Abstract
Background: Precision therapy for lung cancer will require comprehensive genomic testing to identify actionable targets as well as ascertain disease prognosis. RNA-seq is a robust platform that meets these requirements, but microarray-derived prognostic signatures are not optimal for RNA-seq data. Thus, we undertook the first prognostic analysis of lung adenocarcinoma RNA-seq data and generated a prognostic signature.
Methods: Lung adenocarcinoma RNA-seq and clinical data from The Cancer Genome Atlas (TCGA) were divided chronologically into training (n = 255) and validation (n = 157) cohorts. In the training cohort, prognostic association was assessed by univariate Cox analysis. A prognostic signature was built with stepwise multivariable Cox analysis. Outcomes by risk group, stage, and mutation status were analyzed with Kaplan-Meier and multivariable Cox analyses. All the statistical tests were two-sided.
Results: In the training cohort, 96 genes had prognostic association with P values of less than or equal to 1.00x10-4, including five long noncoding RNAs (lncRNAs). Stepwise regression generated a four-gene signature, including one lncRNA. Signature high-risk cases had worse overall survival (OS) in the TCGA validation cohort (hazard ratio [HR] = 3.07, 95% confidence interval [CI] = 2.00 to 14.62) and a University of Michigan institutional cohort (n = 67; HR = 2.05, 95% CI = 1.18 to 4.55), and worse metastasis-free survival in the TCGA validation cohort (HR = 3.05, 95% CI = 2.31 to 13.37). The four-gene prognostic signature also statistically significantly stratified overall survival in important clinical subsets, including stage I (HR = 2.78, 95% CI = 1.91 to 11.13), EGFR wild-type (HR = 3.01, 95% CI = 1.73 to 14.98), and EGFR mutant (HR = 8.99, 95% CI = 62.23 to 141.44). The four-gene prognostic signature also stood out on top when compared with other prognostic signatures.
Conclusions: Here, we present the first RNA-seq prognostic signature for lung adenocarcinoma that can provide a powerful prognostic tool for precision oncology as part of an integrated RNA-seq clinical sequencing program.
Lung cancer is the leading cause of cancer death in the United States, with an estimated 158 000 deaths in 2015, accounting for 27% all cancer deaths and more than colon (∼50 000), breast (∼41 000), and prostate cancer (∼28 000) combined (1). Lung adenocarcinoma is the most common histology, and rates are increasing. Individualized lung cancer therapy primarily consists of tyrosine kinase inhibitors (TKIs) for EGFR- and ALK-altered tumors and, more recently, MET and ROS1 alterations (2). Individualized therapy based on prognostic information has focused on early-stage lung adenocarcinoma, as patients with mediastinal nodal involvement unfortunately have poor outcomes even with high-intensity tri-modality therapy, and five-year survival after surgery alone remains poor at 73% in stage IA and 58% in stage IB (3). Though early-stage patients represent a minority currently, expanded lung cancer screening is likely to increase this proportion (4). Thus, continued efforts to improve management of early-stage lung adenocarcinoma are warranted.
In the genomics era, there have been numerous efforts to use unbiased microarray methods to develop prognostic signatures in lung cancer, dating back to 2002 (5,6). Although there has been some question about the reproducibility and validity in mutation subsets (7), two early-stage lung adenocarcinoma prognostic signatures have been commercialized and are available clinically, though adoption has been slow (8). Logistically, these signatures require a separate clinical test that, along with separate clinical tests for targetable alterations, result in increased cost and specimen handling. Perhaps more importantly, the microarray technology used in those studies is not reflective of the current view of the genome, as it assays only protein-coding genes. In a recent large-scale bioinformatics effort, our group has computationally predicted that long noncoding RNA (lncRNA) genes are several-fold more numerous than protein coding genes, and often cancer- and/or lineage-specific (9). These characteristics make lncRNAs prime biomarker candidates, leading to our characterization of the lncRNA SChLAP1 as a promising single-gene prognostic biomarker in prostate cancer (10). Combining protein- and noncoding genes may prove to increase the robustness of molecular biomarkers.
RNA sequencing (RNA-seq) represents a single comprehensive solution to many of the drawbacks of existing molecular assays. In a single platform, RNA-seq can comprehensively assay for an array of alterations, including point mutations and gene fusions, as well as comprehensively assay gene expression for prognostic and/or predictive genomic signatures and other applications (11). Additionally, exome-capture RNA-seq allows for accurate expression profiling in archival formalin-fixed paraffin-embedded samples (12). In this study, we utilized RNA-seq data from a lung adenocarcinoma cohort to identify a robust prognostic gene signature that can be directly incorporated into an RNA-seq clinical test for prognostic prediction.
Methods
Clinical Cohorts and RNA-Seq and Alteration Data
TCGA lung adenocarcinoma RNA-seq data was downloaded and processed with the Tuxedo pipeline as previously described (13). Briefly, reads were aligned to hg19 with Tophat2 (2.0.4) (14), and FPKM values generated based on the Ensembl v69 assembly (http://www.ensembl.org) (15). As previously described, the validation cohort consisted of 67 lung adenocarcinoma samples that were surgically resected and subjected to RNA-seq at the University of Michigan (5,16). Briefly, size-selected (350 bp) transcriptome libraries were polymerase chain reaction (PCR)–amplified (14 cycles) and analyzed by Agilent 2100 BioAnalyzer (Santa Clara, CA, USA). After paired-end 100 bp sequencing (2 x100 bp) on an Illumina HiSeq 2000 (San Diego, CA, USA), Illumina BaseCall–filtered reads were used and deposited in the Sequence Read Archive (SRA) as SRP048484. EGFR and KRAS nonsilent mutation cases were extracted from the raw mutation data files downloaded from the Broad GDAC FireHose (gdac.broadinstitute.org). ALK fusion cases were identified in the TCGA Fusion gene Data Portal (17). Samples from TCGA data set were divided chronologically into training and validation sets, and we did not find any bias in TCGA test and validation set in case bias analysis.
Evidence Before this Study
Pubmed was searched for articles relating to prognostic signatures in lung adenocarcinoma using the search expression “prognostic [Title/Abstract] AND signature [Title/Abstract] AND lung [Title/Abstract] AND (adenocarcinoma [Title/Abstract] OR non-squamous [Title/Abstract])” with no filters. This search returned over 40 articles, which were reviewed, and reports on 12 prognostic signatures, including two that have been converted into a commercial PCR-based platform, all of which were based on microarray studies. To the original search expression was added “AND (rnaseq[Title/Abstract] OR rna-seq[Title/Abstract] OR rna-sequencing[Title/Abstract] OR rna seq[Title/Abstract] OR rna sequencing[Title/Abstract]),” which returned no results to confirm that no RNA-seq prognostic signatures have been developed in lung adenocarcinoma.
Signature Generation and Statistical Analysis
TCGA clinical data was downloaded from the TCGA data portal and manually curated (Supplementary Tables 1 and 2, available online). Survival data for the validation cohort was collected at the University of Michigan (Supplementary Table 3, available online). TCGA data was partitioned into chronologically consecutive cohorts I (n = 255) and II (n = 157) before and after 2013 as training and validation cohorts, respectively. Univariate and multivariable Cox proportional hazards regression was used to assess association with overall or metastasis-free survival using BRB-ArrayTools (linus.nci.nih.gov/BRB-ArrayTools.html) and SPSS v19 (IBM, Inc., Chicago, IL, USA). Proportional assumptions for Cox proportional hazard model were examined by Kaplan-Meier analysis (for example, low vs high expression), by ensuring two curves do not intersect, and also by scaled Schoenfeld residuals. Benjamini-Hochberg false discovery rate (FDR) multiple hypothesis correction was applied where indicated as such. Hazard ratios (HRs) from univariate Cox regression analysis were used to identify protective (HR < 1) and risky genes (HR > 1). A risk score was calculated by taking into account the expression of gene and correlation coefficient. Kaplan-Meier analysis with log-rank test for difference was performed in GraphPad Prism (La Jolla, CA, USA). Heatmaps were generated in TreeView with z-score normalization within each row (gene). All statistical tests were two-sided. A P value of less than .05 was considered statistically significant.
Gene Signature Analysis
The Oncomine (www.oncomine.com) concept analysis tool was used with all available lung adenocarcinoma studies with default settings (18). PantherDB analysis was performed online (pantherdb.org) (19). Gene set enrichment analysis (GSEA) was performed with the indicated gene sets using phenotype labels “high risk” vs “low risk” (20). Results were exported as the nodes and edges of a concept association network and were visualized using Cytoscape v3.1.1.
Results
Identification of Prognostic Genes
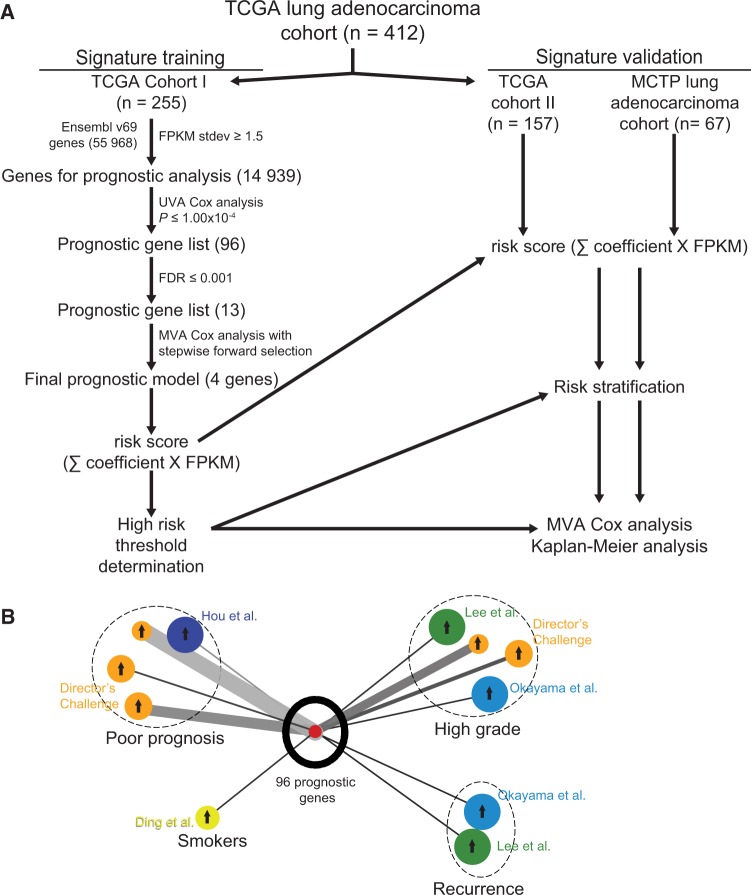
In order to comprehensively analyze the genomic prognostic associations in lung adenocarcinoma, we developed an analysis pipeline (Figure 1A). The Cancer Genome Atlas lung adenocarcinoma cohort (n = 412) was divided chronologically into a training cohort (TCGA cohort I, n = 255) and a validation cohort (TCGA cohort II, n = 157). The demographics of these cohorts were well balanced (Table 1). In TCGA cohort I, we analyzed approximately 15 000 genes with a standard deviation greater than 1.5 FPKM to ensure adequate variance, including approximately 1100 long noncoding RNA genes. Univariate Cox proportional hazards regression analysis showed that 96 genes were statistically significantly correlated, with overall survival at the P value of less than or equal to 1.00x10-4 level, though genes with lower statistical significance may be important as well (Supplementary Table 4, available online). In contrast to prior analyses, which included few or no lncRNAs, we found that five of these top 96 genes were lncRNAs. Oncomine concept analysis of this gene list in lung adenocarcinoma cohorts showed statistically significant positive association with signatures of smoking, high grade, recurrence, and poor prognosis (Figure 1B). An FDR threshold of less than or equal to 0.001 further refined the candidate gene list to 13 genes, including one lncRNA gene (Figure 1A; Supplementary Table 4, available online), to ensure proper algorithm performance for signature generation.
Figure 1.
Identification of prognostic gene signature. A) RNA-seq prognostic analysis and signature generation pipeline. The Cancer Genoma Atlas (TCGA) lung adenocarcinoma cohort was divided chronologically into TCGA cohort I (n = 255) and TCGA cohort II (n = 157). In TCGA cohort I, we filtered the 55 968 Ensembl v69 genes by standard deviation (stdev) greater than or equal to 1.5 fragments per kilobase of transcript per million reads (FPKM). The resulting 14 939 genes were analyzed individually for prognostic significance by univariate Cox proportional hazards models, and 96 genes were statistically significant at the level of P ≤ 1.00x10-4. We further narrowed this gene list to 13 genes with false discovery rates (FDRs) ≤ 0.001 and used multivariable Cox proportional hazards stepwise regression with forward selection to build a prognostic model that included four genes: RHOV, CD109, LINC00941, and FRRS1. This model was used to calculate risk scores for all TCGA cohort I patients by summing the product of model coefficient and FPKM for each gene, and a high risk threshold was chosen. This risk score calculation and high risk threshold was then applied to TCGA cohort II and Michigan Center for Translational Pathology cohort, and prognostic significance was analyzed with multivariable Cox proportional hazards models and Kaplan-Meier analysis. B) Oncomine lung adenocarcinoma signature concept analysis of top 96 prognostic genes. Results were exported as the nodes and edges of a concept association network and visualized using Cytoscape v3.1.1. Oncomine lung adenocarcinoma signatures are color-coded for the cohort in which they were generated, as labeled, and grouped (dashed circles) by the concept of the signature (Poor prognosis, Smokers, High grade, or Recurrence). The size of each signature circle is proportional to the number of genes in the signature, including for the top 96 prognostic genes in our study. Arrows in the center of each signature circle indicate positive (up arrow) or negative (down arrow) correlation with the top 96 prognostic genes, and the width of the connecting line indicates the strength of correlation. FDR = Benjamini-Hochberg false discovery rate; FPKM = fragments per kilobase of transcript per million reads; MCTP = Michigan Center for Translational Pathology; MVA = multivariable; Stdev = standard deviation; TCGA = The Cancer Genome Atlas; UVA = univariate.
Table 1.
Clinical characteristics of the patients*
| Factor | TCGA cohort I | TCGA cohort II | MCTP cohort |
|---|---|---|---|
| No. of patients | 255 | 157 | 67 |
| Age, y, mean (SD) | 65.3 (10) | 65.3 (10.5) | 68.3 (10) |
| Female sex, No. (%) | 134 (52.5) | 88 (56.1) | 36 (53.7) |
| Median survivor follow-up, mo | 19.3 | 20.0 | 33 |
| Smoking history, No. (%) | |||
| Yes | 211 (82.7) | 128 (81.5) | NR |
| No | 35 (13.7) | 24 (15.2) | NR |
| Unknown | 9 (3.5) | 5 (3.2) | NR |
| Stage, No. (%) | |||
| I | 139 (54.5) | 88 (56.1) | 40 (59.7) |
| IA | 63 (24.7) | 55 (35.0) | NR |
| IB | 74 (29.0) | 30 (19.1) | NR |
| II | 59 (23.13) | 40 (25.5) | 13 (19.4) |
| IIA | 25 (9.8) | 19 (12.1) | NR |
| IIB | 34 (13.3) | 21 (13.4) | NR |
| III | 43 (16.9) | 24 (15.2) | 14 (20.9) |
| IIIA | 38 (14.9) | 20 (12.7) | NR |
| IIIB | 5 (1.9) | 4 (2.5) | NR |
| IV | 14 (5.5) | 4 (2.5) | 0 |
*MCTP = Michigan Center for Translational Pathology; NR = not recorded; TCGA = The Cancer Genome Atlas.
Generation and Validation of Prognostic Signature
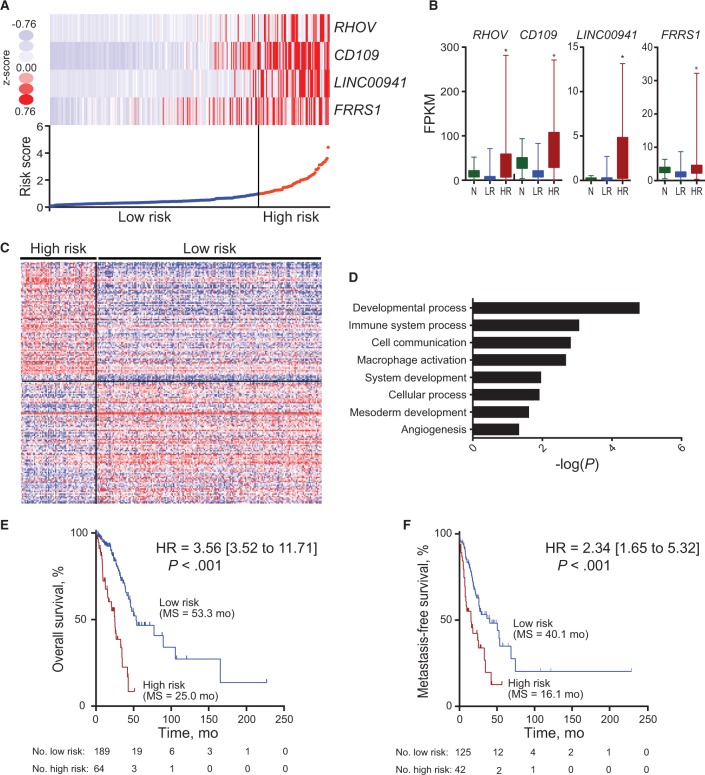
The 13 genes with univariate Cox analysis FDR of less than or equal to 0.001 were used for prognostic signature building using forward conditional stepwise regression with multivariable Cox analysis in the training cohort. This procedure selected a prognostic model containing four genes: protein-coding genes RHOV, CD109, and FRRS1, as well as the lncRNA gene LINC00941 (Supplementary Table 5, available online). A risk score was constructed with the regression coefficients from this model, and a threshold was chosen manually at the 75th percentile (Figure 2A). The four signature genes were statistically significantly overexpressed in high-risk tumors compared with normal lung tissue samples, while there was no difference between low-risk tumors and normal samples (Figure 2B). In order to begin understanding the biology underpinning high-risk tumors, we identified the top 100 genes statistically significantly overexpressed and the top 100 genes underexpressed in high-risk tumors (Figure 2C; Supplementary Table 6, available online). The overexpressed genes are statistically significantly enriched for processes related to cancer biology, including developmental process, immune-related processes, mesoderm development, and angiogenesis (Figure 2D; Supplementary Table 7, available online). Directed GSEA analysis showed that high-risk tumor expression was consistent with previous lung cancer survival–related signatures (Supplementary Figure 1, A and B, available online). Unbiased GSEA analysis showed moderate differentiation to be the top overall signature and an EGFR-related signature to be the top biological signature, consistent with its role in lung cancer (Supplementary Figure 1, C and D, and Supplementary Table 8, available online) (20). High-risk patients, as defined by the four-gene signature-based risk score, had statistically significantly worse overall survival (HR = 3.56, 95% CI = 3.52 to 11.71, P < .001) and metastasis-free survival (HR = 2.34, 95% CI = 1.65 to 5.32, P < .001) in TCGA cohort I independent of age, sex, and stage (Figure 2, E and F; Supplementary Figure 1E and Supplementary Table 9, available online).
Figure 2.
Four-gene prognostic signature biomarker characteristics in The Cancer Genoma Atlast (TCGA) cohort I. A) Four-gene expression and risk score distribution in TCGA cohort I by z-score, with red indicating higher expression and light blue indicating lower expression. The risk scores for all patients in TCGA cohort I are plotted in ascending order and marked as low risk (blue) or high risk (red), as divided by the threshold (vertical black line). B) Fragments per kilobase of transcript per million reads (FPKM) expression distribution of the signature genes in Normal (N), low risk (LR), and high risk (HR) tumors in TCGA cohort I. Bar represents mean, with error bar showing minimum and maximum values. C) Heatmap of the top 200 genes differentially expressed between high and low risk, with red indicating higher expression and blue indicating lower expression. D) Statistically significant Gene Ontology–Slim Biological Processes from PantherDB analysis of the genes differentially overexpressed in high-risk tumors. Full PantherDB results can be found in Supplementary Table 5 (available online). Kaplan-Meier curves of overall survival (E) and metastasis-free survival (F) in TCGA cohort I stratified by four-gene prognostic signature in high and low risk. A two-sided log-rank test was used to calculate hazard ratio (HR). HR, 95% confidence interval, P value, and median survival are shown. *P < .001. FPKM = fragments per kilobase of transcript per million reads; HR = hazard ratio.
Independent Clinical Validation and Signature Comparison
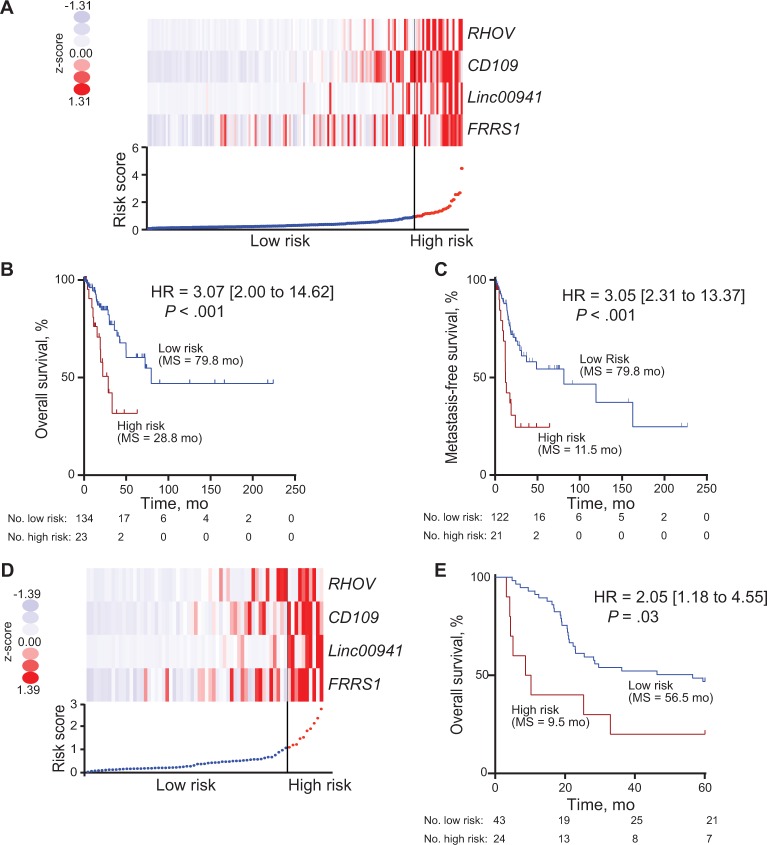
The four-gene prognostic signature was tested in two independent clinical cohorts for validation. Using the same risk score threshold chosen in the TCGA cohort I (Figure 3A), the four-gene prognostic signature risk group statistically significantly stratified the TCGA cohort II for overall survival (HR = 3.07, 95% CI = 2.00 to 14.62, P < .001) and metastasis-free survival (HR = 3.05, 95% CI = 2.31 to 13.37, P < .001) independent of age, sex, and stage (Figure 3B and Table 2; Supplementary Figure 3A, available online). In a second independent institutional MCTP cohort, again using the TCGA cohort I threshold (Figure 3D), the four-gene prognostic signature risk group was also able to statistically significantly stratify patients for overall survival (HR = 2.05, 95% CI = 1.18 to 4.55, P = .03) independent of age, sex, and stage (Figure 3E and Table 3; Supplementary Figure 3B, available online).
Figure 3.
Four-gene prognostic signature biomarker performance in two validation cohorts. A) Four-gene expression and risk score distribution in The Cancer Genome Atlas (TCGA) cohort II by z-score, with red indicating higher expression and light blue indicating lower expression. The risk scores for all patients in TCGA cohort II are plotted in ascending order and marked as low risk (blue) or high risk (red), as divided by the threshold (vertical black line). B and C) Kaplan-Meier curves of overall survival (B) and metastasis-free survival (C) in TCGA cohort II stratified by four-gene prognostic signature high and low risk with log-rank hazard ratio (HR), 95% confidence interval (CI), P value, and median survival. D) Four-gene expression and risk score distribution in the Michigan Center for Translational Pathology (MCTP) cohort by z-score, with red indicating higher expression and light blue indicating lower expression. The risk scores for all patients in MCTP cohort are plotted in ascending order and marked as low risk (blue) or high risk (red), as divided by the threshold (vertical black line). E) Kaplan-Meier curves of overall survival in the MCTP cohort stratified by four-gene prognostic signature in high and low risk. A two-sided log-rank test was used to calculate hazard ratio (HR). HR, 95% CI, P value, and median survival are shown. HR = hazard ratio.
Table 2.
Cox proportional hazards models in TCGA cohort II
| Factor | Univariate |
Multivariable |
||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Stage | 1.61 (1.15 to 2.23) | .004 | 1.54 (1.80 to 2.17) | .02 |
| Sex, female vs male | 0.75 (0.39 to 1.42) | .38 | 0.72 (0.37 to 1.38) | .32 |
| Age | 0.99 (0.96 to 1.01) | .45 | 0.98 (0.95 to 1.01) | .29 |
| Risk score | 1.69 (1.26 to 2.25) | <.001 | 1.67 (1.21 to 2.28) | .001 |
*Two-sided likelihood ratio test. CI = confidence interval; HR = hazard ratio; TCGA = The Cancer Genome Atlas.
Table 3.
Cox proportional hazards models in the MCTP cohort
| Factor | Univariate analysis |
Multivariable analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Stage | 3.12 (2.11 to 4.63) | <.001 | 3.64 (2.39 to 5.54) | <.001 |
| Sex, female vs male | 1.40 (0.74 to 2.65) | .29 | 2.22 (1.08 to 4.53) | .49 |
| Age | 1.01 (0.98 to 1.04) | .54 | 1.01 (0.98 to 1.04) | .03 |
| Risk score | 2.44 (1.49 to 4.00) | <.001 | 3.49 (1.98 to 6.18) | <.001 |
*Two-sided likelihood ratio test. CI = confidence interval; HR = hazard ratio; MTCP = Michigan Center for Translational Pathology.
We compared the four-gene prognostic signature to five other published lung adenocarcinoma prognostic signatures, including a commercial signature, by rederiving a multivariable Cox model using the gene list from each signature due to platform differences (21–25). As might be expected, four out of five signatures were statistically significant on univariate analysis in TCGA cohort I. None of the other five signatures was statistically significant on univariate analysis in both validation cohorts, and these results were typically mirrored in multivariable analysis including age, stage, and sex (Supplementary Figure 2 and Supplementary Table 10, available online).
Validation of Signature in Clinically Important Stage I and Mutation Subsets
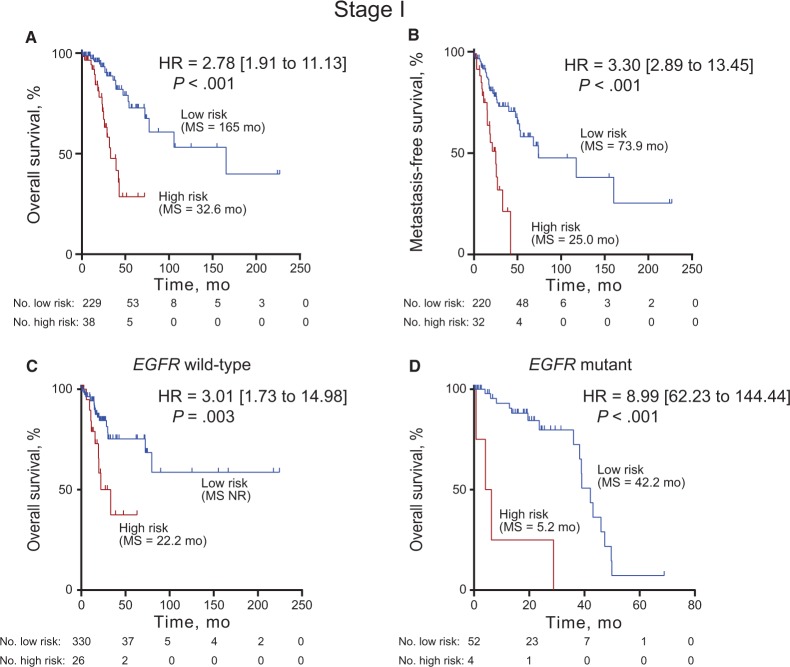
Lung adenocarcinoma prognostic signatures have focused on early-stage patients as a subset where adjuvant treatment decisions might be tailored based on prognosis, as opposed to advanced-stage patients who will have poor outcomes even with full-intensity multimodality therapy. Thus, we evaluated the four-gene prognostic signature in stage I patients from the full TCGA lung adenocarcinoma patients to ensure adequate numbers for analysis (n = 139). The four-gene prognostic signature statistically significantly stratified the stage I patients for both overall survival (HR = 2.78, 95% CI = 1.91 to 11.13, P < .001) and metastasis-free survival (HR = 3.30, 95% CI = 2.89 to 13.45, P < .001) independent of age, sex, and stage (Figures 4, A and B). Additionally, in the full TCGA cohort, the four-gene prognostic signature risk groups statistically significantly stratified stage II and stage III–IV patients (Supplementary Figures 3, C–F, available online). The four-gene prognostic signature also statistically significantly stratified the MCTP cohort stage I patients, though a median threshold was used because of the small number of patients (Supplementary Figure 3G, available online).
Figure 4.
Four-gene prognostic signature biomarker performance in stage I and EGFR mutation subsets. Kaplan-Meier curves with log-rank hazard ratio (HR), 95% confidence interval (CI), P value, and median survival for overall survival (A, C, D) and metastasis-free survival (B) in the stage I cases in The Cancer Genome Atlas (TCGA) cohort I (A, B), EGFR wild-type cases from validation cohort TCGA cohort II (C), and EGFR mutant cases from the full TCGA cohort (D) stratified by four-gene signature into high and low risk. A two-sided log-rank test was used to calculate hazard ratio (HR). HR, 95% CI, P value, and median survival are shown. The hazard ratio and 95% CI for the EGFR mutant analysis could not be accurately calculated due to the small number of patients. HR = hazard ratio.
Given the importance of EGFR and ALK alteration status in lung adenocarcinoma for TKI use and/or prognostication, we analyzed the performance of the four-gene prognostic signature in patient subsets with wild-type or mutant status (Supplementary Tables 11 and 12, available online). In EGFR wild-type patients from TCGA cohort II (131/157, 83%), who would receive chemotherapy if adjuvant therapy were given, the four-gene prognostic signature risk group provided statistically significant overall survival stratification (P = 0.003, HR = 3.01, 95% CI = 1.73 to 14.98) (Figure 4C). EGFR-mutant patients from the full TCGA cohort (56/412, 14%), who might receive an EGFR TKI as adjuvant treatment, were similarly statistically significantly stratified (HR = 8.99, 95% CI = 62.23 to 141.44, P < .001). There is statistically significant enrichment of EGFR mutant cases in the low-risk group from the full TCGA cohort, while KRAS mutant and ALK fusion cases are not statistically significantly different (Supplementary Figure 4A, available online). KRAS mutation status has been reported as a confounder for lung adenocarcinoma prognostic signatures (7). In contrast, the four-gene prognostic signature risk groups statistically significantly stratify KRAS mutant (120/412, 29%) and wild-type KRAS (119/157, 76%) patients for overall survival, as well as patients that are wild-type for ALK, EGFR, and KRAS (Supplementary Figure 4, B–D, available online). Finally, the risk score remained statistically significant (HR = 2.13, 95% CI = 1.79 to 2.52, P < .001) in a multivariable Cox analysis that included EGFR, KRAS, and ALK alteration status (Supplementary Table 13, available online). Interestingly, EGFR mutation status was also statistically significant (HR = 2.03, 95% CI = 1.26 to 3.62, P = .005) in the multivariable analysis.
Discussion
Lung cancer is the leading cause of cancer deaths in the United States, causing 158 000 deaths, with adenocarcinoma histology playing a major role (1). In an effort to bolster clinical tools and biological understanding in lung adenocarcinoma, we present the first RNA-seq prognostic signature. Using a TCGA lung adenocarcinoma cohort subset, we found 96 genes with statistically significant prognostic association, including five lncRNAs. Prognostic model training in this subset selected a four-gene signature, including the lncRNA gene LINC00941, from the top 13 genes, reiterating that lncRNAs are an important class of biomarker candidate genes. The four-gene signature was validated as statistically significantly associated with metastasis-free survival and overall survival in the remaining TCGA lung adenocarcinoma cases, and in an independent institutional cohort. Thus, our four-gene RNA-seq prognostic signature provides biological insights and has potential for rapid incorporation into RNA-seq clinical sequencing programs to tailor management in early-stage lung adenocarcinoma.
Early-stage lung adenocarcinoma is, unfortunately, characterized by statistically significantly worse survival outcomes than many other early-stage cancers, with five-year survival after surgery alone at 73% in stage IA and 58% in stage IB in the most recent 7th TNM staging system (3). Therapy intensification is therefore needed, but patient selection tools have been limited. Based on the Lung Adjuvant Cisplatin Evaluation (LACE) meta-analysis and the CALGB 9633 trial, patients are selected for adjuvant chemotherapy based on clinical criteria (stage IB and tumor ≥4 cm) (26,27), though there is an ongoing trial using a commercial microarray-derived quantitative PCR (qPCR)–based prognostic signature to select patients for adjuvant chemotherapy randomization (28). More sophisticated genomic guidance in the form of mutation identification has been recognized as essential for targeted therapy in the metastatic setting (29); however, outcome variability within mutation cohorts has been underexplored. Furthermore, trials of adjuvant targeted therapy have so far not focused on mutation status, including in the BR19 (30) and RADIANT trials (31). The ongoing ALCHEMIST trial should randomize sufficiently large numbers of early-stage EGFR mutant patients to have a clear indication of benefit, though the results are several years away (32). National basket trials, including NCI-MATCH, NCI-MPACT, and ASCO’s TAPUR, will provide therapeutic response information about targetable alterations that are less prevalent and thus highlight the need for comprehensive parallel genomic alteration testing. These trials are histology-independent but are likely to enroll a large numbers of lung cancer patients.
RNA-seq clinical tools, including prognostic signatures for individualized therapy intensification as presented here, have several key advantages over other platforms. RNA-seq includes noncoding gene expression data that were not represented in the microarrays of past efforts and which the ENCODE Project has demonstrated occupy a majority of the transcribed genome (33). Microarray data have biases and limitations that are improved with RNA-seq, particularly in the detection of low-abundance transcripts (34,35). This advantage of RNA-seq translates into better correlation with qPCR data both in the laboratory and on patient samples and is particularly important for lncRNAs that tend to be differentially expressed but at low absolute abundance (22,35). Continually improving RNA-seq platforms also provide nearly all the genomic alteration information needed, including single-nucleotide variants (SNV)/mutations and gene fusions (36). Moreover, RNA-seq provides comprehensive expression data that will be increasingly important in understanding and predicting therapeutic response in the substantial proportion of tumors that lack a classical targetable alteration (37). Additionally, exome-capture RNA-seq recently published by our group can be used on FFPE samples to mine completed randomized trial samples on the same platform (12). These advantages prompted the Sweden Cancerome Analysis Network – Breast (SCAN-B) clinical sequencing program to focus on RNA-seq; this effort has enrolled nearly 4000 women (∼85% of the primary breast cancer cases in south Sweden) and has collected nearly 3000 tumor samples (38). Though slightly more costly, the major advantages of RNA-seq place it as a highly attractive comprehensive clinical test in precision oncology.
Potential clinical uses of the four-gene signature are driven by its strong prognostic performance in several clinically important settings. As described above, adjuvant therapy for early-stage lung adenocarcinoma is currently driven by a clinical factor of tumor size 4 cm or larger (stage IB). The four-gene signature offers an opportunity for individualized adjuvant therapy based on biological factors, as well comprehensive alteration testing through the RNA-seq platform. For more advanced and metastatic tumors, the four-gene signature offers patient risk stratification for both EGFR mutant and wild-type patients that might be used to intensify EGFR inhibitor therapy in patients at high risk. We also note that the high-risk group identified in our analysis displayed enrichment for genes associated with immune response. It is plausible this would influence the response to immunotherapies that have shown so much promise in recalcitrant disease, including lung adenocarcinoma. The direction of this effect is difficult to predict, as either increased (in high-risk tumors) or decreased (in low-risk tumors) immune response may facilitate either more or less robust immunotherapy response. Clinical integration of the four-gene signature needs to be tested directly, but appears promising from these initial results.
Though the four-gene signature is promising, there are limitations to this initial work. The patient cohorts, including the validation cohorts, were multi-institutional, but retrospective, and therefore these findings must be validated prospectively. Therapy was not randomized or systematic in any way across the cohorts, which limited our ability to test the predictive power of the signature with respect to guidance on specific treatment decisions. Given the low prevalence of mutations, we had limited patient numbers to test the performance of the signature in mutational cohorts. RNA-seq results have some sensitivity to bioinformatics parameters that may vary among clinical sequencing programs and affect the performance of the signature, though validation in the independently collected and processed MCTP cohort demonstrates some robustness to pipeline variations. These limitations can be addressed in future studies.
Our analysis is also likely to provide biological and therapeutic information as well. CD109 is a glycoprotein on the surface of immune and endothelial cells that negatively regulates TGF-beta signaling (39). It is the most studied gene of the four, being nominated as a biomarker and/or therapeutic target in several cancers, including pancreas and breast (40,41). RHOV is an atypical RHO GTPase that has been nominated as upregulated in non–small cell lung cancer in a minor study, and its role in cancer is poorly understood (42). FRRS1 is a cytochrome b561 family iron reductase whose role in cancer is not characterized, though iron metabolism has been studied as a possible therapeutic target (43,44). LINC00941 was identified initially by shotgun cloning, and re-identified by the ENCODE Project, but has not been studied in detail (45). These genes highlight diverse biological processes that may underpin their prognostic significance and provide avenues for further research.
Here, we have performed the first RNA-seq prognostic analysis in lung adenocarcinoma, resulting in an independently validated four-gene prognostic signature that includes an lncRNA, as well as identification of numerous genes with strongly statistically significant prognostic association for further study. Importantly, this four-gene prognostic signature performed well in stage I patients and EGFR-mutant and wild-type cohorts. Thus, this four-gene prognostic signature could be a clinically useful tool easily incorporated into an RNA-seq clinical sequencing program to individualize lung adenocarcinoma therapy.
Funding
Supported in part by the National Cancer Institute Early Detection Research Network (UO1CA113913) and R01CA154365 (to DGB and AMC).
Notes
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
SS, JRE, FYF, RM, SMD, GC, DGB, HJ, and AMC conceived, designed, or planned the study. SS, JRE, RM, SMD, and HJ analyzed the data. SS, JRE, RM, SMD, XC, GC, and DGB acquired data. SS, JRE, FYF, RM, SMD, DGB, HJ, and AMC helped to interpret the results. XC, GC, DGB, and AMC provided study materials or patients. SS and JRE drafted the manuscript. All authors revised and reviewed this work, and all authors had final approval of the submitted manuscript.
All authors declare no conflicts of interest.
Corresponding author confirmation: AMC confirms he had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.Cancer Facts & Figures 2015. In: Atlanta, GA: American Cancer Society; 2015.
- 2.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. [DOI] [PubMed] [Google Scholar]
- 3.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2(8):706–714. [DOI] [PubMed] [Google Scholar]
- 4.Ten Haaf K, van Rosmalen J, de Koning HJ. Lung cancer detectability by test, histology, stage, and gender: Estimates from the NLST and the PLCO trials. Cancer Epidemiol Biomarkers Prev. 2015;24(1):154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. [DOI] [PubMed] [Google Scholar]
- 6.Zhu CQ, Tsao MS. Prognostic markers in lung cancer: Is it ready for prime time? Transl Lung Cancer Res. 2014;3(3):149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starmans MH, Pintilie M, Chan-Seng-Yue M, et al. Integrating RAS status into prognostic signatures for adenocarcinomas of the lung. Clin Cancer Res. 2015;21(6):1477–1486. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y, Bueno R. Commercially available prognostic molecular models in early-stage lung cancer: A review of the Pervenio Lung RS and Myriad myPlan Lung Cancer tests. Expert Rev Mol Diagn. 2015;15(5):589–596. [DOI] [PubMed] [Google Scholar]
- 9.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner JR, Zhao S, Erho N, et al. RNA biomarkers associated with metastatic progression in prostate cancer: A multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damodaran S, Berger MF, Roychowdhury S. Clinical tumor sequencing: Opportunities and challenges for precision cancer medicine. Am Soc Clin Oncol Educ Book. 2015;35:e175–e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieslik M, Chugh R, Wu YM, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015;25:1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balbin OA, Malik R, Dhanasekaran SM, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25(7):1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Pertea G, Trapnell C, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun. 2014;5:5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Garcia W, Zheng S, Sivachenko A, et al. PRADA: Pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30(15):2224–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(Database issue):D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106(8):2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. [DOI] [PubMed] [Google Scholar]
- 23.Kratz JR, Van den Eeden SK, He J, et al. A prognostic assay to identify patients at high risk of mortality despite small, node-negative lung tumors. JAMA. 2012;308(16):1629–1631. [DOI] [PubMed] [Google Scholar]
- 24.Bianchi F, Nuciforo P, Vecchi M, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117(11):3436–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25(35):5562–5569. [DOI] [PubMed] [Google Scholar]
- 26.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26(21):3552–3559. [DOI] [PubMed] [Google Scholar]
- 27.Strauss GM, Herndon JE, 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26(31):5043–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratz JR, Mann MJ, Jablons DM. International trial of adjuvant therapy in high risk stage I non-squamous cell carcinoma identified by a 14-gene prognostic signature. Transl Lung Cancer Res. 2013;2(3):222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. [DOI] [PubMed] [Google Scholar]
- 30.Goss GD, O'Callaghan C, Lorimer I, et al. Gefitinib versus placebo in completely resected non-small-cell lung cancer: Results of the NCIC CTG BR19 study. J Clin Oncol. 2013;31(27):3320–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly K, Altorki NK, Eberhardt WEE, et al. A randomized, double-blind phase 3 trial of adjuvant erlotinib (E) versus placebo (P) following complete tumor resection with or without adjuvant chemotherapy in patients (pts) with stage IB-IIIA EGFR positive (IHC/FISH) non-small cell lung cancer (NSCLC): RADIANT results. J Clin Oncol. 2014;32(15_suppl):7501. [Google Scholar]
- 32.Gerber DE, Oxnard GR, Govindan R. ALCHEMIST: Bringing genomic discovery and targeted therapies to early-stage lung cancer. Clin Pharmacol Ther. 2015;97(5):447–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium EP, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Febbo PG, Kantoff PW. Noise and bias in microarray analysis of tumor specimens. J Clin Oncol. 2006;24(23):3719–3721. [DOI] [PubMed] [Google Scholar]
- 35.Robinson DG, Wang JY, Storey JD. A nested parallel experiment demonstrates differences in intensity-dependence between RNA-seq and microarrays. Nucleic Acids Res. 2015;43(20):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20(12):1479-84. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saal LH, Vallon-Christersson J, Hakkinen J, et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bizet AA, Tran-Khanh N, Saksena A, et al. CD109-mediated degradation of TGF-beta receptors and inhibition of TGF-beta responses involve regulation of SMAD7 and Smurf2 localization and function. J Cell Biochem. 2012;113(1):238–246. [DOI] [PubMed] [Google Scholar]
- 40.Tao J, Li H, Li Q, et al. CD109 is a potential target for triple-negative breast cancer. Tumour Biol. 2014;35(12):12083–12090. [DOI] [PubMed] [Google Scholar]
- 41.Haun RS, Fan CY, Mackintosh SG, et al. CD109 overexpression in pancreatic cancer identified by cell-surface glycoprotein capture. J Proteomics Bioinform. 2014;Suppl 10:S10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepelev MV, Korobko IV. The RHOV gene is overexpressed in human non-small cell lung cancer. Cancer Genet. 2013;206(11):393–397. [DOI] [PubMed] [Google Scholar]
- 43.Vargas JD, Herpers B, McKie AT, et al. Stromal cell-derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim Biophys Acta. 2003;1651(1–2):116–123. [DOI] [PubMed] [Google Scholar]
- 44.Jin Y, Wang L, Qu S, et al. STAMP2 increases oxidative stress and is critical for prostate cancer. EMBO Mol Med. 2015;7(3):315–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.