ABSTRACT
RPGR (retinitis pigmentosa GTPase regulator) is a ciliary protein associated with several forms of inherited retinal degenerative diseases. PDE6D is a ubiquitously expressed prenyl-binding protein and involved in ciliary targeting of prenylated proteins. The current working model for the RPGR function depicts that RPGR acts as a scaffold protein to recruit cargo-loaded PDE6D to primary cilia. Here, we present evidence demonstrating an alternative relationship between RPGR and PDE6D, in which RPGR is a cargo of PDE6D for ciliary targeting. We found that the constitutive isoform of RPGR, which is prenylated, requires prenylation for its ciliary localization. We also found that there are at least two independent ciliary targeting signals in RPGR: one within the N-terminal region that contains the RCC1-like domain and the other near the prenylation site at the C-terminus. Ablation of PDE6D blocked ciliary targeting of RPGR. Our study indicates that prenylated RPGR is one of the cargos of PDE6D for ciliary trafficking and provides insight into the mechanisms by which RPGR is targeted to cilia.
KEY WORDS: Retinitis pigmentosa, Primary cilia, Trafficking, Photoreceptor degeneration, Prenylation
Summary: RPGR is a ciliary protein that functions as a scaffold to recruit cargo-loaded PDE6D to cilia. Our study shows that RPGR is also a cargo of PDE6D.
INTRODUCTION
Protein prenylation is a post-translational lipid modification that adds a farnesyl or geranylgeranyl moiety to the C-terminus of a protein. This post-translational modification mediates protein-membrane and protein-protein interactions, affecting target protein's localization and function. Several proteins (e.g. PDE6 α and β subunits, GRK1, transducin γ subunit, RPGR, and INPP5E) that localize to primary cilia and the photoreceptor outer segment, which is a modified cilium, are prenylated, and mutations in these genes cause photoreceptor degeneration (Huang et al., 1995; Bowes et al., 1990; Yamamoto et al., 1997; Vervoort et al., 2000; Meindl et al., 1996; Bielas et al., 2009; Jacoby et al., 2009; Roepman et al., 1996; McLaughlin et al., 1993). Furthermore, mutations in genes involved in protein prenylation or ciliary trafficking of prenylated proteins (e.g. CHM, RCE1, ARL3, PDE6D, RP2, and ARL13B) also cause retinal degenerations in vertebrates (Seabra et al., 1993; Christiansen et al., 2011; Hanke-Gogokhia et al., 2016; Wright et al., 2016; Thomas et al., 2014; Zhang et al., 2007, 2015; Schwahn et al., 1998; Cantagrel et al., 2008), underscoring the importance of protein prenylation and proper trafficking of prenylated proteins to cilia in photoreceptor function and survival. However, mechanisms by which prenylated proteins are targeted to cilia and the photoreceptor outer segment are not sufficiently understood.
RPGR is a ciliary protein associated with several forms of inherited retinal degenerative diseases (Vervoort et al., 2000; Meindl et al., 1996; Demirci et al., 2002; Shu et al., 2007). Mutations in RPGR account for 50-60% of all X-linked retinitis pigmentosa (RP) cases and up to 20% of all RP cases (Breuer et al., 2002; Churchill et al., 2013; Shu et al., 2007). In addition, certain RPGR mutations are involved in cone-rod dystrophy (CRD) (Demirci et al., 2002). Two major isoforms of RPGR are expressed in vertebrates: RPGRex1-19 and RPGRORF15 (Kirschner et al., 1999; Hong and Li, 2002). The RPGRex1-19 isoform (hereafter RPGR) is constitutively expressed in multiple organs, including the eye, and geranylgeranylated at its C-terminus. This isoform localizes to primary cilia (Gakovic et al., 2011). The RPGRORF15 isoform is specifically expressed in the retina and localizes to the connecting cilium of photoreceptors (Hong et al., 2003; Wright et al., 2011, 2012). Contrary to the constitutive isoform, RPGRORF15 is not geranylgeranylated. How these RPGR isoforms are targeted to primary cilia and to the photoreceptor-connecting cilium is not well understood.
PDE6D (also known as PrBP/δ and PDEδ) is a ubiquitously expressed prenyl-binding protein (Florio et al., 1996; Zhang et al., 2004; Nancy et al., 2002). By binding to prenyl moieties, PDE6D is thought to mediate shuttling of prenylated proteins between membranes (Baehr, 2014; Schmick et al., 2014; Chandra et al., 2012). Recent studies found that PDE6D is involved in ciliary trafficking of INPP5E, a protein linked to Joubert syndrome (Humbert et al., 2012; Bielas et al., 2009; Jacoby et al., 2009; Thomas et al., 2014). PDE6D is also thought to be involved in the outer-segment targeting of PDE6 α/β subunits, transducin γ subunit, and GRK1 in photoreceptors because these proteins either partly mis-localize or are degraded in Pde6d−/− photoreceptors (Zhang et al., 2007).
Previous studies uncovered that RPGR binds to PDE6D through its N-terminal RCC1-like domain (RLD) (Linari et al., 1999; Wätzlich et al., 2013). Based on structural analyses, Wätzlich et al. further proposed that RPGR acts as a scaffold protein to recruit cargo-loaded PDE6D to primary cilia (Wätzlich et al., 2013). More recently, however, an alternative mode of interaction between prenylated RPGR and PDE6D was reported (Lee and Seo, 2015). This study found that PDE6D binds to the C-terminus of RPGR in a prenylation-dependent manner, suggesting that prenylated RPGR could be a cargo of PDE6D in addition to acting as a docking site for cargo-loaded PDE6D. Here, we address this question and report how the prenylated, constitutive isoform of RPGR is targeted to cilia.
RESULTS
Ciliary localization of RPGR is prenylation-dependent
RPGR is previously shown to localize to primary cilia (Gakovic et al., 2011). We validated this finding by immunohistochemistry against endogenous RPGR and knocking down RPGR expression using small interfering RNAs (siRNAs) (Fig. 1A). Although RPGR is detected throughout the cilium, it should be noted that RPGR is significantly enriched within the proximal region of the cilium, which encompasses the transition zone and the proximal end of the ciliary shaft. To dissect the mechanism of RPGR ciliary targeting and identify the minimal element(s) for that, we generated various RPGR deletion and substitution mutant variants with either a FLAG or a GFP tag. These constructs were transfected into IMCD3 cells and their localization was examined (Fig. 1B,C). Consistent with the ciliary localization of endogenous RPGR, FLAG-tagged full-length RPGR (FLAG-RPGR; aa 1-815) showed specific localization to cilia in IMCD3 cells. Similar to endogenous RPGR, FLAG-RPGR exhibited enrichment within the proximal region of the cilium, particularly in low-expressing cells. Then we examined the localization of RPGR deletion mutants. FLAG-RPGR 1-445, 1-704, and 1-811 all showed cytoplasmic localization, while the C-terminal half of RPGR (aa 440-815) exhibited specific ciliary localization but without enrichment near the ciliary base. These data suggest that prenylation of RPGR at the C-terminus may be essential for its ciliary localization and that the C-terminal half of RPGR is sufficient to target it to cilia. To test the requirement of prenylation for RPGR ciliary targeting, we generated a prenylation-incompetent mutant, in which the Cys residue within the CaaX prenylation motif was mutated to Ser (C812S), and examined its localization. As shown in Fig. 1C, this mutation abrogated ciliary localization of RPGR. Therefore, we conclude that prenylation is essential for RPGR ciliary targeting.
Fig. 1.
Localization of RPGR and its mutant variants. (A) Localization of endogenous RPGR in hTERT-RPE1 cells. hTERT-RPE1 cells were transfected with either non-targeting (siCTRL) or human RPGR (siRPGR) siRNAs and the localization of RPGR was probed with anti-RPGR antibodies (green). Cilia and centrosomes (red) were marked with anti-acetylated tubulin and anti-γ-tubulin antibodies. Nuclei were stained with DAPI (blue). (B) Localization of FLAG-tagged RPGR mutant variants in IMCD3 cells. IMCD3 cells were transiently transfected with indicated FLAG-RPGR constructs and localization of RPGR variants were probed with anti-FLAG antibodies (green). Cilia and centrosomes (red) were marked with anti-Arl13b and anti-γ-tubulin antibodies. Numbers denote the amino acid residues included in the construct, while Δ symbolizes a deletion. Nuclei were stained with DAPI (blue). (C) Localization of GFP-tagged RPGR mutant variants. IMCD3 cells were transfected with plasmids expressing indicated GFP-RPGR mutant variants and their localization was examined with anti-GFP antibodies (green). Anti-acetylated tubulin and anti-γ-tubulin antibodies were used to visualize cilia and centrosomes (red). Nuclei were stained with DAPI (blue). Scale bars: 10 µm.
Recently, high affinity binding of PDE6D to prenylated cargos combined with specific release by a ciliary small GTPase ARL3 was suggested as a determinant for ciliary targeting of prenylated proteins (Fansa et al., 2016). Since geranylgeranylated RPGR is predicted to bind to PDE6D with a low-nanomolar range equilibrium dissociation constant (Kd), which is similar to a high-affinity ciliary cargo INPP5E (Fansa et al., 2016, 2015), we tested whether prenylation and high-affinity binding to PDE6D is sufficient for ciliary targeting. To this end, we examined the localization of GFP fused with the last nine amino acids of RPGR (aa 807-815), which is sufficient to bind to PDE6D (Lee and Seo, 2015). As shown in Fig. 1C, GFP-RPGR 807-815 failed to localize to cilia, suggesting that prenylation and high affinity binding to PDE6D is not sufficient for ciliary targeting and that ciliary targeting or retention signals are needed.
Ciliary-targeting signals in RPGR
We then sought to determine additional elements that target RPGR to cilia. While the N-terminal half of RPGR (aa 1-445) failed to localize to cilia, the C-terminal half was sufficient, implying that the ciliary targeting signal(s) are within the latter half. This finding was interesting because it was previously shown that RPGRIP1 was necessary for the connecting cilium localization of RPGR in photoreceptors and that RPGR bound to RPGRIP1 and its structural homolog RPGRIP1L through the RLD (Zhao et al., 2003; Roepman et al., 2000; Boylan and Wright, 2000; Patil et al., 2012; Khanna et al., 2009). To test whether the N-terminal half of RPGR has any role in ciliary localization of RPGR and whether the sequences in the C-terminal half (except for the prenylation motif) are essential for RPGR ciliary targeting, we generated an internal deletion mutant, in which residues between aa 444 and 809 were removed [Δ(445-808)]. It should be noted that the Ser residue at the -3 position (Ser809) relative to the CaaX motif (C is at the 0 position), which is necessary for PDE6D binding (Lee and Seo, 2015), is preserved in this mutant. Interestingly, unlike the full-length RPGR, this deletion mutant [RPGR Δ(445-808)] specifically localized to the ciliary transition zone but was not detected in more distal parts of the cilium (Fig. 1B). In contrast, RPGR 440-815 mutants, which lacked the RLD, were found evenly distributed throughout the cilium with no enrichment at the ciliary base (Fig. 1B,C). These findings suggest that the N-terminal RLD is sufficient to target RPGR to the transition zone as long as RPGR is prenylated (or membrane-anchored). These data also suggest that the RLD in RPGR, presumably by binding to RPGRIP1 and RPGRIP1L (Roepman et al., 2000; Boylan and Wright, 2000; Khanna et al., 2009), induces enrichment of RPGR within the proximal region of the cilium.
To map the ciliary targeting signal(s) within the C-terminal portion of RPGR, we generated a series of deletion mutants with an N-terminal GFP tag (aa 526-815, 699-815, 731-815, 757-815, and 785-815). GFP-RPGR 526-815, 699-815, 731-815, and 757-815 all showed specific localization to cilia (Fig. 1C). GFP-RPGR 785-815 was able to localize to cilia but a considerable amount was found mis-localized to the cytoplasm, indicating that the ciliary targeting signal is partly but not completely compromised. As mentioned above, GFP-RPGR 807-815 was found dispersed throughout the cell. These data indicate that the C-terminal ciliary targeting signal(s) are located between aa 757 and 806. In summary, our data show that RPGR has at least two independent ciliary targeting signals: one within the RLD and the other near the C-terminus.
PDE6D is essential for ciliary targeting of RPGR
Given the requirement of prenylation for RPGR ciliary targeting, we sought to determine the role of PDE6D in the same process. To this end, we generated Pde6d null mutant cell lines using the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology and examined RPGR localization. We designed and tested three single guide RNAs (sgRNAs) targeting mouse Pde6d exons 1 and 2 (Fig. 2; also see Materials and Methods for sgRNA sequences and vector construction). After transient transfection into IMCD3 cells, sgRNAs #1 and #3 effectively induced indels as determined by the T7E1 assay (Fig. 2A). Therefore, we used these two sgRNAs to generate Pde6d null mutant cell lines. Following sequence analyses of the Pde6d locus in individual clones, a total of three independent Pde6d null mutant lines were chosen for further studies (clones #1-2, #1-4, and #3-5; Fig. 2B). The absence of Pde6d proteins in these cell lines was verified by immunoblotting (Fig. 2C).
Fig. 2.
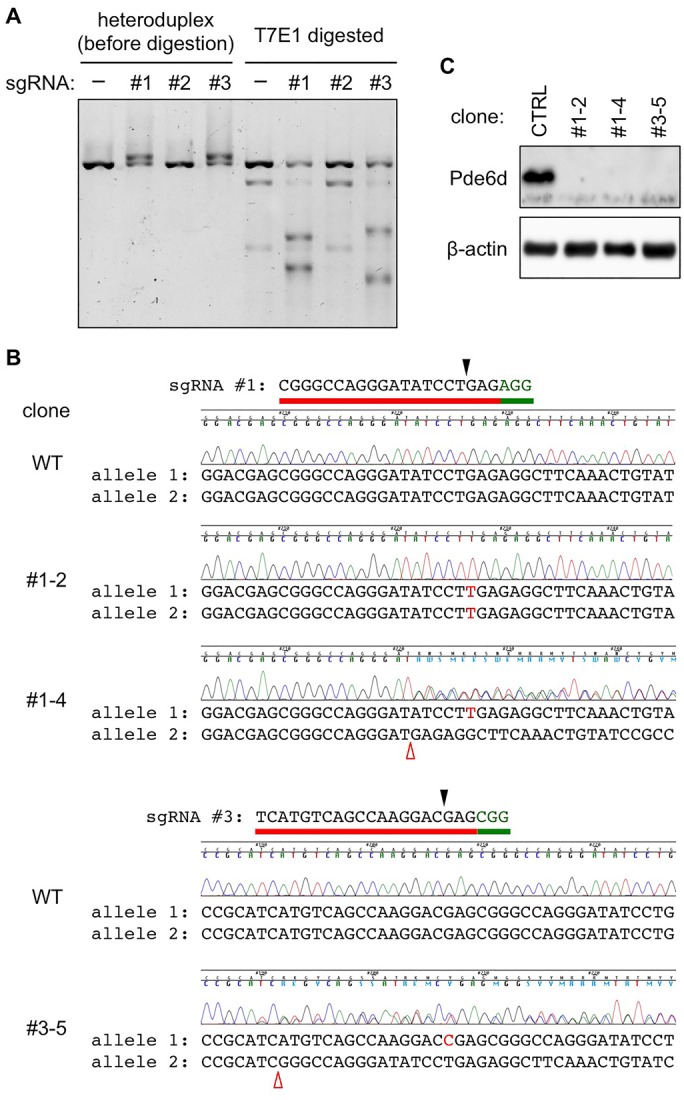
Generation of Pde6d knockout cells. (A) Effective genome editing at Pde6d exon 1 by sgRNAs #1 and #3 in IMCD3 cells. Genomic DNAs were extracted from normal or indicated sgRNA transfected IMCD3 cells and DNA fragments containing Pde6d exon 1 were amplified by PCR (see Materials and Methods for more detail). Heteroduplex DNAs before and after T7 endonuclease I (T7E1) digestion were visualized on a 2% agarose gel. (B) Sequencing results of the target region in normal (WT, wild-type) and selected Pde6d null mutant cell lines. Target site sequences for sgRNAs #1 and #3 are shown above the WT sequence chromatogram. Red bars indicate the sgRNA target sequences, and green bars indicate the protospacer adjacent motif (PAM). Black arrowheads mark the expected double stranded break (DSB) sites. Red open arrowheads denote deletions and inserted nucleotides are written in red upper case letters. Both alleles in clone #1-2 have identical 1-nucleotide (nt) insertions. Alleles in clone #1-4 have a 1-nt insertion and a 5-nt insertion. Alleles in clone #3-5 have a 1-nt insertion and a 19-nt deletion. (C) Immunoblotting against Pde6d verifies the lack of Pde6d proteins in knockout cells. Protein extracts were prepared from the indicated clonal cell lines and the quantity of Pde6d proteins was assessed by immunoblotting. β-actin was used as a loading control.
We first examined whether ciliogenesis is altered by the loss of Pde6d. Normal and Pde6d null mutant IMCD3 cells were cultured in a serum-free medium to induce ciliogenesis and cilium formation was examined by immunocytochemistry using anti-acetylated tubulin and anti-γ-tubulin antibodies. Consistent with the previous observation in human fibroblasts derived from a Joubert syndrome patient with a 42-amino acid in-frame deletion (Thomas et al., 2014) and the relatively normal development of the photoreceptor outer segment in Pde6d−/− mice (Zhang et al., 2007), complete loss of Pde6d function did not cause any ciliogenesis defect (Fig. 3; n>100 in each cell line). These data indicate that Pde6d is not required for ciliogenesis.
Fig. 3.

Normal ciliogenesis in the absence of Pde6d. Cilia formation was examined by indirect immunofluorescence microscopy in (A) normal (WT, wild type) and (B-D) Pde6d null (clones #1-2, #1-4, and #3-5) IMCD3 cell lines. Cells were serum-starved for 48 h and the presence of primary cilium was examined by anti-acetylated tubulin and anti-γ-tubulin antibodies (red). Nuclei were stained with DAPI (blue). Scale bar: 10 µm.
We then investigated the requirement of Pde6d in RPGR ciliary targeting; to this end, we transfected FLAG-RPGR into normal and Pde6d null IMCD3 cells. As shown in Fig. 4, ablation of Pde6d completely abolished RPGR ciliary localization in all three Pde6d null mutant cell lines; no ciliary localization was observed in more than 50 transfected cells in each mutant cell line. Therefore, we conclude that Pde6d is essential for RPGR ciliary targeting.
Fig. 4.
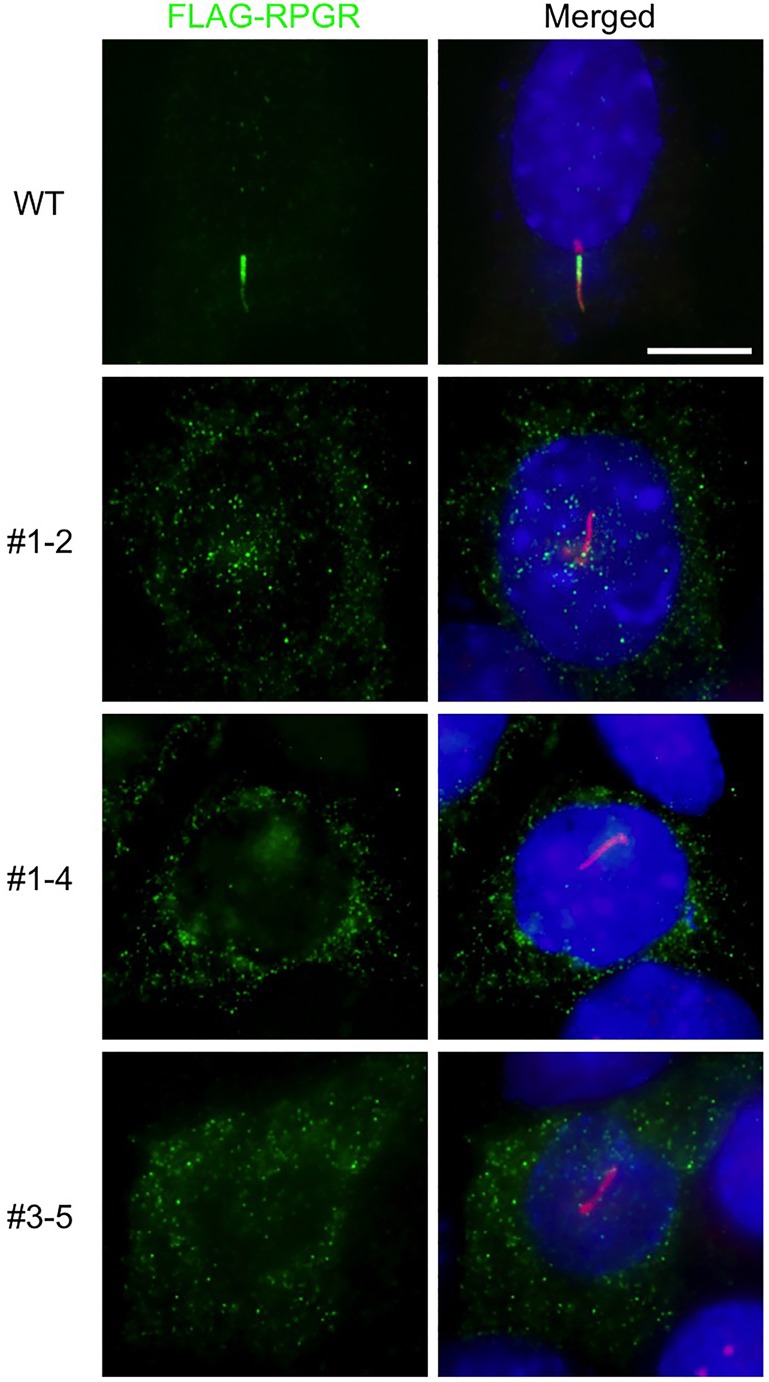
Pde6d is essential for RPGR ciliary targeting. FLAG-RPGR (full-length) was transiently transfected into normal and indicated Pde6d null mutant cell lines and its localization was probed with anti-FLAG antibodies (green). Cilia and centrosomes were marked with anti-Arl13b and anti-γ-tubulin antibodies (red). Nuclei were stained with DAPI (blue). Scale bar: 10 µm.
DISCUSSION
Our study reveals three key findings with respect to the mechanisms of RPGR ciliary targeting: (i) RPGR has at least two independent ciliary targeting signals, (ii) prenylation is essential for RPGR ciliary targeting, and (iii) prenylated RPGR is a cargo of PDE6D. As mentioned in the Introduction, RPGR interacts with PDE6D in two distinct modes (Wätzlich et al., 2013; Linari et al., 1999; Lee and Seo, 2015; Fansa et al., 2015). One of these modes is through its RLD and the other is through its C-terminal prenylation site. Through the RLD-mediated interaction, RPGR was proposed to act as a docking site to recruit prenylated cargo-PDE6D complexes to cilia (Wätzlich et al., 2013; Baehr, 2014). Our study uncovers the significance of the prenylation-dependent interaction between RPGR and PDE6D and demonstrates that prenylated RPGR is a cargo of PDE6D. Our study further suggests that the RLD in RPGR has an additional role in RPGR ciliary localization; it limits or enriches RPGR to the transition zone and the proximal end of the cilium. RPGR binds to RPGRIP1 and RPGRIP1L, which specifically localize to the transition zone in primary cilia and the connecting cilium in photoreceptors, through the RLD (Roepman et al., 2000; Boylan and Wright, 2000; Khanna et al., 2009). Furthermore, Rpgr was found mis-localized in Rpgrip1 knockout photoreceptors (Patil et al., 2012; Zhao et al., 2003). Therefore, we predict that the enrichment of RPGR at the transition zone and the proximal region of the cilium is dependent on the RLD-RPGRIP1/RPGRIP1L interactions.
Based on our findings and previous studies (Baehr, 2014; Wätzlich et al., 2013; Ismail et al., 2011; Zhao et al., 2003; Roepman et al., 2000; Boylan and Wright, 2000; Patil et al., 2012; Khanna et al., 2009), we propose a model for RPGR ciliary targeting (Fig. 5). The constitutive isoform of RPGR is geranylgeranylated upon its synthesis and anchored onto an endomembrane through its geranylgeranyl moiety. The identity of this endomembrane is currently unknown but candidates are endoplasmic reticulum, Golgi, and endosomal membranes. PDE6D is required for the extraction of RPGR from the donor membrane and shuttles it to the plasma membrane of the primary cilium. Although not validated yet, GTP-bound ARL3, a small GTPase that localizes to cilia, is expected to bind to PDE6D and induce the release of RPGR from PDE6D and eventually RPGR membrane anchoring. Interactions with RPGRIP1 and RPGRIP1L in the transition zone prevent RPGR diffusion and limit its localization to the transition zone and the proximal end of the primary cilium. RPGRIP1 and RPGRIP1L may also be involved in the initial entry of the RPGR-PDE6D complex into the cilium.
Fig. 5.
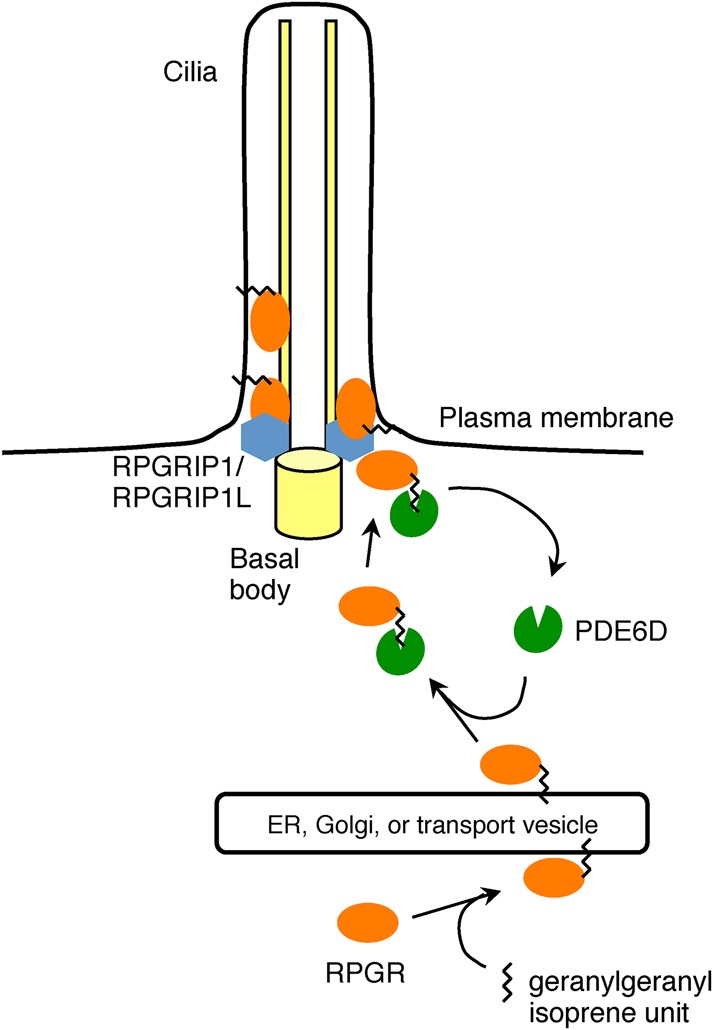
A schematic model for RPGR ciliary targeting.
It should be noted that the prenylation- and PDE6D-dependent ciliary targeting of RPGR is limited to the constitutive isoform of RPGR and that the photoreceptor-specific RPGRORF15 isoform may be targeted to the photoreceptor connecting cilium through a distinct mechanism. This is because RPGRORF15 does not have the CaaX prenylation motif and therefore it is expected not to interact with PDE6D through its C-terminus. Instead, RPGRORF15 has a Glu-rich domain at its C-terminus. Although the function of the Glu-rich domain is currently not well understood, recent studies found that it is glutamylated by tubulin tyrosine ligase like-5 (TTLL5) in photoreceptors (Sun et al., 2016; Rao et al., 2016). Since the RLD has a role in ciliary localization of RPGR but it is not sufficient for its ciliary targeting on its own (i.e. without prenylation), it is tempting to speculate that the Glu-rich domain cooperates with the RLD and contributes to the connecting cilium localization of RPGRORF15 in photoreceptors.
In summary, our study demonstrates that the constitutive isoform of RPGR is targeted to cilia in a prenylation- and PDE6D-dependent manner and, therefore RPGR is a cargo of PDE6D.
MATERIALS AND METHODS
Reagents
Antibodies used in this study and their dilutions for immunocytochemistry are as follows: rabbit polyclonal anti-RPGR (Sigma #HPA001593; 1:100 dilution), mouse monoclonal anti-acetylated tubulin (Proteintech group #66200-1-Ig; 1:2500), mouse monoclonal anti-γ-tubulin (Sigma #T6557; 1:2500), rabbit polyclonal anti-γ-tubulin (Sigma #T3559; 1:2500), mouse monoclonal anti-FLAG (Sigma #F1804; 1:2000), rabbit polyclonal anti-ARL13B (Proteintech group #17711; 1:2000), rabbit monoclonal anti-GFP (Invitrogen #G10362; 1:500), rabbit polyclonal anti-PDE6D (MyBiosources #MBS7005086; 1:1000 for immunoblotting), mouse monoclonal anti-β-actin (Sigma #A1978; 1:10,000 for immunoblotting). Small interfering RNAs (siRNAs) for control (ON-TARGETplus non-targeting siRNA) and human RPGR (ON-TARGETplus SMARTpool) were obtained from GE Dharmacon.
Constructs
The open reading frame encoding full-length RPGRex1-19 was obtained by PCR using the Human Retina Marathon-ready cDNA library (Clontech) and the Expand High Fidelity PCR system (Roche) and cloned into pSS-FS plasmid (Humbert et al., 2012) for N-terminal FLAG tag fusion. For N-terminal GFP fusion, pEGFP-C3 (Clontech) plasmid was used. Deletion and substitution mutagenesis was conducted by PCR and standard molecular biology methodologies. All constructs were sequence-verified.
Immunocytochemistry
Mouse IMCD3 (CRL-2123) and human hTERT-RPE1 (CRL-4000) cells were obtained from American Type Culture Collection (ATCC) and cultured in DMEM/F12 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin/streptomycin (Invitrogen). Transient transfection of plasmid DNAs was conducted with FuGene HD (Promega) and siRNAs were transfected with RNAiMAX (Invitrogen). Cells were seeded on glass coverslips in 24-well plates and transfected with indicated DNAs or siRNAs following the manufacturer's instruction. After 24-48 h of transfection, cells were incubated in a serum-free medium for 24-36 h to induce ciliogenesis. For immunocytochemistry, cells were fixed in 4% paraformaldehyde (PFA) in PBS followed by cold methanol. Samples were blocked with 5% bovine serum albumin (BSA) and 2% normal goat serum in PBST (PBS+0.1% Triton X-100) and incubated with indicated primary antibodies in the blocking buffer for 2 h at room temperature. After washing in PBST, coverslips were incubated with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with Alexa Fluor 488 and 568 (Invitrogen). Coverslips were mounted on VectaShield mounting medium with DAPI (Vector laboratories) and images were taken with Olympus IX71 microscope.
Generation of Pde6d knockout cells
Pde6d knockout IMCD3 cells were generated by using the CRISPR/Cas9 technology (Ran et al., 2013). Three sgRNAs targeting mouse Pde6d exons 1 and 2 were designed using a publicly available sgRNA designer tool (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) (Doench et al., 2016) and cloned into the pX459 plasmid (Ran et al., 2013) (Addgene). Oligonucleotide sequences for sgRNA cloning are as follows: sgRNA #1; 5′-CACCGCGGGCCAGGGATATCCTGAG-3′ and 5′-AAACCTCAGGATATCCCTGGCCCGC-3′, sgRNA #2; 5′-CACCGACCTTCGGGATGCCGAAACA-3′ and 5′-AAACTGTTTCGGCATCCCGAAGGTC-3′, sgRNA #3; 5′-CACCGTCATGTCAGCCAAGGACGAG-3′ and 5′-AAACCTCGTCCTTGGCTGACATGAC-3′. For the initial sgRNA screening, pX459 plasmids with these sgRNAs were transiently transfected into IMCD3 cells for 24 h, and transfected cells were selected by puromycin treatment (0.25 µg/ml) for 2 days. Following expansion in normal growth media, cells were harvested for genomic DNA extraction and Pde6d exons 1 and 2 were PCR-amplified using the following primers and the Universe Hot Start High-Fidelity DNA polymerase (Biotool): for exon 1; 5′-GCTGGCTTGTCATTGGTTTC-3′ and 5′-GGCCTTCAGGACCCTAATTT-3′, for exon 2; 5′-GTTCCAGGACAGGGATTGTT-3′ and 5′-TGGGAAGTGAGAGAAGGAAATG-3′. For T7E1 assays, heteroduplex DNAs were generated by denaturing PCR products at 95°C for 5 min and slowly cooling down to room temperature. Heteroduplex DNAs were digested with T7 endonuclease I (NEB) for 30 min at 37°C. Digested DNAs were loaded on a 2% agarose gel to assess genome editing efficiencies. To generate Pde6d knockout clonal cell lines, IMCD3 cells were transfected with pX459 plasmids with sgRNAs #1 and #3, selected with puromycin as above, and seeded at a low density in 10-cm tissue culture dishes. After 2 weeks of culture, well-separated individual clones were trypsinized and transferred into 24-well plates and expanded. Genomic DNAs were extracted from a subset of cells, and Pde6d exon 1 was PCR amplified using the primers described above and sequenced. Clones with frame-shift mutations were selected and the lack of Pde6d proteins was verified by immunoblotting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
N.D. performed experiments and revised the manuscript. S.S. designed research, performed experiments, and wrote the manuscript.
Funding
This work was supported by a grant from the National Institutes of Health [grant no. R01-EY022616].
References
- Baehr W. (2014). Membrane protein transport in photoreceptors: the function of PDEdelta: the Proctor lecture. Invest. Ophthalmol. Vis. Sci. 55, 8653-8666. 10.1167/iovs.14-16066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas S. L., Silhavy J. L., Brancati F., Kisseleva M. V., Al-Gazali L., Sztriha L., Bayoumi R. A., Zaki M. S., Abdel-Aleem A., Rosti R. O. et al. (2009). Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 41, 1032-1036. 10.1038/ng.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes C., Li T., Danciger M., Baxter L. C., Applebury M. L. and Farber D. B. (1990). Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature 347, 677-680. 10.1038/347677a0 [DOI] [PubMed] [Google Scholar]
- Boylan J. P. and Wright A. F. (2000). Identification of a novel protein interacting with RPGR. Hum. Mol. Genet. 9, 2085-2093. 10.1093/hmg/9.14.2085 [DOI] [PubMed] [Google Scholar]
- Breuer D. K., Yashar B. M., Filippova E., Hiriyanna S., Lyons R. H., Mears A. J., Asaye B., Acar C., Vervoort R., Wright A. F. et al. (2002). A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am. J. Hum. Genet. 70, 1545-1554. 10.1086/340848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V., Silhavy J. L., Bielas S. L., Swistun D., Marsh S. E., Bertrand J. Y., Audollent S., Attié-Bitach T., Holden K. R., Dobyns W. B. et al. (2008). Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 83, 170-179. 10.1016/j.ajhg.2008.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A., Grecco H. E., Pisupati V., Perera D., Cassidy L., Skoulidis F., Ismail S. A., Hedberg C., Hanzal-Bayer M., Venkitaraman A. R. et al. (2012). The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14, 148-158. 10.1038/ncb2394 [DOI] [PubMed] [Google Scholar]
- Christiansen J. R., Kolandaivelu S., Bergo M. O. and Ramamurthy V. (2011). RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc. Natl. Acad. Sci. USA 108, 8862-8866. 10.1073/pnas.1103627108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill J. D., Bowne S. J., Sullivan L. S., Lewis R. A., Wheaton D. K., Birch D. G., Branham K. E., Heckenlively J. R. and Daiger S. P. (2013). Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 54, 1411-1416. 10.1167/iovs.12-11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci F. Y., Rigatti B. W., Wen G., Radak A. L., Mah T. S., Baic C. L., Traboulsi E. I., Alitalo T., Ramser J. and Gorin M. B. (2002). X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am. J. Hum. Genet. 70, 1049-1053. 10.1086/339620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R. et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184-191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fansa E. K., O'reilly N. J., Ismail S. and Wittinghofer A. (2015). The N- and C-terminal ends of RPGR can bind to PDE6delta. EMBO Rep. 16, 1583-1585. 10.15252/embr.201541404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fansa E. K., Kösling S. K., Zent E., Wittinghofer A. and Ismail S. (2016). PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat. Commun. 7, 11366 10.1038/ncomms11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio S. K., Prusti R. K. and Beavo J. A. (1996). Solubilization of membrane-bound rod phosphodiesterase by the rod phosphodiesterase recombinant delta subunit. J. Biol. Chem. 271, 24036-24047. 10.1074/jbc.271.39.24036 [DOI] [PubMed] [Google Scholar]
- Gakovic M., Shu X., Kasioulis I., Carpanini S., Moraga I. and Wright A. F. (2011). The role of RPGR in cilia formation and actin stability. Hum. Mol. Genet. 20, 4840-4850. 10.1093/hmg/ddr423 [DOI] [PubMed] [Google Scholar]
- Hanke-Gogokhia C., Wu Z., Gerstner C. D., Frederick J. M., Zhang H. and Baehr W. (2016). Arf-like protein 3 (ARL3) regulates protein trafficking and ciliogenesis in mouse photoreceptors. J. Biol. Chem. 291, 7142-7155. 10.1074/jbc.M115.710954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D. H. and Li T. (2002). Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Invest. Ophthalmol. Vis. Sci. 43, 3373-3382. [PubMed] [Google Scholar]
- Hong D.-H., Pawlyk B., Sokolov M., Strissel K. J., Yang J., Tulloch B., Wright A. F., Arshavsky V. Y. and Li T. (2003). RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest. Ophthalmol. Vis. Sci. 44, 2413-2421. 10.1167/iovs.02-1206 [DOI] [PubMed] [Google Scholar]
- Huang S. H., Pittler S. J., Huang X., Oliveira L., Berson E. L. and Dryja T. P. (1995). Autosomal recessive retinitis pigmentosa caused by mutations in the alpha subunit of rod cGMP phosphodiesterase. Nat. Genet. 11, 468-471. 10.1038/ng1295-468 [DOI] [PubMed] [Google Scholar]
- Humbert M. C., Weihbrecht K., Searby C. C., Li Y., Pope R. M., Sheffield V. C. and Seo S. (2012). ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA 109, 19691-19696. 10.1073/pnas.1210916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S. A., Chen Y.-X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P. I. H. and Wittinghofer A. (2011). Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942-949. 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- Jacoby M., Cox J. J., Gayral S., Hampshire D. J., Ayub M., Blockmans M., Pernot E., Kisseleva M. V., Compère P., Schiffmann S. N. et al. (2009). INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 41, 1027-1031. 10.1038/ng.427 [DOI] [PubMed] [Google Scholar]
- Khanna H., Davis E. E., Murga-Zamalloa C. A., Estrada-Cuzcano A., Lopez I., den Hollander A. I., Zonneveld M. N., Othman M. I., Waseem N., Chakarova C. F. et al. (2009). A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 41, 739-745. 10.1038/ng.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner R., Rosenberg T., Schultz-Heienbrok R., Lenzner S., Feil S., Roepman R., Cremers F. P., Ropers H.-H. and Berger W. (1999). RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum. Mol. Genet. 8, 1571-1578. 10.1093/hmg/8.8.1571 [DOI] [PubMed] [Google Scholar]
- Lee J.-J. and Seo S. (2015). PDE6D binds to the C-terminus of RPGR in a prenylation-dependent manner. EMBO Rep. 16, 1581-1582. 10.15252/embr.201541220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linari M., Ueffing M., Manson F., Wright A., Meitinger T. and Becker J. (1999). The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 96, 1315-1320. 10.1073/pnas.96.4.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclaughlin M. E., Sandberg M. A., Berson E. L. and Dryja T. P. (1993). Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat. Genet. 4, 130-134. 10.1038/ng0693-130 [DOI] [PubMed] [Google Scholar]
- Meindl A., Dry K., Herrmann K., Manson F., Ciccodicola A., Edgar A., Carvalho M. R. S., Achatz H., Hellebrand H., Lennon A. et al. (1996). A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X–linked retinitis pigmentosa (RP3). Nat. Genet. 13, 35-42. 10.1038/ng0596-35 [DOI] [PubMed] [Google Scholar]
- Nancy V., Callebaut I., El Marjou A. and De Gunzburg J. (2002). The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J. Biol. Chem. 277, 15076-15084. 10.1074/jbc.M109983200 [DOI] [PubMed] [Google Scholar]
- Patil H., Tserentsoodol N., Saha A., Hao Y., Webb M. and Ferreira P. A. (2012). Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell Death Dis. 3, e355 10.1038/cddis.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K. N., Anand M. and Khanna H. (2016). The carboxyl terminal mutational hotspot of the ciliary disease protein RPGRORF15 (retinitis pigmentosa GTPase regulator) is glutamylated in vivo. Biol. Open 5, 424-428. 10.1242/bio.016816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman R., van Duijnhoven G., Rosenberg T., Pinckers A. J. L. G., Bleeker-Wagemakers L. M., Bergen A. A. B., Post J., Beck A., Reinhardt R., Ropers H.-H. et al. (1996). Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum. Mol. Genet. 5, 1035-1041. 10.1093/hmg/5.7.1035 [DOI] [PubMed] [Google Scholar]
- Roepman R., Bernoud-Hubac N., Schick D. E., Maugeri A., Berger W., Ropers H. H., Cremers F. P. M. and Ferreira P. A. (2000). The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet. 9, 2095-2105. 10.1093/hmg/9.14.2095 [DOI] [PubMed] [Google Scholar]
- Schmick M., Vartak N., Papke B., Kovacevic M., Truxius D. C., Rossmannek L. and Bastiaens P. I. (2014). KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 157, 459-471. 10.1016/j.cell.2014.02.051 [DOI] [PubMed] [Google Scholar]
- Schwahn U., Lenzner S., Dong J., Feil S., Hinzmann B., van Duijnhoven G., Kirschner R., Hemberger M., Bergen A. A. B., Rosenberg T. et al. (1998). Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19, 327-332. 10.1038/1214 [DOI] [PubMed] [Google Scholar]
- Seabra M. C., Brown M. S. and Goldstein J. L. (1993). Retinal degeneration in choroideremia: deficiency of rab geranylgeranyl transferase. Science 259, 377-381. 10.1126/science.8380507 [DOI] [PubMed] [Google Scholar]
- Shu X., Black G. C., Rice J. M., Hart-Holden N., Jones A., O'grady A., Ramsden S. and Wright A. F. (2007). RPGR mutation analysis and disease: an update. Hum. Mutat. 28, 322-328. 10.1002/humu.20461 [DOI] [PubMed] [Google Scholar]
- Sun X., Park J. H., Gumerson J., Wu Z., Swaroop A., Qian H., Roll-Mecak A. and Li T. (2016). Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc. Natl. Acad. Sci. USA 113, E2925-E2934. 10.1073/pnas.1523201113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Wright K. J., Le Corre S., Micalizzi A., Romani M., Abhyankar A., Saada J., Perrault I., Amiel J., Litzler J. et al. (2014). A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Hum. Mutat. 35, 137-146. 10.1002/humu.22470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort R., Lennon A., Bird A. C., Tulloch B., Axton R., Miano M. G., Meindl A., Meitinger T., Ciccodicola A. and Wright A. F. (2000). Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 25, 462-466. 10.1038/78182 [DOI] [PubMed] [Google Scholar]
- Wätzlich D., Vetter I., Gotthardt K., Miertzschke M., Chen Y.-X., Wittinghofer A. and Ismail S. (2013). The interplay between RPGR, PDEdelta and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 14, 465-472. 10.1038/embor.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. N., Hong D.-H. and Perkins B. (2011). Misexpression of the constitutive Rpgr(ex1-19) variant leads to severe photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 52, 5189-5201. 10.1167/iovs.11-7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. N., Hong D.-H. and Perkins B. (2012). RpgrORF15 connects to the usher protein network through direct interactions with multiple whirlin isoforms. Invest. Ophthalmol. Vis. Sci. 53, 1519-1529. 10.1167/iovs.11-8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright Z. C., Singh R. K., Alpino R., Goldberg A. F. X., Sokolov M. and Ramamurthy V. (2016). ARL3 regulates trafficking of prenylated phototransduction proteins to the rod outer segment. Hum. Mol. Genet. [Epub ahead of print]. doi:10.1093/hmg/ddw077 10.1093/hmg/ddw077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Sippel K. C., Berson E. L. and Dryja T. P. (1997). Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat. Genet. 15, 175-178. 10.1038/ng0297-175 [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu X.-h., Zhang K., Chen C.-K., Frederick J. M., Prestwich G. D. and Baehr W. (2004). Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J. Biol. Chem. 279, 407-413. 10.1074/jbc.M306559200 [DOI] [PubMed] [Google Scholar]
- Zhang H., Li S., Doan T., Rieke F., Detwiler P. B., Frederick J. M. and Baehr W. (2007). Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl. Acad. Sci. USA 104, 8857-8862. 10.1073/pnas.0701681104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hanke-Gogokhia C., Jiang L., Li X., Wang P., Gerstner C. D., Frederick J. M., Yang Z. and Baehr W. (2015). Mistrafficking of prenylated proteins causes retinitis pigmentosa 2. FASEB J. 29, 932-942. 10.1096/fj.14-257915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hong D.-H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A. and Li T. (2003). The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl. Acad. Sci. USA 100, 3965-3970. 10.1073/pnas.0637349100 [DOI] [PMC free article] [PubMed] [Google Scholar]



