ABSTRACT
Drosophila melanogaster is an excellent model to explore the molecular exchanges that occur between an animal intestine and associated microbes. Previous studies in Drosophila uncovered a sophisticated web of host responses to intestinal bacteria. The outcomes of these responses define critical events in the host, such as the establishment of immune responses, access to nutrients, and the rate of larval development. Despite our steady march towards illuminating the host machinery that responds to bacterial presence in the gut, there are significant gaps in our understanding of the microbial products that influence bacterial association with a fly host. We sequenced and characterized the genomes of three common Drosophila-associated microbes: Lactobacillus plantarum, Lactobacillus brevis and Acetobacter pasteurianus. For each species, we compared the genomes of Drosophila-associated strains to the genomes of strains isolated from alternative sources. We found that environmental Lactobacillus strains readily associated with adult Drosophila and were similar to fly isolates in terms of genome organization. In contrast, we identified a strain of A. pasteurianus that apparently fails to associate with adult Drosophila due to an inability to grow on fly nutrient food. Comparisons between association competent and incompetent A. pasteurianus strains identified a short list of candidate genes that may contribute to survival on fly medium. Many of the gene products unique to fly-associated strains have established roles in the stabilization of host-microbe interactions. These data add to a growing body of literature that examines the microbial perspective of host-microbe relationships.
KEY WORDS: Drosophila, Intestine, Microbiota, Host-microbe
Summary: We examined the genomes of Drosophila-associated bacteria to identify factors that allow survival within the host. These preliminary studies may point at bacterial products that influence host health.
INTRODUCTION
Environmental, microbial, and host factors act at mucosal barriers to establish a unique microclimate that shapes the lives of all participant species (Spor et al., 2011). For example, expression of a host genotype in gastrointestinal tissues works in concert with extrinsic factors to determine microbial associations (Donaldson et al., 2015). The metabolic outputs of the gastrointestinal microbiota influence critical events in the host such as education of immune phenotypes (Hooper et al., 2012; Round and Mazmanian, 2009), development of intestinal structures (Kamada et al., 2013), and access to essential micronutrients (Hacquard et al., 2015). Given the intertwined relationship between host phenotype and microbial genotype, it is of some surprise that hosts often tolerate extensive alterations to their microbiota in response to environmental shifts, such as changes in diet (David et al., 2014). However, alterations to the gastrointestinal microbiota are not invariably without consequence, and intestinal dysbiosis may lead to chronic, debilitating, and occasionally deadly diseases within the host (Belkaid and Hand, 2014; Lee et al., 2011; Schwabe and Jobin, 2013; Wen et al., 2008; Wu et al., 2010). Our appreciation of the holobiont as an intricate network of biochemical and genetic transactions between multiple participants mandates a thorough evaluation of the microbial genomes that shape host physiology. Unfortunately, such studies are tremendously complex in conventional mammalian models due to the size of the microbiome, and also the lack of laboratory techniques for the isolation and manipulation of many mammalian commensals.
The simple invertebrate Drosophila melanogaster is an excellent model holobiont (Buchon et al., 2013; Ma et al., 2015). From a developmental perspective, the Drosophila posterior midgut shares a number of important similarities with the small intestine of more complex mammalian counterparts (Buchon et al., 2013). Both organs are endodermal in origin, and are surrounded by a sheath of mesodermal visceral muscle (Spence et al., 2011; Tepass and Hartenstein, 1994). The mammalian small intestine and Drosophila posterior midgut are maintained by regularly spaced, basal intestinal stem cells that generate transitory progenitor cells (Barker et al., 2008; Jiang and Edgar, 2012; Takashima and Hartenstein, 2012); the non-proliferative enteroblasts of Drosophila; and the transient-amplifying cells of mammals. In both systems, signals along the Notch-Delta axis promote differentiation of transitory progenitors into secretory enteroendocrine cells or absorptive enterocytes (Buchon et al., 2013; Peterson and Artis, 2014). In contrast to mammals, Drosophila lacks specialized basal paneth cells for the release of antimicrobial peptides. Nonetheless, the fly genome encodes antimicrobial peptides that actively contribute to the control of intestinal symbionts and pathogens (Ryu et al., 2008), indicating the release of such factors into the Drosophila intestinal lumen. In both the mammalian small intestine and Drosophila midgut, host factors and biogeography favor association with members of the Lactobacillaceae family (Donaldson et al., 2015; Matos and Leulier, 2014). In return, metabolites from Lactobacilli activate host response pathways that promote intestinal stem cell proliferation and reactive oxygen species generation (Jones et al., 2013). Combined with the genetic accessibility of flies and their suitability for longitudinal studies of large populations in carefully defined environments, these attributes establish Drosophila as an excellent system to decipher the forces that determine genetic interactions within a holobiont (Buchon et al., 2013; Charroux and Royet, 2012; Ferrandon, 2013).
In contrast to conventional vertebrate models, the Drosophila microbiome consists of a small number of aerotolerant bacterial species that are easily isolated and cultured (Broderick and Lemaitre, 2012). The adult Drosophila intestine hosts little to no bacteria immediately after emergence from the pupal case and the microbiotal population grows in number over time (Clark et al., 2015). Several studies established that environmental factors and host genotype influence the diversity of the microbiota (Chandler et al., 2011; Ryu et al., 2008; Wong et al., 2011). It is unclear if bacteria establish stable associations with the host gut, or if they cycle from the intestine to the environment and back (Blum et al., 2013; Broderick et al., 2014). Nonetheless, lab-raised and wild Drosophila frequently associate with representatives of the genii Lactobacillus and Acetobacter. These data suggest that the intestinal lumen of an adult fly favors the survival of specific bacteria, and that such bacteria encode the necessary factors to survive or proliferate within a Drosophila intestine.
Consistent with a long-term association between the fly intestine and specific microbes, many Drosophila phenotypes are influenced by individual Lactobacillus or Acetobacter species. For example, several strains of Lactobacillus plantarum, a common Drosophila-associated microbe, promotes larval development via regulation of the TOR signal transduction pathway and induction of intestinal peptidases (Erkosar et al., 2015; Storelli et al., 2011), while Acetobacter pomorum regulates host insulin growth factor signals to promote development and metabolic homeostasis (Shin et al., 2011). In addition, members of the Acetobacter and Lactobacillus populations regulate levels of essential nutrients in the host (Chaston et al., 2015; Huang and Douglas, 2015; Wong et al., 2014). Combined, these data present a compelling argument that Lactobacilli and Acetobacter are important members of the Drosophila-microbe holobiont.
Despite our advances in the elucidation of Lactobacillus and Acetobacter influences on their Drosophila host, it is unclear if the individual species encode factors that permit survival during passage through the adult Drosophila intestine. We prepared whole genome sequences of three bacterial species that regularly associate with Drosophila – Lactobacillus brevis, Lactobacillus plantarum, and Acetobacter pasteurianus. These sequences included those for a Lactobacillus plantarum strain isolated from our lab-raised flies, and a separate strain isolated from a wild Drosophila melanogaster. For each species, we compared Drosophila-associated bacterial genomes, including ones reported previously, to the genomes of reference strains isolated from non-Drosophila sources. We noted few differences between the genomes of environmental and Drosophila associated Lactobacillus species, and found that environmental Lactobacilli readily established stable associations with a Drosophila host. In contrast, we identified an A. pasteurianus strain that apparently fails to associate with adult Drosophila. In follow-up work, we showed that this particular strain does not survive culture on conventional fly food. Comparisons between the association-competent and incompetent strains of A. pasteurianus uncovered a short list of possible regulators of A. pasteurianus viability on fly food.
RESULTS
The intestine contains structural and chemical barriers that typically inhibit bacterial growth or viability. In response, the intestinal microbiota express factors that overcome host defenses to permit bacterial survival. Here, we examined the genomes of L. brevis, L. plantarum and A. pasteurianus, which are all common members of the Drosophila intestinal community. For each species, we studied whole-genome sequences of bacterial strains that we isolated from adult Drosophila intestines, and compared them to related strains isolated from the environment. Details on the respective genomes characterized in this study are presented in Table 1.
Table 1.
Bacterial strains used in this study
We processed each genome in a similar manner. Where necessary, we used genomic databases to identify the bacterial species of newly sequenced genomes. We then annotated each genome with RAST, used PHAST to scan each genome for intact prophages, and searched for possible CRISPR arrays in the respective genomes. We scrutinized the annotated genomes for functions that might facilitate microbial survival within the intestinal lumen, with a focus on genes involved in signal transduction, transcriptional responses, orchestration of stress responses, or induction of virulence factors. Finally, we compared environmental and Drosophila-associated genomes for each species to identify bacterial factors that are unique to Drosophila-associated genomes. We present the results for each genus below.
Lactobacillus brevis and Lactobacillus plantarum
General genomic features
L. brevis is a common member of the Drosophila intestinal microbiota, and the whole genome sequence of a fly-associated strain, L. brevis EW is available (Kim et al., 2013a). We prepared a whole-genome sequence of an additional L. brevis strain (L. brevis EF) that we isolated from the intestines of wild-type adult Drosophila from our lab. For comparative purposes, we extended our study to include the genome of the environmental ATCC 367 strain. We plated homogenates from flies ten days after feeding a mono-culture of the ATCC 367 isolate and found that L. brevis ATCC 367 retained an association with wild-type adult Drosophila, confirming that the ATCC 367 strain is association-competent (Fig. 1A). Genome-to-genome distance calculations suggest that the Drosophila-associated EW and EF strains are more closely related to each other than to the ATCC 367 strain (Table 2). The genomes of Drosophila-associated strains are also larger than the environmental strain, with approximately 500,000 nucleotides more, and an additional 500 coding sequences (Table 2).
Fig. 1.
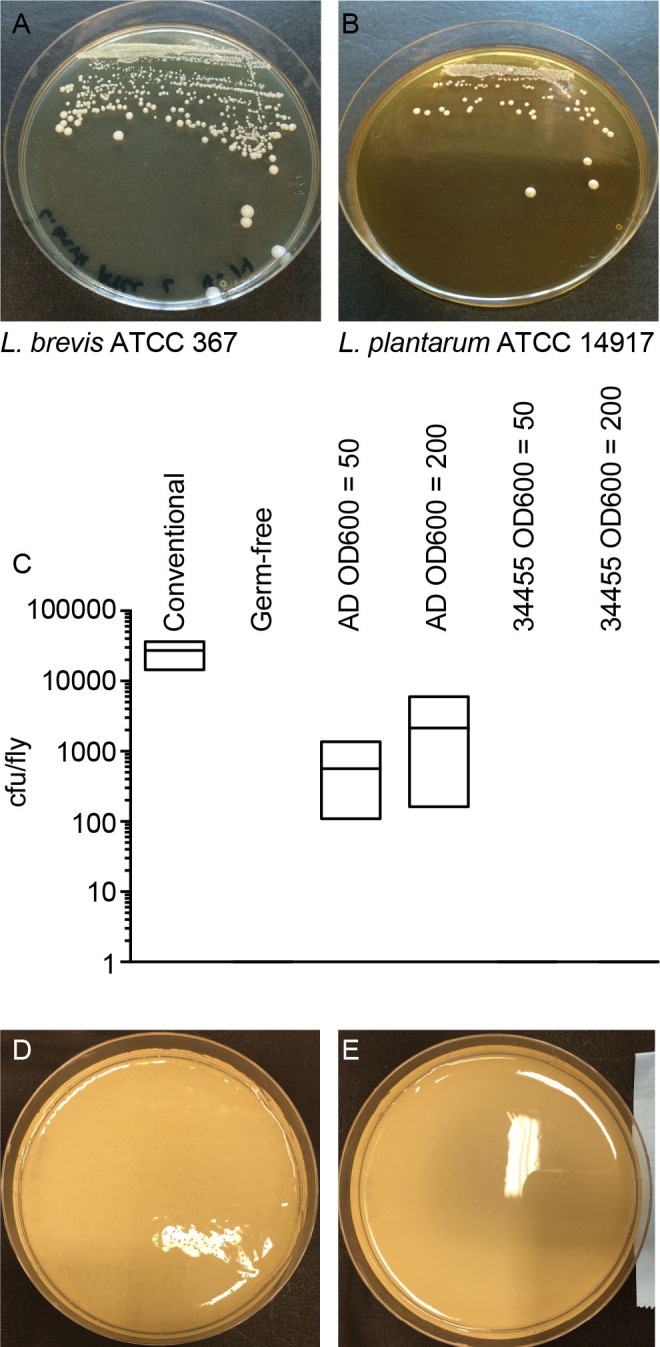
Evaluation of bacterial strain survival. (A,B) Homogenates from gnotobiotic flies mono-associated with L. brevis ATCC 367 (A) and L. plantarum ATCC 14917 (B), 10 days after the initial feeding. Each plate contains the equivalent of 1% of the homogenate of an entire fly. (C) Quantification of A. pasteurianus association with conventionally reared (column 1) flies, germ-free (column 2) flies, gnotobiotic flies that were fed A. pasteurianus strain AD at OD600 of 50 and 200, respectively (columns 3 and 4), or gnotobiotic flies that were fed A. pasteurianus strain ATCC 33445 at OD600 of 50 and 200, respectively (columns 5 and 6). Each column shows the results of three separate measurements, and association was measure as bacterial colony-forming units per fly. (D, E) Liquid cultures A. pasteurianus AD (D) and A. pasteurianus ATCC 33445 (E) were added to fly food, incubated at 29°C for 1 week, rinsed in MRS and re-plated on selective plates.
Table 2.
Details on Lactobacillus genomes described in this study
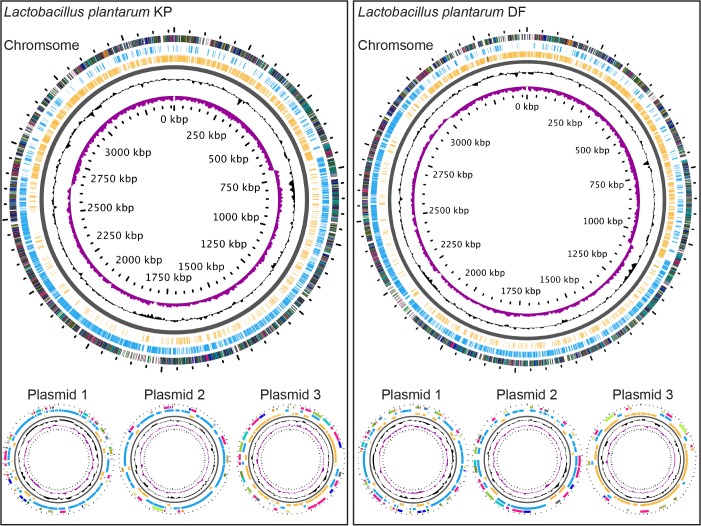
For comparative studies of L. plantarum, we focused on the environmental ATCC 14917 strain. L. plantarum ATCC 14917 was isolated from pickled cabbage. Similar to the ATCC 367 strain of L. brevis, we noticed that the ATCC 14917 strain of L. plantarum remained associated with wild-type Drosophila ten days after feeding (Fig. 1B). We compared the ATCC 14917 strain to four Drosophila-associated genomes: WJL, DMCS_001, DF and KP. WJL and DMCS_001 were isolated from Drosophila raised in geographically separate labs (Kim et al., 2013b; Newell et al., 2014). We isolated the KP strain from the intestines of our lab-raised wild-type strain, and the DF strain from an isofemale wild Drosophila melanogaster line that we captured in Edmonton, Canada in the summer of 2014. The DF and KP genomes encode one chromosome and three closely related plasmids each (Fig. 2). While all five genomes are closely related, genome-to-genome distance calculators suggest a greater degree of identity among the Drosophila-associated KP, DF, WJL and DMCS_001 strains (Table 2). In general, the environmental genome is smaller than the Drosophila-associated genomes, encodes fewer RNAs and coding sequences, and contains fewer phage-associated proteins (Table 2).
Fig. 2.
Illustrations of the genomes for L. plantarum strains KP and DF. GC skew is indicated in purple, and GC content is indicated in black. All positive strand ORFs are shown in blue, and negative strand ORFs are shown in yellow.
Environmental response factors
We then examined genetic regulatory networks within the individual Lactobacillus strains to determine if Drosophila-associated strains encode distinct regulatory components that permit adaptation to the harsh environment of an adult intestine. For these studies, we paid particular attention to two-component systems, transcription factors and additional DNA-binding proteins within the respective genomes. We did not observe substantial differences between Drosophila-associated and environmental genomes for either L. brevis or L. plantarum (Table 2). Likewise, we only observed slight differences between Drosophila-associated and environmental strains when we considered genes dedicated to signal transduction, stress responses, or virulence (Table 2).
Prophages and CRISPR responses
Comparisons between environmental and Drosophila-associated Lactobacillus genomes uncovered a propensity for prophage accumulation within the Drosophila-associated genomes. For example, we detected an average of four intact prophage genomes in Lactobacillus strains isolated from flies, and a maximum of two prophage genomes in environmental strains. The EW and EF L. brevis genomes include four intact temperate prophages, compared to an absence of prophages from the environmental L. brevis strain (Table 2). We found CRISPR sequences that target a common Lactobacillus phage within all three genomes, while the environmental strain encoded a separate CRISPR array that targets a Lactobacillus plasmid (Table 2). These results suggest an ongoing interaction between prophages and CRISPR defenses in the genomes of Drosophila-associated L. brevis strains. Similar to our observations with L. brevis genomes, we observed a greater number of intact prophage genomes in Drosophila-associated L. plantarum strains than in the environmental strain (Table 2). The main difference between the Drosophila-associated brevis and plantarum strains is that the plantarum strains do not appear to encode CRISPR-dependent anti-phage defenses within their genomes.
Function-based comparisons of Drosophila-associated and environmental Lactobacillus strains
In general, the data above suggest very minor differences between the genomes of Drosophila-associated and environmental strains of Lactobacilli. To characterize these differences in greater detail, we performed a function-based comparison of the 185 genes that are common to Drosophila-associated L. brevis genomes, but absent from the environmental strain. This set of 185 genes describes thirteen distinct functional categories, with forty-seven unique roles (Table 2). Unsurprisingly, phage and CRISPR-associated gene products account for two of those categories, and cover eleven of the forty-seven unique roles.
Of the remaining gene products, the dominant functional categories are dedicated to roles that appear suited for survival within an intestine. These include a biochemical cascade that converts α-D-glucose-1-phosphate to dTDP-4-dehydro-6-deoxy-L-mannose, an exopolysaccharide that contributes to prokaryotic survival within a host intestine (Ruas-Madiedo et al., 2006; Zivkovic et al., 2015), and gene products that contribute to the formation of rhamnose-containing glycans, a cell membrane component of acid-fast bacteria that affects several host-microbe interactions, such as adhesion, recognition, and biofilm formation (Martinez et al., 2012).
We also identified gene products within Drosophila-associated L. brevis genomes that facilitate nutrient acquisition from different sources. Bacteria frequently respond to limitations in nutritional environments through activation of the cAMP receptor protein, a transcription factor that we did not identify in the environmental strain of L. brevis, but found in both Drosophila-associated strains. The cAMP receptor protein controls, among other things, the expression of gene products that coordinate metabolism of citrate (Meyer et al., 2001), a function that is also enriched among associated Drosophila-associated L. brevis genomes. In lactic acid bacteria, citrate lyase is activated in acidic environments such as those found in the gut, and increases carbon utilization and energy generation by blocking the inhibitory effects of the Lactobacillus fermentation product lactate (Magni et al., 1999). Finally, we detected an enrichment of genes involved in the transport and degradation of pectin in Drosophila-associated L. brevis genomes. Pectin is an abundant source of energy and carbon for bacteria that grow on plant and vegetable surfaces, and microbial consumption of pectin accelerates the decay of organic matter.
When we looked at the thirty-five genes exclusively observed in the genomes of DF, KP, WJL and DMCS_001 L. plantarum strains, the majority (nineteen) were prophage genes, and an additional five were hypothetical proteins of unknown function. Rather strikingly, several of the remaining genes encode products that actively suppress the growth of competing microbes. These include the PlnMNO operon that encodes a bacteriocin and cognate immunity protein (Diep et al., 1996), and 1,3-propanediol dehydrogenase, an enzyme that converts propane-1,3,-diol to 3-hydroxypropanal. 3-hydroxypropanal, also known as reuterin, is a Lactobacillus reuteri metabolite that exerts broad-spectrum microbicidal effects on intestinal microbes in other animals (Jones and Versalovic, 2009).
Acetobacter pasteurianus
General genomic features
Although Acetobacter frequently associate with Drosophila in the wild and in the lab, we are unaware of any whole-genome sequences of A. pasteurianus strains derived from the intestines of adult Drosophila. To address this shortcoming, we completed a whole-genome sequence of an A. pasteurianus strain (A. pasteurianus AD) that we isolated from the intestines of wild-type Drosophila. For comparative purposes, we examined the available genomic sequences of the NBRC 101655 strain, and the ATCC 33445 strain. Our initial attempts to generate gnotobiotic flies, suggested that the ATCC 33445 strain fails to associate with Drosophila, something we subsequently confirmed (Fig. 1C). These data suggest that the ATCC 33445 isolate is either incapable of survival within the fly gut, or incapable of growth on fly culture medium. To distinguish between these possibilities, we examined the viability of the ATCC 33445 isolate on fly food in the absence of Drosophila. The AD strain isolated from Drosophila survives culture on fly food (Fig. 1D), however, we found that the ATCC 33445 strain failed to do so (Fig. 1E).
The different viability profiles of the different strains prompted us to compare the AD, NBRC 101655, and ATCC 33445 genomes. At first glance, we did not observe substantial differences between the respective genomes. Each genome is approximately 3 MB in length, with similar GC content and similar numbers of RNA, and predicted coding sequences (Table 4). From an evolutionary perspective, A. pasteurianus AD appears more closely related to the NBRC 101655 strain than the ATCC 33445 strain (Table 4). Consistent with a greater evolutionary distance to the ATCC strain, we found that the ATCC 33445 genome encodes 112 unique proteins, while the NBRC 101655 and AD strains share 112 genes that are absent from the ATCC 33445 genome (Fig. 3).
Table 4.
Details on Lactobacillus brevis genomes described in this study
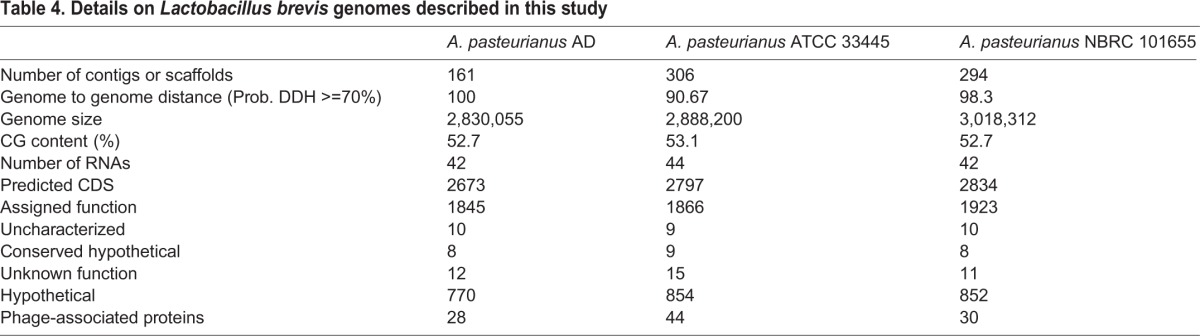
Fig. 3.
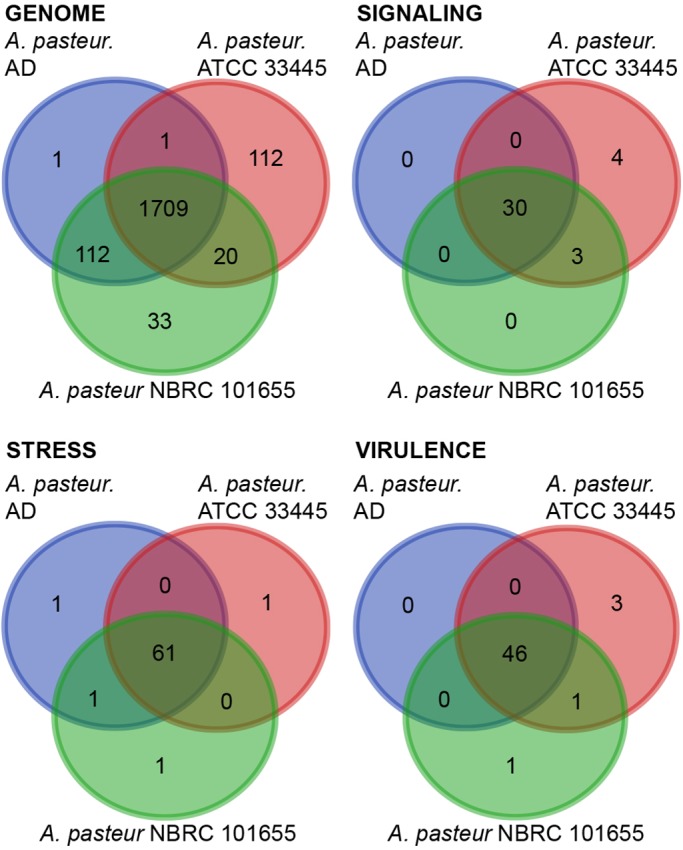
Distribution of unique gene functions in the genomes of A. pasteurianus strains AD, ATCC 33445 and NBRC 101655. All data are based on gene function annotations within RAST and exclude gene products with unknown functions.
Environmental response factors
As with L. brevis, we first compared the Drosophila-associated and environmental genomes for distinctions in gene products that respond to environmental factors. Specifically, we looked at signaling factors stress response factors, and virulence factors (Fig. 3). Across this series of comparisons, the most pronounced differences were commensurate with a closer relationship of the AD strain to the NBRC 101655 strain than to the ATCC 33445 strain. Thus, this admittedly limited comparison does not appear to identify genomic components that readily distinguish Drosophila-associated A. pasteurianus genomes from environmental counterparts. Nonetheless, these functional characterizations uncover differences between the AD and NBRC 101655 A. pasteurianus genomes that survive passage on Drosophila medium, and the ATCC 33445 genome that fails to do so.
Function-based comparisons of individual strains of Acetobacter pasteurianus
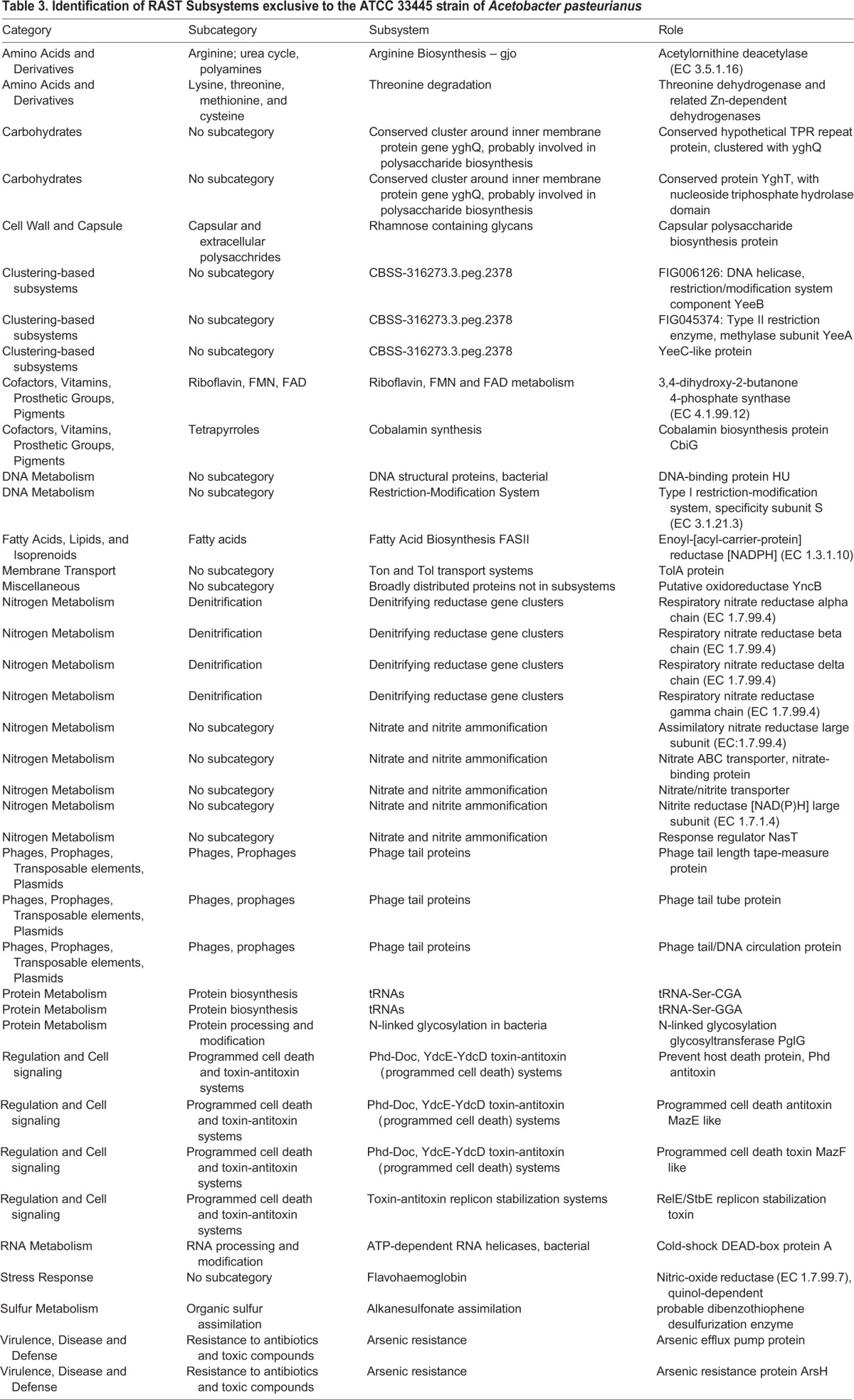
Our fortuitous identification of an environmental A. pasteurianus strain that fails to grow on fly food under experimental conditions that permit growth of all other Lactobacillus and Acetobacter strains tested allowed us to explore A. pasteurianus genomes for factors that may permit survival within a Drosophila-friendly environment. We reasoned that the AD and NBRC 101655 genomes encode biochemical functions absent from ATCC 33445 that permit survival on fly food, or that the ATCC 33445 genome encodes biochemical functions absent from the other strains that prevent survival on fly food. This prompted us to identify biological subsystems shared exclusively by the AD and NBRC 101655 (Table 5), or unique to the ATCC 33445 genome (Table 3).
Table 5.
Identification of RAST Subsystems absent from the ATCC 33445 strain of Acetobacter pasteurianus
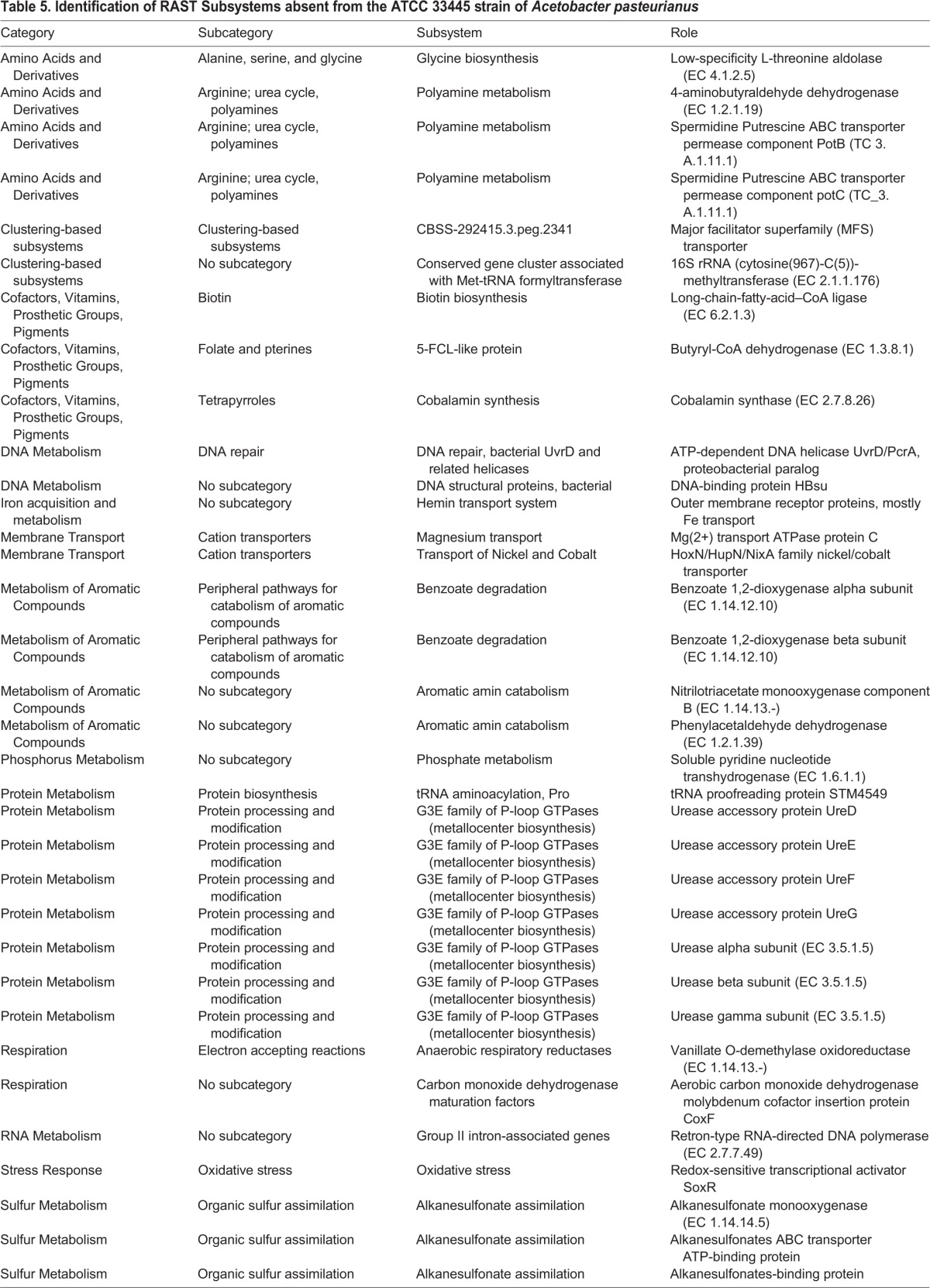
Table 3.
Identification of RAST Subsystems exclusive to the ATCC 33445 strain of Acetobacter pasteurianus
In this comparative analysis, we noted four subsystems exclusive to the AD and NBCR 101655 genomes that may explain their ability to survive on fly food. Both strains encode polyamine metabolism factors that are frequently associated with cellular growth and survival, and have established roles in the formation of biofilms (Di Martino et al., 2013). The association-competent genomes also encode factors necessary for the conversion of urea to ammonium and carbon dioxide. A similar system operates in Helicobacter pylori where it raises the gastric pH to generate a more hospitable environment for microbial survival (Scott et al., 1998). We also detected the redox-sensitive transcriptional activator SoxR in both association-competent genomes. SoxR promotes microbial survival by countering the antibacterial actions of superoxide anions (Imlay, 2015). Finally, we detected several genes that contribute to organic sulfur assimilation in association-competent genomes. These gene products may allow A. pasteurianus AD and NBRC 101655 to use alkanesulfonates as a source of sulfur during sulfate or cysteine starvation and may provide both strains a competitive advantage if sulfur is limiting.
The association-incompetent ATCC 33445 strain also encodes products that may contribute to generation of ammonia. However, the ATCC 33445 strain apparently relies on respiratory nitrate reductase and nitrite reductase to generate ammonia, as well as assimilatory nitrate reductase to access nitrate for metabolic growth. This represents an entirely different strategy to use nitrogen as a fuel for metabolic energy and growth. We also observed two toxin-antitoxin systems unique to the association-incompetent ATCC 33445 genome – an addiction module toxin that ensures propagation of plasmids to progeny cells (Engelberg-Kulka and Glaser, 1999), and a MazE/MazF type toxin-antitoxin (Masuda et al., 1993). The MazE/MazF system induces programmed cell death in prokaryotic cells in response to stressful environments.
DISCUSSION
The last decade witnessed a proliferation of elegant studies that uncovered critical host responses to microbial factors in the Drosophila intestine [reviewed in Buchon et al. (2013)]. Bacterial cues promote larval growth (Shin et al., 2011; Storelli et al., 2011), direct innate immune responses (Broderick et al., 2014; Erkosar et al., 2014), orchestrate the proliferation of intestinal stem cells (Buchon et al., 2009a,b), and regulate the uptake and storage of nutrients (Wong et al., 2014). Despite the importance of the intestinal microbiota for Drosophila health and development, there are gaps in our understanding of the biochemical events that permit bacterial survival within the hostile terrain of a fly intestine. Recent studies identified microbial metabolism and stress response pathways that mediate interactions between intestinal bacterial and their Drosophila host (Chaston et al., 2014; Newell et al., 2014). In this study, we examined the genomes of Drosophila-associated strains of L. brevis, L. plantarum, and A. pasteurianus. We were particularly interested in the identification of candidate bacterial factors that could permit survival within the intestines of adult flies. To this end, we compared fly-associated genomes to environmental strains of the same species. For each species, we observed a small number of genetic pathways that were exclusive to the Drosophila-associated genomes. Many of the Drosophila-associated pathways encode products with established roles in host-microbe interactions, raising the possibility these products may facilitate association of Drosophila with the individual strains.
Caveats
Interpretation of the data presented in this study should be influenced by several important caveats. The experimental design in this study does not distinguish between true colonization of an adult intestine and simple passage through the gut. To date, there are no studies that have identified Lactobacillus strains that fail to associate with the adult intestine of Drosophila. We also observed that environmental strains of L. brevis and L. plantarum form stable associations with Drosophila. The rather indiscriminate associations between flies and Lactobacilli confound attempts to identify fly-specific response factors within a bacterial genome. Indeed, it cannot be excluded that core elements of Lactobacillus genomes are sufficient for survival during transit through a fly intestine. In contrast, we have identified an A. pasteurianus strain that appears incapable of growth on fly food. This strain is a useful starting point for identification of Acetobacter genes that are required for association with Drosophila and we present several potential candidates within this report. As a next step, it is important to perform mutagenesis studies on candidate genes to identify the specific bacterial factors that permit survival within a fly gut lumen. To facilitate such studies, we are developing protocols for genetic manipulation of our lab isolates of Lactobacillus strains. These studies are particularly important given the strain-specific effects of individual Lactobacillus plantarum strains on host phenotypes (Erkosar et al., 2015; Schwarzer et al., 2016; Storelli et al., 2011).
Lactobacilli
For our studies of Lactobacillus genomes, we prepared whole-genome sequences of L. brevis or L. plantarum strains that we isolated from lab-raised wild-type flies, and an L. plantarum strain that we isolated from a wild Drosophila. These genomes formed the cornerstones of a comparative study that included three previously reported Drosophila-associated genomes (Kim et al., 2013a,b; Newell et al., 2014), and the genomes of environmental strains that successfully associate with the intestines of wild-type Drosophila. In this manner, we identified bacterial functions that are unique to the Drosophila-associated genomes of L. brevis and L. plantarum covered in this study. The functions fall into four broad categories: antibacterial, structural, metabolic, and phage-related.
The most striking feature common to all four Drosophila-associated L. plantarum genomes was the presence of broad-spectrum bactericidal factors. For example the DF, KP, WJL and DMCS_001 genomes all encode a complete PnlMNO operon, which encodes a bacteriocin and a corresponding immunity protein (Diep et al., 1996). Bacteriocins are produced by many lactic acid bacteria to kill neighboring bacteria, while the immunity protein protects L. plantarum from collateral damage (Cotter et al., 2005). In addition, the Drosophila-associated L. plantarum genomes encode the enzymatic capacity to generate 3-hydroxypropanal/reuterin, a bacterial toxin expressed by L. reuteri in the gut to suppress the growth of other commensals. Combined, these bactericidal molecules have the potential to counter the growth of competing bacteria inside a Drosophila host, and favor expansion of L. plantarum. The putative competitive advantages conferred by the PnlMNO operon and 3-hydroxypropanal may explain why L. plantarum is frequently reported in studies that characterize the intestinal microbiota of Drosophila.
The Drosophila-associated genomes of L. plantarum and L. brevis also encode structural components that may stabilize associations with their fly host. For example, we detected metabolic pathways for modifications to cell wells that permit host-microbe interactions and biofilm formation. These include the construction of exopolysaccharides by L. brevis and the regulation of sialic acid by L. plantarum. Sialic acid is a comparatively rare microbial metabolite, but has been observed on microbes that associate with deuterostomes. Bacteria use sialic acid as a nutrient, but they also use it to mask detection by host immune responses. While the role of sialic acid in L. plantarum association with Drosophila requires further investigation, we feel that these elements merit consideration as host-microbe interaction factors.
The Drosophila-associated genomes of L. plantarum and brevis also include gene products that may address nutritional requirements. Functional annotation of the respective genomes suggests that these gene products may enhance access to limited resources such as methionine by L. plantarum and utilization of citrate as an energy source by L. brevis. We were particularly struck by the presence of pectin metabolism factors within the genomes of Drosophila-associated strains of L. brevis. Pectin is an excellent source of carbon for bacteria that grow on plants; however, bacterial utilization of pectin accelerates the ripening and decay of the same plants (Abbott and Boraston, 2008). Thus, Drosophila-associated L. brevis genomes express factors that contribute to the decay of organic substrates. We consider this noteworthy, as Drosophila preferably consumes decayed matter as a source of nutrients. The ability of L. brevis to generate meals for their Drosophila host provides a possible explanation for the fact that Drosophila frequently associate with L. brevis. As L. brevis generates palatable meals for fly hosts, we speculate that their chances of association with flies in the wild are greater than those for many other bacteria. This host-microbe relationship is similar to a proposed mechanism for association of Erwinia carotovora with Drosophila in the wild (Basset et al., 2000). Our lab raised fly strains are fed a meal that contains yellow cornmeal, a potential source of pectin, possibly explaining the persistence of pectin metabolism genes in L. brevis strains isolated from flies.
The final difference we noted between environmental and Drosophila-associated Lactobacillus genomes was an accumulation of temperate prophage genomes throughout Drosophila-associated Lactobacilli. Intestinal stresses such as high levels of reactive oxygen species are known to trigger lysogenic induction of temperate prophages (DeMarini and Lawrence, 1992). Thus, it is feasible that bacterial strains that pass though the fly intestine will release and integrate greater numbers of lytic prophages, explaining the increased numbers of prophage genomes in Lactobacillus strains that associate with adult Drosophila.
Acetobacter
In this study, we report the first genome of a Drosophila-associated strain of A. pasteurianus, and identified an A. pasteurianus strain that cannot grow on fly food. Unfortunately, our genomic comparisons are limited by the fact that only one Drosophila-associated genome is available for study. Nonetheless, our study yields a comparatively short list of candidate functions that may regulate growth of Acteobacter on nutrient medium for Drosophila. This list includes bacterial gene products that process nitrogen, and gene products that directly control the induction of cell death in A. pasteurianus. We believe that future characterization of mutations in the respective gene product has the possibility to identify the bacterial factors that control viability of A. pasteurianus on fly food. This approach has considerable potential given the relationship between Acetobacter and Drosophila development.
In summary, our comparative study of bacterial genomes uncovers a short list of possible genetic signatures of association with Drosophila. As many of the gene products have established roles in host-microbe interactions, we propose that these genes include factors that promote the frequent association of Drosophila with Lactobacillus and Acetobacter strains. Future characterization of mutations in the individual products will reveal the relationship between the individual factors and host physiology.
MATERIALS AND METHODS
Drosophila husbandry
All Drosophila assays were performed with virgin w1118 male and female flies raised on standard corn-meal medium (Nutri-Fly Bloomington Formulation, Genesee Scientific) in a humidified incubator at 29°C. To generate germ-free flies, we transferred freshly eclosed (0-16 h old) adult flies to standard medium that we supplemented with an antibiotic cocktail (100 μg/ml ampicillin, 50 μg/ml vancomycin, 100 μg/ml neomycin and 100 μg/ml metronidazole dissolved in ethanol). This mixture has been described previously (Ryu et al., 2008). To generate gnotobiotic flies, we raised adult flies on the antibiotic cocktail for five days, starved flies for two hours, and transferred the flies to a vial containing an autoclaved fly vial cotton plug soaked with the respective bacteria. Bacterial cultures were prepared to OD600 of 50 in 5% sucrose/PBS. Twelve flies per vial were then associated with 1 ml of commensal bacteria suspension on cotton plugs. We fed the flies the bacterial meal for 16 h and transferred the flies to vials of freshly autoclaved food. Flies were raised on the initial vial for one week and transferred to fresh vials weekly thereafter. To test association, we plated fly homogenates on bacterial medium selective for Acetobacter (GYC agar) or Lactobacilli (MRS-agar) every two weeks. For A. pasteurianus, colony forming units were determined by independent quantification of three replicates of five flies/replicate. Flies were sterilized in 50% bleach, 75% ethanol and rinsed in water. Sterilized flies were homogenized in MRS broth (Fluka Analytical) and serial dilutions of the homogenate were plated on GYC agar plates. To test the survival of A. pasteurianus on fly food, bacteria were grown from for 2 days at 29°C with shaking. A bacterial culture of an OD 50 was prepared in 5% sucrose in PBS. From the OD, 50 culture serial dilutions down to 10−7 were prepared. 50 µl of each of the serial dilutions was added to autoclaved fly food. Vials were gently rotated to spread out the bacterial culture. Vials were plugged and incubated at 29°C for one week. Vials were rinsed with 1 ml of MRS and of the 1 ml rinse 50 µl was plated on GYC plates and incubated for 2 days at 29°C. The images shown in panels D and E of Fig. 1 correspond to the 10−3 dilutions.
Bacterial isolation and sequencing
We plated homogenates of 15-day-old adult Drosophila on GYC and MRS culture plates. We found that L. brevis colonies are easily distinguished from L. plantarum colonies on MRS-agar medium. We isolated individual colonies of A. pasteurianus, L. brevis and the KP strain of L. plantarum and grew them statically at 29°C in liquid MRS (L. brevis and L. plantarum), or shaking in liquid (A. pasteurianus). The DF strain of L. plantarum was isolated from a wild, mated isofemale Drosophila melanogaster captured on a rotting strawberry in the kitchen of EF in Edmonton, Canada. Bacterial DNA was isolated with the Microbial DNA Isolation kit from MO BIO Laboratories Inc. (catalog number: 12224-250) according to their instructions. The genomes of L. plantarum strains DF and KP were sequenced and assembled at the McGill University and Génome Québec Innovation Centre on the PacBio platform. The genomes of A. pasteurianus (strain AD) and L. brevis (strain EF) were sequenced at The Applied Genomics Core of the University of Alberta. For the latter genomes, we prepared Nextera XT libraries from the isolated micribial DNA according to Illumina's protocol and sequenced the libraries with using the V3-600 cycle Kit (Illumina). Whole genome sequences were then assembled using Lasergene software (DNASTAR).
Genome assembly and annotation
For each sequencing project, we confirmed the individual species with the SpeciesFinder 1.2 algorithm (Larsen et al., 2014) and calculated genome to genome distances with the genome to genome distance calculator of the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Meier-Kolthoff et al., 2013). We annotated each genome with RAST (Aziz et al., 2008), identified intact prophage genomes with the PHAST server (Zhou et al., 2011) and identified CRISPR arrays with CRISPRFinder (Grissa et al., 2007). We used the CRISPRTarget algorithm to predict CRISP targets for the individual CRISPR arrays (Biswas et al., 2015). To identify regulatory proteins within the respective genomes, we used the P2RP identifier (Barakat et al., 2013). We used the GView tool to generate graphical representations of bacterial genomes (Petkau et al., 2010).
Acknowledgements
Next generation sequencing services were performed by The Applied Genomics Core (TAGC) at the Faculty of Medicine & Dentistry, University of Alberta.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.P., D.F. and A.D. isolated the bacterial strains. K.P. prepared the bacterial genomic DNA for sequencing. E.F. performed the genome evaluations and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by a grant from the Canadian Institutes of Health Research [MOP 77746 to E.F.].
Data availability
This Whole Genome Shotgun project for L. brevis EF and A. pasteurianus AD have been deposited at DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under the accessions LPXV00000000 and LPWU00000000, respectively. The versions described in this paper are version LPXV01000000 and LPWU01000000, respectively. Chromosome1 of L. plantarum KP has been deposited at DDBJ/EMBL/GenBank under the accession CP013749 and plasmids 1-3 for the same strain have been deposited under the accession numbers CP013750, CP013751 and CP013752, respectively. Chromosome1 of L. plantarum DF has been deposited at DDBJ/EMBL/GenBank under the accession CP013753 and plasmids 1-3 for the same strain have been deposited under the accession numbers CP013754, CP013755 and CP013756, respectively.
References
- Abbott D. W. and Boraston A. B. (2008). Structural biology of pectin degradation by Enterobacteriaceae. Microbiol. Mol. Biol. Rev. 72, 301-316. 10.1128/MMBR.00038-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M. et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat M., Ortet P. and Whitworth D. E. (2013). P2RP: a Web-based framework for the identification and analysis of regulatory proteins in prokaryotic genomes. BMC Genomics 14, 269 10.1186/1471-2164-14-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Van De Wetering M. and Clevers H. (2008). The intestinal stem cell. Genes Dev. 22, 1856-1864. 10.1101/gad.1674008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset A., Khush R. S., Braun A., Gardan L., Boccard F., Hoffmann J. A. and Lemaitre B. (2000). The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97, 3376-3381. 10.1073/pnas.97.7.3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y. and Hand T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121-141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A., Fineran P. C. and Brown C. M. (2015). Computational detection of CRISPR/crRNA targets. Methods Mol. Biol. 1311, 77-89. 10.1007/978-1-4939-2687-9_5 [DOI] [PubMed] [Google Scholar]
- Blum J. E., Fischer C. N., Miles J. and Handelsman J. (2013). Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. MBio 4, e00860-e00813 10.1128/mbio.00860-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick N. A. and Lemaitre B. (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307-321. 10.4161/gmic.19896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick N. A., Buchon N. and Lemaitre B. (2014). Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio 5, e01117-e01114 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Chakrabarti S. and Lemaitre B. (2009a). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333-2344. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Poidevin M., Pradervand S. and Lemaitre B. (2009b). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200-211. 10.1016/j.chom.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A. and Lemaitre B. (2013). Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol. 11, 615-626. 10.1038/nrmicro3074 [DOI] [PubMed] [Google Scholar]
- Chandler J. A., Lang J. M., Bhatnagar S., Eisen J. A. and Kopp A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7, e1002272 10.1371/journal.pgen.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B. and Royet J. (2012). Gut-microbiota interactions in non-mammals: what can we learn from Drosophila? Semin. Immunol. 24, 17-24. 10.1016/j.smim.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Chaston J. M., Newell P. D. and Douglas A. E. (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 5, e01631-e01614 10.1128/mBio.01631-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J. M., Dobson A. J., Newell P. D. and Douglas A. E. (2015). Host genetic control of the microbiota mediates Drosophila nutritional phenotype. Appl. Environ. Microbiol. 82, 671-679. 10.1128/AEM.03301-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. I., Salazar A., Yamada R., Fitz-Gibbon S., Morselli M., Alcaraz J., Rana A., Rera M., Pellegrini M., Ja W. W. et al. (2015). Distinct shifts in microbiota composition during drosophila aging impair intestinal function and drive mortality. Cell Rep. 12, 1656-1667. 10.1016/j.celrep.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Hill C. and Ross R. P. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777-788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A. et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559-563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarini D. M. and Lawrence B. K. (1992). Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat. Res. 267, 1-17. 10.1016/0027-5107(92)90106-C [DOI] [PubMed] [Google Scholar]
- Di Martino M. L., Campilongo R., Casalino M., Micheli G., Colonna B. and Prosseda G. (2013). Polyamines: emerging players in bacteria–host interactions. Int. J. Med. Microbiol. 303, 484-491. 10.1016/j.ijmm.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Diep D. B., Havarstein L. S. and Nes I. F. (1996). Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178, 4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G. P., Lee S. M. and Mazmanian S. K. (2015). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20-32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H. and Glaser G. (1999). Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53, 43-70. 10.1146/annurev.micro.53.1.43 [DOI] [PubMed] [Google Scholar]
- Erkosar B., Defaye A., Bozonnet N., Puthier D., Royet J. and Leulier F. (2014). Drosophila microbiota modulates host metabolic gene expression via IMD/NF-kappaB signaling. PLoS ONE 9, e94729 10.1371/journal.pone.0094729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkosar B., Storelli G., Mitchell M., Bozonnet L., Bozonnet N. and Leulier F. (2015). Pathogen virulence impedes mutualist-mediated enhancement of host juvenile growth via inhibition of protein digestion. Cell Host Microbe 18, 445-455. 10.1016/j.chom.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D. (2013). The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr. Opin. Immunol. 25, 59-70. 10.1016/j.coi.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G. and Pourcel C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52-W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard S., Garrido-Oter R., González A., Spaepen S., Ackermann G., Lebeis S., Mchardy A. C., Dangl J. L., Knight R., Ley R. et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603-616. 10.1016/j.chom.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Littman D. R. and Macpherson A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268-1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-H. and Douglas A. E. (2015). Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol. Lett. 11, 20150469 10.1098/rsbl.2015.0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2015). Transcription Factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 69, 93-108. 10.1146/annurev-micro-091014-104322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. and Edgar B. A. (2012). Intestinal stem cell function in Drosophila and mice. Curr. Opin. Genet. Dev. 22, 354-360. 10.1016/j.gde.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E. and Versalovic J. (2009). Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 9, 35 10.1186/1471-2180-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. M., Luo L., Ardita C. S., Richardson A. N., Kwon Y. M., Mercante J. W., Alam A., Gates C. L., Wu H., Swanson P. A. et al. (2013). Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 32, 3017-3028. 10.1038/emboj.2013.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Seo S.-U., Chen G. Y. and Núñez G. (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321-335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Kim E.-K., Park Y. M., Lee O. Y. and Lee W.-J. (2013a). Draft genome sequence of Lactobacillus brevis strain EW, a Drosophila gut pathobiont. Genome Announc. 1, e00938-e00913 10.1128/genomea.00938-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-K., Park Y. M., Lee O. Y. and Lee W.-J. (2013b). Draft genome sequence of Lactobacillus plantarum strain WJL, a Drosophila gut symbiont. Genome Announc. 1, e00937-e00913 10.1128/genomea.00937-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Lukjancenko O., Saputra D., Rasmussen S., Hasman H., Sicheritz-Ponten T., Aarestrup F. M., Ussery D. W. and Lund O. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529-1539. 10.1128/JCM.02981-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Menezes J. S., Umesaki Y. and Mazmanian S. K. (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 108 Suppl. 1, 4615-4622. 10.1073/pnas.1000082107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Storelli G., Mitchell M. and Leulier F. (2015). Studying host-microbiota mutualism in Drosophila: Harnessing the power of gnotobiotic flies. Biomed. J. 38, 285-293. 10.4103/2319-4170.158620 [DOI] [PubMed] [Google Scholar]
- Magni C., De Mendoza D., Konings W. N. and Lolkema J. S. (1999). Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J. Bacteriol. 181, 1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K., Slesarev A., Wolf Y., Sorokin A., Mirkin B., Koonin E., Pavlov A., Pavlova N., Karamychev V., Polouchine N. et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103, 15611-15616. 10.1073/pnas.0607117103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez V., Ingwers M., Smith J., Glushka J., Yang T. and Bar-Peled M. (2012). Biosynthesis of UDP-4-keto-6-deoxyglucose and UDP-rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana. J. Biol. Chem. 287, 879-892. 10.1074/jbc.M111.287367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Miyakawa K., Nishimura Y. and Ohtsubo E. (1993). chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175, 6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos R. C. and Leulier F. (2014). Lactobacilli-Host mutualism: “learning on the fly”. Microb. Cell Fact. 13 Suppl. 1, S6 10.1186/1475-2859-13-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsutani M., Hirakawa H., Saichana N., Soemphol W., Yakushi T. and Matsushita K. (2012). Genome-wide phylogenetic analysis of differences in thermotolerance among closely related Acetobacter pasteurianus strains. Microbiology 158, 229-239. 10.1099/mic.0.052134-0 [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H.-P. and Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Dimroth P. and Bott M. (2001). Catabolite repression of the citrate fermentation genes in Klebsiella pneumoniae: evidence for involvement of the cyclic AMP receptor protein. J. Bacteriol. 183, 5248-5256. 10.1128/JB.183.18.5248-5256.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Chaston J. M., Wang Y., Winans N. J., Sannino D. R., Wong A. C. N., Dobson A. J., Kagle J. and Douglas A. E. (2014). In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Front. Microbiol. 5, 576 10.3389/fmicb.2014.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orla-Jensen, S. (1919) The Lactic Acid Bacteria. Copenhagen: Ejnar. Fred. Host & Son. [Google Scholar]
- Peterson L. W. and Artis D. (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141-153. 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- Petkau A., Stuart-Edwards M., Stothard P. and Van Domselaar G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125-3126. 10.1093/bioinformatics/btq588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L. and Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313-323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas-Madiedo P., Gueimonde M., Margolles A., De Los Reyes-Gavilan C. G. and Salminen S. (2006). Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J. Food Prot. 69, 2011-2015. [DOI] [PubMed] [Google Scholar]
- Ryu J.-H., Kim S.-H., Lee H.-Y., Bai J. Y., Nam Y.-D., Bae J.-W., Lee D. G., Shin S. C., Ha E.-M. and Lee W.-J. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777-782. 10.1126/science.1149357 [DOI] [PubMed] [Google Scholar]
- Schwabe R. F. and Jobin C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13, 800-812. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M. E., Balmand S., Hudcovic T., Heddi A. et al. (2016). Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854-857. 10.1126/science.aad8588 [DOI] [PubMed] [Google Scholar]
- Scott D. R., Weeks D., Hong C., Postius S., Melchers K. and Sachs G. (1998). The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114, 58-70. 10.1016/S0016-5085(98)70633-X [DOI] [PubMed] [Google Scholar]
- Shin S. C., Kim S.-H., You H., Kim B., Kim A. C., Lee K.-A., Yoon J.-H., Ryu J.-H. and Lee W.-J. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670-674. 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- Spence J. R., Lauf R. and Shroyer N. F. (2011). Vertebrate intestinal endoderm development. Dev. Dyn. 240, 501-520. 10.1002/dvdy.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A., Koren O. and Ley R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279-290. 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- Storelli G., Defaye A., Erkosar B., Hols P., Royet J. and Leulier F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403-414. 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Takashima S. and Hartenstein V. (2012). Genetic control of intestinal stem cell specification and development: a comparative view. Stem Cell Rev. 8, 597-608. 10.1007/s12015-012-9351-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. and Hartenstein V. (1994). Epithelium formation in the Drosophila midgut depends on the interaction of endoderm and mesoderm. Development 120, 579-590. [DOI] [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A. et al. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109-1113. 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. N. A., Ng P. and Douglas A. E. (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889-1900. 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C.-N., Dobson A. J. and Douglas A. E. (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 217, 1894-1901. 10.1242/jeb.101725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-J., Ivanov I. I., Darce J., Hattori K., Shima T., Umesaki Y., Littman D. R., Benoist C. and Mathis D. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815-827. 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J. and Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347-W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic M., Miljkovic M., Ruas-Madiedo P., Strahinic I., Tolinacki M., Golic N. and Kojic M. (2015). Exopolysaccharide production and ropy phenotype are determined by two gene clusters in putative probiotic strain Lactobacillus paraplantarum BGCG11. Appl. Environ. Microbiol. 81, 1387-1396. 10.1128/AEM.03028-14 [DOI] [PMC free article] [PubMed] [Google Scholar]