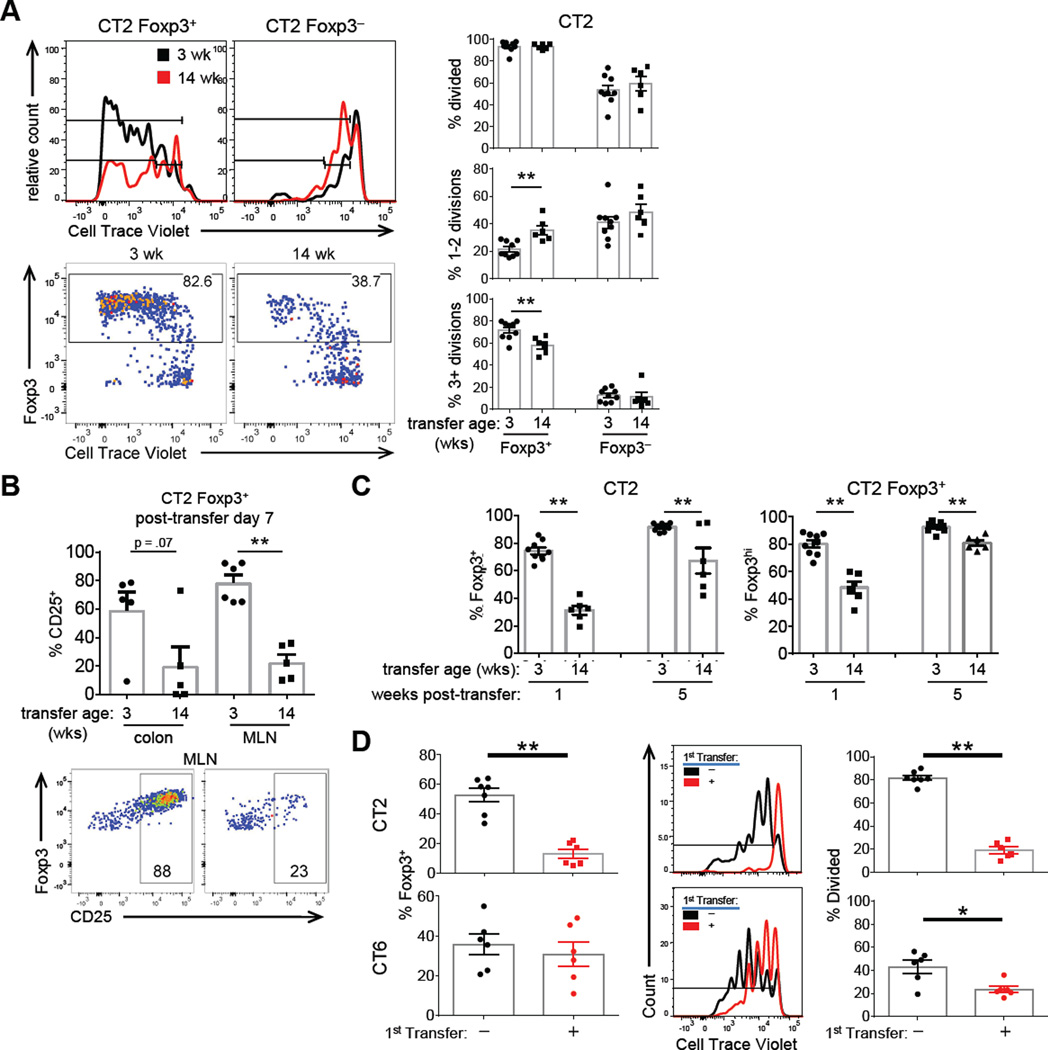
pTreg selection efficiency increased from 1 to 3 wk old mice (Figure 1A), but decreased in 8–14-week-old adult mice (Figure 1A, right; S5A). CT2 cells also proliferated less in older mice (Figure 5A), and had a relative increase of Foxp3int CD25lo cells amongst the CT2 Foxp3+ population (Figure 5B–C). Yet, CT2 cells become mostly Foxp3+ by 5 wks after transfer (Figure 5C), suggesting that older mice are kinetically slower in their ability to support pTreg cell selection.
Figure 5. Effects of host age on Foxp3 induction and maintenance.
(A–C) Treg cells generated in older mice show lower proliferation, Foxp3, and CD25 expression. Naïve CTV labelled CT2 Tg cells were injected into 3 or 14 wk old mice. CT2 dMLN cells were assessed for Foxp3 expression and cell division at 1 wk (A), with representative FACS plots on the left, and cell divisions summarized on the right (expt=2+, includes data in Figure 1A,2C). CD25 expression is shown in (B). Foxp3 percentage and expression level at 1 and 5 wks after transfer is summarized in (C) (expt=2+, n≥2). (D) Pre-existing cells can inhibit subsequent pTreg selection. 3–4 wk old littermates were injected with PBS or naïve CT2 or CT6 cells. One wk later, congenically marked naïve CTV labelled CT2/CT6 cells was injected into all hosts. 4 days later, cells from the second transfer were analyzed for Foxp3 expression and CTV dilution (expt=2, n=2–4). See also Figure S5.
One possible explanation is decreased bacteria in older mice. However, in vitro stimulation with fecal antigens from 3–14 wk old mice caused equivalent CD25 upregulation (Figure S5B), suggesting similar antigen concentration in the lumen. Another possibility is the Treg niche is already filled with endogenous cells, leading to decreased conversion of naïve cells. Consistent with this hypothesis, we found that pre-existing CT2 cells decrease the efficiency of Treg cell selection after a second transfer of naïve CT2 cells (Figure 5D). Pre-existing CT6 cells decreased proliferation, but not Foxp3-induction of newly transferred naïve CT6 cells (Figure 5D), consistent with the milder effect of older age on CT6 pTreg generation (Figure 1A, right). Although these data do not exclude other possibilities such as changes in the consortia resulting in antigen-independent effects (e.g. SCFA or other metabolites), they suggest inhibition by pre-existing antigen-specific Treg cells is one possible mechanism for the decreased Foxp3-induction and proliferation seen in older 14 wk old mice (Figure 1A, 5D).
TGFβ is not a “master specifying” factor for pTreg cell generation
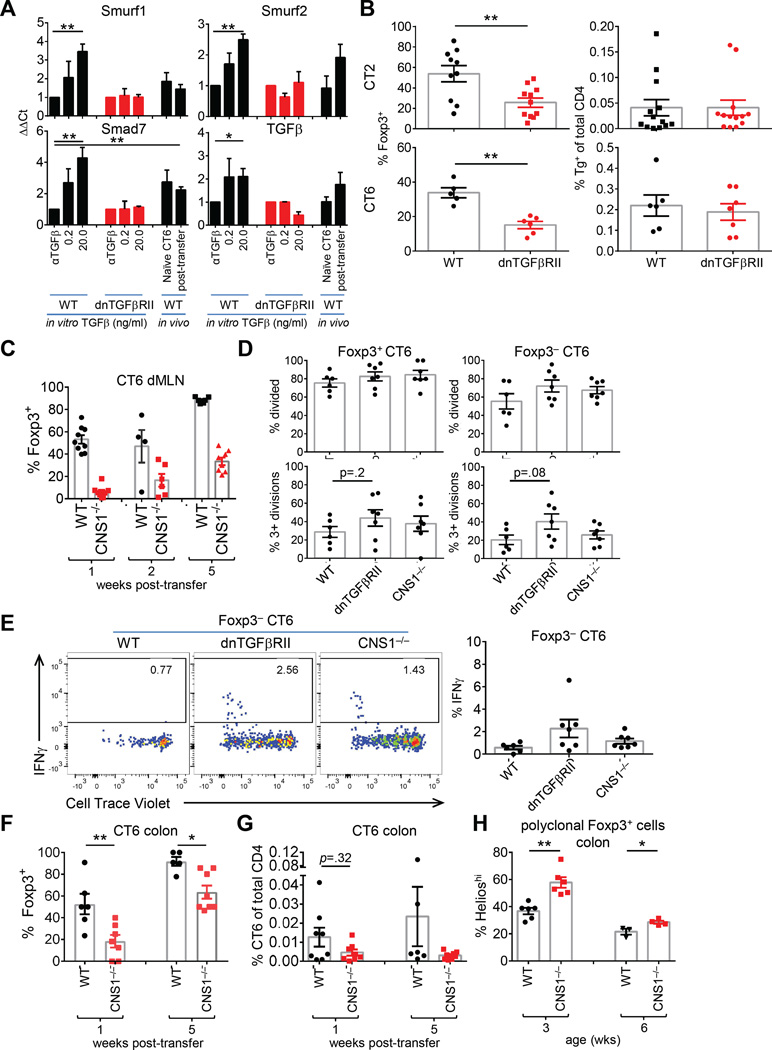
TGFβ is generally accepted as a critical signal for pTreg cell selection in the gut (Konkel and Chen, 2011). However, in vivo studies analyzing the role of TGFβ in Treg cell development to intestinal commensal antigens have not been performed. To address this, we bred the CT6 TCR Tg line to CD4-cre Tgfbr2fl/fl mice, and observed a partial decrease in Treg cell selection in vivo (Figure S6A). Although we observed the reported lethality in the non-TCR Tg littermates around 3–4 wks of age (Marie et al., 2005), substantial TGFβ-dependent Foxp3 induction still occurred in vitro (Figure S6B), suggesting that CD4-Cre Tgfbr2fl/fl incompletely abrogated TGFβ signaling or affected TGFβ signaling differently than dnTGFβRII (Ishigame et al., 2013).
We therefore bred CT2 and CT6 to the dnTGFβRII Tg line, which diminishes in vivo pTreg cell generation by 20-fold (Kretschmer et al., 2005). We confirmed that dnTGFβRII inhibits by 100–1000 fold the ability of exogenous TGFβ to induce Foxp3 in vitro (Figure S6C) and blocked the in vitro induction of TGFβ-responsive genes (Figure 6A) (Feuerer et al., 2010; Hill et al., 2008). Consistent with notion that gut T cells receive TGFβ signals in vivo, we found that a subset of TGFβ responsive genes were upregulated in CT6 cells (Figure 6A).
Figure 6. Partial reduction of Treg cell selection with impaired TGFβ receptor signaling and CNS1-deficiency.
(A) Analysis of TGFβ dependent genes. For in vitro experiments, sorted naïve polyclonal T cells were stimulated with anti-CD3/CD28 with the indicated TGFβ condition. After 2 days, TGFβ-responsive genes (Table S1) were analyzed by qPCR. For in vivo experiments, 1.5×105 naïve CT6 cells were injected into WT mice. After 2 days, CD25+Foxp3− cells were sorted and gene expression assessed by qPCR. Data represent fold-change over in vitro αTGFβ condition (in vitro expt=2, n=1; in vivo expt=5, n=1 for naïve and n=1–2 post-transfer of 4–5 pooled mice). (B) Treg cell selection with impaired TGFβ receptor signaling. Naive CT2 (5×104) or CT6 (2×105) cells on a WT or dnTGFβRII background were injected into 3–4 wk old mice, and analyzed 1 wk later. Data show frequency of Foxp3+ cells amongst CT2/CT6 Tg cells in the dMLN (left), and the percentage of Tg cells in the total CD4+ population (right). (C) Impaired Treg cell selection in CNS1-deficient CT6 cells. Naïve CT6 Tg cells (WT or CNS1−/−) were injected into 3–4 wk old mice. Cells from the dMLN were analyzed at the indicated time point (expt=2–3, n=2–4). (D) Analysis of proliferation. The percentage of divided cells with the indicated mutations is shown (expt=2, n=2–4 pools of 2 mice). (E) Assessment of IFNγ expression. Naïve CT6 Tg cells (2×105) (WT, dnTGFβRII or CNS1−/−) were stained with CTV and injected into 3–4 wk old hosts. 1 wk later, cells were stimulated with PMA/Ionomycin for intracellular staining. Representative plots are shown (left) and summarized (right) (expt=2, n=2–4 pools of 2 mice). (F–G) CNS1-independent Treg cell selection of CT6 cells. (F) Naïve WT or CNS1−/− CT6 Tg cells were injected into 3–4 wk old mice, and the colons analyzed 1 or 5 wks later for the frequency of Foxp3+ CT6 cells (F) and of CT6 cells amongst the total CD4+ T cell population (G) (expt=3, n=1–3). (H) Helios expression in polyclonal colonic CNS1−/− Treg cells with age. CNS1−/− mice were cross-fostered to limit variability in microbiota (expt=2–3, n=1–3). See also Figure S6–7.
Contrary to the in vitro inhibition of Foxp3 (Figure S6C), dnTGFβRII only led to a relatively modest 50% reduction of CT6 Foxp3+ cells in vivo at 3 or 7 days post-transfer in comparison to WT CT6 cells (Figure 6B, S6D). We did not find evidence that hyper-expansion of a small fraction of Foxp3+ dnTGFβRII cells was responsible for the higher than expected Foxp3+ frequency (Figure S6E). As the dnTGFβRII is a hypomorph (Figure S6C), we cannot exclude the possibility that low levels of TGFβ signals are sufficient for Foxp3 induction in vivo. However, our data suggest that blockade of TGFβ signals by >100 fold has a disproportionately small effect on pTreg generation in vivo compared with in vitro, arguing that TGFβ is unlikely to be the singular, or “master,” signal that specifies a naïve T cell to upregulate Foxp3 in vivo.
CNS1 is important for initial, but not late, peripheral Treg cell selection
TGFβ mediates its effects via the activation of SMAD transcription factors which can bind to the promoter and CNS1 enhancer region in the Foxp3 locus (Schlenner et al., 2012; Zheng et al., 2010). We therefore asked whether CNS1-deficiency would phenocopy the inhibition of TGFβ receptor signaling on Treg cell selection. However, CNS1-deletion had a much greater effect on Treg cell selection of naïve CT6 cells (Figure 6C), consistent with prior reports (Josefowicz et al., 2012; Zheng et al., 2010).
The reduction of Foxp3+ cells was not due to effects on T cell activation, as there were no differences in proliferation among the Foxp3+ or Foxp3− cells lacking CNS1 (Figure 6D). There was a trend towards increased proliferation of both Foxp3+ and Foxp3− dnTGFβRII cells (Figure 6D). Despite this, few of the Foxp3− dnTGFβRII or CNS1−/− CT6 cells developed the ability to produce IFNγ or IL-17 in 1 wk (Figure 6E, S7A). None of the Foxp3+ cells expressed IFNγ, and the frequency of CT6 Tg cells remained the same (Figure S7A). We did note a decreased frequency of CNS1−/− CT6 cells in the colon, suggesting that the block in Treg cell selection may result in decreased cell survival or trafficking (Figure 6F–G). Thus, these data suggest that CNS1 is essential for the rapid induction of Foxp3 in response to intestinal commensal antigens.
Unexpectedly, a substantial proportion of CNS1−/− CT6 cells express Foxp3 by 2 wks increasing by 5 wks post transfer (Figure 6C,F), suggesting that CNS1 is not absolutely required for pTreg selection. Consistent with the CT6 data, polyclonal T cells in CNS1−/− mice showed increased Helioshi tTreg cell frequencies at 3, but not 6, wks of age (Figure 6H, S7B). Thus, it appears CNS1 primarily affects the kinetics of Foxp3 induction without completely altering cell fate.
Notch2-dependent Dendritic Cells are necessary for optimal pTreg cell generation
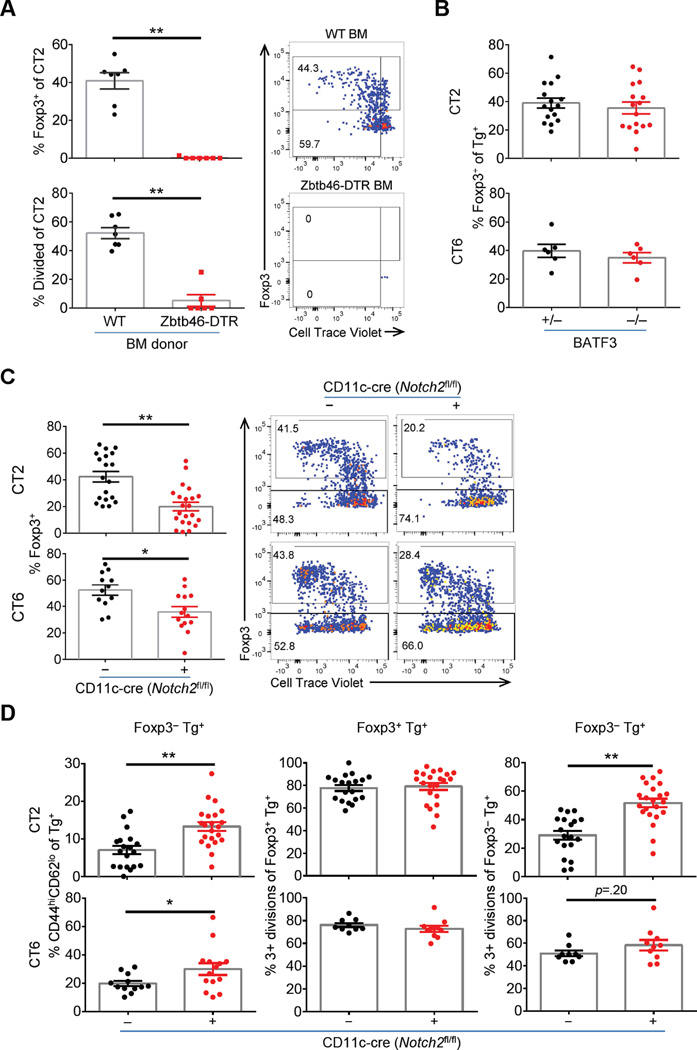
To determine whether DCs are primarily responsible for activating commensal specific T cells, we used bone marrow chimeras with Zbtb46-DTR to deplete all conventional DCs (Meredith et al., 2012). We tested CT2 cells as they show more efficient Treg selection (Figure 1A), and found conversion and proliferation of CT2 was markedly blocked with DT depletion of DCs (Figure 7A).
Figure 7. Role of Notch2-dependent DCs in Foxp3 induction to commensal antigens.
(A) Conventional DCs cells are necessary for activation of CT2. Bone marrow chimeras with WT or Zbtb46-DTR donors were generated as described in Supplemental Materials. CTV labelled naïve CT2 cells were injected into DC-depleted mice and analyzed after 1 wk (expt=2–3, n=2–5). (B) Loss of CD103+CD11b− DCs does not decrease pTreg cells. Naïve Tg cells were injected into BATF3+/− or −/− littermates and analyzed at 1 wk for Foxp3 (expt=2–3, n=1–3). (C–D) Notch2 dependent DCs are involved in pTreg cell generation. Naïve Tg cells were stained with CTV and injected into Notch2cKO mice. Foxp3 expression and representative FACS plots (C), and the summary of effector cell frequency (CD44hiCD62Llo) and cell divisions (D) are shown for dMLN cells 1 wk after transfer (expt=4, n=2–5).
In the gut, CD103+ DCs are reported to be the primary APC subset that induces pTreg cell selection in the MLN (Coombes et al., 2007; Sun et al., 2007). However, their role in pTreg generation to commensal bacterial antigens has not been tested in vivo. To address the role of CD103+ DCs, we used BATF3−/− mice to ablate CD103+CD11b− DCs (Hildner et al., 2008) and CD11c-cre Notch2fl/fl (Notch2cKO) mice to ablate CD103+CD11b+ DCs (Satpathy et al., 2013). Whereas the frequency of CT2/CT6 Foxp3+ cells appeared unaffected in BATF3−/− mice, we observed significant decreases in Treg cell selection in Notch2cKO hosts (Figure 7B–C). In Notch2cKO hosts, the frequency of Foxp3−CD44hiCD62Llo effector Tg cells and proliferation of Foxp3− Tg cells was increased (Figure 7D). We interpret this to indicate that Notch2-deficiency in DCs does not affect the overall antigen presentation and T cell activation of commensal-antigen reactive T cells, but rather skews the balance of differentiation from the Treg cell subset.
Discussion
Treg cell-mediated immune tolerance to commensal antigens is crucial for intestinal homeostasis. Using Tg lines that express TCRs derived from colonic Treg cells, we have characterized the kinetics, anatomy, and molecular mechanisms of Treg cell selection to commensal antigens during homeostasis in lymphoreplete animals. We make the following observations: (1) generation of Treg cells can be the dominant outcome of naïve T cell activation to commensal antigens; (2) this process is first seen in the distal mesenteric lymph node; (3) the acquisition of effector cytokines such as IL-10 occurs post-Foxp3 expression; (4) TGFβ does not appear to be the “master specifying” factor for Treg versus effector cell differentiation; (5) the CNS1-region in Foxp3 affects the kinetics of pTreg cell selection, but does not abrogate it; and (6) Notch2-deficiency in DCs skews T cell differentiation from Treg to effector cell subsets.
We found that pTreg differentiation from naïve T cells in response to commensal antigens was unexpectedly rapid and efficient, in contrast with previous studies (Gottschalk et al., 2010; Kretschmer et al., 2005; Weissler et al., 2015), which found pTreg cells in the least divided population, suggesting that they were a byproduct of the immune response. This difference may arise from the models used. First, we used activation in a mucosal tissue which may favor Treg cell selection (Sun et al., 2007). Second, our model utilizes endogenous antigen presentation of commensal antigens, rather than transient exogenous administration. Finally, the TCR affinity/avidity of CT2/CT6 for their antigens are unknown, and may fall within a range favorable for pTreg cell generation (Gottschalk et al., 2010). Though the response to food antigens may differ, a previous study also observed proliferation of Treg cells in OTII TCR Tg cells, though with a lower Foxp3+ frequency, in response to oral OVA (Hadis et al., 2011). Thus, our data show that naïve T cell responses can be dominated by the generation of pTreg cells.
We found the efficiency of Treg cell selection has a monophasic dependence on age with a peak around weaning, consistent with literature showing that early host-commensal interactions are important for immune homeostasis in the gut (Keeney et al., 2014; Stefka et al., 2014). The low, if any, pTreg development in young mice appeared related to the microbiota, which undergoes marked changes around the time of weaning (Pantoja-Feliciano et al., 2013). Another possibility which our studies do not address is that changes in the consortia of commensal species also affect pTreg cell generation via factors such as SCFA that are independent of TCR specificity (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). Additionally, it is possible that in older mice pre-existing adaptive responses to commensal bacteria may limit antigen access to the immune system (Peterson et al., 2007). We also observed that pre-existing T cell responses can limit naïve CT2/CT6 cell activation and CT2 Treg cell generation. In addition to these non-mutually exclusive possibilities, Treg cell selection may be affected by other changes in microbial, mucosal or immune physiology in very young or adult mice.
Analysis of early time points after T cell transfer allowed the identification of the primary site of T cell activation for CT2/CT6 to the dMLN. Prior mapping studies revealed that this lymph node drains the cecum and proximal colon (Mowat and Agace, 2014). Thus, these data suggest a pathway for commensal antigen uptake in the colon, perhaps via bacteria outer membrane vesicles (Hickey et al., 2015) or colonic goblet cell-associated passages (Knoop et al., 2015).
The TCR Tg transfer model also allows study of specific molecular and cellular events affecting commensal bacteria-specific T cells within a normal intestinal environment. For example, it has been postulated that TGFβ is a crucial cytokine for pTreg cell selection in the gut (Konkel and Chen, 2011). We were therefore surprised that dnTGFβRII showed a relatively modest (50%) effect on early Treg cell selection, as it was previously reported to markedly (90%) inhibit Treg cell selection in a different TCR Tg model (Kretschmer et al., 2005). Although dnTGFβRII is a hypomorph, our data showing a >100-fold blockade of TGFβ effects on Foxp3-induction in in vitro assays versus a 2-fold reduction in Foxp3+ cells in vivo suggest that either (1) TGFβ levels in vivo are well over 100-fold greater than that necessary for Treg cell specification; or that (2) TGFβ is not a singular master specifying factor for Treg cell selection in vivo. However, the first scenario, that TGFβ levels are excessively high in vivo, seems unlikely based on our analysis of TGFβ-dependent gene expression, as well as previous transcriptome analysis showing that the signature of TGFβ responsive genes was not enriched in Treg cells from the lamina propria versus other peripheral organs (Feuerer et al., 2010). Thus, our data support the hypothesis that TGFβ may not be the singular “master” specifying factor for pTreg cell selection in the gut, and suggests the involvement of additional factors.
One target downstream of TGFβ-dependent transcription factors is the CNS1 region in the Foxp3 locus (Zheng et al., 2010). Mice deficient in this region develop spontaneous colitis (Josefowicz et al., 2012) and those lacking the SMAD binding site in this region show decreased Treg cells in the gut (Schlenner et al., 2012). However, the colitis develops only in older mice, and was not initially observed on a mixed genetic background. Consistent with this data, we found a striking defect in initial, but not late, Treg cell selection, which may explain why CNS1-mice show less pathology than might be predicted if pTreg cell selection was completely abrogated (Haribhai et al., 2011).
Finally, our studies using BATF3−/− and NotchcKO mice suggest that CD103+ CD11b+ DCs play a role in Treg/Teffector cell selection in the gut, contrary to reports that removal of this DC subset in several models (IRF4, human Langerin-DTA, Notch2) does not affect total Treg cell numbers, but does lower Th17 cell numbers in the intestines (Lewis et al., 2011; Persson et al., 2013; Satpathy et al., 2013; Welty et al., 2013). The discrepancy could be due to the analysis of steady state Treg cell numbers, which may be less sensitive than a kinetic analysis of pTreg generation. Future studies are needed to investigate which DC subsets present to commensal-specific T cells, and the mechanisms by which APCs influence the effector-regulatory cell fate decision.
In summary, the use of TCR Tg lines expressing Treg TCRs has allowed us to evaluate the process of pTreg cell selection to commensal antigens, providing insight regarding the kinetics, efficiency, geography, molecular and cellular requirements. Future studies will be required to understand the molecular mechanism determining Treg versus effector cell differentiation, which may facilitate the development of treatments for diseases of disordered gut tolerance such as inflammatory bowel disease.
Experimental Procedures
Mice
CT6 (Lathrop et al., 2011) and CT2 Tg mice were generated as described (Bautista et al., 2009), with microinjection into B6 × 129 fertilized eggs and backcrossed 5+ generations to B6 background Foxp3IRES-GFP (Lin et al., 2007) and Rag1−/− mice obtained from Jackson Labs. Additional mice are described in supplemental material.
T cell transfers into normal hosts
Naïve (CD44lo or CD25−CD44loCD62Lhi) CD4+Foxp3− T cells were FACS purified from the MLN and spleen from CD45.2 Foxp3IRES-GFP Rag1−/− TCR Tg mice. 5×104 (typical, up to 5×105) cells were injected intraperitoneal (1 or 2 wk old) or retro-orbital (older hosts) into congenic CD45.1 Foxp3gfp mice. The entire colon lamina propria, small intestine lamina propria, Peyer’s patches, half of the spleen, mesenteric and peripheral lymph nodes were harvested at varying times and analyzed by flow cytometry. Transferred cells were identified as CD4+CD45.2+CD45.1−Vα2+. Data from colon samples with ≥10 TCR Tg cells were used, except in cases where the experimental condition typically resulted in <10 cells (e.g. Figures S1G, 2A, and 7A). In order to recover enough cells for qPCR or intracellular cytokine staining, mice in Figures 6A, 6D–E, and S7A were injected with additional cells (1.5–2×105).
Statistical analysis
Graphpad Prism v6 was used for used for statistical and graphical analysis. Student’s t-test was used for between-subjects analyses. Benjamini-Hochberg false discovery rate correction (padj) was used on Mann-Whitney U calculations for phyla and class comparisons (p.adjust, R v3.2). Unless otherwise indicated: each dot represents data from an individual host; bars indicate mean ± SEM. *p < .05, **p < .005, Student’s t-test. Experiments (expt) are performed independently with n replicates per experiment.
Additional procedures in supplemental procedures.
Supplementary Material
Acknowledgments
We thank Brian Kim (Wash.U.), A.Y. Rudensky (MSKCC), K. Hogquist (U. Minn.), and T. Egawa (Wash.U.), for critical reading of the manuscript and advice; A. Rudensky (MSK), and CT Weaver (UAB) for gifts of mice. Experimental support was provided by the Facility of the Rheumatic Diseases Core Center. C.S.H. is supported by NIH R01 DK094995, R21 AI097535, the CCFA, and the Burroughs Wellcome Fund. C.R.N is supported by NIH R01 AI106302 and University of Chicago Digestive Diseases Research Core Center P30 DK42086.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.M.N., T.F., C.R.N., and C.S.H. conceived of the project and designed the experiments; K.M.N., J.N.C., T.L.A., T.F., and E.R.G. performed the experiments; and K.M.N. and C.S.H. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Deroos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013 doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista JL, Lio CW, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh CS. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, Bry L, Kraj P, Kisielow P, Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei K-j, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+ CD25− naive T cells to CD4+ CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. Journal of Experimental Medicine. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmunity reviews. 2013;12:661–665. doi: 10.1016/j.autrev.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Elson CO, Cong Y. Host-microbiota interactions in inflammatory bowel disease. Gut microbes. 2012;3:332–344. doi: 10.4161/gmic.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk RA, Corse E, Allison JP. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol. 2012;188:976–980. doi: 10.4049/jimmunol.1102964. [DOI] [PubMed] [Google Scholar]
- Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Muller W, Sparwasser T, Forster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B, Ziegelbauer J, Yassai M, Li SH, Relland LM, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011;35:109–122. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigame H, Mosaheb MM, Sanjabi S, Flavell RA. Truncated form of TGF-betaRII, but not its absence, induces memory CD8+ T cell expansion and lymphoproliferative disorder in mice. J Immunol. 2013;190:6340–6350. doi: 10.4049/jimmunol.1300397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annual review of microbiology. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 2015;8:198–210. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel JE, Chen W. Balancing acts: the role of TGF-beta in the mucosal immune system. Trends in molecular medicine. 2011;17:668–676. doi: 10.1016/j.molmed.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von Boehmer H. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MW, Shen S, Lafaille JJ. TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J Exp Med. 2009;206:2121–2130. doi: 10.1084/jem.20091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, Klinakis A, Charo IF, Jung S, Gommerman JL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, Chatila TA. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-beta1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J Exp Med. 2005;201:1061–1067. doi: 10.1084/jem.20042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3(+) and Foxp3(−) precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011 doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. The ISME journal. 2013;7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, Kc W, Wu X, Thomas SR, Lee WL, Turkoz M, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J Exp Med. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, et al. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissler KA, Garcia V, Kropf E, Aitken M, Bedoya F, Wolf AI, Erikson J, Caton AJ. Distinct modes of antigen presentation promote the formation, differentiation, and activity of foxp3+ regulatory T cells in vivo. J Immunol. 2015;194:3784–3797. doi: 10.4049/jimmunol.1402960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welty NE, Staley C, Ghilardi N, Sadowsky MJ, Igyarto BZ, Kaplan DH. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J Exp Med. 2013;210:2011–2024. doi: 10.1084/jem.20130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, Forster R, Pabst O. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. S1711–S1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.