Abstract
Background and Aims
Large indels are commonly identified in patients but are not detectable by routine Sanger sequencing and panel sequencing. We specially designed a multi-gene panel that could simultaneously test known large indels in addition to ordinary variants, and reported the diagnostic yield in patients with intrahepatic cholestasis.
Methods
The panel contains 61 genes associated with cholestasis and 25 known recurrent large indels. The amplicon library was sequenced on Ion PGM system. Sequencing data were analyzed using a routine data analysis protocol and an internal program encoded for large indels test simultaneously. The validation phase was performed using 54 patients with known genetic diagnosis, including 5 with large insertions. At implement phase, 141 patients with intrahepatic cholestasis were evaluated.
Results
At validation phase, 99.6% of the variations identified by Sanger sequencing could be detected by panel sequencing. Following the routine protocol, 99.8% of false positives could be filtered and 98.8% of retained variations were true positives. Large insertions in the 5 patients with known genetic diagnosis could be correctly detected using the internal program. At implementation phase, 96.9% of the retained variations, following the routine protocol, were confirmed to be true. Twenty-nine patients received a potential genetic diagnosis when panel sequencing data were analyzed using the routine protocol. Two additional patients, who were found to harbor large insertions in SLC25A13, obtained a potential genetic diagnosis when sequencing data were further analyzed using the internal program. A total of 31 (22.0%) patients obtained a potential genetic diagnosis. Nine different genetic disorders were diagnosed, and citrin deficiency was the commonest.
Conclusion
Specially designed multi-gene panel can correctly detect large indels simultaneously. By using it, we assigned a potential genetic diagnosis to 22.0% of patients with intrahepatic cholestasis.
Introduction
Cholestasis results from impairment of bile acid biosynthesis, bile secretion and excretion [1]. The etiology is diverse, and includes a range of genetic defects that represent a collection of disorders [2]. The pathophysiology of cholestasis is complex, and depends on the specific genetic defects. Although subtle clinical and biochemical differences exist, these genetic disorders are difficult to be differentiated based on clinical and routine laboratory findings [3–4]. Meanwhile, patients with same genetic defects can present different clinical phenotypes [5–6]. Therefore, genetic tests are extremely important for the establishment of a clear genetic diagnosis, and hence for the initiation of tailored treatment and genetic counseling [3,7].
Quite often, several candidate genes have to be evaluated in clinic practice, because the differential diagnoses are numerous [2]. If the candidate genes are tested by Sanger sequencing, the process is time-consuming and expensive. Multi-gene panel, a time and cost efficient alternative to Sanger sequencing, can screen multiple candidate genes simultaneously [8], and is increasingly used for diagnostic evaluation of patients with intrahepatic cholestasis in children [2,9–10]. However, large insertions/deletions (indels) can neither be detected by routine Sanger sequencing [6,11], nor by routine multi-gene panel sequencing [2,10]. Furthermore, some special large indels, i.e. IVS16ins3kb and IVS4ins6kb in SLC25A13, even can’t be detected by copy number variation (CNV) analysis, but have high frequency in patients and contribute substantively to disease burden [12–13].
To facilitate genetic diagnosis, we specially designed a multi-gene panel that could not only sequence the coding exons of 61 cholestasis-related genes, but also test 25 known recurrent large indels (>150bp) in the genes. In this study, we first validated our system using 54 patients with known genetic diagnosis, then evaluated 141 consecutive patients with intrahepatic cholestasis using this panel and reported the diagnostic yield.
Methods
Multi-Gene Panel Design
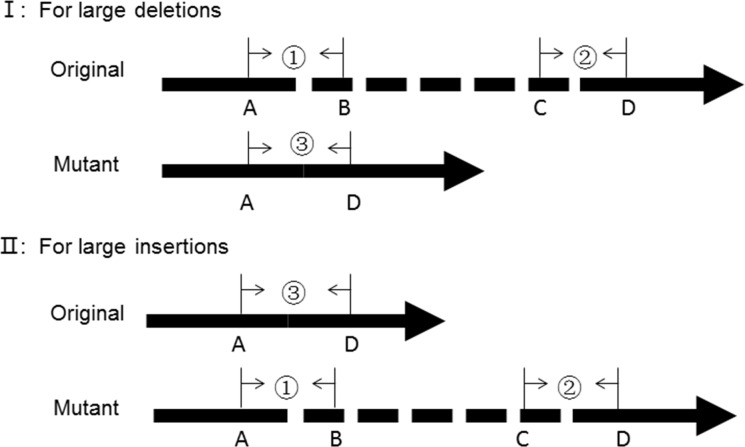
Cholestasis was defined as direct bilirubin (DB) >20% of the total bilirubin (TB) if TB >5 mg/dL or DB >1 mg/dL if TB <5 mg/dL [10]. A custom AmpliSeq panel was designed to cover the target regions, including the coding DNA sequence and at least 5bp of flanking intronic regions, of 61 genes associated with cholestasis (S1 Table). Among the 61 genes, 25 were known intrahepatic cholestasis disease-causing genes [14–15]. The remaining 36 included potential candidate genes and genes for differential diagnoses. In addition, 25 related known large indels in these genes were also included in this panel (S2 Table). Three pair primers were designed for each large indel, and yield three amplicons (AP1, AP2 and AP3) covering the original allele and mutant allele (Fig 1). An internal program was encoded to detect the three amplicons. The implication of these amplicons was summarized in Table 1.
Fig 1. Schematic diagram of primer design for large indels.
①, ②, ③ were three pair primers designed for each large indel, and yield three amplicons AP1, AP2, AP3 respectively. BC was the inserted or deleted sequence.
Table 1. The implication of amplicons for large indels detection.
| AP1 | AP2 | AP3 | Deletion | Insertion |
|---|---|---|---|---|
| + | + | + | Heterozygous | Heterozygous |
| + | + | - | Normal | Homozygous |
| - | - | + | Homozygous | Normal |
AP1, AP2, AP3, three amplicons yield from primers designed for each large indel
Subjects and Experiment Design
At validation phase, 54 cholestatic patients, who had received a clear genetic diagnosis by Sanger sequencing, were re-evaluated using this panel. Among them, 5 patients harbored large insertions in SLC25A13, and were diagnosed with neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD). Both panel sequencing data and Sanger sequencing results of the 54 patients were used to optimize parameter settings of the data analysis protocol that helped to retain true positives and filter false positives effectively.
At implement phase, 141 consecutive patients with intrahepatic cholestasis were evaluated from April 2015 to November 2015. The patients were referred to the Center for Pediatric Liver Disease of the Children’s Hospital of Fudan University and the Department of Pediatrics, Jinshan Hospital of Fudan University. Following a reported extensive workup [16–17], other causes were excluded, including infections, drug-induced, metabolic and surgical causes. Cytomegalovirus (CMV) infection was considered if serum immunoglobulin M (IgM) or pp65 antigenemia or urinary CMV-DNA was positive [18]. CMV infection was not excluded for its high prevalence in Chinese infants [19]. The 141 patients included 92 boys and 49 girls. The age ranged from 1 month to 17 years old when panel sequencing was ordered.
This study was approved by the ethics committee of Jinshan Hospital of Fudan University and Children’s Hospital of Fudan University. Written informed consent was obtained from guardian/their parents. Patients’ information was de-identified prior to analysis.
Library Construction, Enrichment and Sequencing
Library construction, enrichment and sequencing were performed according to the manufacturer’s instructions (Life Technologies, USA). Briefly, targets were amplified by a multiple polymerase chain reaction assay, and ligated to Ion Xpress barcode adapters. Then, libraries were purified and normalized to ~100pM. Template-positive Ion Sphere Particles were prepared and enriched. 8 samples were loaded per 316 v2 chip. Sequencing was performed on the Ion Torrent Personal Genome machine (Ion PGM) system.
Data Analysis and Variations Classification
Torrent Suite, the Ion Torrent platform-specific pipeline software, was used to process raw data. Human genome reference sequence (hg19) was used for the reference. Variation calling was performed using settings [Homopolymer Indel Balance ≤0.5, and Frequency ≤50%] by NextGene software (version 2.3.3). For large indels test, panel sequencing data were further analyzed using the internal program encoded for known large indels test.
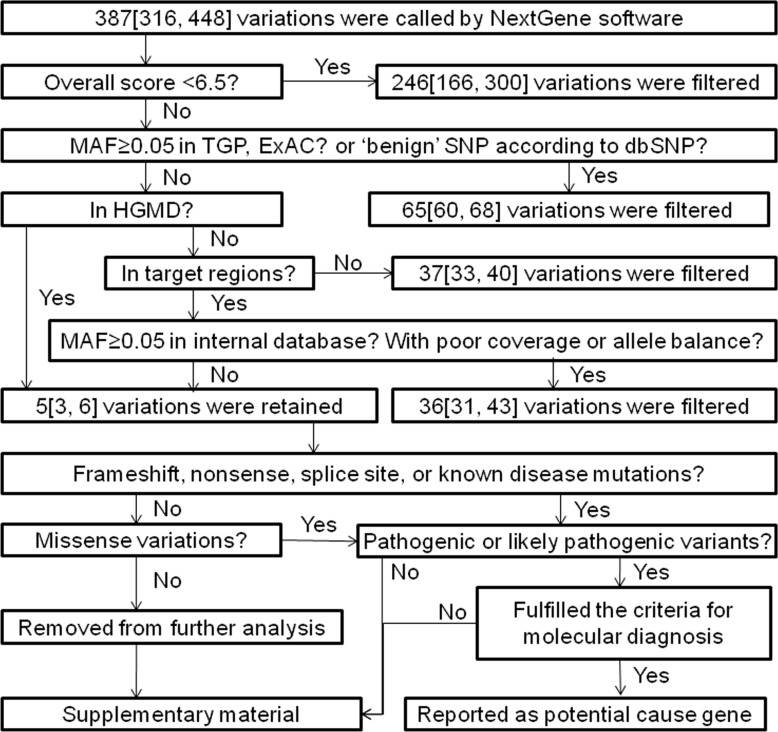
Variations with suboptimal overall scores (OS) were filtered as false positives (Fig 2). Variations with minor allele frequency (MAF) ≥0.05 according to 1000 Genomes Project (TGP) or Exome Aggregation Consortium (ExAC) and benign single nucleotide polymorphism (SNP) according to dbSNP were considered benign and filtered [20]. Of the remaining variations, mutations in Human Gene Mutation Database (HGMD) were retained [21], while variations beyond 5bp from exon boundaries, and variations with MAF ≥0.05 in internal database or poor coverage (<20x) or poor allele balance were discarded.
Fig 2. The optimized protocol for panel sequencing data analysis.
The data from 195 patients were presented as median [P25, P75]. MAF, minor allele frequency; TGP, 1000 Genomes Project; ExAC, Exome Aggregation Consortium; dbSNP, Single Nucleotide Polymorphism database; HGMD, Human Gene Mutation Database.
Of the retained variations, frameshift, nonsense, canonical splice site variants, previous reported mutations and missense variations were analyzed further, while synonymous variations and variations beyond the canonical splice site were removed from further analysis. The prediction of missense variations was performed by in silico predictors (Polyphen 2, MutationTaster and SIFT). The pathogenicity was assessed according to the standards and guidelines for interpretation of sequence variants [20].
Sanger Sequencing and Electrophoresis
Pathogenic and likely pathogenic variants of interest were confirmed by directly sequencing the affected exons from the patients and the parents using Sanger sequencing. Primer sequences and PCR conditions were available on request. Purified PCR products were directly sequenced on an ABI Prism 3500 Genetic Analyzer. Large indels were amplified by long and accurate-PCR (LA-PCR). LA-PCR products were confirmed by electrophoresis.
Statistical Analysis
Statistic analysis was done using SPSS version 17.0 software (University of Chicago, Chicago, IL, United States). Data were expressed as mean±SD for normality, or median [P25, P75] for non-normality. Comparisons of two means or two medians were done by using two independent samples t-test or nonparametric Mann-Whitney test respectively. P<0.05 was considered significant.
Results
Performance of Ion PGM Sequencing
A total of 1278 pair primers were designed to cover the target regions of the 61 cholestasis associated genes, including 886 coding exons. The numbers of total reads and mapped reads, and the average depth of coverage were similar between patients at validation phase and at implementation phase (P >0.05, Table 2). Few exons were found to have bases with poor coverage (<20x) in patients at implementation phase, and the range was 13~41 (1.5%~4.6% of the 886 exons) while it was 23~43 (2.6%~4.9% of the 886 exons) for patients at validation phase.
Table 2. The performance of Ion PGM sequencing.
| At validation phase(N = 54) | At implementation phase(N = 141) | |
|---|---|---|
| Total reads | 36.5k±7.1k | 39.0k±7.2k |
| Mapped reads | 35.4k±6.7k | 37.6k±7.0k |
| Average depth of coverage | 277.2±52.6 | 294.2±54.5 |
| Exons with poor coverage (<20x) | 32.0 [29.0, 35.0] | 28.0 [23.0, 32.0] * |
| Total called variations | 417.0±80.3 | 366.4±103.3 * |
| Variations with OS ≥6.5 | 142.0 [136.0, 156.0] | 139.5 [133.0, 151.0] |
| Retained variations | 5.0 [4.0, 7.0] | 4.0 [3.0, 5.0] * |
| Mutations per sample | 2.0 [1.0, 2.0] | 0.0 [0.0, 1.0] * |
* Patients at validation phrase vs. patients at implement phrase, P <0.05
Validation of Detection Efficiency
In the target regions of the 54 patients with known genetic diagnosis, Sanger sequencing identified 225 variations, including 11 distinct indels and 84 different substitutions. Of them, 224 (99.6%) were detected by panel sequencing with one missing for low coverage (5×). Additional 420 variations were identified by panel sequencing in the same regions, but not detected by Sanger sequencing. These variations were regarded as false positives, and the majority (99.3%) was indels. Tree false substitutions were identified and had OS<6.5. The features of false and true positives were summarized in Table 3. Following a data analysis protocol (Fig 2), 99.8% of false positives could be filtered. Of the 86 retained variations, 85 (98.8%) were true positives. The remaining one false positive (1.2%) was 1bp deletion.
Table 3. Features of false and true positives.
| True positives | False positives | |
|---|---|---|
| Total | 224 | 420 |
| Substitutions | 213 (95.1%) | 3 (0.7%) |
| Indels | 11 (4.9%) | 417 (99.3%) |
| Overall score (OS) | ||
| OS ≥6.5 | 224 (100.0%) | 36 (8.6%) |
| OS >12.0 | 217 (96.9%) | 24 (5.7%) |
| Original/mutant allele ratio <2.5:1 | 224 (100.0%) | 127 (30.2%) |
| Filtered variations § | 139 (62.1%) | 419 (99.8%) |
| OS <6.5 (false positives) | 0 (0.0%) | 384 (91.4%) |
| Benign variations ‡ | 129 (57.6%) | 0 (0.0%) |
| Discarded variations † | 10 (4.5%) | 35 (8.3%) |
§ Filtered variations: 558 variations detected by panel sequencing were filtered following the protocol shown in Fig 2.
‡ Benign variations were defined as variations with MAF ≥0.05 in TGP and ExAC, or variations classified as benign in dbSNP.
† Discarded variations included variations beyond the target regions, and variations with MAF ≥0.05 in internal database or poor coverage or poor allele balance.
Re-evaluation of Patients with Known Genetic Diagnosis
In the 54 patients with known genetic diagnosis, 74 pathogenic or likely pathogenic variants were detected by Sanger sequencing. Following the data analyzing protocol in Fig 2, about 5 (range: 1~11) variations were retained by panel sequencing per sample (Table 2). The retained variations contained all the 74 pathogenic or likely pathogenic variants detected by Sanger sequencing. Then, panel sequencing data were further analyzed using the internal program encoded for large indels test. Large insertions were correctly detected in the 5 NICCD patients (Table 4). Therefore, by combination of the two data analysis methods, all known short genetic variants (≤50bp) and large indels, were detected successfully.
Table 4. Information of large indels identified in 5 citrin deficiency patients.
| Patient | Gene | Mutation | Zygosity | NextGene | In-program ‡ |
|---|---|---|---|---|---|
| NO.1 | SLC25A13 | c.329-18_329-17ins6057bp | Homozygous | Not | Yes |
| NO.2 | SLC25A13 | c.1751-5_1751-4ins3kb | Homozygous | Not | Yes |
| NO.3 | SLC25A13 | c.1751-5_1751-4ins3kb | Heterozygous | Not | Yes |
| SLC25A13 | c.1638_1660dup | Heterozygous | Yes | Not | |
| NO.4 | SLC25A13 | c.1751-5_1751-4ins3kb | Homozygous | Not | Yes |
| NO.5 | SLC25A13 | c.1402C>T | Heterozygous | Yes | Not |
| SLC25A13 | c.1751-5_1751-4ins3kb | Heterozygous | Not | Yes |
‡ In-program, internal program encoded by our team to detect known large indels.
Evaluation of Patients without a Previous Genetic Diagnosis
Panel sequencing data of the 141 patients with intrahepatic cholestasis were analyzed using the same protocol (Fig 2). About 4 (range: 0~11) variations were retained per sample (Table 2). A total of 127 retained variations, including 110 substitutions and 17 indels, were chosen to validate by Sanger sequencing. Among them, 123 (96.9%; 123/127) were confirmed to be true, including 110 substitutions and 13 indels. Fourteen filtered variations, including 1 substitution with OS<6.5, 1 deletion with MAF>0.05 in internal database and 11 indels with OS>6.5 (range: 7.1~18.3) but original/mutant allele ratios ≥2.5:1, were confirmed to be false. Additional variations were not identified in target regions of the 138 affected exons evaluated by Sanger sequencing.
Pathogenic or likely pathogenic variants were identified in 59 (41.8%; 59/141) patients. Twenty-nine patients obtained a potential genetic diagnosis (Table 5). Twenty-three patients were diagnosed with autosomal recessive (AR) disorders and 6 had autosomal dominant (AD) disorders. Nine distinct genetic disorders were diagnosed, including 4 seen only once. For the 29 patients, a total of 36 different mutations were identified in causal genes. The 36 mutations included 2 deletions, 2 insertions, 5 nonsense, 7 canonical splice sites and 20 missense mutations. Of the 20 missense mutations, 6 were novel and predicted to be damaging (S3 Table).
Table 5. The spectrum of genetic disorders diagnosed by panel sequencing.
| Patient | Gene | Nucleotide change | Amino acid change |
|---|---|---|---|
| 1 | SLC25A13 | c.1177+1G>A/c.1177+1G>A | -/- |
| 2 | SLC25A13 | c.1095delT/c.1157G>T | p. F365LfsX43/p.G386V |
| 3 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 4 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 5 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 6 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 7 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 8 | SLC25A13 | c.851_854del/c.851_854del | p.M285PfsX2 /p.M285PfsX2 |
| 9 | ABCC2 | c.2302C>T/c.4024T>C | p.R768W/p.S1342P |
| 10 | ABCC2 | c.632+2_632+5del/c.4238_4239dup | -/p.H1414LfsX18 |
| 11 | ABCC2 | c.3825C>G/c.4146+1G>T | p.Y1275X/- |
| 12 | ABCC2 | c.2366C>T/c.2366C>T | p.S789F/p.S789F |
| 13 | ABCC2 | c.1963C>T/c.2153A>G | p.R655X/p.N718S |
| 14 | ABCC2 | c.1281T>G/c.4025C>A | p.D427E/p.S1342Y |
| 15 | ABCC2 | c.2224G>A/c.4025C>A | p.D742N/p.S1342Y |
| 16 | JAG1 | c.133G>T/- | p.V45L/- |
| 17 | JAG1 | c.133G>T/- | p.V45L/- |
| 18 | JAG1 | c.133G>T/- | p.V45L/- |
| 19 | JAG1 | c.463G>C/- | p.A155P/- |
| 20 | JAG1 | c.2698C>T/- | p.R900X/- |
| 21 | ABCB11 | c.1550G>A/c.908+1G>T | p.R517H/- |
| 22 | ABCB11 | c.3691C>T/c.872T>C | p.R1231W/p.V291A |
| 23 | ABCB11 | c.2197C>T/c.1489C>T | p.Q733X/p.Q497X |
| 24 | CYP27A1 | c.379C>T/c.1263+1G>A | p.R127W/- |
| 25 | CYP27A1 | c.379C>T/c.1214G>A | p.R127W/p.R405Q |
| 27 | CFTR | c.214G>A/c.650A>G+c.3406G>A | p.A72T/p.E217G+ p.A1136T |
| 27 | GALT | c.377+2dup/c.377+2dup | -/- |
| 28 | NOTCH2 | c.6027+1G>A/- | -/- |
| 29 | NPC1 | c.1421C>T/c.2728G>A | p.P474L/p.G910S |
Novel mutations are shown in bold.
Large Indels Test Facilitates Genetic diagnosis
Sequencing data of the 141 patients were analyzed using the internal program encoded for large indels test. Large insertions were identified in SLC25A13 in two patients, and were confirmed to be true by LA-PCR. Consequently, two additional patients obtained a potential genetic diagnosis, and were diagnosed with NICCD (one with c.1640_1641ins23bp/IVS4ins6kb and the other with c.851_854del/IVS16ins3kb). Hence, the rate of positive genetic diagnosis was 22.0%. The top 4 common genetic disorders: NICCD (32.3%), Dubin-Johnson syndrome (DJS, 22.6%), Alagille syndrome (19.4%), PFIC2/BRIC2 (9.8%).
Discussion
Multiple specific genetic defects can cause intrahepatic cholestasis. It is still challenging to assign a molecular diagnosis because the differential diagnoses are numerous [2]. To ease the process of diagnosis, we designed a multi-gene panel that contained 61 genes associated with cholestasis and 25 related known large indels. We demonstrated that this panel was very practical, and that the ability of large indels detection could further facilitate genetic diagnosis. Using this panel, we assigned a potential molecular diagnosis to 22.0% of patients with intrahepatic cholestasis.
Poor coverage was one of the important causes for missing true positives [2]. In this study, more than 95.0% of the coding exons had desired coverage. Hence, bases with poor coverage (<20x) were much less than 5.0%. This might account for that 99.6% of known variations were successfully detected. However, numerous false positives were also produced during sequencing. The majority was indels, only a few (0.7%) were substitutions. At validation phase, all false substitutions had OS <6.5, while all true substitutions had OS ≥6.5. At implementation phase, all retained substitutions with OS ≥6.5 were confirmed to be true, while filtered substitutions with OS <6.5 were confirmed to be false. Hence, we inferred that OS ≥6.5 was reliable for true substitutions. It was different from previous researches [22–23]. According to our data, 3.1% of true positives would be missed if OS >12.0 was set as a cut-off value. Furthermore, 8.6% of false indels also had OS ≥6.5, even >12.0. Therefore, the retained variations also contained a few false indels. To filter these false indels effectively, additional parameters were needed, i.e. original/mutant allele ratio, as described in this study.
Similar presentation, symptoms and management of patients with intrahepatic cholestasis hindered genetic diagnosis [3–4]. Multi-gene panel overcame the complexity of candidate gene approach, and had advantages in evaluating not only patients with atypical presentations, but also clinical diagnoses associated with multiple candidate genes, and patients lacking a genetic diagnosis despite extensive Sanger sequencing [10,20,24]. Using a multi-gene panel, 9 distinct genetic disorders were diagnosed in this study. 4 were seen only once, and most were first reported in Chinese child patients with intrahepatic cholestasis. This indicated that this panel was very powerful for the assignment of genetic diagnosis. However, we failed to include all causal genes of cholestasis in this panel, especial those reported recently [25].This might be one of the reasons for that the majority (78.0%) of the patients still lacked a genetic diagnosis.
Large indels were commonly identified in patients [13], but failed to be detected by using routine Sanger sequencing and routine multi-gene panel sequencing [2]. Multiplex ligation-dependent probe amplification (MLPA) analysis, genomic microarrays, and other CNV analysis tools based on routine next generation sequencing (NGS) data were used to identify large indels characterized by DNA copy number loss/gain [10,26–27]. However, some special large indels, i.e. a retrotransposal insertion IVS16ins3kb that the inserted sequence was an antisense strand of complementary DNA (cDNA) processed from C6orf68 and a transposal insertion IVS4ins6kb in SLC25A13 [6,28], could not be detected by routine CNV analysis. These large indels had high frequency in patients, and were identified in 26.0% of NICCD patents and accounted for 14.3% of total mutant alleles [29]. The detection of these large indels could facilitate the diagnosis and improve the diagnostic efficiency. However, other labor-intensive and cost-expensive molecular tools were needed to identify them [11]. Using our design, we could simultaneously test these special large indels in addition to ordinary variants in a multi-gene panel. Our data demonstrated that this design was very practical to detect known large indels with definite chromosomal location. However, this panel failed to enroll other known large indels without definite chromosomal location, and failed to identify novel large indels. By using this panel, 22.0% patients with intrahepatic cholestasis obtained a potential genetic diagnosis, including two harboring large insertions in SLC25A13. Large indels were only identified in NICCD patients, it might attribute to that large indels had high frequency in SLC25A13 and that NICCD was common in Asian [10,29]. Therefore, we suggest that SLC25A13 should be compulsory after CFTR, which causes the most common AR genetic disorder in white population [30], in every panel for children with intrahepatic cholestasis. This is the first report to our knowledge that using panel sequencing to test known large indels in addition to small sequencing changes at one assay without CNV analysis. Without a doubt, the design of this panel can be used for other panels that involve known large indels, especial those failed to be detected by CNV analysis.
Conclusion
We specially designed a multi-gene panel that contained 61 genes associated with cholestasis and 25 related known large indels. We demonstrated that this panel was practical and powerful in evaluation of patients with intrahepatic cholestasis. Using this panel, we assigned a potential genetic diagnosis to 22.0% of patients with intrahepatic cholestasis, including two patients with large indels.
Supporting Information
(DOCX)
(DOC)
(DOC)
(DOC)
Acknowledgments
The authors would like to express our gratitude to patients and their families for their participation, and to Dr. Bansal S (King's College Hospital, UK) for editing the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by National Natural Science Foundation of China (NO. 81361128006) to Jian-She Wang, and Shanghai Municipal Commission of Health and Family Planning (NO.2013-27) to Jian-She Wang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zollner G, Trauner M. Mechanisms of cholestasis. Clin Liver Dis. 2008;12:1–26. 10.1016/j.cld.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 2.Herbst SM, Schirmer S, Posovszky C, Jochum F, Rödl T, Schroeder JA, et al. Taking the next step forward-Diagnosing inherited infantile cholestatic disorders with next generation sequencing. Mol Cell Probes. 2015;29:291–8. 10.1016/j.mcp.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Aronow BJ, Jegga AG, Wang N, Miethke A, Mourya R, et al. Novel resequencing chip customized to diagnose mutations in patients with inherited syndromes of intrahepatic cholestasis. Gastroenterology. 2007;132:119–26. 10.1053/j.gastro.2006.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalaf R, Phen C, Karjoo S, Wilsey M. Cholestasis beyond the Neonatal and Infancy Periods. Pediatr Gastroenterol Hepatol Nutr. 2016;19:1–11. 10.5223/pghn.2016.19.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordo-Gilart R, Andueza S, Hierro L, Martínez-Fernández P, D'Agostino D, Jara P, et al. Functional analysis of ABCB4 mutations relates clinical outcomes of progressive familial intrahepatic cholestasis type 3 to the degree of MDR3 floppase activity. Gut. 2015;64:147–55. 10.1136/gutjnl-2014-306896 [DOI] [PubMed] [Google Scholar]
- 6.Song YZ, Zhang ZH, Lin WX, Zhao XJ, Deng M, Ma YL, et al. SLC25A13 gene analysis in citrin deficiency: sixteen novel mutations in Asian patients, and the mutation distribution in a large pediatric cohort in China. PLoS One. 2013;8:e74544 10.1371/journal.pone.0074544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. 10.1056/NEJMoa1306555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinrichs JW, van Blokland WT, Moons MJ, Radersma RD, Radersma-van Loon JH, de Voijs CM, et al. Comparison of next-generation sequencing and mutation-specific platforms in clinical practice. Am J Clin Pathol. 2015; 143:573–8. 10.1309/AJCP40XETVYAMJPY [DOI] [PubMed] [Google Scholar]
- 9.Matte U, Mourya R, Miethke A, Liu C, Kauffmann G, Moyer K, et al. Analysis of gene mutations in children with cholestasis of undefined etiology. J Pediatr Gastroenterol Nutr. 2010;51:488–93. 10.1097/MPG.0b013e3181dffe8f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Togawa T, Sugiura T, Ito K, Endo T, Aoyama K, Ohashi K, et al. Molecular genetic dissection and neonatal/infantile intrahepatic cholestasis using targeted next-generation sequencing. J Pediatr. 2016;171:171–7. 10.1016/j.jpeds.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 11.Zheng QQ, Zhang ZH, Zeng HS, Lin WX, Yang HW, Yin ZN, et al. Identification of a Large SLC25A13 Deletion via Sophisticated Molecular Analyses Using Peripheral Blood Lymphocytes in an Infant with Neonatal Intrahepatic Cholestasis Caused by Citrin Deficiency (NICCD): A Clinical and Molecular Study. Biomed Res Int. 2016;2016:4124263 10.1155/2016/4124263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang ZH, Lin WX, Deng M, Zhao ST, Zeng HS, Chen FP, et al. Clinical, molecular and functional investigation on an infant with neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD). PLoS One. 2014;9:e89267 10.1371/journal.pone.0089267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009; 360:599–605. 10.1056/NEJMoa0805392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balistreri WF, Bezerra JA, Jansen P, Karpen SJ, Shneider BL, Suchy FJ. Intrahepatic Cholestasis: Summary of an American Association for the Study of Liver Diseases Single-Topic Conference. Hepatology. 2005;42:222–35. 10.1002/hep.20729 [DOI] [PubMed] [Google Scholar]
- 15.McKiernan P. Metabolic liver disease. Clin Res Hepatol Gastroenterol. 2012;36:287–90. 10.1016/j.clinre.2012.03.028 [DOI] [PubMed] [Google Scholar]
- 16.Wang JS, Tan N, Dhawan A. Significance of low or normal serum gamma glutamyl transferase level in infants with idiopathic neonatal hepatitis. Eur J Pediatr. 2006;165:795–801. 10.1007/s00431-006-0175-3 [DOI] [PubMed] [Google Scholar]
- 17.Fang LJ, Wang XH, Knisely AS, Yu H, Lu Y, Liu LY, et al. Chinese children with chronic intrahepatic cholestasis and high γ-glutamyl transpeptidase: clinical features and association with ABCB4 mutations. J Pediatr Gastroenterol Nutr. 2012;55:150–6. 10.1097/MPG.0b013e31824ef36f [DOI] [PubMed] [Google Scholar]
- 18.Wang NL, Li LT, Wu BB, Gong JY, Abuduxikuer K, Li G, et al. The Features of GGT in Patients with ATP8B1 or ABCB11 Deficiency Improve the Diagnostic Efficiency. PLoS One. 2016,11:e0153114 10.1371/journal.pone.0153114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang MH, Hsu HC, Lee CY, Wang TR, Kao CL. Neonatal hepatitis: A follow-up study. J Pediatr Gastroenterol Nutr. 1987;6:203–7. 10.1097/00005176-198703000-00006 [DOI] [PubMed] [Google Scholar]
- 20.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009; 1:13 10.1186/gm13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarabeux J, Zeitouni B, Moncoutier V, Tenreiro H, Abidallah K, Lair S, et al. Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–41. 10.1038/ejhg.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck J, Pittman A, Adamson G, Campbell T, Kenny J, Houlden H, et al. Validation of next-generation sequencing technologies in genetic diagnosis of dementia. Neurobiol Aging. 2014;35:261–5. 10.1016/j.neurobiolaging.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 24.Buckley RH. Variable phenotypic expression of mutations in genes of the immune system. J Clin Invest. 2005;115:2974–6. 10.1172/JCI26956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Ospina N, Potter CJ, Xiao R, Manickam K, Kim MS, Kang HK et al. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat Commun. 2016;7:10713 10.1038/ncomms10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Dong J, Wang X, Guo H, Wang H, Zhao J, et al. JAG1 Mutation Spectrum and Origin in Chinese Children with Clinical Features of Alagille Syndrome. PLoS One. 2015;10:e0130355 10.1371/journal.pone.0130355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearney HM1, Thorland EC, Brown KK, Quintero-Rivera F, South ST; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5. 10.1097/GIM.0b013e3182217a3a [DOI] [PubMed] [Google Scholar]
- 28.Tabata A, Sheng JS, Ushikai M, Song YZ, Gao HZ, Lu YB, et al. Identification of 13 novel mutations including a retrotransposal insertion in SLC25A13 gene and frequency of 30 mutations found in patients with citrin deficiency. J Hum Genet. 2008;53:534–45. 10.1007/s10038-008-0282-2 [DOI] [PubMed] [Google Scholar]
- 29.Lin WX, Zeng HS, Zhang ZH, Mao M, Zheng QQ, Zhao ST, et al. Molecular diagnosis of pediatric patients with citrin deficiency in China: SLC25A13 mutation spectrum and the geographic distribution. Sci Rep. 2016;6:29732 10.1038/srep29732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaiuolo R, Sangiuolo F, Bombieri C, Bonizzato A, Cardillo G, Raia V, et al. Epidemiology and a novel procedure for large scale analysis of CFTR rearrangements in classic and atypical CF patients: a multicentric Italian study. J Cyst Fibros. 2008;7:347–51. 10.1016/j.jcf.2007.12.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.