Abstract
Background
Despite major advances in lung cancer treatment, early detection remains the most promising way of improving outcomes. To detect lung cancer in earlier stages, many serum biomarkers have been tested. Unfortunately, no single biomarker can reliably detect lung cancer. We combined a set of 2 tumor markers and 4 inflammatory or metabolic markers and tried to validate the diagnostic performance in lung cancer.
Methods
We collected serum samples from 355 lung cancer patients and 590 control subjects and divided them into training and validation datasets. After measuring serum levels of 6 biomarkers (human epididymis secretory protein 4 [HE4], carcinoembryonic antigen [CEA], regulated on activation, normal T cell expressed and secreted [RANTES], apolipoprotein A2 [ApoA2], transthyretin [TTR], and secretory vascular cell adhesion molecule-1 [sVCAM-1]), we tested various sets of biomarkers for their diagnostic performance in lung cancer.
Results
In a training dataset, the area under the curve (AUC) values were 0.821 for HE4, 0.753 for CEA, 0.858 for RANTES, 0.867 for ApoA2, 0.830 for TTR, and 0.552 for sVCAM-1. A model using all 6 biomarkers and age yielded an AUC value of 0.986 and sensitivity of 93.2% (cutoff at specificity 94%). Applying this model to the validation dataset showed similar results. The AUC value of the model was 0.988, with sensitivity of 93.33% and specificity of 92.00% at the same cutoff point used in the validation dataset. Analyses by stages and histologic subtypes all yielded similar results.
Conclusions
Combining multiple tumor and systemic inflammatory markers proved to be a valid strategy in the diagnosis of lung cancer.
Keywords: Lung neoplasms, Biomarkers
INTRODUCTION
Despite notable advances in diagnosis and treatment, lung cancer remains the most lethal cancer in most industrialized countries. More people in the United States die from lung cancer than any other type of cancer. In 2013, 150,000 people in the United States died from lung cancer. Moreover, lung cancer accounted for 19.4% of cancer deaths in adults worldwide.
Much effort has been focused on diagnosing lung cancer at earlier stages. For example, the National Lung Screening Trial showed that low-dose computed tomography (CT) screening of high-risk groups reduced lung cancer mortality by 20% compared to those who have been screened with plain chest radiography.1 Many other screening trials have been conducted or are likely to confirm this result.
Although CT screening for lung cancer is promising, it has some limitations. It is too costly to be used in mass screening and the radiologic exposure is a problem. Accordingly, most CT lung cancer screening trials included only high-risk individuals. The incidence of lung cancer among non-smokers, however, is increasing, especially in Asian populations.1–3
Serum biomarkers, on the other hand, are relatively inexpensive and repeatable. They impose no biological hazards. Many tumor markers and various potential biomarkers have been tested for this purpose, but few showed sufficient diagnostic performance to be used alone. 4,5
Systemic inflammatory markers have also been tested for use in early detection of malignancies.6–8 These operate on the premise that cancers cause extensive inflammatory activation and metabolic derangement.9 However, there are still no convincing data to suggest that inflammatory or metabolic markers can reliably detect early stage cancers.
We devised a set of 6 biomarkers consisting of 2 tumor markers and 4 inflammatory markers. The aim of this study was to validate their diagnostic value in lung cancer.
MATERIALS AND METHODS
1. Cohorts and serum samples
Serum samples from patients with lung cancer were obtained from the Korea Regional Biobank of Keimyung University Dongsan Medical Center (DSMC), Korea and the Pulmonary Division of the Department of Internal Medicine of Asan Medical Center (AMC), Korea. Serum samples from healthy donors were obtained from the Healthcare Promotion Center in the Department of Family Medicine, Seoul National University Hospital (SNUH), Korea. All samples used in this study were IRB-approved (2013-08-012, DSMC; H-1308-075-514 and H-1308-076-514, SNUH). The cancer samples were obtained before any therapeutic approaches performed. The samples were stored at −80°C or in liquid nitrogen until analysed. A total of 355 lung cancer patients (242 from AMC, 113 from DSMC) and 590 healthy control serum were used for this study. Additional 38 samples of small cell lung cancer (SCLC) were collected from AMC and one sample from DSMC. Total of 39 SCLC were submitted for testing the diagnostic performance in the cases of SCLC, using the algorithm obtained through this study.
2. Measurement of serum protein markers
Initially, thirteen biomarkers were measured and screened for this study as follows and details in Materials and Methods were described in supplement data.
Human epididymis secretory protein 4 (HE4) and leucine rich alpha-2-glycoprotein 1 (LRG-1) were measured by ELISA. Briefly, standard proteins, capture and biotinylated detect antibodies for human HE4 (XEMA Co. Ltd., Moscow, Russia) and human LRG1 (R&D systems, Minneapolis, MN, USA) were used.
Multiplexed serum immunoassay of regulated on activation, normal T cell expressed and secreted (RANTES) and secretory vascular cell adhesion molecule-1 (sVCAM-1) were performed using the xMAP technology platform with Magplex-bead array (Luminex Corp. Austin, TX, USA). Coupling and biotinylated detect antibodies of both biomarkers were purchased from R&D systems. Calibration standards were obtained from PeproTech (RANTES; Rocky Hill, NJ, USA) and R&D systems (sVCAM-1).
Apolipoprotein A (ApoA) 1, ApoA2, and beta-2 microglobulin (B2M) were measured on the Clinical Analyzer 7080 (Hitachi Medical Corp., Tokyo, Japan) by immunoturbidimetric method; cancer antigen (CA) 125, CA 19-9, carcinoembryonic antigen (CEA), and cytokeratin 19 fragment (CYFRA21.1) were measured on the Cobas e601 (Hoffmann-La Roche AG., Basel, Switzerland) using electrochemiluminescent detection; and C-reactive protein (CRP) and transthyretin (TTR) were measured on the BN2 System (Siemens AG., Berlin, Germany) by means of immunonephelometry according to the manufacturer’s instructions.
3. Statistical methods
Before analysis using various classification methods, biomarker experimental values were logarithm-transformed to base 10. Since log10 (HE4) and age were highly correlated (Pearson’s correlation, 0.50; P < 0.001), the residual of log10 (HE4) predicted linearly by age was used instead of log10 (HE4). Unless otherwise specified in this paper, the transformed biomarkers were used. If raw values were used, “raw” would be specified.
Various classification methods with linear decision boundaries, logistic regression, linear discriminant analysis, support vector machine, and lasso penalty logistic regression, were considered. The area under the receiver operating characteristic (ROC) curve (AUC) was used to compare the power of combinations of markers or individual methods.
As an initial step, the feature selection process was performed using training data. In this step, 6 biomarkers and age from among 13 biomarkers, age, and gender were carefully selected: HE4, CEA, ApoA2, transthyretin (TTR), sVCAM-1, RANTES, and age were chosen, and ApoA1, B2M, CA 125, CA 19-9, CRP, CYFRA21-1, LRG1, and gender were removed. First, the chi-square test and Student’s t-test were used to evaluate the significance of 13 biomarkers, age, and gender. In this step, gender (X-squared = 1.03, degrees of freedom [df] = 1, P = 0.314) and CA 19-9 (t = −1.075, df = 401.249, P = 0.2829) were not significant at a significance level of 0.05. Second, by removing 6 biomarkers, performance was maintained similar to that using 12 biomarkers and age. The range of AUCs using 12 biomarkers and age was from 0.987 (logistic) to 0.988 (support vector machines [SVM]), which made little difference to the range of AUCs (from 0.986 [logistic] to 0.988 [SVM]) using 6 selected biomarkers and age, but labor and material costs were reduced.
In the second step, using 6 selected biomarkers and age, the training data were used to develop non-SCLC (NSCLC) predictive models, and the test data were used to assess the accuracies of the models developed. The AUCs were compared to determine the best predictive methods. All calculations were performed using the R program package (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
1. Cohort of patients with lung cancer and controls
Total 355 lung cancer patients and 590 controls were included in this study. The clinical and demographic characteristics of lung cancer patients and controls used in this study are presented in Table 1. Five hundreds and ninety control samples from SNUH were divided into the training sets (515 controls) and the test (i.e., validation) sets (75 controls). One hundred and thirteen NSCLC samples from DSMC were used only in training sets and 242 NSCLC samples from AMC were divided into the training sets (167 cases) and the test sets (75 cases).
Table 1.
Clinical and demographic characteristics of the lung cancer cases and controls used in the training set and test set
| Variable | Training set | Test set |
|---|---|---|
| Cases | 280 | 75 |
| Age (yr) | 66 (25–82) | 64 (38–83) |
| Gender, female/male | 108/172 | 31/44 |
| TNM stage | ||
| 1 | 145 | 17 |
| 2 | 31 | 11 |
| 3 | 44 | 18 |
| 4 | 60 | 29 |
| Histology | ||
| Adenocarcinoma | 181 | 49 |
| Squamous cell carcinoma | 87 | 22 |
| Large cell carcinoma | 4 | - |
| Neuroendocrine carcinoma | 2 | - |
| Others | 6* | 4† |
| Controls | 515 | 75 |
| Age (yr) | 56 (38–79) | 56 (39–77) |
| Gender, female/male | 219/296 | 28/47 |
Values are presented as number only or median (range).
Other histology in training set includes mucoepidermoid carcinoma (n = 1), non-small cell lung cancer (NSCLC), not otherwise specified (n = 1), sarcomatoid carcinoma (n = 2), synchronous squamous cell carcinoma and adenocarcinoma (n = 1), and adenosquamous carcinoma (n = 1).
Other histology in test set includes NSCLC, not otherwise specified (n = 2), combined small cell carcinoma and adenocarcinoma (n = 1), sarcomatoid carcinoma with adenocarcinoma (n = 1), and sarcomatoid carcinoma combined with squamous cell carcinoma (n = 1).
2. Selection of six serum biomarkers and age from thirteen biomarkers and demographic features
Supplement 1 shows raw expression levels of each biomarker of cases and controls from both training and test datasets before logarithmic transformation.
The feature selection process was performed using training. Six biomarkers and age were carefully selected: HE4, CEA, ApoA2, TTR, sVCAM-1, RANTES, and age. First, the Student’s t-test were used to evaluate the significance of 13 biomarkers, age, and gender. In this step, CA 19-9 (t = −1.075, df = 401.249, P = 0.2829) were not significant under significant level 0.05. Second, we considered the residuals of each variables predicted by linear combination of the other variables using linear regression, and no significant effect was remained in the removed biomarkers to discriminate between NSCLC cases and controls. Again, the Student’s t-test was used to test the significance of each residuals. ApoA1, B2M, CA 125, CA 19-9, CRP, CYFRA21-1, LRG1 and gender were not significant under significant level 0.05 (Supplement 2-1). To review this result, we also considered the residuals of each removed variables predicted by linear combination of the selected variables using linear regression. Again, no significant effect was remained in the removed biomarkers under significant level 0.05 (Supplement 2-2). In other words, the things that the removed variables could explain but selected variables could not were not significant to function as a classifier for NSLC cases and controls. Moreover, by removing 6 biomarkers, performance was maintained similar to that by using 12 biomarkers and age. The range of AUCs using 12 biomarkers and age was from 0.987 (logistic) to 0.988 (SVM) which makes little difference to the range of AUCs which was 0.986 to 0.988 (Supplement 3-1) using selected 6 biomarkers and age, but with reduced labor and material costs.
3. Comparing the classification methods
Using selected 6 biomarkers and age, the training data was used to develop the NSCLC predictive models and the test data was used to assess the accuracies of the models developed. The AUCs were compared to determine the most predictive methods. Among the various statistical methods, logistic regression shows the highest prediction that the AUC value was 0.988 in test data (Supplement 3-2). All subsequent analysis was performed using the logistic regression.
4. Predictive value of biomarkers in training set
AUC values of each biomarker are presented in Table 2. The marker that showed the highest AUC value was ApoA2 (AUC value 0.867), and the lowest was sVCAM-1 (AUC value 0.867).
Table 2.
Comparison of AUC values of training data set for single/combined marker(s) using logistic regression
| Marker | AUC | Probability | Sensitivity at 94% specificity (%) | Cutoff at 94% specificity |
|---|---|---|---|---|
| HE4 | 0.821 | 5.04E-51 | 51.43 | 0.1094199 |
| CEA | 0.753 | 1.78E-32 | 33.21 | 0.6434527 |
| RANTES | 0.858 | 1.02E-62 | 2.86 | 5.076444 |
| ApoA2 | 0.867 | 4.98E-66 | 0.71 | 5.60206 |
| TTR | 0.83 | 1.20E-53 | 1.43 | 5.630428 |
| sVCAM-1 | 0.552 | 0.0074418 | 6.07 | 3.161243 |
| CEA, HE4 | 0.855 | 6.12E-62 | 55.00 | 0.5850375 |
| RANTES, ApoA1, TTR, sVCAM-1 | 0.964 | 2.88E-104 | 84.29 | 0.4779614 |
| CEA, HE4, RANTES, ApoA2, TTR, sVCAM-1 | 0.983 | 2.03E-112 | 92.50 | 0.357278 |
| CEA, HE4, RANTES, ApoA2, TTR, sVCAM-1, age | 0.986 | 3.68E-114 | 93.21 | 0.3700928 |
AUC, the area under the curve; HE4, human epididymis secretory protein 4; CEA, carcinoembryonic antigen; RANTES, regulated on activation, normal T cell expressed and secreted; ApoA2, apolipoprotein A2; TTR, transthyretin; sVCAM-1, secretory vascular cell adhesion molecule-1.
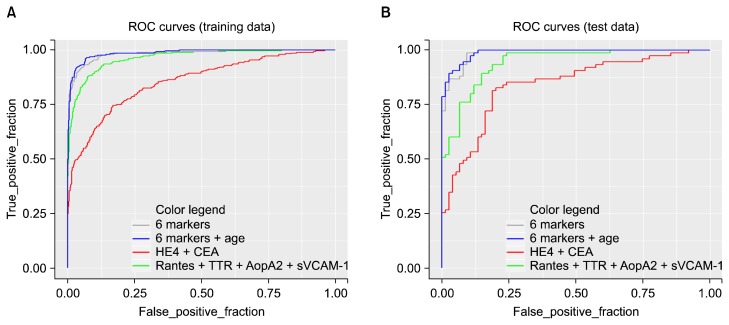
At first we used two tumor markers (CEA, HE4) together. Using these two tumor markers yielded AUC value of 0.855. Then we put four inflammatory markers (RANTES, ApoA1, TTR, sVCAM-1) together in a model. It yielded AUC value of 0.964. When using all six biomarkers (CEA, HE4, ANTES, ApoA1, TTR, sVCAM-1), AUC value increased to 0.983. When age as a clinical variable was included in the model, AUC value further increased to 0.986. Figure 1 shows ROC curves of the training set using each models. Sensitivity to discriminate lung cancer from control was 55% for tumor markers only (CEA, HE4), 84% for inflammatory markers (RANTES, ApoA2, TTR, sVCAM-1), 92.5% for all six markers, and 93.21% for six markers and age (cutoff at 94.17% specificity).
Figure 1.
Receiver operating characteristic (ROC) curve. (A) training and (B) test data set. HE4, human epididymis secretory protein 4; CEA, carcinoembryonic antigen; RANTES, regulated on activation, normal T cell expressed and secreted; TTR, transthyretin; ApoA2, apolipoprotein A2; sVCAM-1, secretory vascular cell adhesion molecule-1.
The same analyses were done by tumor stages (early vs. late, stage 1 vs. 2 vs. 3 vs. 4). But there was no significant difference according to tumor stages (Supplement 4-1, 4-2).
5. Predictive value of biomarkers in test (validation) set
Table 3 presents AUC values of each biomarker in test set. The biomarker of highest AUC value was RANTES (AUC value 0.829) and the one of the lowest was sVCAM-1 (AUC value 0.698) again.
Table 3.
Comparison of AUC values of test data set for single/combined marker(s) using logistic regression
| Marker | AUC | Probability | Cutoff from training | Specificity at the cutoff (%) | Sensitivity at the cutoff (%) |
|---|---|---|---|---|---|
| HE4 | 0.842 | 2.25E-13 | 0.1094199 | 89.33 | 57.53 |
| CEA | 0.681 | 6.60E-05 | 0.6434527 | 94.67 | 28.77 |
| RANTES | 0.829 | 1.83E-12 | 5.076444 | 92.00 | 0.00 |
| ApoA2 | 0.828 | 1.97E-12 | 5.60206 | 90.67 | 1.37 |
| TTR | 0.791 | 3.89E-10 | 5.630428 | 92.00 | 4.11 |
| sVCAM-1 | 0.698 | 1.48E-05 | 3.161243 | 82.67 | 5.48 |
| CEA, HE4 | 0.841 | 2.66E-13 | 0.5850375 | 86.67 | 57.33 |
| RANTES, ApoA1, TTR, sVCAM-1 | 0.942 | 4.47E-21 | 0.4779614 | 89.33 | 77.33 |
| CEA, HE4, RANTES, ApoA2, TTR, sVCAM-1 | 0.985 | 5.26E-25 | 0.357278 | 90.67 | 94.67 |
| CEA, HE4, RANTES, ApoA2, TTR, sVCAM-1, age | 0.988 | 2.71E-25 | 0.3700928 | 93.33 | 92.00 |
AUC, the area under the curve; HE4, human epididymis secretory protein 4; CEA, carcinoembryonic antigen; RANTES, regulated on activation, normal T cell expressed and secreted; ApoA2, apolipoprotein A2; TTR, transthyretin; sVCAM-1, secretory vascular cell adhesion molecule-1.
When the model developed in training set was applied to test set, the AUC value of the two tumor markers (CEA, HE4) was 0.841 and that of the four inflammatory markers (RANTES, ApoA1, TTR, sVCAM-1) was 0.942. When using a model including all six biomarkers (CEA, HE4, ANTES, ApoA1, TTR, sVCAM-1) of the training set, the AUC value of test set was 0.985. The value went up to 0.988 when the model including age was used (Table 3 and Fig. 2). Sensitivity to discriminate lung cancer from control was 87% for tumor markers only (CEA, HE4), 89% for inflammatory markers (RANTES, ApoA2, TTR, sVCAM-1), 91% for all six markers, and 93.3% for six markers and age at the same cutoff point.
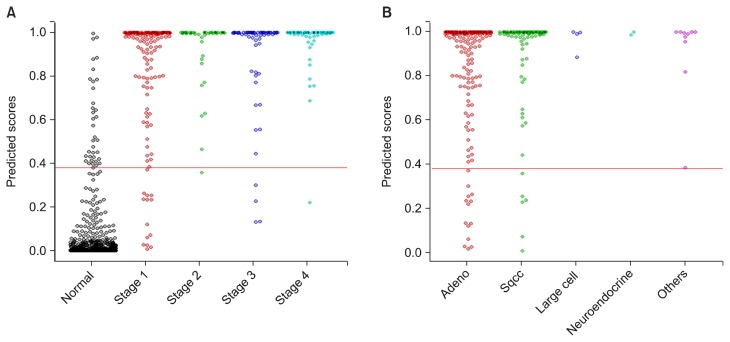
Figure 2.
Distribution of predicted scores in combined data set. Data were combined according to (A) the stage and (B) histologic subtype using 10-fold validation. Sqcc, squamous cell carcinoma.
When analysed the test set by tumor stage, we could acquire the similar results again (Supplement 5-1, 5-2).
6. Diagnostic utility of biomarkers in combined training and test set
We merged the two datasets together, and the AUC value was calculated in the same way as calculating the AUC of the training set. The diagnostic performance of our model in the combined dataset by tumor stage is shown in Figure 2 (Supplement 6). The AUC values were between 0.981 to 0.998 across all tumor stages.
When analysed by histologic subtypes, our model also showed stable diagnostic performance across all histologic subtypes (Supplement 6).
7. Diagnostic utility of biomarkers in small cell lung cancer
We used 39 samples of SCLC patients to test the diagnostic performance of our model in SCLC. Thirty-eight samples of them (97%) were determined to have cancer using our model developed in NSCLC patients.
DISCUSSION
To our knowledge, this is the first study to develop a diagnostic model for lung cancer using both tumor markers and systemic inflammatory markers together. Our strategy combining tumor markers and inflammatory biomarkers into a single model achieved high sensitivity and specificity across all stages and histologic subtypes of NSCLC.
We chose 2 tumor markers, CEA and HE4. CEA has long been widely used in clinical practice. However, its sensitivity in diagnosing lung cancer is only modest when used alone.10 On the other hand, HE4 is a relatively new and promising biomarker for ovarian cancer.11–13 Although its biological function is still unknown, the blood level of HE4 has been reported to be associated with other cancers, including endometrial cancer, breast cancer, and lung cancer.14–17
In one meta-analysis including 715 lung cancer cases and 549 controls, HE4 showed 72% sensitivity and 85% specificity.18 In one recent study, HE4 showed better sensitivity and specificity than CEA, neuron-specific enolase, CYFRA21-1, and progastrin-releasing peptide.14 Moreover, it was reported that the serum levels of HE4 were associated with treatment response and prognosis in NSCLC patients.19,20 We used both CEA and HE4 in a single model, and the AUC values of the model with the 2 tumor markers were 0.855 in the training set, and 0.841 in the validation set.
The fact that patients with cancers have enhanced systemic inflammation and metabolic derangement is well known. Many systemic inflammatory or metabolic markers have been found to be associated with cancers, but they were seldom useful in clinical practice due to relatively low sensitivity and specificity.
We used systemic inflammatory or metabolic markers such as RANTES, ApoA2, TTR, and sVCAM-1, based on our previous studies,21,22 and in accordance with previously reported data. There are many studies reporting the association of apolipoproteins and lung cancer.23,24 TTR is also reported to be expressed in lung cancer tissue,25 and helps differentiate lung cancer from lung infection. Soluble intercellular adhesion molecule-1 levels reportedly have diagnostic and prognostic value in lung cancer patients.26–28
Our model maintains sensitivity even in early stage patients, which is very important for use as a screening tool. The sensitivity of our model in stage 1 patients was 88.9% in the training set and 82.4% in the validation set. Moreover, the diagnostic performance of our model was stable across all subtypes of NSCLC. This might be related to the fact that our model has both tumor markers that could reflect tumor volume and activity as well as systemic inflammatory markers that could be associated with systemic response to the tumor. The model seems to be effective in SCLC patients, but this needs further confirmation.
In summary, we devised a set of biomarkers consisting of 2 tumor markers and 4 inflammatory markers. These showed excellent diagnostic performance both in a training set and a validation set, and remained sensitive even in early stage lung cancer patients and across all subtypes of lung cancer. The model has the potential to be useful as a screening test for lung cancer.
Supplementary Materials
ACKNOWLEDGMENTS
Authors are deeply appreciated for Professor Chang Min Choi, Department of Pulmonary and Critical Care Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, for the supplying lung cancer serum samples used in this study.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Supplementary Data can be found with this article online at http://www.jcpjournal.org/, and at http://dx.doi.org/10.15430/ JCP.2016.21.3.187
REFERENCES
- 1.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quaife SL, Ruparel M, Beeken RJ, McEwen A, Isitt J, Nolan G, et al. The Lung Screen Uptake Trial (LSUT): protocol for a randomised controlled demonstration lung cancer screening pilot testing a targeted invitation strategy for high risk and ‘hard-to-reach’ patients. BMC Cancer. 2016;16:281. doi: 10.1186/s12885-016-2316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patz EF, Jr, Greco E, Gatsonis C, Pinsky P, Kramer BS, Aberle DR. Lung cancer incidence and mortality in national lung screening trial participants who underwent low-dose CT prevalence screening: a retrospective cohort analysis of a randomised, multicentre, diagnostic screening trial. Lancet Oncol. 2016;17:590–9. doi: 10.1016/S1470-2045(15)00621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuca LE, Coy ED, Alarcón MA, Fernández A, Aristizábal FA. Cytotoxic effect of some natural compounds isolated from Lauraceae plants and synthetic derivatives. Biomedica. 2011;31:335–43. doi: 10.1590/S0120-41572011000300006. [DOI] [PubMed] [Google Scholar]
- 5.Zhao W, Yu H, Han Z, Gao N, Xue J, Wang Y. Clinical significance of joint detection of serum CEA, SCCA, and bFGF in the diagnosis of lung cancer. Int J Clin Exp Pathol. 2015;8:9506–11. [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigues D, Monteiro M, Jerónimo C, Henrique R, Belo L, Bastos MdL, et al. Renal cell carcinoma: a critical analysis of metabolomic biomarkers emerging from current model systems. Transl Res. 2016 doi: 10.1016/j.trsl.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Zhou JC, Guo JF, Teng RY, Wang QC, Wang J, Wei Q, et al. New utility of an old marker: serum low-density lipoprotein predicts histopathological response of neoadjuvant chemotherapy in locally advanced gastric cancer. Onco Targets Ther. 2016;9:5041–7. doi: 10.2147/OTT.S97061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funk CM, Honjoh S, Rodriguez AV, Cirelli C, Tononi G. Local slow waves in superficial layers of primary cortical areas during REM sleep. Curr Biol. 2016;26:396–403. doi: 10.1016/j.cub.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aminzadeh S, Vidali S, Sperl W, Kofler B, Feichtinger RG. Energy metabolism in neuroblastoma and Wilms tumor. Transl Pediatr. 2015;4:20–32. doi: 10.3978/j.issn.2224-4336.2015.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang-Chun F, Min F, Di Z, Yan-Chun H. Retrospective study to determine diagnostic utility of 6 commonly used lung cancer bio-markers among Han and Uygur population in Xinjiang Uygur autonomous region of People’s Republic of China. Medicine (Baltimore) 2016;95:e3568. doi: 10.1097/MD.0000000000003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diavatis S, Papanikolaou A. Level of HE4 is correlated with diagnosis of struma ovarii: a case report. Am J Case Rep. 2016;17:459–61. doi: 10.12659/AJCR.897158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho HY, Park SH, Park YH, Kim HB, Kang JB, Hong SH, et al. Comparison of HE4, CA125, and risk of ovarian malignancy algorithm in the prediction of ovarian cancer in Korean women. J Korean Med Sci. 2015;30:1777–83. doi: 10.3346/jkms.2015.30.12.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehtinen L, Vesterkvist P, Roering P, Korpela T, Hattara L, Kaipio K, et al. REG4 is highly expressed in mucinous ovarian cancer: a potential novel serum biomarker. PLoS One. 2016;11:e0151590. doi: 10.1371/journal.pone.0151590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Q, Liu M, Zhou N, Liu L, Song X. Serum human epididymis protein 4 (HE4) may be a better tumor marker in early lung cancer. Clin Chim Acta. 2016;455:102–6. doi: 10.1016/j.cca.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Gündüz UR, Gunaldi M, Isiksacan N, Gündüz S, Okuturlar Y, Kocoglu H. A new marker for breast cancer diagnosis, human epididymis protein 4: a preliminary study. Mol Clin Oncol. 2016;5:355–60. doi: 10.3892/mco.2016.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li BT, Lou E, Hsu M, Yu HA, Naidoo J, Zauderer MG, et al. Serum biomarkers associated with clinical outcomes fail to predict brain metastases in patients with stage IV non-small cell lung cancers. PLoS One. 2016;11:e0146063. doi: 10.1371/journal.pone.0146063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang QF, Zhou ZW, Ji HB, Pan WH, Sun MZ. Value of serum marker HE4 in pulmonary carcinoma diagnosis. Int J Clin Exp Med. 2015;8:19014–21. [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng D, Sun Y, He H. The diagnostic accuracy of HE4 in lung cancer: a meta-analysis. Dis Markers. 2015;2015:352670. doi: 10.1155/2015/352670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan WG, Hao YZ, Xu DH, Wang P, Zhou YL, Ma LB. Serum human epididymis protein 4 is associated with the treatment response of concurrent chemoradiotherapy and prognosis in patients with locally advanced non-small cell lung cancer. Clin Transl Oncol. 2016;18:375–80. doi: 10.1007/s12094-015-1375-y. [DOI] [PubMed] [Google Scholar]
- 20.Lamy PJ, Plassot C, Pujol JL. Serum HE4: an independent prognostic factor in non-small cell lung cancer. PLoS One. 2015;10:e0128836. doi: 10.1371/journal.pone.0128836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HJ, Kim YT, Park PJ, Shin YS, Kang KN, Kim Y, et al. A novel detection method of non-small cell lung cancer using multiplexed bead-based serum biomarker profiling. J Thorac Cardiovasc Surg. 2012;143:421–7. doi: 10.1016/j.jtcvs.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 22.Kim BK, Lee JW, Park PJ, Shin YS, Lee WY, Lee KA, et al. The multiplex bead array approach to identifying serum biomarkers associated with breast cancer. Breast Cancer Res. 2009;11:R22. doi: 10.1186/bcr2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgquist S, Butt T, Almgren P, Shiffman D, Stocks T, Orho-Melander M, et al. Apolipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138:2648–56. doi: 10.1002/ijc.30013. [DOI] [PubMed] [Google Scholar]
- 24.Chandler PD, Song Y, Lin J, Zhang S, Sesso HD, Mora S, et al. Lipid biomarkers and long-term risk of cancer in the women’s health study. Am J Clin Nutr. 2016;103:1397–407. doi: 10.3945/ajcn.115.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao S, Sun S, Xiao X, He D, Liu L. Selective expression of transthyretin in subtypes of lung cancer. J Mol Histol. 2016;47:239–47. doi: 10.1007/s10735-016-9666-3. [DOI] [PubMed] [Google Scholar]
- 26.Kotteas EA, Gkiozos I, Tsagkouli S, Bastas A, Ntanos I, Saif MW, et al. Soluble ICAM-1 levels in small-cell lung cancer: prognostic value for survival and predictive significance for response during chemotherapy. Med Oncol. 2013;30:662. doi: 10.1007/s12032-013-0662-0. [DOI] [PubMed] [Google Scholar]
- 27.Guney N, Soydinc HO, Derin D, Tas F, Camlica H, Duranyildiz D, et al. Serum levels of intercellular adhesion molecule ICAM-1 and E-selectin in advanced stage non-small cell lung cancer. Med Oncol. 2008;25:194–200. doi: 10.1007/s12032-007-9026-y. [DOI] [PubMed] [Google Scholar]
- 28.Kotteas EA, Boulas P, Gkiozos I, Tsagkouli S, Tsoukalas G, Syrigos KN. The intercellular cell adhesion molecule-1 (icam-1) in lung cancer: implications for disease progression and prognosis. Anticancer Res. 2014;34:4665–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.