Abstract
MicroRNAs (miRs) are small single-stranded noncoding RNA that function in RNA silencing and post-transcriptional regulation of gene expression. An increasing number of studies have shown that miRs play an important role in tumorigenesis, and understanding the regulatory mechanism of miRs in this gene regulatory network will help elucidate the complex biological processes at play during malignancy. Despite advances, determination of miR–target interactions (MTIs) and identification of functional modules composed of miRs and their specific targets remain a challenge. A large amount of data generated by high-throughput methods from various sources are available to investigate MTIs. The development of data-driven tools to harness these multi-dimensional data has resulted in significant progress over the past decade. In parallel, large-scale cancer genomic projects are allowing new insights into the commonalities and disparities of miR–target regulation across cancers. In the first half of this review, we explore methods for identification of pairwise MTIs, and in the second half, we explore computational tools for discovery of miR-regulatory modules in a cancer-specific and pan-cancer context. We highlight strengths and limitations of each of these tools as a practical guide for the computational biologists.
Keywords: data integration, transcriptional regulation, microRNA networks
Introduction
MicroRNAs (miRs) are small, single-stranded, noncoding RNAs (~20–22 nucleotides) that regulate target gene expression by binding to complementary sites (seed sequences) in the messenger RNA (mRNA) target gene.1 This interaction, mediated by the miR-induced silencing complex (miR-RISC), reduces the stability and translational rate of the mRNA target.2,3 These miRs are predicted to target one-third of all genes in the genome, where each miR is expected to target hundreds of transcripts.4,5 As the number of published miR sequences continues to increase with small RNA deep sequencing experiments,6 the biological implications of miRs as modulators of post-transcriptional regulation expand. As of April 2016, there are 1,881 miR sequences in the human genome annotated in mirBase (http://www.mirbase.org), the primary miR sequence repository. However, the functions of only a subset of these miRs have been experimentally determined. To date, >300 cancer-related miRs and 829 target genes from >25 cancer tissues have been collected in OncomiRDB, a manually curated database of cancer-related miRs with direct experimental evidence.7 As miRs mainly regulate function through their targets, elucidating the miR–target interactions (MTIs) is vital for functional characterization of miRs. Therefore, much progress has been made over the past decade to develop high-throughput experimental and computational methods for MTI identification.
Relevance of miR biology to cancer studies
Recent studies have found that miR oncogenes (oncomiRs) and miR tumor suppressors tend to regulate tumor suppressors and oncogenes, respectively.8–11 Tumors that depend on over-expression of oncomiRs are said to demonstrate “oncomiR addiction,”12 for example, mice overexpressing miR-21 were found to contract pre-B malignant lymphoid-like phenotype tumors, and inactivation of this oncomiR resulted in complete tumor regression.13 Dysregulation of miR–gene networks has been shown to play an important role in tumor initiation and progression.14 The dysregulation of miR expression in cancer can be attributed to (i) DNA point mutations, (ii) epigenetic mechanisms (eg, DNA methylation), (iii) alterations of chromosomes (eg, deletions or amplifications of miR genes), and (iv) changes in the machinery responsible for miR processing.14
While global expression of miRs is usually repressed in cancer,15 a pan-cancer co-regulated “superfamily” of upregulated oncomiRs co-targeting critical tumor suppressors has been identified.11 Several miRs have been shown to inhibit metastasis through negative regulation of the epithelial–mesenchymal transition (EMT) and stemness pathways.16 Yang et al.17 found that miR-506 functioned as a potent EMT inhibitor in an orthotopic mouse model of ovarian cancer. Delivery of miR-506 via lipid-based nanoparticles to the tumor resulted in reduced tumor growth. Therapeutic inhibition of oncomiRs using antisense oligomers (called antimiRs) has also been shown to reduce tumor growth.12 These recent studies have established the role for miRs as “druggable targets” with vast potential for anti-cancer therapies.
In addition to novel targets for cancer therapy, miR expression profiles have the potential to play an important role in the diagnosis and management of patients with cancer. Numerous studies in multiple cancer types found miR expression profiles that serve as diagnostic and prognostic biomarkers. MiRs detected in various bodily fluids have been found to originate from cancer cells secreting exosomal vesicles (exosomes).18 High-quality miR samples have been extracted from a variety of sources, including plasma/serum, urine, and formalin-fixed, paraffin-embedded (FFPE) tissues.19 MiR expression profiles from FFPEs have been shown to determine the tissue of origin from metastatic tumors.20,21 Remarkably, miRs remain largely intact in bodily fluids and FFPE tissues in contrast to most mRNAs.22,23 The resistance of miRs to degradation from RNases and severe conditions has been attributed to their small size, encapsulation by lipid vesicles (eg, exosomes or apoptotic bodies), and association with RNA binding proteins.24,25 Russo et al.26 created a manually curated database of extracellular circulating miRs called miRandola (http://atlas.dmi.unict.it/mirandola/browse.php) containing 581 miRs from 21 types of samples. This website allows users to efficiently review the literature on circulating miRs that have been studied as biomarkers in cancer and various other diseases. For example, a recent study by Razzak et al.27 found that expression profiling of three miRs (miR-21, miR-210, and miR-372) in sputum from patients with early stage non-small cell lung cancer (NSCLC) and cancer-free controls detected NSCLC with 67% sensitivity and 90% specificity. Upregulation of miR-21 in the biopsies of NSCLC lung tumors has also been associated with poor prognosis.28,29 Using a matched analysis of miR and gene expression from cancer tissue samples, a number of studies have identified cancer subtype-specific miR-regulatory networks that serve as network biomarkers for colorectal and breast cancers.30–33
Framework for elucidating MiR-regulatory modules from multi-dimensional omics data
The rapidly increasing availability of transcriptome-wide matched miR and gene expression data via microarray and more recently RNA- sequencing technologies has greatly expanded our understanding of miR–gene regulation and interactions. The interactions between miRs and their target genes form networks, which much like gene–gene interaction (GGI) networks, are understood to consist of modules in which co-expressed miRs have a greater tendency to be functionally associated with miRs within the same module than to those outside the module.34–36 Groups of miRs coordinately regulate sets of targets forming miR-regulatory modules (MRMs), which function to control different biological processes.37,38 Recent studies have found that the modular organization and co-expression of miRs are dysregulated in cancer.35,39 However, the targets of most miRs remain unknown and the complex regulatory mechanisms of MRM are an open area of research.
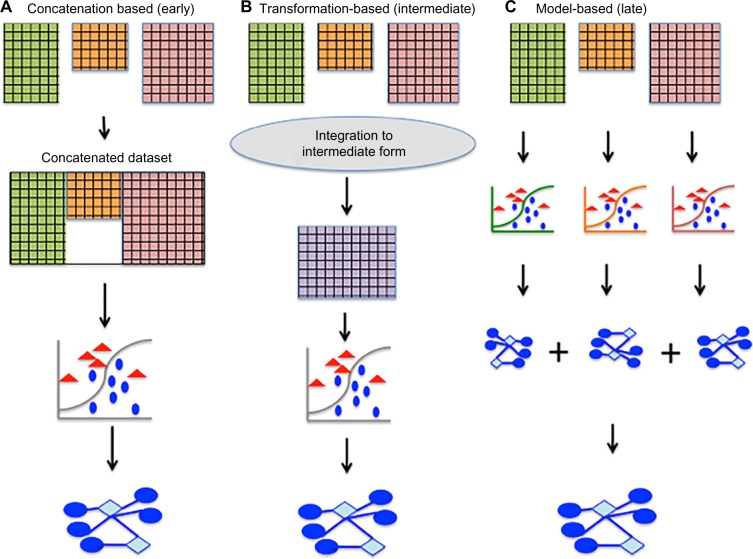
Integration of multiple types of molecular data in a simultaneous analysis, termed multi- or meta-dimensional analysis,40 is fundamental for discovery of MRMs. Multi-dimensional analysis is broadly classified into three categories40,41: (i) concatenation based (or early integration), (ii) model based (or late or decision integration), and (iii) transformation based (or intermediate integration); the schema is depicted in Figure 1. Concatenation-based (or early) integration combines multiple pre-processed molecular data matrices into one larger data matrix before constructing a model. Early integration requires an appropriate method to combine the different data types into one model. This task is often challenging, given that different data types often have differing properties and scales.42 Taskesen et al.43 found that gene expression and DNA-methylation profiles combined in an early integration strategy improved prediction of leukemia subtypes versus analysis of datasets individually. In model-based integration, the models are generated from each of the different data types and fused into a final model. For example, Kim et al.44 developed an integrative method that combined variables from models derived from each genomic data type to generate a final model to predict survival in ovarian cancer.
Figure 1.
Simultaneous integration of multiple data types: classification of approaches into three categories. (A) Concatenation-based (early) integration of multiple preprocessed molecular data matrices into one larger data matrix before constructing a model. (B) Transformation-based (intermediate) integration combines the multiple data types after transforming each type into an intermediate form before generating a model. (C) Model-based (late) integration creates models for each of the different data types and then combines each model into a final model for analysis. Modified from Ritchie et al.40
Transformation-based (or intermediate or partial) integration combines multiple data types after transforming each type into an intermediate form, such as a graph or matrix, before generating a model.40 The advantage of transformation-based integration over early integration is that when each type of data is transformed into an appropriate intermediate form, the data type-specific properties of each dataset are preserved without information loss.45 The transformation-based integration strategy is most frequently applied for identification of MTIs and MRM.42,46 Typically, miR–gene correlation matrix and sequence-based target prediction scores are integrated by transformation into binary matrices or bipartite graphs (discussed in the “Methods for inferring MRMs in a single cancer-type via network approaches” and “Methods for inferring pan-cancer MTIs and MRMs via joint analysis of sample data” sections).42,47,48 An alternative integration strategy developed by Zhang et al.49 used a joint matrix factorization technique to project multiple data types onto a common coordinate system to detect profiles that are highly (anti-) correlated (discussed in the “Methods for inferring MRMs in a single cancer-type via network approaches” section).
Integration of miR and gene expression data with sequence-based target prediction has been shown to improve prediction of MTIs.50–52 Integration of other high-throughput molecular layers, including methylomic and proteomic data, can also improve MRM discovery.49,53,54 Over the past decade, there has been a tremendous increase in the amount of publically available genomic, transcriptomic, methylomic, proteomic, and clinical data for many types of cancer.55 This has provided a major opportunity for researchers to study genetic and molecular abnormalities across human cancer types to discover pan-cancer commonalities as well as cancer (sub)type-specific features. The Cancer Genome Atlas (TCGA) project,56 a large systematic cancer genomics project, provides clinical, transcriptomic, and genomic data for >33 cancer types and subtypes. Initiated in 2006, TCGA has currently characterized tissues from matched tumor and healthy tissues from 11,000 patients, making it an invaluable resource for multi-dimensional omics projects. The rapidly increasing abundance of multi-dimensional omics data has been met with novel computational methods, providing significant advances in our understanding of genomic and epigenetic drivers of cancer. More specifically, the TCGA project has allowed researchers to develop new approaches to identify both cancer (sub)type-specific and pan-cancer MRM. However, discovery of MRM presents a number of challenges and potential pitfalls to the computational biologists. In the following sections, we highlight research progress that has addressed these challenges to identify MTIs and MRMs in cancer datasets.
Recent reviews of integration of multiple omic data40,45,57 and integrative analysis of cancer data58–60 are available and are outside the scope of this review article. While transcription factors (TFs) and miRs can jointly regulate target expression in MRM in the form of feed-forward and feedback loops,61 the involvement of TFs in MRM networks is also beyond the scope of this review. In the next section, we provide a brief overview of sequence-based target prediction algorithms and recent high-throughput experimental target identification methods.
Mir–Target Interactions
Overview of sequence-based target prediction and recent high-throughput experimental methods
Sequence-based miR–target prediction
A number of bioinformatics algorithms for predicting miR recognition sites within transcripts have been developed using knowledge from experimentally validated target sites. Early experimental studies found that a primary determinant of target specificity was perfect complementarity (canonical site) at the 5′ end of the miR “seed region” at positions 2–7.1,2,62 Therefore, these algorithms initially were primarily focused on sequence complementarity between the seed region of miR and the 3′ untranslated region (UTR) of the putative target.62,63 However, given the large number of randomly occurring six nucleotide sequences in a 3′ UTR of a gene, perfect seed match itself is a poor predictor of miR regulation.64–66
Several studies have proposed that target sites in which the pairing between miR seed and mRNA does not completely match (termed non-canonical sites) also confer regulatory effect.67,68 A recent study by Agarwal et al.69 found that while miRs bind to non-canonical sites, there was no detectable repression based on mRNA stability or translation using multiple cell types. This finding has supported the focus on canonical binding sites by sequence-based target prediction programs. Other algorithms consider additional data, including mRNA secondary structure and target accessibility70,71; however, this also results in a large number of predictions with many false positives (FPs).2 Many algorithms, including TargetScan69 and TargetRank,62 use evolutionary conservation of the target site to select predicted targets based on conservation to reduce FP predictions. However, approximately 20% of functional target sites are not conserved between mammals, and conservation is further decreased in a step-wise manner in larger taxonomic groups,72 indicating that sensitivity of target prediction decreases with higher conservation thresholds.65
The growing number of experimental MTI data has prompted more recent use of machine learning (ML) algorithms to train classifiers directly on the experimental data. For example, miRSVR exploits mRNA expression data from miR transfection experiments to train an ML algorithm to predict MTIs.64 The miRSVR scoring model for predicted MTIs is calibrated to correspond linearly with the probability of downregulation of the target, providing a meaningful guide for selecting a score cutoff. Agarwal et al.69 recently compared 17 sequence-based target prediction algorithms and found that the number of potential MTIs varied greatly, reflecting the varied strategies of these algorithms. Their analysis found that TargetScan version 7 performed significantly better than the existing models and was as good as recent high-throughput experimental approaches to identify effective target sites. Table 1 summarizes the target prediction algorithms discussed earlier. Sequence-based miR target prediction algorithms have been comprehensively reviewed elsewhere.73
Table 1.
List of software, websites, and references to methods for sequence-based miR target prediction and method for inferring miR–target relationships using paired expression profiles of miRs and genes in single-cancer datasets.
| SEQUENCE BASED METHODS FOR miR TARGET PREDICTION (PREDICTING WHETHER A GIVEN mRNA IS TARGETED BY A miR) | ||
|---|---|---|
| METHOD/REFERENCE/SOFTWARE | DATA TYPES | COMMENTS |
|
TargetRank Nielsen et al.62 http://hollywood.mit.edu/targetrank/ |
*mRNA sequence | *scoring system based on sequence complementarity and conservation |
|
miRanda Enright et al.114 |
*mRNA sequence | *sequence complementarity and estimated minimum free energy *source code in C freely available |
|
TargetScan (Version 7) Agarwal et al.69 http://www.targetscan.org/vert_71/ |
*mRNA sequence | *scoring system based on 14 features found to be informative of binding efficacy using a regression model |
|
STarMir Rennie et al.70 http://sfold.wadsworth.org/cgi-bin/starmir.pl |
*mRNA sequence | *model for binding site predictions trained on miR binding sites from CLIP data |
|
miRanda-miRSVR Betel et al.64 http://www.microrna.org/microrna/home.do |
*mRNA sequence | *regression model trained on miRanda predicted target site features and miR transfection data to predict target site binding efficacy |
| METHODS FOR INFERRING miR–TARGET RELATIONSHIPS USING PAIRED MIR AND GENE EXPRESSION PROFILES IN SINGLE-CANCER DATASETS | ||
|---|---|---|
| METHOD | DATA TYPES | COMMENTS |
|
Correlation coefficient based methods Peng et al.47 |
*miR & gene expression *sequence predicted targets |
*Proposed permutation based method to estimate FDR of MTIs |
|
MLR Yang et al.17 |
*miR & gene expression *sequence predicted targets *CNA *PM |
*models gene expression by a linear combination of all miR expression profiles (adjusting for epigenetic and genomic effects) |
|
LASSO Lu et al.52 |
*miR & gene expression | *models gene expression given multiple potentially competing miRs |
|
Elastic net regression Sass et al.82 |
*miR & gene expression | *found superior performance for identification of experimentally validated MTIs versus LASSO and PCC |
|
Causal inference (IDA) Le et al.88 |
*miR & gene expression | *ensemble of LASSO, PCC, and IDA detected more MTIs than any single method |
|
Maximal information content (MIC) Le et al.88 |
*miR & gene expression | *mutual information based method to detect linear and non-linear associations between two variables |
|
GenmiR++ Huang et al.51 |
*miR & gene expression *sequence predicted targets |
*Bayesian inference method for MTI prediction |
Abbreviations: FDR, false discovery rate; MTI, miR–target interactions; UTR, untranslated region; LASSO, least absolute shrinkage and selection operator; PM, DNa promoter methylation; CNA, copy number abnormalities; PCC, Pearson’s correlation coefficient; IDA, interventional calculus when the directed acyclic graph is absent; CLIP, crosslinking and immunoprecipitation.
High-throughput experimental methods for miR–target identification
High-quality experimentally derived training data are generally required to improve sequence-based target prediction performance.74 High-throughput methods such as those employing crosslinking and immunoprecipitation (CLIP) are an important class of capture-based methods for detection of direct miR–target binding events associated with the Argonaute protein (Ago).75,76 Argonaute high-throughput sequencing of RNAs isolated by CLIP simultaneously sequences Ago-miR and Ago-mRNA binding sites to identify interaction sites between miR–target pairs.76 One limitation of this approach is that miR–target complexes are dissociated prior to sequencing, requiring the target sequence in each miR–target pair to be inferred computationally, which is prone to error.
Recently a method for producing ligation of the miR–target pair called crosslinking, ligation, and sequencing of hybrids (CLASH) has shown to be more robust than CLIP for identification of miR target sites.67 The former method is similar to CLIP, but adds a ligation step between the miR and target, allowing direct characterization of the chimeric or hybrid miR–target to unambiguously identify the miR bound at a specific target site. A novel finding from the CLASH analysis was the detection of strongly overrepresented motifs in the interaction sites of several miRNAs, suggesting that individual miRs systematically differ in their binding site modes. Although this likely affects the response of RISC to miR–target binding, it is unclear how it impacts in vivo function of MTIs.67
While CLASH holds much promise, at the present time, this method has a very low yield with only ~2% of the reads obtained in an experiment corresponding to miR–target chimeras. Thus, further improvements to CLASH will be needed before comprehensive mapping of MTIs will be possible.77 As each cell line has a different miR expression profile, the cell line used in an experimental analysis will yield different sets of MTIs than other cell lines or disease conditions. For example, an miR with low expression in a cancer (sub)type profiled using CLASH may not be detected, whereas this miR may have high expression in another (sub) type of cancer. Therefore, studies have integrated knowledge of MTIs from one cell line or condition identified by CLIP and CLASH with miR and gene expression profiles from the cell line or condition of interest, yielding condition-specific MTIs. For example, StarBase, a database of 108 CLIP- and CLASH-based datasets from 37 studies, integrates TCGA pan-cancer expression data with CLIP-seq data to provide MTI pan-cancer predictions.78 Recent comprehensive reviews of experimental target prediction methods are available elsewhere.75,77
While high-throughput experimental (in vitro) approaches are being increasingly performed, they currently suffer from several disadvantages compared to in silico predicted MTIs (based on sequence based and/or structural stability): 1) the number of experimentally derived MTIs remains far fewer than the predicted MTIs79 and 2) the tissue specificity of the experimental method may exclude availability of cancer (sub) types of interest to the investigator. Therefore, the most common approach to facilitate inference of cancer-specific MTIs involves profiling paired miR and gene expression datasets. Integrating condition-specific (dynamic) expression profiles with in silico predicted MTI datasets (static) datasets (as discussed in the next section) has been shown to improve MTI prediction.80 Several recent studies have demonstrated better MTI prediction in cancer datasets by integrating CLIP data with sequence-based target predictions.11,81 In the next section, we discuss commonly applied methods to integrate paired miR and gene expression with in silico and in vitro data types for multi-dimensional analysis to identify significant MTIs.
Methods for inferring MTIs
The integration of matched miR and gene expression data with sequence-based target prediction has been shown to significantly improve the quality of the identified MTIs.50,80 While high-throughput measurement of miR and gene expression has become relatively straightforward, their joint integration for detecting high-confidence interacting miR–mRNA pairs is more challenging.80,82 A number of approaches have been used to quantify the statistical significance for association between an miR and its target using their expression measurements. These approaches include correlation and mutual information-based methods,47 multiple linear regression (MLR) models,17 partial least squares (PLS) regression,83 and regularized least-squares regression models.52 A PLS model extracts coefficients (miRs) that explain the maximum variance in the dependent variable (gene expression) by ensuring good fit of the model.80 A regularized least-squares model also ensures good model fit and adds an extra term to prevent model overfitting (discussed in the “Regularized least squares” section). GenMiR++, the first developed target prediction algorithm that integrated miR and gene expression data with sequence-based target predictions, applies a Bayesian inference to score potential targets.51 In the following section, we survey the major classes of statistical approaches for MTI detection in paired miR and gene expression data from cancer samples.
Correlation coefficient-based methods
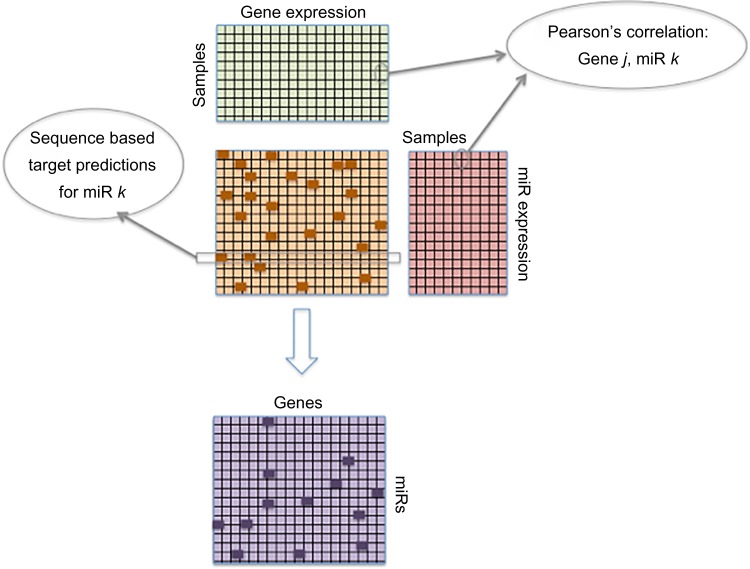
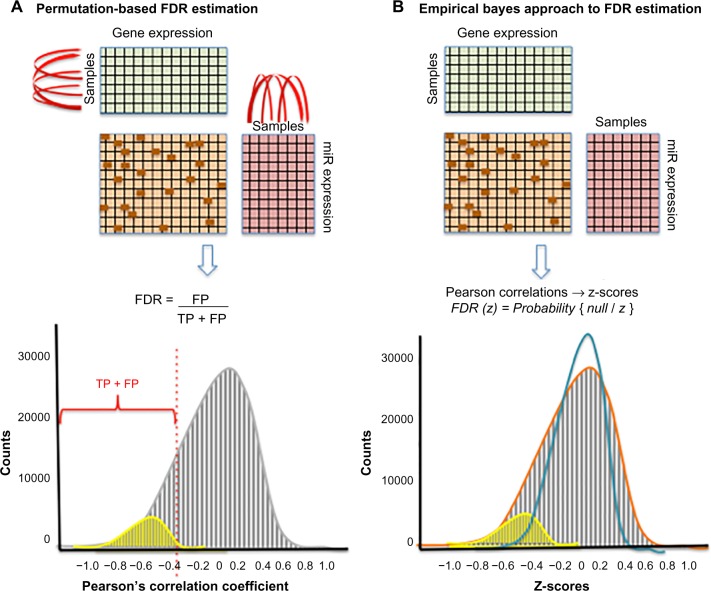
The principle of assuming that the expression levels of miRs and target mRNAs are negatively correlated is commonly used to detect MTIs.47,48,84 These methods typically select potential miR–target pairs that (i) are negatively correlated above some statistical significance threshold and (ii) have been identified to interact using sequence-based target prediction or experimental methods (Fig. 2). The large number of miR–target correlations calculated necessitates estimation of the false discovery rate (FDR), defined as the number of FP divided by the number of FP and true positives (TPs). Peng et al.47 proposed a permutation-based method to estimate the FDR of miR–target correlations at a given statistical threshold (Fig. 3A). The FDR was defined as the ratio of the number of correlated miR–mRNA pairs above a given threshold (eg, Pearson’s r < −0.5 and P < 0.01) in a randomly permuted dataset (ie, FP) to the number of pairs above the threshold in the original dataset (ie, TP and FP). To generate the randomly permutated miR–mRNA datasets, the sample labels for miR and mRNA were randomly swapped such that the samples in the random miR datasets did not correspond to the samples in the random mRNA datasets. This process was repeated 100 times, and a median value of FDR was selected. Using this approach, the authors were able to ascertain that Pearson’s r < −0.55 with P-value < 0.01 was associated with an approximately 5% FDR. An alternative method to estimate FDR using empirical Bayes (EB) is discussed in the “Bayesian models” section (Fig. 3B).
Figure 2.
Inferring MTIs by integrating match miR–gene expression profiles and sequence-based target prediction data. Sequence predicted targets from a pre-selected database (orange matrix) depicted as a binary matrix (indicating the presence or absence of miR–target pairs, as dark orange or light orange boxes, respectively). Expression profiles from matched gene (green matrix) and miR (red matrix) microarrays are correlated using the PCC and input into the purple matrix. MTIs with PCC above a selected threshold and present in the sequence-based target prediction database are indicated as dark purple boxes, while all other pairs as light purple boxes.
Figure 3.
Methods for estimation of FDR from MTI data. (A) Permutation-based method to estimate FDR at a given statistical threshold. Randomly permuted datasets are generated to estimate the proportion of FPs above a given threshold. To generate randomly permuted datasets, the sample labels for miR and gene expression matrices are randomly swapped such that samples in the random miR datasets do not correspond to samples in the random gene datasets (swapping of sample labels indicated by red arching arrows above miR and above gene expression matrices). PCC is calculated for each miR–gene pair in the sequence-based target prediction matrix (dark orange boxes above). The distribution of PCC values is depicted as a gray histogram (below). PCC values below a threshold of −0.4 (vertical dashed red line) represent FP in the permuted data (gray). PCC values below this threshold using the original (unpermuted dataset) represent TPs (yellow region in the histogram). The FDR is calculated as the number of FP divided by the number of FP and TP. (B) EB estimation of FDR using local FDR. PCC values for all miR–gene pairs within the sequence-based target prediction matrix are calculated and transformed into z-scores. The z-scores corresponding to false (null) interactions are depicted as a histogram in gray (below), and TPs are shown in yellow within the histogram. The empirical null distribution (orange curve) and theoretical null distribution (blue curve) are shown overlapping the histogram. Note that the theoretical null is too narrow for the data, whereas the empirical null (determined using local FDR) is a better fit for the data.
For miR and gene expression profiles from heterogeneous conditions such as multiple (sub)types of cancer samples or cancer and healthy controls, correlations are often calculated for each condition separately rather than over the data as a whole.81,85,86 The rationale for this approach is that each cancer type may have differences in dysregulation of MTIs. Farazi et al.81 found significant differences in the medians of the correlation distributions for miRs and their predicted targets comparing between three subtypes of breast cancer. They also detected distinct correlations between the expression of specific miR families and their predicted targets among the three cancer subtypes, providing a rationale for performing separate MTI analysis based on the subtype. Importantly, the top-ranked miRs according to regulatory activity did not necessarily overlap with the top-ranked miRs according to association with tumor subphenotype. This disconnection becomes especially relevant for validation and development of future miR-based therapies.
MLR and regularized least-squares models
MLR models
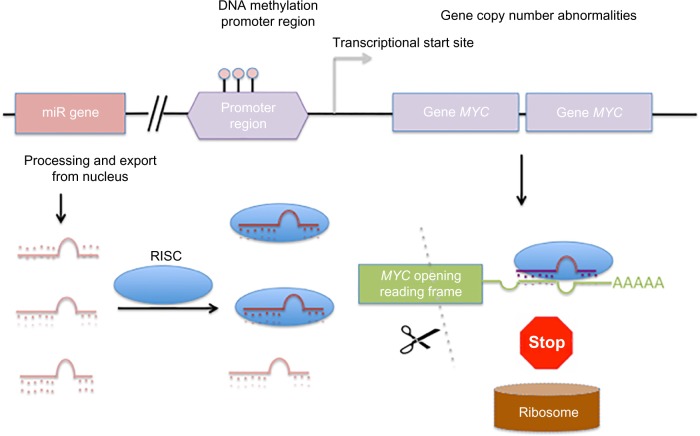
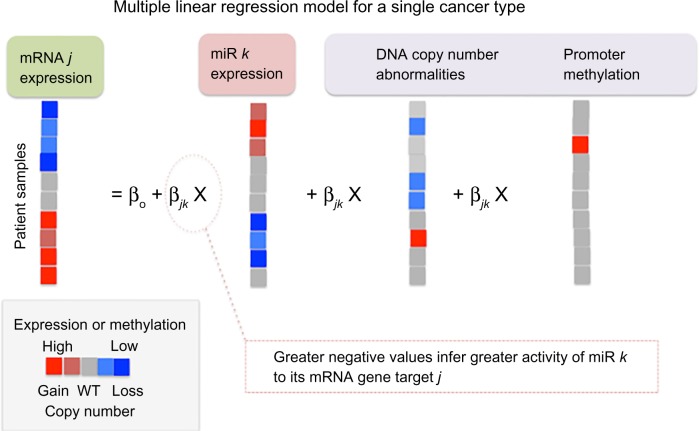
Whereas correlation-based approaches consider only the pairwise miR–gene expression, MLR models gene expression by a linear combination of all miR expression profiles targeting the gene. Furthermore, other epigenetic or genomic factors can also be modeled such that gene expression is the response (dependent) variable and the transcriptional and epigenetic regulators are the independent variables in the models (Fig. 4). Integrating gene expression with associated alterations in genomic, epigenetic, and miR expression has been undertaken in several studies to identify molecular drivers of cancer (sub) types. Yang et al.17 applied an MLR model to analyze gene expression of each gene in a mesenchymal signature of ovarian cancer. The putative regulatory factors of each gene selected for analysis were the associated DNA copy number abnormalities (CNA), promoter methylation (PM), and miR expression level, which were used as independent variables in the regression model. Based on this analysis, the investigators detected a set of 219 genes predicted to be targeted by 19 miRs in an miR–mRNA network. These genes could be used to distinguish the mesenchymal subgroup of ovarian cancer from other ovarian cancer subtypes.
Figure 4.
Factors controlling gene expression in cancer. The expression of the oncogene MYC is regulated by miRs gene expression (pink square, left), the availability of the miR-RISC (shown in blue), DNA-methylation promoter region (purple hexagon), and gene copy number abnormalities (duplicated copies of MYC shown as purple squares). MiR loading to RISC leads to targeting the 3′ UTR of the MYC mRNA, where it is degraded and translation by the ribosome is inhibited.
Regularized least squares
Least absolute shrinkage and selection operator
The least absolute shrinkage and selection operator (LASSO) algorithm performs variable selection procedure by estimating linear regression coefficients through L1-constrained least squares. This L1 constraint (penalty) tends to result in regression coefficients that are exactly zero, thereby imposing sparsity on feature selection, making the models more interpretable. Several investigators have argued that LASSO avoids overfitting in the presence of noisy expression data and a large number of explanatory variables, leading to better prediction accuracy.53,87 Li et al.54 found LASSO with negative coefficient constraint as the best performing method for the joint analysis of miR and mRNA expression data compared to Pearson’s correlation-based methods and Genmir++ (a Bayesian learning method to infer the probabilities of targets)51 using a database of experimentally verified MTIs as a reference.
MLR with elastic net penalty
While LASSO models multiple potentially competing miRs,52 several investigators82,86 have noted that it fails to account for co-expression between miRs targeting the same gene. The sparse solution (L1 penalty) implemented in LASSO selects one representative miR from each correlated group of miRs, disregarding other potentially biologically relevant co-expressed miRs within the group. In contrast, the ridge regression model maintains predictors in the model by using an L2 penalty in which the coefficients are small but non-zero. However, this approach does not facilitate feature selection. The elastic net penalty overcomes the drawbacks of LASSO and ridge regression by combining L1 and L2 penalties in order to account for co-expression among miRs while simultaneously performing feature selection. Compared to Pearson’s correlation and LASSO, elastic net regression in combination with negativity constraint coefficients was applied to a head and neck cancer dataset and was found to identify MTIs with greater enrichment for experimentally validated MTIs.82
Ensemble of computational methods
Le et al.88 investigated the performance of eight methods for joint analysis of miR–target expression and prediction methods using datasets from multiple cancer types. The computational methods compared were (i) Pearson’s correlation coefficient, (ii) MIC (maximal information coefficient, a mutual information based method to detect linear and non-linear associations between two variables),89 (iii) z-score (a network inference method to estimate the effects of gene knockout experiments),90 (iv) IDA (interventional calculus when the directed acyclic graph is absent, a causal inference method),91 (v) ProMISe (probabilistic MiR–mRNA interaction signature, a method that estimates the probability of a gene to be a target of an miR by taking competition between genes and between miRs into account),92 (vi) GenMiR++,51 (vii) LASSO, and (viii) elastic net regression methods.
An ensemble of three methods (Pearson, IDA, and LASSO) obtained more targets than any single method and identified targets with enhanced functional enrichment. From this, we can infer that different classes of computational methods tend to identify unique sets of validated targets and therefore each has its own merits.91 It is important to note that this analysis did not incorporate sequence-based target predictions. Thus, the investigators hypothesized that the poor performance by GenMiR++ and ProMISe, two methods originally designed to incorporate sequence-based predicted targets, may have been due to the use of miR and gene expression data alone. Le et al.93 provided a software pipeline integrating the above computational methods in the R package miRLAB that is freely available on Bioconductor at http://bioconductor.org/packages/miRLAB/.
The statistical approaches described in this section quantify the association between any given single miR and single gene (refer “Correlation coefficient-based methods” section) or associations between multiple (co-expressed) miRs and a single gene (refer “MLR and regularized least-squares models” section). These approaches are summarized in Table 1. In the next section, we explore methods for identifying associations between co-expressed miRs and groups of gene targets in a network of “many-to-many” miRs and genes termed an MRM. As is discussed in the following, a greater understanding of the (patho)biological roles of miRs and their targets can be gained by identifying MRM compared to single MTI analysis.
Mir-Regulatory Modules
Methods for inferring MRMs in a single-cancer type via network approaches
A number of methods have been developed to study MRM. Most approaches aim to identify groups of co-expressed miRs and their inversely expressed targets by integrating paired miR and gene expression profiles with sequence-based predicted MTIs. The methods discussed in this section implement analysis of a single condition (eg, cancer type versus some reference). Here, we discuss three distinct learning frameworks to elucidate MRM: biclique enumeration, matrix factorization, and Bayesian networks (BNs). The methods that have been designed for analysis of more than two conditions (eg, pan-cancer analysis) are discussed in the “Methods for inferring pan-cancer MTIs and MRMs via joint analysis of sample data” section. The methods for inferring MRM in cancer and pan-cancer datasets are summarized in Table 2.
Table 2.
List of software, websites, and references to methods for inferring MRMs in single-cancer or pan-cancer datasets.
| METHODS FOR INFERRING MRMs IN SINGLE-CANCER DATASETS | ||
|---|---|---|
| METHOD/REFERENCE/SOFTWARE | DATA TYPES | COMMENTS |
|
Maximal Biclique enumeration Peng et al.47 Software upon request |
*Sequence predicted targets *miR & gene expression (DE) |
*Maximal biclique enumeration method to sensitive to noise in the data *frequently often produces MRM with only 1 miR |
|
Hypergraph Kim et al.100 Software upon request |
*miR & gene expression *cancer stage |
*Hyperedges (MRMs) weighted by discriminative ability to predict cancer stage |
|
Matrix Factorization Zhang et al.49,96 SNMNMF http://zhoulab.usc.edu/SNMNMF/ |
*Sequence predicted targets *miR & gene expression *GGI *DNA-protein interaction |
*MRM had greater enrichment for GO terms compared to Peng et al.47 *requires estimation of pre-defined number of modules *the solution is often not unique *omits MRM regulated by a single type of regulator |
|
Matrix Factorization Yang and Michailidis101 iNMF https://github.com/yangzi4/iNMF |
*miR & gene expression *PM *CNA |
*MRM detection more robust to noisy datasets than Zhang et al.49 |
|
Bayesian network Jin and Lee105 Software upon request |
*miR & gene expression (DE) *GGI data |
*Condition-specific analysis *MRM had greater enrichment for GO terms compared to Zhang et al.96 |
|
Bayesian network Liu et al.104 Software upon request |
*Sequence predicted targets *miR & gene expression (DE) |
*Condition-specific analysis *Top interactions under each condition determined then merged in final network |
|
Bayesian network Zacher et al.33 birta https://www.bioconductor.org/packages/release/bioc/html/birta.html |
*Sequence predicted targets *miR & gene expression (DE) |
*Condition-specific analysis *Detects miRs with regulatory activity differing between two conditions and their gene targets *Does not detect MRM |
| METHODS FOR INFERRING miR–TARGET RELATIONSHIPS AND MRMS IN PAN-CANCER DATASETS | ||
|---|---|---|
| METHOD | DATA TYPES | COMMENTS |
|
MLR Jacobsen et al.8 |
*miR & gene expression *sequence predicted targets *PM *CNA |
*Inferred MTIs for 11 cancer types to derive high-confidence network *Results available in the CancerMiner website http://cancerminer.org |
|
LASSO Setty et al.87 RegulatorInference https://bitbucket.org/leslielab/regulatorinference/ |
*miR & gene expression *CNA *sequence predicted targets |
*Group LASSO used to learn regression models of all samples simultaneously to identify common and subtype specific miRs associated with gene expression |
| LASSO Le and Bar-Joseph53 Software upon request |
*miR & gene expression *sequence predicted targets *GGI |
*Dependent on quality of GGI data *Number of modules must be determined in advance (fixed for each cancer (sub)type) |
|
Empirical Bayes Chen et al.86 MCMG http://bioinformatics.med.yale.edu/group |
*miR & gene expression | *Prioritizes MTIs by sharing information between cancers (joint posterior estimation). *Higher precision than Pearson correlation-based and LASSO approaches for identification of MTIs across multiple cancer types |
|
Empirical Bayes Li et al.112 PanMiRa http://www.cs.utoronto.ca/~yueli/PanMiRa.html |
*miR & gene expression *CNA *PM |
*Integrates MLR approach of Jacobsen et al.8 (accounting for biases due to PM and CNA) with the joint posterior estimation method of Chen et al.86 for pan-cancer analysis |
Notes: The names of packages available in R are italicized where available. Methods with data types that input DE miRs and genes are indicated.
Abbreviations: MTI, miR–target interaction; NMF, non-negative matrix factorization; LASSO, least absolute shrinkage and selection operator; PM, DNa promoter methylation; CNA, copy number abnormalities; BN, Bayesian network.
Bipartite graphs and biclique enumeration approach
A bipartite graph (network) consists of two sets of nodes (in this case, miR and target) and a set of edges connecting the nodes (in this case, edges represent association strength between miR and target).36 A putative module (in this case, MRM) in the bipartite network has been postulated to correspond to a biclique, a special type of bipartite network where every node in the first set (miRs) is connected to every node in the second set (target genes).47 A biclique is called a maximal (complete) biclique if it is not contained in a larger biclique. To perform enumeration of the maximal bipartite cliques (EBC) within the bipartite network representing putative MRMs, the module input consensus algorithm (MICA),94 the most efficient algorithm for finding bicliques, is often used.47 Zhang et al.95 argued that MICA is designed for general graphs and unable to take advantage of the bipartite structure. Therefore, the investigators developed the maximal biclique enumeration algorithm, which outperformed MICA.95
Peng et al.47 developed one of the earliest approaches to identify MRM using a maximal biclique method on MTIs discovered in a multi-step approach. In the first step, pairwise Pearson’s correlation between the differentially expressed (DE) miRs and genes across all samples was preformed to identify putative MTIs. The MTIs were selected if they exceeded a predefined FDR threshold (discussed in the “Correlation coefficient-based methods” section). Then the MTIs were further selected if present in a set of sequence-based target predictions, resulting in a binary matrix of MTIs. This matrix of selected MTIs was graphically represented as a bipartite network. Using MICA, maximal bicliques (ie, putative MRMs) were identified, which comprised between one and three miRs per MRM.
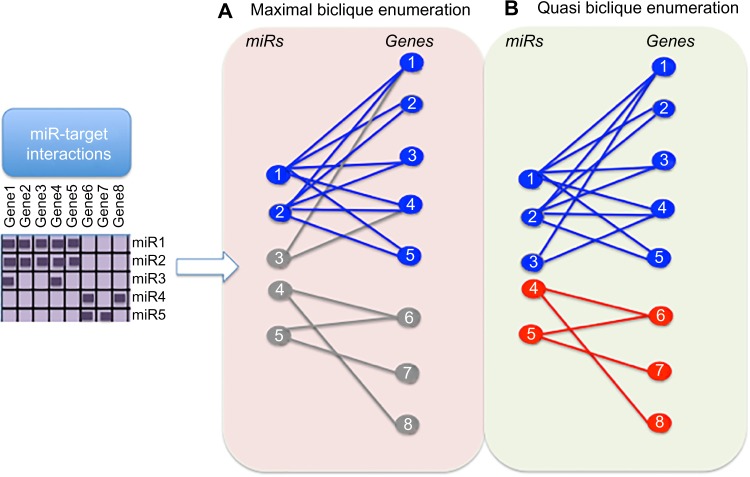
Maximal biclique-based methods to discover putative MRM have been argued to be too sensitive to noise in the data and frequently produce MRM with a high level of redundancy and only a single miR that cannot be used to explore miR combinatorial regulation.95,96 Missing subsets (false negatives) or erroneous (FP) MTIs may have an adverse effect on the quality of the maximal bicliques detected. Furthermore, searching only for maximal bicliques may be too restrictive as they are defined by an all-to-all relation between miRs and targets within the MRM.97 To add flexibility to MRM detection, several studies98,99 have applied quasi-bicliques, which exhibit a most-to-most interaction between miRs and genes within the MRM (Fig. 5).97 Quasi-bicliques allow all nodes in the bipartite network (miRs and genes) to accommodate missing interactions up to some user-determined level. Veksler-Lublinsky et al.99 found that MRMs discovered using the quasi-biclique method more significantly identified MTIs than a maximal biclique approach.
Figure 5.
Comparison of maximal biclique and quasi-biclique enumeration of bipartite graph for identification of MRMs. A matrix of miRs and genes (purple) indicating known MTIs (dark purple boxes) is represented as a bipartite graph with miRs shown as circles on the left (labeled 1–5) and genes shown on the right (labeled 1–8) in a) and b). MTIs are depicted with an edge (straight line) connecting the miR and gene. (A) Maximal biclique enumeration (all-to-all interactions): miRs and genes within a maximal biclique (representing an MRM) are shown in blue, whereas those excluded from a maximal biclique are shown in gray. Unlike miR-1 and miR-2, miR-3 is not connected to all genes within the biclique and is therefore excluded. Likewise, miR 4 and miR 5 do not form a maximal biclique and are excluded. (B) Quasi-biclique enumeration (most-to-most interactions): quasi-biclique enumeration allows some missing interactions within the biclique; therefore, miR-3 is included with miR-1 and miR-2 (blue, above); miR-4 and miR-5 form a biclique (red, below). Thus, in this toy example, quasi-biclique method is more sensitive to detecting MRM than maximal biclique enumeration.
Kim et al.100 have argued that the bi-relationships modeled between miRs and targets using bicliques are unsuitable for complex genetic interactions because information is lost. Instead, they applied hypergraphs to generalize the concept of an edge between nodes to a hyperedge by which more than two variables could be connected simultaneously. As the weight of a hyperedge reflects the strength of higher order dependency among variables, it was hypothesized that each hyperedge potentially behaves as a gene module. To model prostate cancer stage-specific MRM networks, the investigators developed a hypergraph model with each hyperedge represented by miR and gene expression for each cancer stage. Using a learning hypergraph model, hyperedges having high discriminative capacity between cancer stages were selected The investigators identified putative prostate cancer stage- specific MRMs; however, it is unclear whether the hypergraph structure improves MRM discovery over the maximal biclique approach utilized by Peng et al.47
Matrix factorization approach
An alternative strategy to the biclique approach was implemented by Zhang et al.96 by integrating miR and target expression profiles using non-negative matrix factorization (NMF) to identify putative MRM. The NMF method decomposes a matrix to find two smaller (lower rank) non-negative matrices, allowing substructures to be readily identified within the data. This method is similar to principal component analysis, another unsupervised matrix decomposition technique, except that it employs a constraint of non-negativity instead of orthogonality.101 Zhang et al.96 extended the NMF method by simultaneously factoring two variable types (expression profiles for miRs and genes) into a common basis matrix and two lower rank matrices. The decomposed matrices were then used to identify MRM with each miR or gene permitted to be assigned to multiple MRMs. Sparsity was induced via L1 penalty, and they named their algorithm the sparse network-regularized multiple NMF (SNMNMF) technique. To guide the optimization process, the investigators incorporated sequence-based predicted target data and GGI using a semi-supervised learning framework to define constraints for MRM. Using a multiplicative interactive procedure, the model is learned until convergence to a local optimum.
Comparing NMF to EBC methods using the TCGA ovarian cancer data, only 19% of the modules identified by EBC were enriched for at least one gene ontology (GO) biological term compared to 53% of the modules identified by NMF, indicating that this method performed better than EBC. The limitations of the NMF approach include the requirement for a pre-defined number of modules in order to perform the matrix factorization (which may be difficult to estimate empirically), the solution is often not unique, and it does not enforce a negative correlation constraint. A negativity constraint is implemented by other MRM methods to account for the fact that since miRs mainly repress mRNA target expression levels, positively correlated MTIs are likely to be FP.3,82
An additional limitation of the conventional NMF method is that it only permits two types of genomic data (eg, miR and gene expression) to be analyzed. Therefore, Zhang et al.49 implemented a joint NMF (jNMF) semi-supervised framework for the integrative analysis of more than two types of genomic data to identify groups of methylomic markers, miRs, and genes in putative modules in ovarian cancer datasets. Yang and Michailidis101 have noted that the jNMF method is not different from NMF, and as a consequence, jNMF does not distinguish between different data sources in the integrative analysis, which can be problematic for analyzing heterogeneous data. Thus, the investigators developed an integrative NMF (iNMF) method that applied a novel tuning selection procedure that allows the model to adapt to the level of heterogeneity among the datasets. The iNMF method was found to be more robust to heterogeneous noise across the data sources than jNMF for the detection of true modules. These matrix factorization approaches exemplify transformation-based integration of multi-dimensional datatypes (refer “Framework for elucidating MiR-regulatory modules (MRM) from multi-dimensional omics data” section).
BN models
A BN is a graphical model based on probabilistic relationships among the measured variables,102 which can be applied to miR and mRNA expression datasets to discover MRM.103,104 Inference of the BN structure (topology) involves searching among the possible networks and then scoring these structures. Two nodes are expected to be connected to each other if one node (ie, miR expression) affects another (eg, gene target expression). If the search space (number of potentially interacting variables) is not sufficiently restricted, the process of learning a BN can become very computationally time consuming, as all possible networks will be formed.103 Therefore, most BN approaches rely on constraints to decrease the search space to provide a more compact representation of probability distributions. Prior information such as predicted target data can be incorporated to improve network construction.
After the topology of the BN is determined, the strength of the relationship between any two nodes is quantified using conditional probabilities (network parameters). Thus, the expression values (activities) of miRs and genes are represented as nodes, and the dependence between nodes (regulatory interactions) is represented by the edges within the network.
Jin and Lee105 proposed a method to integrate miR–gene expression data with GGI data using the TCGA ovarian cancer dataset as an alternative to the NMF method developed by Zhang et al.49 To constrain the search space, DE genes along with labeled samples were input into a biclustering algorithm. Clustering has been previously demonstrated to aid in the inference of BN by reducing the parameter space and avoiding highly correlated gene profiles from inhibiting interaction inference.106 The interaction of sample and gene expression profiles was modeled as a bipartite graph and generated subgraphs, termed gene-sample modules (GSMs). Within each GSM, the expression trend was similar for most genes among the subset of selected samples.
Since previous studies have shown that not all the genes in cancer-related pathways undergo expression changes, the investigators expanded the GSMs using GGI data by including genes that are significantly correlated with at least one gene in a GSM. Next, to discover MRMs, a BN model was used to estimate dependencies between expression values of miRs and genes in the GSM using Bayes information criterion (BIC). The BIC is a score function used to assess the degree to which the BN explains the data (ie, whether it provides a “good” structure).102 The search space is constrained to the miRs whose correlation coefficient values for genes in a given GSM are in the top T%. The investigators found that the average number of enriched pathways in modules using this method was larger than that in the NMF approach by Zhang et al.96 described in the “Matrix factorization approach” section.
Liu et al.104 were the first to model separate BN structures in two different conditions (eg, cancer and normal) in order to effectively identify both strong and subtle MTIs. For each condition, the BN learning was performed using the miR and gene expression as input, initially generating separate BN models for each condition. The BN learning iteratively evaluated the MTIs, removing MTIs from the initial structure using the expression data and retaining only high-confidence MTIs, with the goal of selecting the structure that best fits the data based on a scoring function. To avoid statistically insignificant results and overfitting with small sample sizes, a bootstrapping step (sampling with replacement) was added to achieve reliable inference and integration of the BN models from the two conditions. To significantly reduce the search space, the BN was assumed to have a bipartite graph topology, and sequence-based predicted targets were further used to constraint the initial network. Furthermore, miR and gene expression profiles were discretized into a binary status: “up” or “down” regulated. This condition known as “splitting” approach, initially used to generate separate models, was shown to capture complex MTIs from the cancer and control samples. Using datasets from epithelial and mesenchymal cell lines, their method named Bayesian networks splitting and averaging was found to identify more co-regulated targets by multiple miRs compared to conventional BN that did not separate the conditions. This example of intermediate integration40 learned after combining the sample types was also found to result in improved performance over concatenation-based integration by Gevaert et al.107 for integration of clinical and gene expression data.
Zacher et al.33 formulated a BN integrating miR and gene expression to infer miR activities in a specific condition versus a reference condition (eg, controls). In their model, if the activity of certain miRs changes between conditions, a shift in the expression value of the targets is expected. For example, an miR is considered to be active in a condition if it is upregulated and its gene targets are downregulated compared to the reference condition. While TF-target activity can also be inferred using this approach, we will focus on the use of this application for miR–target discovery. The investigators applied a Bayesian linear regression, where the expression level of each gene in each condition was determined by a linear combination of miR activities. Then a score was used to rank the degree to which miRs explained the observed differential gene expression, where a score close to 1 indicates that the corresponding miR is essential (ie, high miR activity) for explaining the differential gene expression in the condition versus reference. The miR activity states are inferred using Markov Chain Monte Carlo sampling. The model swaps the activity states of any two miRs with opposite activity states sampled from the posterior distributions. To reduce the size of the potential network, the investigators (i) input only DE miRs and genes between the two conditions, (ii) limited MTIs to those present in experimental or sequence-based predicted MTIs datasets, and (iii) required that the target genes for two miRs exhibit a minimal overlapping similarity (ie, 0.8). While this method identifies miRs (and their targets) with condition-specific activity, it does not directly identify MRMs in contrast to the other BN methods described earlier.
The investigators applied their method to ovarian cancer data from TCGA. They separated patients into “early” or “late” relapse (>1 year) and discovered 12 miRs active in the “early” but not in the “late” relapse patients, with target genes of 11 of the 12 miRs directly associated with relapse-free survival times. This method named Bayesian influence of regulation of transcriptional activity is freely available through the R package birta on Bioconductor at https://www.bioconductor.org/packages/release/bioc/html/birta.html.
These BN methods represent examples of transformation-based (intermediate) integration.40,57 While the methods described earlier facilitate MTI or MRM discovery using datasets from a single-cancer type (or subtype), other recent works have investigated methods for inferring MTIs and MRMs across many types of cancers, which are discussed in the next section.
Methods for inferring pan-cancer MTIs and MRMs via joint analysis of sample data
The recent availability of miR and gene expression across multiple cancer types has spurred investigators to develop approaches to compare the similarities and differences of MTIs and MRMs across cancer types. Recurrent MTIs across cancer types are hypothesized to play important roles in tumorigenesis.8 Pan-cancer analysis has identified convergent cancer subtypes composed of several distinct cancer types providing novel prognostic and therapeutic insights.108 Joint analysis of multiple cancer types to infer pan-cancer MTIs and MRMs presents an additional set of challenges in comparison to single-cancer analysis. These challenges include systematic confounding biases owing to differences in study size, sample heterogeneity (eg, tumor purity), experimental platforms, differences in miR expression across cancers, and other technical factors. Thus, pan-cancer analysis requires robust algorithms to detect key features (MTIs) in the setting of noisy multi-dimensional datasets. While early efforts addressed these challenges by performing analysis of each cancer individually before combining these results,8 more recent studies have developed probabilistic approaches to large-scale pan-cancer analysis as discussed in the following.
MLR and regularized least-squares models
Multiple linear regression. Jacobsen et al.8 analyzed associations between miR and gene expression for individual cancer types using an MLR model that was fit to the gene expression, taking into account biases due to copy number abnormalities (CNA) and PM at the gene locus (Fig. 6). The linear coefficients for each miR in the MLR model indicate the magnitude of association between the miR–target pairs for each cancer type, where the greater (negative) value of the coefficient infers greater miR activity on the target gene. This method more accurately evaluated miR–target expression association in the presence of CNA and PM abnormalities that influence gene expression. To evaluate the relative strength of association between given miR and gene in at least half of the cancer types while avoiding potential biases in cancer datasets (such as sample size), the authors developed a rank-based, statistic, association RECurrence (REC), under the null hypothesis that no association exists between the miR–target pair in any of the cancer types. Jacobsen et al.8 applied a model-based (late integration) approach40,57 (refer “Framework for elucidating MiR-regulatory modules from multi-dimensional omics data” section) that applies an MLR model to each cancer type individually and combines the results (ranks) into a pan-cancer REC score.
Figure 6.
MLR of mRNA expression in a single-cancer type to identify miR–mRNA associations. mRNA expression from each sample is modeled as the response variable (left), and miR expression, DNA copy number abnormalities (CNA), and level of PM are the independent variables. The MLR evaluates pairwise associations between mRNA j and miR k while controlling for the confounding variables CNA and PM. The greater the negative value of βjk in the MLR, the greater the miR–mRNA association. The degrees of PM and mRNA and miR expression are shown as a heat plot (with increased, unchanged, and decreased levels shown in red, gray, and blue, respectively). CNA are expressed as an increase in the number of copies (gain), no change in copy number (wild type), or loss of copies of the gene, shown in red, gray, and blue, respectively. Modified from Jacobsen et al.8
Using this approach, the investigators identified 143 miR–target pairs (40 miRs and 72 mRNA targets) having both strong negative REC score and exceeding sequence-based predicted target thresholds (using TargetScan and miRanda scores). The 40 miRs in the pan-cancer network were more likely to be dysregulated by genetic and epigenetic alterations – a common property of cancer driver genes – than 180 other miRs expressed across all cancer types. The results of this analysis are available in the CancerMiner website (http://cancerminer.org).
Regularized regression (LASSO)
Setty et al.87 applied a LASSO model using CNA, miR binding site counts in mRNA 3′ UTR, and TF binding site counts in the gene promoter as covariates to explain gene expression changes (tumor versus normal) from subtypes of glioblastoma multiforme. First, LASSO was applied on a sample-by-sample basis to identify key direct regulators (miR and TF) that account for DE genes in glioblastoma multiforme relative to normal brain samples. A second LASSO model used a multi-task learning framework (group LASSO) to learn regression models of all samples simultaneously. Multi-task learning seeks to perform individual tasks (eg, selecting MTIs in subtypes), while exploiting the relationships between several learning tasks to increase the power of the search, thereby improving model performance. Group LASSO, an extension of LASSO, was developed for multi-task learning to select a common subset of features among tasks.109 For both LASSO models, Setty et al.87 used a feature-scoring scheme to determine significant regulators for common and subtype-specific gene expression. This scoring scheme estimated increase in total residual error in the models when an miR was excluded in order to determine its degree of influence in predicting gene expression changes. Using this method, common and subtype-specific miR and TFs were identified. This software is available in the R package RegulatorInference (https://bitbucket.org/leslielab/regulatorinference/).
Le and Bar-Joseph53 developed a probabilistic regression model called protein interaction-based miR modules (PIMiM) to discover MRMs in specific cancers (conditions) and across multiple cancers. The goal of this approach was to determine MRMs that explain gene expression as a function of the miR expression and the set of proteins the genes interact with. This module-based method assigned miRs and predicted targets to one of K modules (where K was a predetermined number). A regularized probabilistic regression model was learned in which gene expression data were regressed to the expression data of the miRs in the assigned modules. Their algorithm used sequence-based target prediction and GGI data as constraints. The cancer-specific approach used an L1-norm to encourage sparser modules and additional terms to reward sequence-predicted MTIs and GGI pairs to the same network.
PMiM was also found to detect MRMs with higher functional enrichment than the matrix factorization method by Zhang et al.96 when PIMiM was applied to the ovarian cancer dataset from TCGA. The investigators also found that using the prior knowledge from GGI datasets greatly increased precision and recall of 115 miRs known to participate in ovarian cancer. A potential disadvantage of supervised MRM identification methods such as PMiM is that the MRMs identified naturally tend toward the reference (in this case, GGI data). Yang and Michaelidis101 have argued that publically available GGI datasets are prone to accumulated errors and oversimplification, and therefore, supervised MRM methods depend on the accuracy of these databases. Indeed, a recent evaluation of Mirsynergy,54 a LASSO and clustering-based MRM algorithm that is also dependent on GGI data, was found to produce module structures that were highly dependent on initial clustering of miRs and the GGI data.110 Moreover, supervised methods are less likely to select new candidates based on the existing data and therefore are at a disadvantage for novel discovery.101
To integrate multiple types of cancers, PMiM uses a group LASSO to regularize the models over multiple cancer (sub)types using an L1/L2 penalty to encourage miRs and genes to be assigned to the same modules across cancer types. One limitation of this approach is that the total number of modules must be determined in advance and is fixed for each cancer (sub)type. The group selection approaches implemented by Setty et al.87 and Le and Bar-Joseph53 are examples of model-based (late) integration methods (refer “Framework for elucidating MiR-regulatory modules from multidimensional omics data” section) in which different models are learned simultaneously across cancer types, increasing the power to detect pan-cancer MTIs compared to traditional approaches that integrate parameters after each cancer has been separately modeled.
Bayesian models
Chen et al.86 demonstrated the advantage of joint analysis of multiple cancers using an EB method over a single-cancer analysis for identification of MTIs. The underlying goal of their method was to integrate MTI commonalities among cancers by explicitly borrowing information among the cancer types to increase the power of detecting TP and reducing FP MTIs, respectively. It was recognized that different cancer types have distinct sets of MTIs, and therefore, the investigators expected that only a fraction of the MTIs would be shared between cancers. Additionally, cancers that were more closely related (eg, ovarian and breast cancers) were expected to share a higher degree of common MTIs than distantly related cancers. Thus, similarity between two cancers was quantified by the fraction of shared MTIs.
In the first stage, Pearson’s correlation coefficients (PCCs) for all miR–mRNA pairs were expressed as z-scores (derived from Fisher transformation), which approximately followed a normal distribution, where increasingly negative z-scores were more likely to represent MTIs. Next, to correct for multiple hypothesis testing, a variant of the Benjamini–Hochberg FDR, termed local FDR, was applied to the set of all z-scores.111 The local FDR method estimated the empirical null distribution (histogram of z-scores) using maximum likelihood, where the central peak of the distribution mainly consisted of null cases (non-interacting pairs) and the (negative) tail tended to contain non-null cases (interacting pairs with negative z-scores). Thus, the FDR could be determined at any given z-score threshold; a lower (absolute) z-score threshold resulted in a greater sensitivity at the cost of a higher FDR and vice-versa (Fig. 3B). Using samples from glioblastoma multiforme and ovarian cancer, the investigators estimated that ~10% of miR–mRNA pairs may interact.
In the next stage, the estimated similarity between cancers based on the fraction of shared MTIs was calculated. Then the estimated similarity among cancers and the estimated MTI probability in individual cancers (obtained from local FDR) were combined to derive the posterior marginal probability of MTIs between miR–target pairs. The prior probabilities that measure cancer similarity were estimated from the observed data using an iterative updating procedure. Unlike the local FDR approach for single-cancer analysis, the pan-cancer approach was shown to change the order of z-scores, thereby re-prioritizing candidate MTIs by sharing information between cancers. This method called joint analysis of multiple cancers for miR–gene interactions (MCMG) was shown to have a higher precision than Pearson’s correlation-based and LASSO approaches for identification of MTIs in a dataset of multiple cancer types. The MCMG method is implemented in R and available for download (http://bioinformatics.med.yale.edu/group).
Li and Zhang112 developed a pan-cancer analysis method called pan-cancer miRNA–target associations (PanMiRa) that directly infers the posterior distribution of the pan-cancer MTIs and accounts for potential genomic confounders. Unlike the MCMG approach that predicts cancer-specific MTIs by borrowing information from other cancers, PanMiRa aims to infer recurrent MTIs across all cancers while taking into account biases due to CNA and PM. Similar to Jacobsen et al.8, PanMiRa applies an MLR model for individual cancers with gene expression as the response (dependent) variable and miR expression, CNA, and PM as the independent variables (Fig. 6). The coefficient in the MLR model for miR expression (denoting relationship between miR k gene i in cancer type d) is converted to a t-statistic, which is subsequently transformed into z-scores. To reduce FP, only the interactions with negative z-scores in at least 75% of the cancer types were retained. Next, the investigators exploited the empirical distribution approach of the local FDR method applied to the z-scores to estimate the joint posterior of the true interactions across the pan-cancer types via an EB algorithm. The miRs and targets involved in pan-cancer interactions detected using PanMiRa were significantly enriched for known oncomiRs and oncogenes, respectively. The investigators also found a significantly higher enrichment number of MTIs detected by CLASH when comparing PanMiRa to the recurrence association method by Jacobsen et al.8 and randomly shuffled posteriors. The source code is freely available in R (http://www.cs.utoronto.ca/~yueli/PanMiRa.html).
Conclusion and Outlook
Dysregulation of miR activity is increasingly being recognized as a pivotal factor in the development and progression of cancer. Expression profiling of miRs within tumors and those secreted from tumors as circulating miRs detected in bodily fluids have great potential for diagnostic and prognostic biomarkers in many types of cancer. miRs possess several characteristics making them particularly advantageous as therapeutic targets. Their small size and resistance to degradation make delivery of miRs to the tumor site relatively achievable.
To detect miRs with therapeutic potential in cancer, the investigators have sought to identify these miRs acting as central drivers of cancer within miR-regulatory networks. These networks are composed of subsets of densely interconnected co-expressed miRs, and their overlapping targets are termed as MRM. As MRM are composed of many MTIs, MRM identification is dependent on the quality of the MTIs. Much progress has been made over the past decade in experimental high-throughput methods to detect MTIs, and this knowledge has served to improve in silico sequence-based target predictions. Integration of matched miR and gene expression profiles from cancer samples with in silico and/or experimental MTIs is instrumental for most MRM detection methods.
The general challenges for MRM detection are the complexity of the regulatory networks, the large number of FPs generated from computational analysis, and the frequently small sample sizes available. With massive multi-omic projects such as TCGA, the latter issue is abating within the cancer field. Furthermore, as our knowledge of miR biology and regulatory mechanisms continues to augment, it is expected that computational tools being developed or refined will reflect this biological reality. Our understanding of the complexities of post-transcriptional regulation by miR will continue to be refined. While mRNA destabilization has been found to be the dominant form of miR-mediated repression in endogenous genes, translational repression has been reported to be the major mode for reporter genes. Some investigators have identified miRs capable of repressing translation in one cellular context while inducing translation upregulation in another,113 challenging basic assumptions regarding miR-regulatory roles. Moreover, advances in our understanding of miR–miR interactions as well as TF–miR interactions are currently being applied to develop increasingly sophisticated network analyses to reflect the complexity of post-transcriptional regulation.
In this review, we have highlighted major classes of statistical and computational approaches to identify MTIs and MRM in individual cancer and pan-cancer analyses. Future developments in the MRM tool are anticipated to improve integration of multiple heterogeneous omics datasets across multiple types of cancer types. Exploiting improved integration of these omic datasets and the information borrowed from many cancer types is expected to facilitate novel discovery and prioritization of MRM. Specifically, MRM that encompass the interplay between miRs and TFs for gene co-regulation will help to further elucidate the miR-regulatory mechanisms. Knowledge of these important regulatory layers will be vital for the development of effective therapeutic interventions in cancer and other challenging diseases.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1293 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: CJW. Contributed to the writing of the manuscript: PH, CCdS. Agree with manuscript results and conclusions: CJW, PH, JB, CCdS. Jointly developed the structure and arguments for the paper: CJW, PH. Made critical revisions and approved final version: CJW, JB, PH, CCdS. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Gu J, Wang T, Ding Z. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics. 2014;30(15):2237–8. doi: 10.1093/bioinformatics/btu155. [DOI] [PubMed] [Google Scholar]
- 8.Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat Struct Mol Biol. 2013;20(11):1325–32. doi: 10.1038/nsmb.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64(5):311–36. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One. 2010;5(9):e13067. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton MP, Rajapakshe K, Hartig SM, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–10. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 14.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15(6):546–54. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Sun Y, Hu L, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23(2):186–99. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22(1):34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 21.Ferracin M, Pedriali M, Veronese A, et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1–2):85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo F, Di Bella S, Bonnici V, et al. A knowledge base for the discovery of function, diagnostic potential and drug effects on cellular and extracellular miRNAs. BMC Genomics. 2014;15(suppl 3):S4. doi: 10.1186/1471-2164-15-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razzak R, Bedard EL, Kim JO, et al. MicroRNA expression profiling of sputum for the detection of early and locally advanced non-small-cell lung cancer: a prospective case-control study. Curr Oncol. 2016;23(2):e86–94. doi: 10.3747/co.23.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian L, Shan W, Zhang Y, Lv X, Li X, Wei C. Up-regulation of miR-21 expression predicate advanced clinicopathological features and poor prognosis in patients with non-small cell lung cancer. Pathol Oncol Res. 2016;22(1):161–7. doi: 10.1007/s12253-015-9979-7. [DOI] [PubMed] [Google Scholar]
- 29.Ma X-L, Liu L, Liu X-X, et al. Prognostic role of MicroRNA-21 in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2012;13(5):2329–34. doi: 10.7314/apjcp.2012.13.5.2329. [DOI] [PubMed] [Google Scholar]
- 30.Cantini L, Isella C, Petti C, et al. MicroRNA–mRNA interactions underlying colorectal cancer molecular subtypes. Nat Commun. 2015;6:8878. doi: 10.1038/ncomms9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deyati A, Bagewadi S, Senger P, Hofmann-Apitius M, Novac N. Systems approach for the selection of micro-RNAs as therapeutic biomarkers of anti-EGFR monoclonal antibody treatment in colorectal cancer. Sci Rep. 2015;5:8013. doi: 10.1038/srep08013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D, Xia H, Li ZY, Hua L, Li L. Identification of novel breast cancer subtype-specific biomarkers by integrating genomics analysis of DNA copy number aberrations and miRNA-mRNA dual expression profiling. Biomed Res Int. 2015;2015:746970. doi: 10.1155/2015/746970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zacher B, Abnaof K, Gade S, Younesi E, Tresch A, Frohlich H. Joint Bayesian inference of condition-specific miRNA and transcription factor activities from combined gene and microRNA expression data. Bioinformatics. 2012;28(13):1714–20. doi: 10.1093/bioinformatics/bts257. [DOI] [PubMed] [Google Scholar]
- 34.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144(6):986–98. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Li CX, Lv JY, et al. Prioritizing candidate disease miRNAs by topological features in the miRNA target-dysregulated network: case study of prostate cancer. Mol Cancer Ther. 2011;10(10):1857–66. doi: 10.1158/1535-7163.MCT-11-0055. [DOI] [PubMed] [Google Scholar]
- 36.Uhlmann S, Mannsperger H, Zhang JD, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Zhang J, Yuan X, et al. Detecting pan-cancer conserved microRNA modules from microRNA expression profiles across multiple cancers. Mol Biosyst. 2015;11(8):2227–37. doi: 10.1039/c5mb00257e. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Y, Xu C, Guan J, et al. Discovering dysfunction of multiple microRNAs cooperation in disease by a conserved microRNA co-expression network. PLoS One. 2012;7(2):e32201. doi: 10.1371/journal.pone.0032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin CC, Mitra R, Cheng F, Zhao Z. A cross-cancer differential co-expression network reveals microRNA-regulated oncogenic functional modules. Mol Biosyst. 2015;11(12):3244–52. doi: 10.1039/c5mb00443h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16(2):85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 41.Hamid JS, Hu P, Roslin NM, Ling V, Greenwood CM, Beyene J. Data integration in genetics and genomics: methods and challenges. Hum Genomics Proteomics 2009. 2009;1(1):1–12. doi: 10.4061/2009/869093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gade S, Porzelius C, Falth M, et al. Graph based fusion of miRNA and mRNA expression data improves clinical outcome prediction in prostate cancer. BMC Bioinformatics. 2011;12:488. doi: 10.1186/1471-2105-12-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taskesen E, Babaei S, Reinders MM, de Ridder J. Integration of gene expression and DNA-methylation profiles improves molecular subtype classification in acute myeloid leukemia. BMC Bioinformatics. 2015;16(suppl 4):S5. doi: 10.1186/1471-2105-16-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Li R, Dudek SM, Ritchie MD. ATHENA: identifying interactions between different levels of genomic data associated with cancer clinical outcomes using grammatical evolution neural network. BioData Min. 2013;6(1):23. doi: 10.1186/1756-0381-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gligorijevic V, Przulj N. Methods for biological data integration: perspectives and challenges. J R Soc Interface. 2015;12(112):1–19. doi: 10.1098/rsif.2015.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, Joung JG, Sohn KA, et al. Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J Am Med Inform Assoc. 2015;22(1):109–20. doi: 10.1136/amiajnl-2013-002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng X, Li Y, Walters KA, et al. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu J, Tang W, Du P, et al. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012;6:68. doi: 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Liu CC, Li W, Shen H, Laird PW, Zhou XJ. Discovery of multidimensional modules by integrative analysis of cancer genomic data. Nucleic Acids Res. 2012;40(19):9379–91. doi: 10.1093/nar/gks725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer SU, Sass S, Mueller NS, et al. Integrative analysis of MicroRNA and mRNA data reveals an orchestrated function of MicroRNAs in skeletal myocyte differentiation in response to TNF-alpha or IGF1. PLoS One. 2015;10(8):e0135284. doi: 10.1371/journal.pone.0135284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang JC, Morris QD, Frey BJ. Bayesian inference of MicroRNA targets from sequence and expression data. J Comput Biol. 2007;14(5):550–63. doi: 10.1089/cmb.2007.R002. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y, Zhou Y, Qu W, Deng M, Zhang C. A Lasso regression model for the construction of microRNA-target regulatory networks. Bioinformatics. 2011;27(17):2406–13. doi: 10.1093/bioinformatics/btr410. [DOI] [PubMed] [Google Scholar]
- 53.Le HS, Bar-Joseph Z. Integrating sequence, expression and interaction data to determine condition-specific miRNA regulation. Bioinformatics. 2013;29(13):i89–97. doi: 10.1093/bioinformatics/btt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Liang C, Wong KC, Luo J, Zhang Z. Mirsynergy: detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics. 2014;30(18):2627–35. doi: 10.1093/bioinformatics/btu373. [DOI] [PubMed] [Google Scholar]
- 55.Kannan L, Ramos M, Re A, et al. Public data and open source tools for multi-assay genomic investigation of disease. Brief Bioinform. 2016;17(4):603–15. doi: 10.1093/bib/bbv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh C, Hu P, Batt J, Santos C. Microarray meta-analysis and cross-platform normalization: integrative genomics for robust biomarker discovery. Microarrays. 2015;4(3):389–406. doi: 10.3390/microarrays4030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kristensen VN, Lingjaerde OC, Russnes HG, Vollan HK, Frigessi A, Borresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014;14(5):299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Q, Shi X, Xie Y, Huang J, Shia B, Ma S. Combining multidimensional genomic measurements for predicting cancer prognosis: observations from TCGA. Brief Bioinform. 2015;16(2):291–303. doi: 10.1093/bib/bbu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei Y. Integrative analyses of cancer data: a review from a statistical perspective. Cancer Inform. 2015;14(suppl 2):173–81. doi: 10.4137/CIN.S17303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang HM, Kuang S, Xiong X, Gao T, Liu C, Guo AY. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief Bioinform. 2015;16(1):45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- 62.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13(11):1894–910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellwanger DC, Buttner FA, Mewes HW, Stumpflen V. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27(10):1346–50. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13(9):849–51. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 67.Helwak A, Tollervey D. Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH) Nat Protoc. 2014;9(3):711–28. doi: 10.1038/nprot.2014.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19(3):321–7. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:1817–21. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rennie W, Liu C, Carmack CS, et al. STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014;42(Web Server issue):W114–8. doi: 10.1093/nar/gku376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 72.Xu J, Zhang R, Shen Y, Liu G, Lu X, Wu CI. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res. 2013;23(11):1810–6. doi: 10.1101/gr.148916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dweep H, Sticht C, Gretz N. In-silico algorithms for the screening of possible microRNA binding sites and their interactions. Curr Genomics. 2013;14(2):127–36. doi: 10.2174/1389202911314020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-Ligation studies. Bioinformatics. 2016;32(9):1316–22. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steinkraus BR, Toegel M, Fulga TA. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdiscip Rev Dev Biol. 2016;5(3):311–62. doi: 10.1002/wdev.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions – beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 78.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 80.Muniategui A, Pey J, Planes FJ, Rubio A. Joint analysis of miRNA and mRNA expression data. Brief Bioinform. 2013;14(3):263–78. doi: 10.1093/bib/bbs028. [DOI] [PubMed] [Google Scholar]
- 81.Farazi TA, Ten Hoeve JJ, Brown M, et al. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol. 2014;15(1):R9. doi: 10.1186/gb-2014-15-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sass S, Pitea A, Unger K, Hess J, Mueller NS, Theis FJ. MicroRNA-target network inference and local network enrichment analysis identify two microRNA clusters with distinct functions in head and neck squamous cell carcinoma. Int J Mol Sci. 2015;16(12):30204–22. doi: 10.3390/ijms161226230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Gill R, Cooper NG, Yoo JK, Datta S. Modeling microRNA-mRNA interactions using PLS regression in human colon cancer. BMC Med Genomics. 2011;4:44. doi: 10.1186/1755-8794-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W, Edwards A, Fan W, Flemington EK, Zhang K. miRNA-mRNA correlation-network modules in human prostate cancer and the differences between primary and metastatic tumor subtypes. PLoS One. 2012;7(6):e40130. doi: 10.1371/journal.pone.0040130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hecker N, Stephan C, Mollenkopf HJ, Jung K, Preissner R, Meyer HA. A new algorithm for integrated analysis of miRNA-mRNA interactions based on individual classification reveals insights into bladder cancer. PLoS One. 2013;8(5):e64543. doi: 10.1371/journal.pone.0064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen X, Slack FJ, Zhao H. Joint analysis of expression profiles from multiple cancers improves the identification of microRNA-gene interactions. Bioinformatics. 2013;29(17):2137–45. doi: 10.1093/bioinformatics/btt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Setty M, Helmy K, Khan AA, et al. Inferring transcriptional and microRNA-mediated regulatory programs in glioblastoma. Mol Syst Biol. 2012;8:605. doi: 10.1038/msb.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le TD, Zhang J, Liu L, Li J. Ensemble methods for MiRNA target prediction from expression data. PLoS One. 2015;10(6):e0131627. doi: 10.1371/journal.pone.0131627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Jia S, Huang H, Qiu J, Zhou C. A novel algorithm for the precise calculation of the maximal information coefficient. Sci Rep. 2014;4:6662. doi: 10.1038/srep06662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prill RJ, Marbach D, Saez-Rodriguez J, et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS One. 2010;5(2):e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang J, Le TD, Liu L, et al. Identifying direct miRNA-mRNA causal regulatory relationships in heterogeneous data. J Biomed Inform. 2014;52:438–47. doi: 10.1016/j.jbi.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 92.Li Y, Liang C, Wong KC, Jin K, Zhang Z. Inferring probabilistic miRNA-mRNA interaction signatures in cancers: a role-switch approach. Nucleic Acids Res. 2014;42(9):e76. doi: 10.1093/nar/gku182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le TD, Zhang J, Liu L, Liu H, Li J. miRLAB: an R Based Dry Lab for exploring miRNA-mRNA regulatory relationships. PLoS One. 2015;10(12):e0145386. doi: 10.1371/journal.pone.0145386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alexe G, Alexe S, Crama Y, Foldes S, Hammer PL, Simeone B. Consensus algorithms for the generation of all maximal bicliques. Discrete Appl Math. 2004;145(1):11–21. [Google Scholar]
- 95.Zhang Y, Phillips CA, Rogers GL, Baker EJ, Chesler EJ, Langston MA. On finding bicliques in bipartite graphs: a novel algorithm and its application to the integration of diverse biological data types. BMC Bioinformatics. 2014;15:110. doi: 10.1186/1471-2105-15-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang S, Li Q, Liu J, Zhou XJ. A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics. 2011;27(13):i401–9. doi: 10.1093/bioinformatics/btr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sim K, Li J, Gopalkrishnan V, Liu G. Mining maximal quasi-bicliques: Novel algorithm and applications in the stock market and protein networks. Stat Anal Data Min. 2009;2(4):255–73. [Google Scholar]
- 98.Guzzi PH, Veltri P, Cannataro M. Unraveling multiple miRNA-mRNA associations through a graph-based approach; Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine; ACM conference held in Orlando. Association for Computing Machinery (ACM); 2012. pp. 649–54. [Google Scholar]
- 99.Veksler-Lublinsky I, Shemer-Avni Y, Meiri E, Bentwich Z, Kedem K, Ziv-Ukelson M. Finding quasi-modules of human and viral miRNAs: a case study of human cytomegalovirus (HCMV) BMC Bioinformatics. 2012;13:322. doi: 10.1186/1471-2105-13-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim SJ, Ha JW, Zhang BT. Constructing higher-order miRNA-mRNA interaction networks in prostate cancer via hypergraph-based learning. BMC Syst Biol. 2013;7:47. doi: 10.1186/1752-0509-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Z, Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2016;32(1):1–8. doi: 10.1093/bioinformatics/btv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. A primer on learning in Bayesian networks for computational biology. PLoS Comput Biol. 2007;3(8):e129. doi: 10.1371/journal.pcbi.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Le TD, Liu L, Liu B, et al. Inferring microRNA and transcription factor regulatory networks in heterogeneous data. BMC Bioinformatics. 2013;14:92. doi: 10.1186/1471-2105-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu B, Li J, Tsykin A, Liu L, Gaur AB, Goodall GJ. Exploring complex miRNA-mRNA interactions with Bayesian networks by splitting-averaging strategy. BMC Bioinformatics. 2009;10:408. doi: 10.1186/1471-2105-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jin D, Lee H. A computational approach to identifying gene-microRNA modules in cancer. PLoS Comput Biol. 2015;11(1):e1004042. doi: 10.1371/journal.pcbi.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Godsey B. Improved inference of gene regulatory networks through integrated Bayesian clustering and dynamic modeling of time-course expression data. PLoS One. 2013;8(7):e68358. doi: 10.1371/journal.pone.0068358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gevaert O, De Smet F, Timmerman D, Moreau Y, De Moor B. Predicting the prognosis of breast cancer by integrating clinical and microarray data with Bayesian networks. Bioinformatics. 2006;22(14):e184–90. doi: 10.1093/bioinformatics/btl230. [DOI] [PubMed] [Google Scholar]
- 108.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meier L, Van De Geer S, Bühlmann P. The group lasso for logistic regression. J R Stat Soc Series B Stat Methodol. 2008;70(1):53–71. [Google Scholar]
- 110.Masud Karim SM, Liu L, Le TD, Li J. Identification of miRNA-mRNA regulatory modules by exploring collective group relationships. BMC Genomics. 2016;17(suppl 1):7. doi: 10.1186/s12864-015-2300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Efron B. Microarrays, empirical Bayes and the Two-Groups Model. Stat Sci. 2008;23(1):1–22. [Google Scholar]
- 112.Li Y, Zhang Z. Potential microRNA-mediated oncogenic intercellular communication revealed by pan-cancer analysis. Sci Rep. 2014;4:7097. doi: 10.1038/srep07097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 114.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]