Abstract
Cancers often evade immune surveillance by adopting peripheral tissue–tolerance mechanisms, such as the expression of programmed cell death ligand 1 (PD-L1), the inhibition of which results in potent antitumor immunity. Here, we show that cyclin-dependent kinase 5 (Cdk5), a serine-threonine kinase that is highly active in postmitotic neurons and in many cancers, allows medulloblastoma (MB) to evade immune elimination. Interferon-γ (IFN-γ)-induced PD-L1 up-regulation on MB requires Cdk5, and disruption of Cdk5 expression in a mouse model of MB results in potent CD4+ T cell–mediated tumor rejection. Loss of Cdk5 results in persistent expression of the PD-L1 transcriptional repressors, the interferon regulatory factors IRF2 and IRF2BP2, which likely leads to reduced PD-L1 expression on tumors. Our finding highlights a central role for Cdk5 in immune checkpoint regulation by tumor cells.
Cyclin-dependent kinase 5 (Cdk5) is a non-stereotypical Cdk whose activity depends on coactivators, p35 and/or p39. A proline-directed serine-threonine kinase (1), Cdk5 is essential in central nervous system (CNS) development (2, 3). Cdk5 also contributes to angiogenesis, apoptosis, myogenesis, vesicular transport, and senescence in nonneuronal cells, including tumors (4–6), which makes Cdk5 a potential therapeutic target in cancers (7–9). We explored whether Cdk5 plays a role in medulloblastoma (MB), a common malignant pediatric CNS tumor.
MB cell lines and clinical specimens expressed Cdk5, p35, and p39 (Fig. 1A and fig. S1A). Cdk5-specific kinase activity could be abolished in vitro by roscovitine, a nonselective inhibitor against Cdks 1,2,5,7, and 9 (fig. S1B) (10). To interrogate Cdk5-specific functions, we disrupted Cdk5 in wild-type murine MB cells (MM1WT) by short hairpin-mediated RNA interference (MM1 shCdk5) and clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9–targeted mutation (MM1 crCdk5), with nontargeting constructs as controls (MM1 shNS and MM1 crNeg). A reduction in Cdk5 was confirmed at the transcript (fig. S1C) and protein levels (fig. S1D). In vitro, there were no significant differences in cell proliferation among all constructs (fig. S1, E and F) (1).
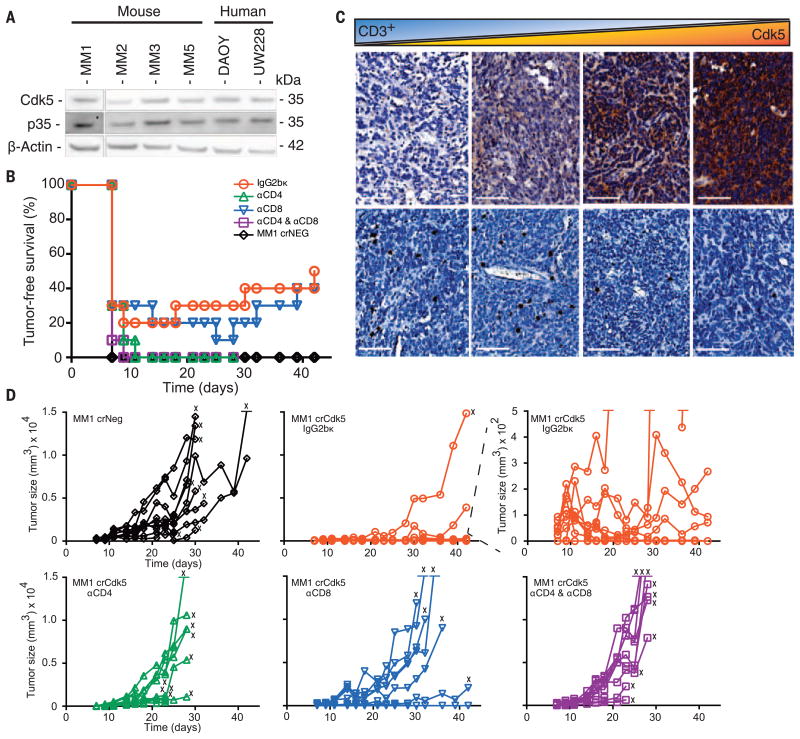
Fig. 1. Targeted deletion of Cdk5 in MB results in rejection by CD4+ Tcells.
(A) Cdk5 and p35 proteins are expressed in murine and human MB cell lines in vitro. (B) TFS in C57BL/6J mice injected with MM1 crNeg or crCdk5 cells with various depleting antibodies (n = 10 per group). (C) Immunohistochemistry of six clinical MB samples reveals an inverse correlation between tumor Cdk5 expression (top) and CD3+ T cell infiltration (bottom). Pearson correlation = −0.91 (fig. S2D). Scale bars, 100 μm. (D) Tumor-growth kinetics for individual animals in each group from (B). The top right-hand graph shows the center one with an expanded scale. X indicates that an animal was killed because of tumor size or ulceration.
To assess MB growth in vivo, 5 × 104 Cdk5-deficient or control cells were inoculated subcutaneously (s.c.) into the flanks of immunodeficient mice. All mice developed comparable-sized tumors by day 14 (fig. S2, A to C). However, 78 to 50% of C57BL/6 mice injected s.c. with Cdk5-deficient MB cells showed tumor-free survival (TFS) at 19 and 42 days, whereas mice injected with WT and control tumors exhibited 0 and 7% TFS after 19 days, respectively (Fig. 1B and fig. S3A). Mice injected with Cdk5-deficient MB cells developed significantly smaller tumors (0.02 ± 0.04 g) than mice injected with WT (0.91 ± 0.39 g) or NS (0.51 ± 0.21 g) cells (fig. S3B). These data suggest a T cell-dependent rejection mechanism of Cdk5-deficient MM1 cells. This interpretation is supported by the observation that Cdk5 expression inversely correlated with T cell infiltration in human MB (Fig. 1C and fig. S2D).
To identify T cell populations mediating this potent rejection, we depleted CD8+ T cells, CD4+ T cells, or both subsets in mice inoculated with MM1 crCdk5 or crNeg cells (5 × 104 s.c). By day 11,100% of mice injected with MM1 crNeg and 80% of mice receiving MM1 crCdk5 developed measurable tumors (Fig. 1B), although MM1 crNeg tumors were 8 times the size of MM1 crCdk5 tumors (808.8 ± 382.1 versus 101.1 ± 92.9 mm3) (Fig. 1D). Depletion with CD4-specific (αCD4) antibody alone or with both αCD4 and αCD8 antibodies resulted in 100% MM1 crCdk5 tumor incidence accompanied by rapid tumor growth, whereas CD8 depletion alone yielded 30% TFS, similar to isotype control (Fig. 1D). Among mice receiving isotype antibody, three of eight crCdk5 tumor outgrowths regressed starting on day 17, whereas three of nine crCdk5 tumor outgrowths among mice depleted of CD8+ T cells regressed starting on day 25; these outgrowths contributed to a total TFS of 50 and 40%, respectively (Fig. 1, B and D). Tumors harvested from MM1 crCdk5-bearing mice remained Cdk5− without evidence of Cdk5+ escape (fig. S4A). Similar results were seen in mice receiving MM1 shCdk5 and shNS inoculations, with a dependency on CD4+ T cells for tumor rejection (fig. S3C). Cdk5-deficient tumors also grew aggressively in mice deficient in major histocompatibility complex class II (MHC-II) (fig. S3D). Finally, 60 to 75% of mice that rejected Cdk5-deficient tumors remained tumor free after rechallenge with a lethal dose of MM1 WT cells (fig. S3E). Collectively, these studies point to a CD4+ T cell–dependent rejection of Cdk5-deficient tumors with robust antitumor immune memory generation.
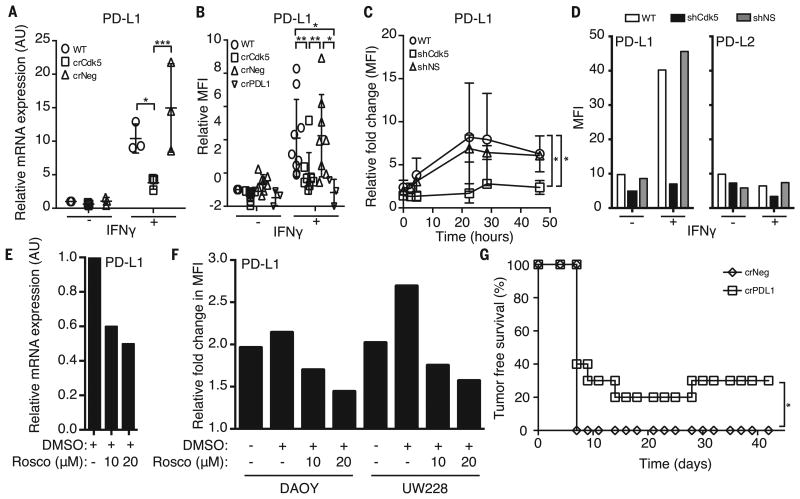
Interferon-γ (IFN-γ) is a major CD4+ T cell effector cytokine (11) and was abundant in Cdk5-deficient tumor mass (fig. S5A). IFN-γ induces p35 (12), which results in enhanced Cdk5 activity (fig. S5B). IFN-γ is known to induce PD-L1 (13), whose expression on infiltrating immune cells is evidence of an ongoing intratumoral immune response (14). We examined whether disruption of Cdk5 expression in MB impaired PD-L1 induction in response to IFN-γ stimulation. We analyzed human tumor databases and found a cooccurrence of Cdk5 and PD-L1 mRNA expression in many tumor types (fig. S6). In Cdk5-deficient MM1, we observed a 37.58 ± 14.28% reduction in basal PD-L1 mRNA level (Fig. 2A). Note that Cdk5-deficient MM1 cells exhibited a blunted PD-L1 up-regulation in response to IFN-γ stimulation in vitro (Fig. 2, A and B, and fig. S4B). Other IFN-γ–responsive proteins, such as MHC H-2Kb and H-2Db (fig. S7A), were not significantly affected in the Cdk5-deficient tumors, which indicated that a global disruption of the IFN-γ receptor (IFNGR) signaling was not responsible for failed PD-L1 up-regulation or enhanced immune sensitivity. Disrupting Cdk5 in rhabdomyosarcoma also led to a blunted IFN-γ-induced PD-L1 up-regulation (fig. S4, C to E), which indicated that the link between Cdk5 and PD-L1 regulation by IFN-γ is not MB-specific. Twenty-four hours after IFN-γ exposure, surface PD-L1 expression reached a peak of 8.2- and 6.8-fold above baseline in WT and NS cells, respectively (Fig. 2C), whereas Cdk5-deficient cells only up-regulated PD-L1 2.8-fold, so it reached a peak level similar to the basal levels in unstimulated WT and NS controls. The blunted response to IFN-γ is specific for PD-L1 but not PD-L2 (Fig. 2D). To further corroborate the link between Cdk5 and PD-L1 synthesis, we treated MM1 WT cells with roscovitine and observed a dose-dependent decrease in PD-L1 transcripts (Fig. 2E). In vitro treatment of human MB with roscovitine also diminished surface PD-L1 up-regulation with IFN-γ in a dose-dependent manner (Fig. 2F). Finally, to establish a functional link between PD-L1 and in vivo rejection of Cdk5-deficient MM1, we disrupted the PD-L1 gene (CD274) in MM1 cells (MM1 crPDL1). Similar to MM1 crCdk5 experiments, 30% of mice inoculated with MM1 crPDL1 remained tumor-free for more than 4 weeks (Figs. 2G and 1B).
Fig. 2. Disruption of either Cdk5 gene expression or Cdk5 activity suppresses PD-L1 expression that cannot be overcome with IFN-γ stimulation in both human and murine MBs.
(A) In vitro mRNA expression in arbitrary units (AU) of PD-L1 by MM1 WT, crCdk5, and crNeg cells with or without 24 hours of IFN-γ stimulation. Values represent the average of three biological replicates ± SD. (B) In vitro PD-L1 surface staining of MM1 WT, crCdk5, crNeg, and crPDL1 cells with or without 24 hours of IFN-γ stimulation. Values represent the average mean fluorescence intensity (MFI) ± SD compared with unstimulated MM1 WT cells over seven or eight replicates. (C) Fold change of surface PD-L1 expression in MM1 WT, shCdk5, and shNS cells over the course of 48 hours of IFN-γ stimulation. (D) MFI of PD-L1 and PD-L2 expressed in MM1 WT, shCdk5, and shNS cells. (E) PD-L1 mRNA expression in MM1 WT cells when treated with roscovitine and stimulated with IFN-γ for 24 hours. AU relative to untreated. DMSO, dimethyl sulfoxide. (F) DAOY and UW228 human MB lines treated with roscovitine and stimulated with IFN-γ for 24 hours. MFI relative to untreated samples. (G) TFS of MM1 crNeg and crPDL1 injected mice over 36 days (n = 10 mice per group). *P < 0.05; **P < 0.01; ***P < 0.001. Significance was determined by two-way ANOVA with Bonferoni posttest (A) and (B), Student's t test (C), or log-rank test (G). Representative experiments are shown in (D) and (E).
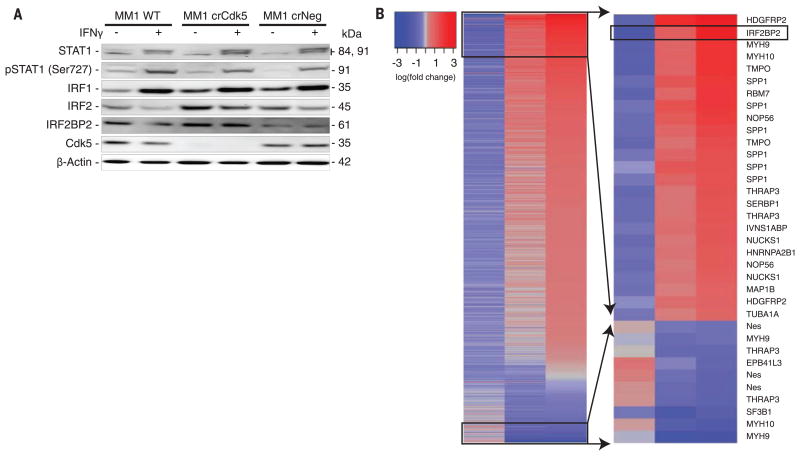
Next, we interrogated the IFNGR signaling pathway. Western blot analysis of various MM1 cells failed to show differences, after IFN-γ exposure, in STAT1, STAT2, or STAT3 (members of the family of signal transducers and activators of transcription) (Fig. 3A and fig. S7B), in agreement with the robust MHC class I induction in Cdk5-deficient MM1 cells (fig. S7A). To further dissect this STAT1-independent signaling, we examined interferon regulatory factor-1 (IRF1) and interferon regulatory factor-2 (IRF2), which are implicated as positive and negative regulators of PD-L1 transcription, respectively (13, 15). IRF1 protein was rapidly induced by IFN-γ and remained elevated for up to 48 hours regardless of Cdk5 expression (Fig. 3A and fig. S7C). We observed a rapid loss of the PD-L1 transcription repressor, IRF2, in WT and crNeg cells. In contrast, IRF2 and its corepressor IRF2BP2 (16) were elevated at baseline in Cdk5-deficient cells and persisted for up to 48 hours after IFN-γ exposure (Fig. 3A and fig. S7C). This protein expression difference cannot be accounted for at the transcriptional level (fig. S7D). Phosphoproteomic analysis identified 77 distinct phosphopeptides in the shCdk5 versus WT or shNS screen (tables S1 and S2), and 798 phosphopeptides in the crCdk5 versus WT or crNeg screen (tables S3 and S4). Between these two data sets, 22 common proteins were differentially phosphorylated in Cdk5-deficient cells, with IRF2BP2 among the highest phosphorylated peptide species (Fig. 3B).
Fig. 3. Cdk5 gene silencing alters the IFN-γ signaling pathway and is associated with hyperphosphorylation of IRF2BP2.
(A) IFN-γ stimulation of MM1 WT, crCdk5, and crNeg cells for 24 hours. IFNGR downstream mediators STAT1, phosphorylated (p)STAT1, IRF1, IRF2, and IRF2BP2 were assayed. (B) Global quantitative phosphoproteomic analysis (left) of MM1 WT, crCdk5, and crNeg cells shows a change in phosphorylation status of 35 different phosphopeptides found in 18 of the 22 identified proteins (right). Twelve proteins exhibit only increased phosphorylation, three exhibit only decreased phosphorylation, and three have both increased and decreased phosphorylation sites. Phosphoproteomic analysis of three biologic repeats of each cell line. Boxes on the left indicate peptides identified from the highest and lowest phosphorylated species that are magnified on the right.
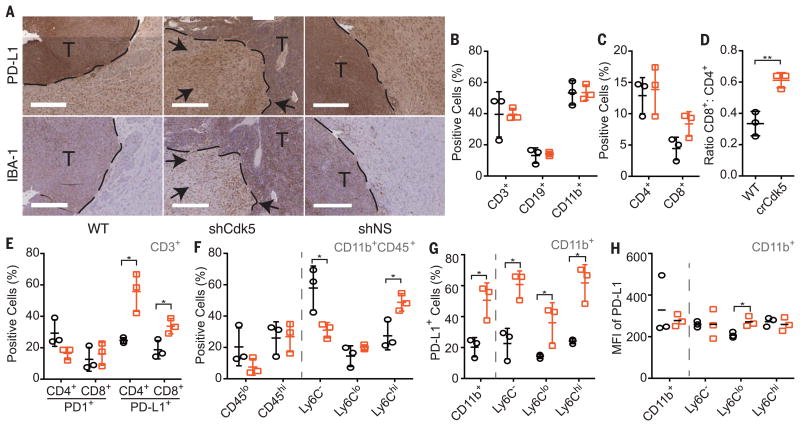
Finally, we introduced Cdk5-deficient MM1 cells orthotopically into C57BL/6 mice. Gross inspection revealed a 50% tumor incidence in mice injected with Cdk5-deficient MM1, mirroring s.c. tumors. In contrast, 100% of mice injected with WT or NS MM1 cells developed gross brain tumors by day 14 (fig. S8A). Intracranial (i.c.) Cdk5-deficient tumor outgrowth remained devoid of Cdk5 expression without the emergence of a Cdk5+ escape variant (fig. S8B). Histological analysis showed increased accumulation of IBA-1+ cells, which marks microglia and infiltrating monocytes, and PD-L1+ staining in the Cdk5-deficient MM1 tumor margin and surrounding stroma (Fig. 4A). Immune cell composition analysis showed a modest increase in CD3+ T cells, similar to that shown by immunohistochemical data (Fig. 1C and fig. S8, C and D). However, the percentages of CD3+ cells were equivalent in crCdk5 and WT tumor samples by flow cytometry (Fig. 4B). Cdk5-deficient tumors elicited an increased ratio of CD8+ to CD4+ T cell infiltrate, lower PD-1 expression in CD4+ T cells, and higher PD-L1 expression in both T cell subsets (Fig. 4, C to E). Although CD8+ T cells are not the primary antitumor effector cells in this model, their increased recruitment likely reflects an overall inflammatory tumor milieu as evidenced by increased PD-L1 expression and overall tissue IFN-γ levels (Fig. 1B and figs. S3C, S5A, and S9, A to E). The myeloid infiltrate in i.c. tumors shifted from a Ly6C− to a Ly6Chi population with an increased percentage of PD-L1+ cells in bulk CD11b+ cells and in each Ly6C subset (Fig. 4, F to H), accompanied by a decrease in the percentage of microglia (CD11b+CD45lo) (Fig. 4F). The Ly6Clo subset expressed ah gher density of surface PD-L1 in the Cdk5-deficien tumors (Fig. 4H). Again, this finding was recapi ulated in s.c. tumors, which showed a significan increase in the percentage of PD-L1+ immune cells, with a trend toward increased density of PD-L1 staining in the crCdk5 tumor microenvironment (fig. S9, F to I). The observed increase in PD-L1+ populations and staining density aligns with histologic analyses (Fig. 4A and fig. S9B), which suggests a state of global immune activation in response to ongoing IFN-γ stimulation. This finding is in good agreement with reports showing increased PD-L1+ immune cells in MB stroma undergoing active immune checkpoint blockade (17).
Fig. 4. Orthotopic Cdk5-deficient tumors exhibit increased PD-L1 staining, CD4+ tumor–infiltrating lymphocytes (TILs), and accumulating infiltrates of CD11b+ populations.
(A) Tumors extracted 14 days postinoculation from MM1 WT, shCdk5, or shNS mice stained for PD-L1 expression. Dashed line represents margin between tumor (T) and stroma. Black arrows point to increased PD-L1+ and IBA-1+ cells in the tumor stroma. Scale bars, 400 μm. (B and C) Fluorescence-activated cell sorting (FACS) analysis of MM1 WT (black circle) and MM1 crCdk5 (orange square) tumor infiltrate by percentage of cell type. (D) Ratio of total CD8+:CD4+ cell infiltrate. (E) FACS analysis of the percentage of PD-1+ or PD-L1+ cells in the CD4+ or CD8+ populations. (F) FACS analysis of the percentage of myeloid cells in tumor infiltrate based on differential CD45 staining (left) or Ly6C staining among CD11b+CD45+ cells (right). (G) Percent of total CD11b+ population (left) and subpopulations (right) present in tumor infiltrate that express PD-L1. (H) MFI of PD-L1 expression among CD11b+ total population (left) and subpopulations (right). (B), (C), (D), (E), (F), and (G) were graphed as means ± SD. (H) was graphed as individual MFI with mean indicated. n = 9 per group. Each data point represents pooled samples from three mice. *P < 0.05; **P < 0.01. Significance was determined using the Student's t test.
Here, we showed that Cdk5 disruption sensitizes MB to CD4+ T cell-dependent rejection via posttranslational modification of IRF2BP2, which increases IRF2 and IRF2BP2 abundance and sustains PD-L1 transcriptional repression after IFN-γ stimulation. Downstream IFN-γ signaling induces interferon-stimulated genes, including IRF1 (18), which activates secondary-response genes, including PD-L1 (13,19,20). IRF2 acts as a repressor that competes with IRF1 for binding to the same promoter element (15). Constitutively present, IRF2 is up-regulated in response to either type I IFNs or IRF1 (15,20) and provides a negative-feedback loop by binding to its own promoter to block transcription (15). The prolonged half-life of IRF2 (8 hours) relative to IRF1 (0.5 hours) provides a mechanism for IRF2 antagonism (20). IRF2BP2 was recently identified as a corepressor with IRF2 (16), and low IRF2BP2 expression was correlated with high PD-L1 expression in breast cancer (21). Our data provide a direct link between disruption of Cdk5 activity and IRF2BP2 hyperphosphorylation at sites that are distinct from previously described sites that affect nuclear localization, vascular endothelial growth factor A, or MHC-I expression (22–24), which suggests that Cdk5 either directly or indirectly inhibits other kinase(s) that phosphorylate IRF2BP2 (fig. S10).
PD-L1 and PD-1 play a critical role in tumor immune evasion, with ∼30% of tumors responding to immune checkpoint blockade (25,26). High Cdk5 expression correlates with worse clinical outcome in multiple cancers (fig. S11). In our studies, both Cdk5- and PD-L1–deficient MB cells exhibit similar TFS (Figs. 1B and 2G). More CD4+ T cells with lower PD-1 expression were found in the Cdk5-deficient CNS tumors, whereas CD11b+ cells accumulate in larger quantities with higher PD-L1+ expression (Fig. 4, C to G). Myeloid PD-L1 up-regulation may be a response to overall increased IFN-γ (13,27). Alternatively, these cells may play a distinct role modulating infiltrating T cell function, which are present in most human MB specimens (28). Last, as Cdk5 directly phosphorylates MYC on Ser62 (29), it remains to be determined whether Cdk5 plays a role in MYC-regulated PD-L1 expression (30).
Supplementary Material
Fig S1. Kinase active Cdk5 is expressed by murine and human MB cell lines and deletion of Cdk5 does not alter proliferation. (A) Genomic analysis from the UCSC cancer genome browser demonstrates that Cdk5, p35, and p39 are highly expressed in pediatric MB (4 patients) and neuroblastoma (NB) (39 patients) samples (above black line) compared to normal brain tissue (below black line). (B) Cdk5 kinase assay activity is detected in human MB (UW228, DAOY) and mouse MB (MM1) cell lines, with lysates from Cdk5−/− and Cdk5+/+ embryonic brains as negative and positive controls, respectively (Black Bars). The non-selective Cdk5 inhibitor, Roscovitine (10 μM), abrogates this kinase activity (Gray bars). (C) mRNA expression of Cdk5 transcript in MM1 WT (white), shCdk5 (black), or shNS (gray). (D) MM1 shCdk5 and MM1 crCdk5 are deficient in Cdk5 protein expression. (E) Proliferation curves of MM1 WT (square), shCdk5 (triangle), and shNS (circle) in media with 10% serum (black lines) or 1% serum (gray lines) over 72 hours (F) Proliferation curves of MM1 WT (white), crCdk5 (black), crNeg (gray) in media containing 10% serum over 72 hours. Data represent mean ±SD from biologic triplicates.
Fig S2. Similar subcutaneous (s.c.) growth kinetics of crCdk5 MM1 and crNeg MM1 in immune-compromised mice. (A) In vivo, growth kinetics of MM1 crCdk5 (circle) and crNeg (square) in NSG mice (n = 10/group). Points were graphed as the average size ± SD. (B) TFS of MM1 crCdk5 (circle) and crNeg (square) in NSG mice. (C) Tumor weights of MM1 crCdk5 (circle) and crNeg (square) in NSG mice. The solid line represents the average weight ± SD. (D) Inverse correlation between CD3+ cell count per HPF and Cdk5 staining intensity of clinical MB tissue samples as revealed by IHC analysis in Fig. 1D. Significance was determined using a t test (C).
Fig S3. In vivo growth kinetics of MM1 shCdk5 reveals thymic dependent tumor rejection upon s.c. injection. (A) TFS of MM1 WT (diamond, n = 13), shCdk5 (circle, n = 17), and shNS in B6 mice (triangle, n = 7). Survival curves were calculated from 3 independent experiments. (B) Tumor weights of MM1 WT (triangles, n = 13) and shCdk5 (circles, n = 13) and shNS (squares, n = 7). The solid line represents the average weight ± SD. (C) TFS of B6 mice injected s.c. with MM1 shNS (diamond, n = 10) or MM1 shCdk5 (circle) with injection of αCD4 (triangle), αCD8 (inverted triangle), or αCD4 and αCD8 (square) antibodies (n = 15/group). (D) Tumor size measurement of MHC-II KO (circle) and MHC-I KO (square) mice injected s.c. with Cdk5-deficient MM1 (n = 10/group). (E) TFS of B6 mice re-challenged with MM1 WT at least 60 days post-initial injection of MM1 shCdk5 (square) or MM1 crCdk5 (circle) (n = 5/group). **P < 0.01; ***P < 0.001 Significance was determined by log rank test (A, C, E) or t test (B, D).
Fig S4. A link between Cdk5 and PD-L1 expressions in human and murine tumors. (A) Cdk5 expression of cells cultured from subcutaneous tumor outgrowths isolated from animals initially injected with MM1 WT (n = 4), crCdk5 (n = 4), or crNeg (n = 6). (B) PD-L1 expression on primary cell cultures of MM1 crNeg, crCdk5, or outgrowth of crCdk5 tumors. Cells were stimulated with IFNγ for 24 hours (black) and compared to isotype staining (dotted line) and un-stimulated cells (gray). Histogram is a representative staining from three separate experiments. (C) PD-L1 mRNA expression by RMS 76.9 WT (white), shCdk5 (black), and shNS (gray) with or without 24 hours of IFNγ stimulation. (D) Surface PD-L1 staining of RMS 76.9 shNS (gray) and shCdk5 (black) with or without 24 hours of IFNγ stimulation. Values represent the average MFI ± SD compared to unstimulated RMS 76.9 shNS over three independent experiments. (E) Surface PD-L1 staining of SJCRH30 WT (white) and crCdk5 (black) with or without 24 hours of IFNγ stimulation in vitro. Values represent average MFI ± SD compared to unstimulated WT over three independent experiments. *P<0.05; **P<0.005. Significance was determined by two-way analysis of variance (ANOVA) (D) and (E).
Fig S5. Disruption of Cdk5 gene expression results in more IFNγ in the tumor bed and decreased Cdk5 activity in response to IFNγ. (A) In vivo IFNγ mRNA expression in tumor extracts from i.c. inoculations (n = 3 mice/group; ± SD). (B) Cdk5 kinase activity in MM1 WT, shCdk5, and shNS over the course of 24-hour stimulation with IFNγ (black) and the addition of 10 μM Roscovitine (gray). EB: embryonic brain.
Fig S6. Co-occurrence of upregulated Cdk5 and PD-L1 expression in human cancer samples. TCGA provisional data sets for all available cancer types were analyzed for Cdk5 and PD-L1 (CD274) mRNA expression through cBioPortal (http://www.cbioportal.org/). (A) Relative mRNA expression values of samples with existing RNA sequencing analysis (RNA Seq V2) were downloaded and analyzed with microsoft excel. The average log2 mRNA expression across each cancer type examined was individually graphed for Cdk5 and PD-L1. (B) Co-occurrence of elevated Cdk5 and PD-L1 gene expression is observed in several cancer types. mRNA expression z-scores of RNA Seq V2 RSEM from available tumors were analyzed for significance, their log odds ratio, and type of association. Representative cancer types showing co-occurrence of elevated Cdk5 and PD-L1 transcript levels are displayed.
Fig S7. Silencing Cdk5 in MB alters IRF-2 expression but does not affect other IFN signaling pathways. (A) Surface expression of MHC-I is upregulated on MM1 WT, crCdk5, and crNeg. Cells were stimulated with IFNγ for 24 hours (black) and compared to isotype staining (dotted line) and unstimulated cells (gray). Histogram is a representative staining from three separate experiments. (B) 24-hour stimulation of MM1 WT, crCdk5, and crNeg cells. Downstream IFNγ pathway elements STAT3, phosphorylated (p)STAT3, and STAT2 were assayed. (C) IFNγ stimulation of MM1 WT, crCdk5, and crNeg over the course of 48 hours. IFNγR downstream mediators IRF1, and IRF2 were assayed. (D) In vitro mRNA analysis of IRF-1, IRF-2, and IRF2BP2 present in MM1 WT (white), crCdk5 (black), and crNeg (gray) after treatment with IFNγ for 24 hours. Significance was determined using a 2-Way ANOVA (D).
Fig S8. CD3+ cell infiltrate and diminished growth rates associate with a decrease in tumor incidence following orthotopic administration of Cdk5-deficient MM1 cells. (A) Tumor incidence of MM1 WT, shCdk5, and shNS 14 days after i.c. tumor inoculation. (B) Western blot analysis for Cdk5 in cells cultured from B6 mice tumor outgrowths initially inoculated with MM1 WT (n = 2) and MM1 crCdk5 (n = 3). (C) IHC of tumor injected i.c. with MM1 WT, shCdk5, and shNS (scale bar = 200 μm). (D) Quantification of intratumoral CD3+ cells in MM1 WT (n = 3), shCdk5 (n = 3), and shNS (n = 4) tumors identified via IHC per mm2 of tumor tissue. Solid line represents the average ± SD. Significance was determined using a Student's t test (D).
Fig S9. Subcutaneous Cdk5 deficient tumors characterized by increased stromal PD-L1 staining, CD4+ tumor infiltrating lymphocytes (TIL), and infiltrating CD11b+ populations. (A) In vivo IFNγ mRNA expression in tumor extracts from subcutaneous inoculations (n = 3 mice/group; ± SD). (B) S.c. tumors extracted 14 days post-inoculation from MM1 shCdk5 or shNS stained for F4/80, PD-L1, and PD-1 expression (Scale bars = 2mm). (C), (D) FACS analysis of MM1 WT (black circle) and MM1 crCdk5 (red square) tumor infiltrate by percentage of cell type. (E) Ratio of total CD8+:CD4+ cell infiltrate. (F) FACS analysis of the percentage of PD-1+ or PD-L1+ cells in the CD4+ or CD8+ populations. (G) FACS analysis of the percentage of myeloid cells in tumor infiltrate based on differential CD45 staining (left) or Ly6C staining among CD11b+CD45+ cells (right). (H) Percent of total CD11b+ population (left) and sub-populations (right) present in tumor infiltrate that express PD-L1. (I) MFI of PD-L1 expression between total (left) and sub-populations (right) of CD11b+ cells. C thru H were graphed as the mean ± SD. (I) was graphed as individual MFI with mean indicated. n = 9/group. Each data point represents pooled samples from 3 mice. *P < 0.05 Significance was determined by Student's t test.
Fig S10. Putative mechanism by which Cdk5 controls PD-L1 expression. (A) When Cdk5 WT cells are stimulated with IFNγ, signaling through the JAK/STAT pathway stimulates IRF-1 driven transcription of PD-L1. Additionally, IFNγ stimulates p35 expression and thereby increases Cdk5 activity. Phosphorylation of unknown kinase(s) by Cdk5 inhibits kinase function, resulting in a reduced abundance of the IRF2/IRF2BP2 repressor complex. (B) In Cdk5 deficient cells, unknown kinase(s) hyper-phosphorylate IRF2BP2, prolong half-life of IRF2/IRF2BP2 repressor complex despite intact JAK/STAT/IRF-1 signaling, ultimately leading to decreased PD-L1 expression. IRF-E: IRF-binding element.
Fig. S11 Higher Cdk5 correlates with adverse clinical outcomes in human cancers. PrognoScan database-based Kaplan-Meier analysis of six independent patient cohorts including CNS and peripheral tumors that were analyzed for the effects of higher levels of Cdk5 gene expression (red) compared to low Cdk5 levels (blue) on overall survival, disease free survival, or metastasis free survival (http://www.prognoscan.org/). High levels of Cdk5 are associated with reduced (A) overall survival in a cohort of 74 glioma patients with astrocytoma (n = 8), GBM (n = 50), mixed glioma (n = 7), and oligodendroglioma (n = 9) (GSE 4412-GPL96); (B) overall survival in 50 patients with anaplastic oligodendroglioma (n = 22) and GBM (n = 28) (MGH-glioma); (C) overall survival of cutaneous melanoma patients (n = 38) (GSE19234); (D) distant metastasis free survival in uveal melanoma patients (n = 63) (GSE22138); (E) overall survival in a cohort of 104 patients with lung adenocarcinoma (GSE68465); and (F) disease free survival in a cohort of 249 breast cancer patients (GSE4922-GPL96). All p-values represent the corrected p-value.
Table S1. shCdk5-All phosphoproteomics data
Table S2. shCdk5-Significant phosphoproteomics data
Table S3. crCdk5-All phosphoproteomics data
Table S4. crCdk5-Significant phosphoproteomics data
Acknowledgments
We thank F. Scrimieri, G. Valdivieso, A. Awadallah, A. Kresak, D. Schlatzer, and the Case Western Reserve University Center for Proteomics for technical assistance; M. Couce for constructing human MB tissue microarray; and M. Chieppa, D. Askew, and B. Benson for careful review of the manuscript. The data reported in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by NIH F31CA196265 (R.D.D.), NIH T32GM007250 (R.D.D. and S.M.C.), NIH T32AI089474 (S.M.C.), NIH R01GM086550 (D.W.A.), NIH R01CA154656 (A.Y.H.), NIH R21CA181875 (A.Y.H. and J.J.L.), NIH R01HL111682 (A.Y.H. and J.J.L.), NIH P30CA043703 (A.Y.H. and A.P.), Wolstein Research Scholarship (R.D.D.), St. Baldrick's Foundation (A.P., J.J.L., and A.Y.H.), Hyundai “Hope-on-Wheels” Program (A.P., D.S.S., and A.Y.H.), Marc Joseph Fund (A.Y.H.), Alex's Lemonade Stand Foundation (A.P. and A.Y.H.), the Angie Fowler Adolescent and Young Adult Cancer Research Initiative at the Case Comprehensive Cancer Center (A.P., J.J.L., D.S.S., S.A., and A.Y.H.), and Theresia G. and Stuart F. Kline Family Foundation in Pediatric Oncology (A.Y.H.). This work was supported by the Clinical and Translational Science Collaborative of Cleveland UL1TR000439 (A.P.), and KL2TR000440 (S.A.) from the National Center for Advancing Translational Sciences (NCATS) component of the NIH.
Footnotes
Authors declare no competing financial interests. All phosphoproteomics data are available in the supplementary materials.
Other Supplementary Material for this manuscript includes the following: (available at www.sciencemag.org/content/353/6296/399/suppl/DC1)
Contributor Information
Alex Y. Huang, Email: ayh3@case.edu.
Agne Petrosiute, Email: axp125@case.edu.
References and Notes
- 1.Dhavan R, Tsai LH. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 2.Ohshima T, et al. Proc Natl Acad Sci USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Utreras E, Futatsugi A, Pareek TK, Kulkarni AB. Drug Discov Today Ther Strateg. 2009;6:105–111. doi: 10.1016/j.ddstr.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contreras-Vallejos E, Utreras E, Gonzalez-Billault C. Cell Signal. 2012;24:44–52. doi: 10.1016/j.cellsig.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Arif A. Biochem Pharmacol. 2012;84:985–993. doi: 10.1016/j.bcp.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Pareek TK, et al. J Exp Med. 2010;207:2507–2519. doi: 10.1084/jem.20100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu FN, et al. J Biol Chem. 2011;286:33141–33149. doi: 10.1074/jbc.M111.252080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann G, et al. Cancer Res. 2010;70:4460–4469. doi: 10.1158/0008-5472.CAN-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu R, et al. Proc Natl Acad Sci USA. 2008;105:7570–7575. doi: 10.1073/pnas.0712306105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil HS, Mitev V, Vlaykova T, Cavicchi L, Zhelev N. J Biotechnol. 2015;202:40–49. doi: 10.1016/j.jbiotec.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Cantor H. Cancer Immunol Res. 2014;2:91–98. doi: 10.1158/2326-6066.CIR-13-0216. [DOI] [PubMed] [Google Scholar]
- 12.Song JH, et al. J Biol Chem. 2005;280:12896–12901. doi: 10.1074/jbc.M412139200. [DOI] [PubMed] [Google Scholar]
- 13.Lee SJ, et al. FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 14.Taube JM, et al. Sci Transl Med. 2012;4:27ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada H, et al. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 16.Childs KS, Goodbourn S. Nucleic Acids Res. 2003;31:3016–3026. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pham CD, et al. Clin Cancer Res. 2016;22:582–595. doi: 10.1158/1078-0432.CCR-15-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platanias LC. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 19.Yao S, et al. Mucosal Immunol. 2015;8:746–759. doi: 10.1038/mi.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi T, Takaoka A. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 21.Soliman H, Khalil F, Antonia S. PLOS ONE. 2014;9:e88557. doi: 10.1371/journal.pone.0088557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng AC, et al. PLOS ONE. 2011;6:e24100. doi: 10.1371/journal.pone.0024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng AC, et al. FASEB J. 2010;24:4825–4834. doi: 10.1096/fj.10-167049. [DOI] [PubMed] [Google Scholar]
- 24.Jarosinski KW, Massa PT. J Neuroimmunol. 2002;122:74–84. doi: 10.1016/s0165-5728(01)00467-2. [DOI] [PubMed] [Google Scholar]
- 25.Pardoll DM. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou W, Chen L. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, et al. Blood. 2007;110:296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 28.Salsman VS, et al. PLOS ONE. 2011;6:e20267. doi: 10.1371/journal.pone.0020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo HR, Kim J, Bae S, Soh JW, Lee YS. J Biol Chem. 2008;283:15601–15610. doi: 10.1074/jbc.M800987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casey SC, et al. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Kinase active Cdk5 is expressed by murine and human MB cell lines and deletion of Cdk5 does not alter proliferation. (A) Genomic analysis from the UCSC cancer genome browser demonstrates that Cdk5, p35, and p39 are highly expressed in pediatric MB (4 patients) and neuroblastoma (NB) (39 patients) samples (above black line) compared to normal brain tissue (below black line). (B) Cdk5 kinase assay activity is detected in human MB (UW228, DAOY) and mouse MB (MM1) cell lines, with lysates from Cdk5−/− and Cdk5+/+ embryonic brains as negative and positive controls, respectively (Black Bars). The non-selective Cdk5 inhibitor, Roscovitine (10 μM), abrogates this kinase activity (Gray bars). (C) mRNA expression of Cdk5 transcript in MM1 WT (white), shCdk5 (black), or shNS (gray). (D) MM1 shCdk5 and MM1 crCdk5 are deficient in Cdk5 protein expression. (E) Proliferation curves of MM1 WT (square), shCdk5 (triangle), and shNS (circle) in media with 10% serum (black lines) or 1% serum (gray lines) over 72 hours (F) Proliferation curves of MM1 WT (white), crCdk5 (black), crNeg (gray) in media containing 10% serum over 72 hours. Data represent mean ±SD from biologic triplicates.
Fig S2. Similar subcutaneous (s.c.) growth kinetics of crCdk5 MM1 and crNeg MM1 in immune-compromised mice. (A) In vivo, growth kinetics of MM1 crCdk5 (circle) and crNeg (square) in NSG mice (n = 10/group). Points were graphed as the average size ± SD. (B) TFS of MM1 crCdk5 (circle) and crNeg (square) in NSG mice. (C) Tumor weights of MM1 crCdk5 (circle) and crNeg (square) in NSG mice. The solid line represents the average weight ± SD. (D) Inverse correlation between CD3+ cell count per HPF and Cdk5 staining intensity of clinical MB tissue samples as revealed by IHC analysis in Fig. 1D. Significance was determined using a t test (C).
Fig S3. In vivo growth kinetics of MM1 shCdk5 reveals thymic dependent tumor rejection upon s.c. injection. (A) TFS of MM1 WT (diamond, n = 13), shCdk5 (circle, n = 17), and shNS in B6 mice (triangle, n = 7). Survival curves were calculated from 3 independent experiments. (B) Tumor weights of MM1 WT (triangles, n = 13) and shCdk5 (circles, n = 13) and shNS (squares, n = 7). The solid line represents the average weight ± SD. (C) TFS of B6 mice injected s.c. with MM1 shNS (diamond, n = 10) or MM1 shCdk5 (circle) with injection of αCD4 (triangle), αCD8 (inverted triangle), or αCD4 and αCD8 (square) antibodies (n = 15/group). (D) Tumor size measurement of MHC-II KO (circle) and MHC-I KO (square) mice injected s.c. with Cdk5-deficient MM1 (n = 10/group). (E) TFS of B6 mice re-challenged with MM1 WT at least 60 days post-initial injection of MM1 shCdk5 (square) or MM1 crCdk5 (circle) (n = 5/group). **P < 0.01; ***P < 0.001 Significance was determined by log rank test (A, C, E) or t test (B, D).
Fig S4. A link between Cdk5 and PD-L1 expressions in human and murine tumors. (A) Cdk5 expression of cells cultured from subcutaneous tumor outgrowths isolated from animals initially injected with MM1 WT (n = 4), crCdk5 (n = 4), or crNeg (n = 6). (B) PD-L1 expression on primary cell cultures of MM1 crNeg, crCdk5, or outgrowth of crCdk5 tumors. Cells were stimulated with IFNγ for 24 hours (black) and compared to isotype staining (dotted line) and un-stimulated cells (gray). Histogram is a representative staining from three separate experiments. (C) PD-L1 mRNA expression by RMS 76.9 WT (white), shCdk5 (black), and shNS (gray) with or without 24 hours of IFNγ stimulation. (D) Surface PD-L1 staining of RMS 76.9 shNS (gray) and shCdk5 (black) with or without 24 hours of IFNγ stimulation. Values represent the average MFI ± SD compared to unstimulated RMS 76.9 shNS over three independent experiments. (E) Surface PD-L1 staining of SJCRH30 WT (white) and crCdk5 (black) with or without 24 hours of IFNγ stimulation in vitro. Values represent average MFI ± SD compared to unstimulated WT over three independent experiments. *P<0.05; **P<0.005. Significance was determined by two-way analysis of variance (ANOVA) (D) and (E).
Fig S5. Disruption of Cdk5 gene expression results in more IFNγ in the tumor bed and decreased Cdk5 activity in response to IFNγ. (A) In vivo IFNγ mRNA expression in tumor extracts from i.c. inoculations (n = 3 mice/group; ± SD). (B) Cdk5 kinase activity in MM1 WT, shCdk5, and shNS over the course of 24-hour stimulation with IFNγ (black) and the addition of 10 μM Roscovitine (gray). EB: embryonic brain.
Fig S6. Co-occurrence of upregulated Cdk5 and PD-L1 expression in human cancer samples. TCGA provisional data sets for all available cancer types were analyzed for Cdk5 and PD-L1 (CD274) mRNA expression through cBioPortal (http://www.cbioportal.org/). (A) Relative mRNA expression values of samples with existing RNA sequencing analysis (RNA Seq V2) were downloaded and analyzed with microsoft excel. The average log2 mRNA expression across each cancer type examined was individually graphed for Cdk5 and PD-L1. (B) Co-occurrence of elevated Cdk5 and PD-L1 gene expression is observed in several cancer types. mRNA expression z-scores of RNA Seq V2 RSEM from available tumors were analyzed for significance, their log odds ratio, and type of association. Representative cancer types showing co-occurrence of elevated Cdk5 and PD-L1 transcript levels are displayed.
Fig S7. Silencing Cdk5 in MB alters IRF-2 expression but does not affect other IFN signaling pathways. (A) Surface expression of MHC-I is upregulated on MM1 WT, crCdk5, and crNeg. Cells were stimulated with IFNγ for 24 hours (black) and compared to isotype staining (dotted line) and unstimulated cells (gray). Histogram is a representative staining from three separate experiments. (B) 24-hour stimulation of MM1 WT, crCdk5, and crNeg cells. Downstream IFNγ pathway elements STAT3, phosphorylated (p)STAT3, and STAT2 were assayed. (C) IFNγ stimulation of MM1 WT, crCdk5, and crNeg over the course of 48 hours. IFNγR downstream mediators IRF1, and IRF2 were assayed. (D) In vitro mRNA analysis of IRF-1, IRF-2, and IRF2BP2 present in MM1 WT (white), crCdk5 (black), and crNeg (gray) after treatment with IFNγ for 24 hours. Significance was determined using a 2-Way ANOVA (D).
Fig S8. CD3+ cell infiltrate and diminished growth rates associate with a decrease in tumor incidence following orthotopic administration of Cdk5-deficient MM1 cells. (A) Tumor incidence of MM1 WT, shCdk5, and shNS 14 days after i.c. tumor inoculation. (B) Western blot analysis for Cdk5 in cells cultured from B6 mice tumor outgrowths initially inoculated with MM1 WT (n = 2) and MM1 crCdk5 (n = 3). (C) IHC of tumor injected i.c. with MM1 WT, shCdk5, and shNS (scale bar = 200 μm). (D) Quantification of intratumoral CD3+ cells in MM1 WT (n = 3), shCdk5 (n = 3), and shNS (n = 4) tumors identified via IHC per mm2 of tumor tissue. Solid line represents the average ± SD. Significance was determined using a Student's t test (D).
Fig S9. Subcutaneous Cdk5 deficient tumors characterized by increased stromal PD-L1 staining, CD4+ tumor infiltrating lymphocytes (TIL), and infiltrating CD11b+ populations. (A) In vivo IFNγ mRNA expression in tumor extracts from subcutaneous inoculations (n = 3 mice/group; ± SD). (B) S.c. tumors extracted 14 days post-inoculation from MM1 shCdk5 or shNS stained for F4/80, PD-L1, and PD-1 expression (Scale bars = 2mm). (C), (D) FACS analysis of MM1 WT (black circle) and MM1 crCdk5 (red square) tumor infiltrate by percentage of cell type. (E) Ratio of total CD8+:CD4+ cell infiltrate. (F) FACS analysis of the percentage of PD-1+ or PD-L1+ cells in the CD4+ or CD8+ populations. (G) FACS analysis of the percentage of myeloid cells in tumor infiltrate based on differential CD45 staining (left) or Ly6C staining among CD11b+CD45+ cells (right). (H) Percent of total CD11b+ population (left) and sub-populations (right) present in tumor infiltrate that express PD-L1. (I) MFI of PD-L1 expression between total (left) and sub-populations (right) of CD11b+ cells. C thru H were graphed as the mean ± SD. (I) was graphed as individual MFI with mean indicated. n = 9/group. Each data point represents pooled samples from 3 mice. *P < 0.05 Significance was determined by Student's t test.
Fig S10. Putative mechanism by which Cdk5 controls PD-L1 expression. (A) When Cdk5 WT cells are stimulated with IFNγ, signaling through the JAK/STAT pathway stimulates IRF-1 driven transcription of PD-L1. Additionally, IFNγ stimulates p35 expression and thereby increases Cdk5 activity. Phosphorylation of unknown kinase(s) by Cdk5 inhibits kinase function, resulting in a reduced abundance of the IRF2/IRF2BP2 repressor complex. (B) In Cdk5 deficient cells, unknown kinase(s) hyper-phosphorylate IRF2BP2, prolong half-life of IRF2/IRF2BP2 repressor complex despite intact JAK/STAT/IRF-1 signaling, ultimately leading to decreased PD-L1 expression. IRF-E: IRF-binding element.
Fig. S11 Higher Cdk5 correlates with adverse clinical outcomes in human cancers. PrognoScan database-based Kaplan-Meier analysis of six independent patient cohorts including CNS and peripheral tumors that were analyzed for the effects of higher levels of Cdk5 gene expression (red) compared to low Cdk5 levels (blue) on overall survival, disease free survival, or metastasis free survival (http://www.prognoscan.org/). High levels of Cdk5 are associated with reduced (A) overall survival in a cohort of 74 glioma patients with astrocytoma (n = 8), GBM (n = 50), mixed glioma (n = 7), and oligodendroglioma (n = 9) (GSE 4412-GPL96); (B) overall survival in 50 patients with anaplastic oligodendroglioma (n = 22) and GBM (n = 28) (MGH-glioma); (C) overall survival of cutaneous melanoma patients (n = 38) (GSE19234); (D) distant metastasis free survival in uveal melanoma patients (n = 63) (GSE22138); (E) overall survival in a cohort of 104 patients with lung adenocarcinoma (GSE68465); and (F) disease free survival in a cohort of 249 breast cancer patients (GSE4922-GPL96). All p-values represent the corrected p-value.
Table S1. shCdk5-All phosphoproteomics data
Table S2. shCdk5-Significant phosphoproteomics data
Table S3. crCdk5-All phosphoproteomics data
Table S4. crCdk5-Significant phosphoproteomics data