Abstract
Targeted proteomics technique has emerged as a powerful protein quantification tool in systems biology, biomedical research, and increasing for clinical applications. The most widely used targeted proteomics approach, selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM), can be used for quantification of cellular signaling networks and preclinical verification of candidate protein biomarkers. As an extension to our previous review on advances in SRM sensitivity herein we review recent advances in the method and technology for further enhancing SRM sensitivity (from 2012 to present), and highlighting its broad biomedical applications in human bodily fluids, tissue and cell lines. Furthermore, we also review two recently introduced targeted proteomics approaches, parallel reaction monitoring (PRM) and data-independent acquisition (DIA) with targeted data extraction on fast scanning high-resolution accurate-mass (HR/AM) instruments. Such HR/AM targeted quantification with monitoring all target product ions addresses SRM limitations effectively in specificity and multiplexing; whereas when compared to SRM, PRM and DIA are still in the infancy with a limited number of applications. Thus, for HR/AM targeted quantification we focus our discussion on method development, data processing and analysis, and its advantages and limitations in targeted proteomics. Finally, general perspectives on the potential of achieving both high sensitivity and high sample throughput for large-scale quantification of hundreds of target proteins are discussed.
Keywords: Biomarker, DIA, PRISM, PRM, Signaling pathway, SRM, Technology
1 Introduction
Mass spectrometry (MS)-based proteomics is a promising technology in characterization of human proteome at a genome scale [1–3]. For example, MS-based global proteomics allows achieving genome-scale proteome coverage and quantitative changes of thousands of proteins and their posttranslational modifications (PTMs) in response to perturbation when combined with labeling strategies [4, 5]. It has been mature and routinely used in systems biology for measuring proteome changes during signal transduction [6, 7] and biomedical research for discovering novel candidate protein biomarkers [8, 9]. However, such global measurements generally lack quantification precision and have inherently poor reproducibility for low-abundance proteins due to the stochastic selection of precursor ions for MS/MS fragmentation by the data-dependent acquisition (DDA) mode [10, 11]. Thus, DDA-based measurements are not suitable for reliable validation of candidate protein biomarkers because most informative biomarkers are typically low abundance proteins and the human proteome has a wide dynamic range in concentrations (e.g., >10 orders of magnitude in human plasma or serum, where serum is similar to plasma in composition but exclude clotting factors of blood [12]).
With significant advances in MS instrumentation thousands of candidate protein biomarkers have been generated from proteomics studies [13, 14], but sadly none of them has been successfully translated into an FDA-approved clinical test [15, 16]. One important reason is the lack of robust protein quantification tools for enabling precise measurements of such numerous candidate protein biomarkers simultaneously and effectively in large sample sets (e.g., ~1000 patient samples). Traditional antibody-based ELISA has a limited multiplexing power in biomarker validation (in general measuring one or several protein biomarker at a time) and the antibodies for new biomarker proteins are often not available, especially for variant proteins and PTMs [17, 18]. Targeted proteomics has been emerging as a powerful protein quantification technology in terms of reproducibility, multiplexing capability and quantification accuracy [17,19,20].
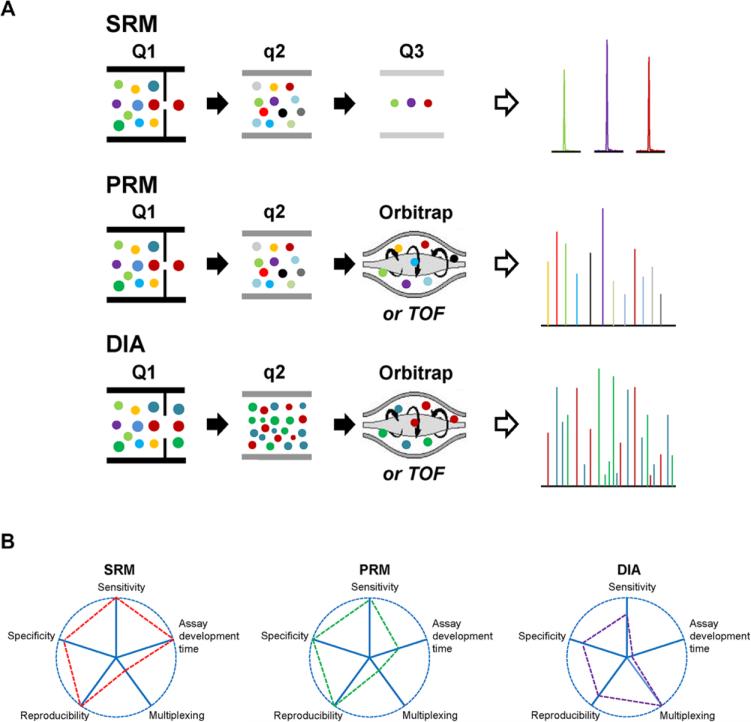
We can readily group three types of targeted proteomics approaches: selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM) [17,21]; parallel reaction monitoring (PRM) [22,23]; and data-independent acquisition (DIA) combined with targeted data extraction of the MS/MS spectra (e.g., sequential windowed acquisition of all theoretical product ion mass spectra, termed SWATH [11]). SRM is a classic targeted proteomics approach performed on a triple quadrupole mass spectrometer (QqQ MS) [17]. It exploits the unique features of QqQ with two levels of mass selection (i.e., Q1 and Q3 isolation of precursor ions and their product ions, respectively) and a relatively long dwell time (in general 10 ms per pair of precursor/product ions) over a narrow m/z window (± 0.02 m/z) (Fig. 1A), which result in significantly improved selectivity and sensitivity, at least one to two orders of magnitude higher than full scan global proteomics analysis [17,21]. Thus, conceptually SRM is similar to Western blotting [24]. More recently, hybrid mass spectrometers with the substitution of the third quadrupole with a high-resolution and accurate-mass (HR/AM) mass analyzer, such as the quadrupole-Orbitrap (i.e., Q Exactive) [22,23] and quadrupole-time of flight (e.g., the ‘Triple-TOF’) MS [25] instruments, were developed and operated in the PRM mode for targeted MS quantification (Fig. 1A). In PRM, the isolation and fragmentation of peptide precursors are very similar to those in SRM. The only difference for PRM is that full MS/MS spectra are acquired for each precursor (i.e., the parallel detection of all product ions) in the HR/AM mass analyzer; while in SRM only predefined product ions (i.e., transitions) are monitored by the low-resolution quadrupole mass analyzer. Thus, in complex biological samples PRM can provide higher selectivity than SRM for a better discrimination of the product ion signal of target peptides from the co-eluting interferences [26]. With the use of internal standards both SRM and PRM enable reproducible accurate quantification of target proteins across many samples (Fig. 1B). However, both of them suffer from the limitations in the scale of quantification (i.e., multiplexing capability), typically ~500 peptides/125 proteins per a single SRM [21] or PRM analysis [27] for reliable quantification (assuming that each protein has two surrogate peptides with light and heavy versions per peptide). Increasing the number of target peptides requires the tight control of peptide LC elution time for accommodating more transitions or peptides in a narrow time window. Furthermore, with the scale of quantification increasing, the sensitivity could drop significantly, especially for PRM whose sensitivity is inversely proportional to the degree of multiplexing [28].
Figure 1.
(A) Schematic diagrams of typical SRM, PRM, and DIA. In SRM, the precursor ion of a specific peptide is selected in Q1, and then transmitted into q2 for fragmentation; specific product ions from the target peptide (typically three product ions) are selected in Q3 for detection. In PRM, the first two steps are the same as SRM; whereas in the last step all fragment ions from the target peptide are monitored by HR/AM mass analyzer. In DIA, all peptide ions within a defined mass-to-charge (m/z) window are isolated in Q1, then fragmented together in q2, and the highly complex mixture of all product ions are monitored by HR/AM mass analyzer; the analysis is repeated as MS progresses the full m/z range in a stepwise fashion. (B) Targeted quantification performance comparisons of SRM, PRM, and DIA. In the charts each analytical variable is represented by a spoke (the radial handle projecting from the center). The length of a spoke indicates the magnitude of the variables. PRM has similar multiplexing capability as SRM but with higher specificity due to the use of HR/AM mass analyzer to monitor the product ions. Note that the sensitivity comparison between SRM and PRM is based on quantification of the relatively small number of target peptides (e.g., ~50 peptides) in a single analysis.
To alleviate the limitations in multiplexing, DIA-based targeted quantification (e.g., SWATH [11]) was recently introduced for proteome-wide quantification of target proteins of interest. This method consists of high specificity DIA for generating product ion maps of all detectable precursors and targeted data analysis with the use of the SRM concept for data extraction [11]. In a DIA analysis, a set of wide precursor acquisition windows are predefined to cover the whole m/z range of proteolytic peptides. All peptides within a defined mass-to-charge (m/z) window are fragmented and MS records a complete high accuracy product ion spectrum for each detectable peptide (Fig. 1A). Thus, DIA provides highly multiplexed product ion spectra for all the detectable peptides over the LC elution time, and requires more elaborate data processing and interpretation. A few software tools have been developed (e.g., OpenSWATH [29]) but DIA data analysis is still challenging. DIA-based targeted quantification is performed using a targeted data extraction strategy based on the utilization of a priori information (e.g., peptide retention time and product ion intensity) from spectral libraries for confident identification of target peptides of interest in the DIA product ion map, as well as using the most intense product ions for peptide quantification [29,30]. Because of its unbiased, broad range of precursor ion selection and fragmentation, DIA-based targeted quantification could potentially lead to a paradigm shift in targeted proteomics from small-scale to proteome-wide quantification in complex samples with good reproducibility and high accuracy (Fig. 1B). But when compared to SRM or PRM with only focusing on a small number of predefined target peptides, DIA-based targeted quantification has somewhat lower sensitivity, specificity, and reproducibility because of the much shorter dwell time for each individual peptide, a wider precursor isolation window, as well as the lack of using internal standards to correct MS run-to-run variability [11]. For example, SRM was demonstrated to offer at least 10-fold higher sensitivity than DIA-based targeted quantification [11].
In this review we provide an overview of recent advances in targeted proteomics and its broad applications in human bodily fluids, tissue and cell lines, including (i) recent advances in SRM sensitivity and its applications to biomedical research and systems biology (from 2012 to present), which expand our previous review article in advancing SRM sensitivity (covering the time period of 2002 to late 2011) [17], (ii) new development in targeted MS/MS quantification on fast hybrid MS operated in the PRM mode, (iii) DIA-based targeted proteomics for proteome-wide quantification of target proteins, and (iv) future perspectives in targeted proteomics for reliable validation of candidate protein biomarkers in a high throughput manner.
2 Recent advances in SRM sensitivity and its application
In principle, MS sensitivity is governed by the ability to provide sufficient target analyte ions for the MS detection and the overall resolving power of MS mass analyzers for a specific measurement of analyte signal in the presence of background/interference. Thus, enhancing the MS-based targeted proteomics sensitivity can be built upon the following three aspects: frond-end (sample preparation, fractionation, and LC separations), interface (ion sampling and ion transmission) and back-end (the resolving power of the mass analyzer for removal of co-eluting interferences) [17]. Compared to other targeted proteomics platforms SRM has been well documented with demonstrated performance and robustness both within and across laboratories and instrument platforms [31–34]. Therefore, the implementation of the front-end methods to improve the targeted proteomics sensitivity has been primarily evaluated on the SRM platform. However, in general all the front-end methods are equally applicable with other targeted proteomics platforms.
2.1 Affinity enrichment
2.1.1 Antibody
Antibody-based immunoaffinity enrichment coupled to SRM (i.e., immuno-SRM) has emerged as a promising technology for sensitive precise quantification of target proteins in complex matrices [17]. For example, it was applied for quantification of mutant RAS (G12D) at LOD of 12 amol (i.e., 0.25 pg) or 240 amol/mg of total protein in patient tumors by anti-RAS protein antibody at the protein-level enrichment [35] and FGF15 at LOD of 0.1 ng/mL in mouse plasma by anti-peptide antibody at the peptide-level enrichment (i.e., SISCAPA, stable isotope standard and capture by the anti-peptide antibody) [36]. Recently, the development of immuno-SRM has been primarily focused on multiplexing [37], assay sensitivity [35, 38], and rapid cost-effective generation of antibodies [39, 40]. For example, the sequential enrichment of target peptides was used for increasing the multiplexing capability. After enrichment with one set of anti-peptide antibodies the next set of anti-peptide antibodies was immediately added to the same incubation plate and repeated in an identical manner for the sequential capture of the target peptides. Immuno-SRM assays allow concurrently measuring up to 50 peptides at a single step enrichment with the similar data quality as a 10-plex configuration with five groups of antibodies in a sequential fashion (e.g., peptide recovery, assay reproducibility and accuracy) but with slightly lower sensitivity [37]. Further increasing the levels of multiplexing has been recognized as challenging because nonspecifically bound matrix components, likely on the surface of the stationary media will be increased [37]. This can lead to decreasing the S/N ratio of target peptides and lowering SRM sensitivity. For rapid generation of anti-peptide antibodies at low cost, Whiteaker et al. have recently evaluated the enrichment efficiency of replacing traditional monoclonal or polyclonal antibodies with recombinant antibody fragments (Fabs) by spiking exogenous target peptides into human plasma [39]. They concluded that at the peptide level high-affinity Fab-based SRM provided similar sensitivity (LODs of 0.3-2.9 ng/mL in human plasma) as monoclonal antibody-based SRM with similar reproducibility (~10% of average coefficient of variation (CV) at the LOD for both methods) by evaluation of three proteins (i.e., osteopontin, E-selection, and ADAM17). This suggests that high-affinity recombinant Fabs can be applied in peptide enrichment immuno-SRM assays. In another study they have shown the feasibility of using multiplex immuno-SRM assays that are originally developed for unmodified peptides to quantify the pharmacodynamics of their corresponding phosphorylated peptides because those anti-peptide antibodies can concurrently enrich those phosphorylated peptides and SRM can easily distinguish the phosphorylated and unmodified forms of a given peptide [40]. A 69-plex peptide-level immuno-SRM assay targeting the DNA damage response network can provide sensitivity at a median LOQ of 2.0 fmol/mg and reproducibility with a median interassay variability of 10% CV for measuring endogenous phosphopeptides. This shows the potential of using immuno-SRM for rapid and precise quantification of cell signaling networks.
To further increase immuno-SRM sensitivity, tandem affinity enrichment approaches have recently been employed for highly specific enrichment of target peptides to quantify extremely low-abundance proteins in human blood (Table 1). With the use of the two-step sequential enrichment method (e.g., isolation of human β-nerve growth factor (β-NGF) protein using anti-NGF antibody from patient serum, followed by peptide immunoaffinity capture of the predefined surrogate peptide from the digested β-NGF protein) to generate a highly enriched peptide sample [41], LC-SRM can reliably measure the total β-NGF at concentration levels of 7.03–450 pg/mL in human serum with <15% interassay CV. Such sensitivity is at least 10-fold higher than that provided by regular immuno-SRM with single antibody-based enrichment. The other tandem affinity enrichment approach was to combine immunoaffinity depletion (e.g., plasma albumin depletion to remove the most abundant albumin protein that accounts for ~50% of the total plasma protein mass [42] or IgY14 depletion [43] to remove the 14 high abundance proteins that account for ~95% of the total plasma protein mass) for reducing the dynamic range of protein concentration and anti-peptide antibody enrichment for effectively enriching target peptides. The combination of plasma albumin depletion and affinity enrichment of the unique epitope peptide from TnI by anti-TnI mAb-coated mircoparticles [42] was demonstrated to significantly improve SRM sensitivity for measuring TnI, a low abundant protein in human plasma. This method was demonstrated to have low ng/mL (i.e., ~4 ng/mL) sensitivity and high reproducibility within 6% CV. Another example is to quantify low-abundance serum transferrin receptor (sTfR) [43]. Coupling the IgY14 depletion to SRM can only reliably quantify sTfR at a concentration of ≥1000 ng/mL. However, with anti-peptide affinity enrichment, ~10-fold enhancement in SRM sensitivity can be achieved (i.e., the LOQ of 100 ng/mL for IgY14-SISCAPA-SRM).
Table 1.
A survey of recent advances in front-end strategies (2012-present) for enhancing SRM sensitivity of detection and quantification of target proteins in blood plasma or serum (with starting volume for sample analysis) as well as in human cell or tissue (with the description written in italics)
| Strategy | Volumea) | Proteinsb) | LOD | LOQ | CV | Ref. |
|---|---|---|---|---|---|---|
| SISCAPA | 30 μL | 1c) | 0.1 ng/mL | [36] | ||
| 10 μL | 3 | 0.3-3 ng/mL | 4–15% | [39] | ||
| 69 d) | 1–46 fmol/mg e) | 1–400 fmol/mg e) | 2–39% | [40] | ||
| Protein enrichment | 1 | 240 amol/mg e) | 280 amol/mg e) | 4-20% | [35] | |
| Protein enrichment, SISCAPA | 600 μL | 1f) | ~10-450 pg/mL | <15% | [41] | |
| Depletion, SISCAPA | 35 μL | 1 | ~4 ng/mLg) | 1-6% | [42] | |
| 1 | 100 ng/mLh) | ≤15% | [43] | |||
| MSIA (protein enrichment) | 1-1000 μLi) | 14 | 20 pg/mL-300 μg/mL | ≤20% | [38] | |
| 75 μL | 1 | ~20 ng/mL | <25% | [44] | ||
| Gel extraction | 4j) | 80–1100 fmol/mg e) | ~14% | [56] | ||
| NPE-Gel extraction k) | 10 | ~250 copies/nucleus e) | <15% | [54] | ||
| Protein precipitation | 7 μL | 5 | 50–1250 ng/mL | <15% | [55] | |
| Depletion, C18-SCX StageTip | 800 μL | 3j) | 75-130 amol/mLg) | 150-320 amol/mLg) | <20% | [57] |
| PRISM | ~2 μL | 3 | <0.1-1 ng/mL | 0.5-5 ng/mL | ~10% | [58] |
| ~2 μL | 1j) | ~0.6 ng/mL | <20% | [63] | ||
| 7 j) | 0.5-5 fmol/mg e) | 2–50 fmol/mg e) | <15% | [60] | ||
| Depletion, PRISM | 10 μL | 4 | ≤0.05 ng/mLh) | 0.05-0.1 ng/mLh) | ~10% | [18] |
| 10 μL | 1 | ~130 pg/mLh) | <10% | [59] | ||
| μRPLCl) | 30 μLm) | 2n) | 19 ng/mL-36 μg/mL | [64] | ||
| 2D RPLC | 30 μLm) | 31n) | 0.5-10 ng/mL | ~10% | [64] | |
| 41n) | >10 ng/mL | ~10% | [64] | |||
| RP-HILIC | 100 μL | 1 | 1 ng/mL | 6-8% | [68] | |
| LG | ~2 μL | 2 | ~5 ng/mL | ~10 ng/mL | ~7% | [69] |
| 16 | ~0.5-13 ng/mLo) | ~1-39 ng/mLo) | <10% | [70] |
The starting volume of human plasma/serum for SRM quantification.
The number of target proteins in plasma or serum.
Mouse plasma.
Peptides including phosphopeptides.
Cells or tissues.
β-NGF neuropeptide.
Plasma albumin depletion.
IgY14 depleted human serum.
Plasma volume depending on the abundance of target proteins.
Peptides.
Nuclear protein extraction followed by gel extraction.
One dimensional microflow RPLC separation followed by SRM measurement.
30 μL of human plasma used for both the 1D and 2D parallel experiments.
2 undetected proteins by 2D RPLC-SRM but with detection by 1D RPLC-SRM; 31 and 41 detected proteins by 2D RPLC-SRM but without detection by 1D RPLC-SRM.
MARS14 depleted human plasma.
Besides the affinity enrichment at the peptide level, mass spectrometric immuno assay (MSIA)-SRM [38], in which immunoenrichment is performed at the protein level on a monolithic microcolumn activated with an anti-protein antibody rather than magnetic beads and fixed in a pipette tip, was developed for sensitive multiplexed quantification of target proteins ranging in concentration from pg/mL to mg/mL (Table 1). When compared to other immunoenrichment methods, MSIA has less non-specific binding and is flexible with a wide range of sample volumes, thus MSIA allows rapid and quantitative enrichment of very low abundance (pg/mL) plasma proteins from a large volume (e.g., 1 mL) of human plasma. MSIA-SRM assays also demonstrated high sensitivity (within published clinical ranges for target proteins or peptides). Very recently MSIA-SRM assay was used for sensitive multiplexed quantification of several forms of circulating PCSK9 in human plasma [44]. The average LOQ of MSIA-SRM is ~20.8 ng/mL for PCSK9 in human plasma, which is below the clinical range. This sensitive PCSK9 MSIA-SRM assay revealed novel relationships between PCSK9 and metabolic phenotypes.
2.1.2 Aptamer
Single-stranded oligonucleotide (ssDNA) termed “aptamer” can be used for molecular detection in many screening platforms (e.g., the SOMAscan assay [45]). Similar to antibodies, high-quality aptamers have a strong binding affinity to target proteins with a dissociation constant (Kd) of ~10−9-10−10 and can form stable aptamer-protein complexes. With the aptamers serving as both folded protein-binding entities with defined shapes and unique nucleotide sequences recognizable by specific hybridization probes, the target protein concentrations can be reliably determined by measuring their corresponding aptamer concentrations using a quantitative DNA microarray. Thus, DNA aptamers have been widely used as affinity reagents for identifying and validating protein biomarkers in complex fluids [45–52]. However, the use of the aptamers to enrich the target proteins for SRM analysis has never been reported until recently. An aptamer enrichment technique coupled to SRM was developed for absolute quantification of activated ReIA in cytokine-stimulated eukaryotic cells [53]. The isolated aptamer P028F4 was demonstrated to have a strong binding affinity (Kd: 6.4 × 10−10) with the activated form of ReIA. Compared to direct SRM quantification of ReIA and associated proteins from crude cell extracts, aptamer-based enrichment dramatically reduced the sample complexity and the co-eluting interference, and thus significantly improved the S/N ratio of endogenous ReIA (~36-fold enhancement), quantification accuracy and reproducibility [53]. ReIA aptamer-SRM assay provided quantitative estimates of the number of activated ReIA molecules within a cell, which was in good agreement with previous Western blot experiments but with higher accuracy. ReIA aptamer-SRM is the first example of coupling of aptamer enrichment of target proteins to SRM-based targeted quantification. Aptamer-SRM is a promising alternative to immuno-SRM for reliable quantification of low-abundance target proteins in complex protein mixtures because of the demonstrated higher multiplexing capability of aptamer assays.
2.1.3 Other enrichment strategies
Other enrichment strategies (e.g., subcellular fractionation [54] and protein precipitation [55]) have been incorporated into SRM workflow for reducing sample complexity to enhance SRM sensitivity. Following the sequential protein extraction, in which the nuclear proteins were extracted from the whole cell lysate, the resulting protein mixture was then separated by SDS-PAGE, and the transcription factor (TF) protein bands were excised from the gel, low-abundance TFs were significantly enriched. Regular LC-SRM enabled sensitive reproducible quantification of the enriched TFs at ~250 copies per nucleus in mammalian cells [54]. This sensitive SRM method has been used for absolute quantification of the dynamic change of TFs during cellular differentiation. A single gel electrophoresis (Ge)-based enrichment method has recently been used for enriching KRAS at a low molecular weight protein fraction prior to SRM analysis. GeLC-SRM was demonstrated for reproducible quantification of KRAS mutant variants (14% CV for processing replicates) in a panel of cancer cell lines [56]. Next it was applied for robust sensitive quantification of mutant KRAS in pancreatic benign and cancer subjects at concentrations of 0.08–1.1 fmol/μg protein (Table 1). Recently, protein precipitation with acetonitrile was used to extract low mass proteins and to remove medium and high mass proteins from human blood. This step also precipitated the high mass and high abundance plasma albumin and immunoglobulins, and thus significantly reducing the sample complexity. The enriched low mass target proteins were then measured by SRM [55]. This simple, low-cost SRM method was demonstrated to reliably quantify five low mass biomarker proteins at a medium to low abundance (56.1–20 547.0 ng/mL) in 40 individual blood samples. Protein precipitation was also integrated into multistep sample purifications for highly efficient enrichment of target peptides. With the application of the combined plasma albumin depletion to remove the highest abundance albumin, acetonitrile protein precipitation to remove plasma proteins, and C18-SCX multi-StageTip to remove unnecessary peptides from pretreated plasma by using appropriate buffer solvents [57], low-abundance APL1β peptides, a surrogate marker for Alzheimer's disease, in human plasma were significantly enriched and can be sensitively quantified by SRM without immunoaffinity enrichment. The quantifiable absolute concentration of APL1β peptides was identified as several hundred amol/mL, which is probably the lowest detection level of endogenous plasma peptides as claimed by the authors.
2.2 High-resolution liquid chromatography separations
2.2.1 PRISM
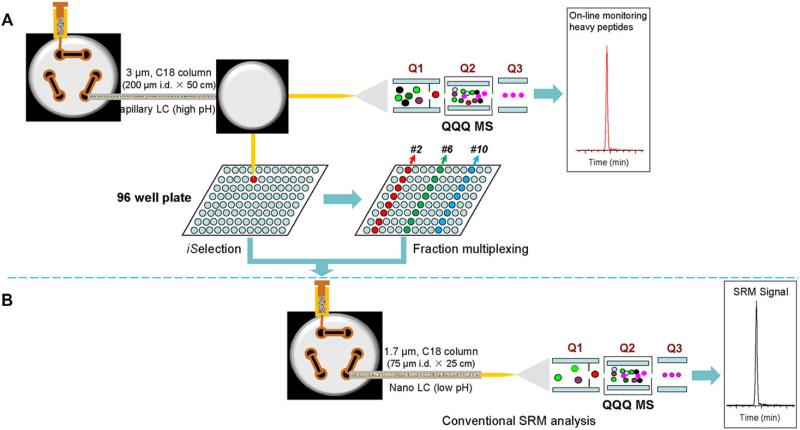
Similar to antibody-based immunoassays, affinity reagent-based SRM has significant potential for high-throughput studies, but it shares similar limitations in terms of availability of affinity reagents for new proteins or PTMs and multiplexing capability [17]. To address this issue, we recently developed an antibody-free chromatography-based technique termed PRISM (short for high-pressure, high-resolution separations with intelligent selection and multiplexing) that effectively enriches target peptides for sensitive LC-SRM analysis [18]. In PRISM, high pH reversed-phase capillary LC (RP cLC) separation is used to fractionate a peptide mixture into either 96- or 384-fractions, thus enriching target peptides by isolating them into specific fractions (Fig. 2). Selection of target fractions of interest is accomplished by online SRM monitoring of heavy isotope-labeled internal standards during sample fractionation. The accurate elution profiles of the internal standards allow precise determination of the locations of target peptides in the 96- or 384-well plate because the light and heavy forms of individual peptides are chemically identical so they co-elute during PRISM fractionation. This enables selection of the most informative target fractions for downstream LC-SRM analysis. Furthermore, a limited number of target fractions eluted at different times during the first-dimension separation can be multiplexed to further increase the overall analytical throughput because the high-and low-pH RP cLC are partially orthogonal. Therefore, only a small number of target fractions need to be analyzed, which effectively addresses the concern on the analytical throughput.
Figure 2.
Schematic diagram of the PRISM-SRM workflow reproduced from Shi et al. [18] with permission. (A) PRISM workflow. Peptide sample (typically 25 μg) spiked with internal standard (IS) heavy peptides was injected and separated by a high-resolution reversed-phase capillary LC (cLC) system using high-pH mobile phases. The eluent from the cLC column at a flow rate of 2.2 μL/min was split into two flowing streams via a Tee union (the split ratio of flow rates is 1:10): a small fraction (9%) of the column eluent went to a triple quadrupole mass spectrometer for on-line SRM monitoring IS peptides; a large fraction (91%) of the column eluent was automatically collected every minute into a 96-well plate during a ~100-min LC run. The specific target peptide fractions were either selected based on the same elution times of IS being monitored by the on-line SRM (iSelection) or multiplexed. For example, 96 fractions collected along with the first-dimension LC separation can be multiplexed into 12 fractions by combining 8 fractions from the first-dimension LC separation into 1 fraction for downstream LC-SRM analyses, such as pooling fractions 2, 14, 26, 38, 50, 62, 74, and 86 (marked in red color) into one sample (#2) for the second-dimension LC-SRM. (B) Conventional LC-SRM workflow. Following iSelection, a target peptide fraction was either directly subjected to nanoLC-SRM with 4 μL of sample per injection (~50 ng of peptides on the nanoLC column) or multiplexed with other target fractions with a final volume of 20 μL before nanoLC-SRM analysis.
Combined with IgY14 immunoaffinity depletion (i.e., the removal of the 14 most abundant proteins), PRISM-SRM enables accurate and reproducible quantification of plasma proteins at the 50–100 pg/mL range [18], and without depletion, in the sub-ng/mL to low ng/mL range [58]. Compared to regular LC-SRM analysis, PRISM provides ≥ 200-fold enhancement in SRM sensitivity. Since its development, PRISM-SRM has been serving as a key enabling technology for rapid preclinical verification of low-abundance protein biomarkers in human bodily fluids and tissues. For example, PRISM-SRM was applied to reliably measure candidate biomarker AGR2 at low pg/mL levels in prostate cancer patient urine and ~100 pg/mL levels in human serum when combined with the IgY14 depletion [59]. We also applied PRISM-SRM to measure TMPRSS2-ERG fusion protein products in prostate cancer cell lines and tumors [60]. PRISM-SRM enabled to confidently detect six unique ERG peptides in both TMPRSS2-ERG positive cell lines and tissues with the LOQ ranged from 2 to 50 amol/μg of total protein. Systematic comparison of three analytical platforms, ELISA, Western blotting, and PRISM assays, has shown that PRISM-SRM having a 20 pg detection limit is the most sensitive among the three platforms [61]. Very recently, we also applied PRISM-SRM for direct, site-specific quantification of the dynamics of PTMs (e.g., ERK phosphorylation dynamics and stoichiometry) [62]. PRISM-SRM was demonstrated to provide a level of sensitivity of ~1000 molecules per cell in quantifying ERK phosphorylation from as little as 25 μg tryptic peptides. Besides our broad applications PRISM-SRM was also applied by others to precisely quantify the total Aβ level in human plasma for avoiding the underestimation of Aβ by antibody-based assays [63]. This is the first study for measuring the plasma Aβ level by LC-SRM without antibody-based affinity enrichment.
Besides its application in PRISM fractionation, high pH reversed phase separation was also used to generate a limited number of fractions (e.g., 13 fractions) with pooling neighboring LC fractions that contained the same target peptides [64]. The neighboring fraction pooling enhances overall SRM signal but with the compromise of concurrently increasing the potential co-eluting interferences because the high-resolution reversed phase separation was not fully utilized. The partial orthogonality between the high pH and low pH RP separations only originates from the utilization of the difference in the ionic nature of peptides. Thus, 2D RPLC-SRM is very similar to SCX-RPLC-SRM because in both cases a limited number of continuous fractions were collected for LC-SRM analysis and the overall orthogonality between 2D RPLC and SCX-RP is similar [65–67]. 2D RPLC-SRM was demonstrated for enabling quantification of 31 proteins below 10 ng/mL and 41 proteins above 10 ng/mL [64] (Table 1).
Another example of using two dimensional LC to enhance SRM sensitivity is interfacing RP and hydrophilic interaction LC (HILIC) separations through an anionic cartridge [68]. RPHILIC-SRM was demonstrated for enabling quantification of PSA at 1 ng/mL level without any front-end immunoaffinity depletion. However, this 2D-LC-SRM method has two limitations, lacking multiplexing power (i.e., one target peptide at a time) and effectively trapping the selected target peptide on the anion exchange cartridge often requiring optimization of the buffer condition.
2.2.2 Long gradient (LG) separations
As an alternative to multidimensional sample fractionation strategies we recently evaluated the performance of the long-gradient separations coupled with SRM (LG-SRM) for targeted protein quantification [69]. Direct comparison of LG-SRM (5 h gradient) and regular LC-SRM (45 min gradient) showed that the long-gradient separations significantly reduced background interference levels and provided an 8- to 100-fold improvement in LOQ for target proteins in human female serum. LG-SRM enables reliable quantification of plasma proteins at ~10 ng/mL levels in non-depleted human plasma, which is comparable to most low-resolution fractionation-based SRM methods in the overall sensitivity [17, 65]. Later a similar study for systematic evaluation of LG-SRM performance has also been conducted by Carr's group [70]. They concluded that LG-SRM could provide low ng/mL LOQs for proteins in the IgY14 depleted plasma when the optimal LC conditions were used. Such sensitivity is very consistent with our results of ~10 ng/mL LOQs in non-depleted human plasma because the IgY14 depletion resulted in removing the 14 most abundant proteins in human plasma [69] that constitute 90–95% of the total protein mass (i.e., 10-fold sample enrichment) [71]. Besides the improved sensitivity, LG-SRM potentially offers much higher multiplexing capacity than conventional LC-SRM due to an increase in average peak widths (~3-fold) for a 300-min gradient compared to a 45-min gradient [69]. This allows a QqQ to use ~3 times longer duty cycle for monitoring more transitions (i.e., ~3 times more proteins) in a given elution window without affecting the quality of quantification (e.g., the sufficient data points across the LC peak). Furthermore, when compared to LC fractionation-based SRM, LG-SRM has several advantages in terms of sample throughput (one analysis per sample without multiple fraction runs), minute amounts of starting materials (e.g., ~4 μg), simple implementation, and easy operation [69,70].
2.3 Broad applications in biomedical research and systems biology
With the recent advances in SRM technology, front-end sample preparation, and bioinformatics tools (e.g., Skyline [30], QuaSAR [72], and Qualis-SIS [73]), SRM-based targeted proteomics technique is nearly mature for routinely reliable quantification of 10–100 s of target proteins in complex biological samples [17, 19]. In recent years studies for systematic evaluation of SRM assays and their performance have been conducted, such as developing a system suitability protocol for quantitative assessment of intra- and inter-laboratory instrument performance in LC-SRM analysis [34, 74], large-scale development of standardized SRM assays [33, 75], inter-laboratory evaluation of SISCAPA-SRM [32], and defining three tiers of SRM assays using a “fit-for-purpose” approach [76]. In 2012 targeted proteomics was selected as Method of the Year in Nature Methods [77]. SRM-based targeted proteomics merits have been well documented in multiple reviews [17, 19, 20, 78] and commentary reports [24, 79, 80]. SRM has been broadly applied in systems biology and biomedical research fields for reliable protein quantification. Compared to antibody-based assays SRM assays have several advantages in terms of multiplexing, detection specificity, and the assay development time and success rate for new target proteins [17, 80]. Thus, it may be time to “turn the tables” in new target protein quantification with the more frequent use of SRM assays [24] because it is considerably easier to develop reliable SRM assay with high specificity [17, 81, 82] and SRM assay has been proved to have an excellent correlations with analytically validated ELISA [18,59,83–85].
2.3.1 Biomedical research
As SRM is mature with distinct advantages over antibody-based assay, in recent years a number of papers have been published on verification of protein biomarkers with SRM in different types of diseases (e.g., prostate cancer [59–61, [86]], lung cancer [87–92], colorectal cancer [93–95], pancreatic cancer [96,97], cardiovascular disease [84], Parkinson disease [98–101], Down syndrome [102–104], type 1 diabetes [8], and other rare diseases [105–109]) (Table 2). The candidate protein biomarkers are generated either from comparative proteomics (or genomics) discovery studies or from previous literature reports. These candidate protein biomarkers are then measured in a medium or large matched set of clinical cohorts. For example, to identify lung cancer biomarkers in blood that are shed or secreted from lung tumor cells, comparative proteomics studies of protein expression levels between lung cancer tumor cells and normal lung cells have been conducted [91]. In combination with literature-resource biomarkers 388 protein candidates were selected for validation and performance assessment. SRM assays were applied in a three-site discovery study (n = 143) on plasma samples from patients with benign and stage IA lung cancer, producing a 13-protein classifier. The classifier was validated on an independent set of plasma samples (n = 104), exhibiting a negative predictive value of 90%. It provides a new metric, independent of current diagnostic risk factors, for assessing the molecular status of a lung nodule. Furthermore, to expedite SRM-based protein biomarker verification, SRM assays for most cancer-associated proteins were recently developed, and their sensitivity, reproducibility, quantitation dynamic range, and detectability were systematically evaluated in human bodily fluids (e.g., human plasma/serum [110–112], urine [110], CSF [113]). For example, SRM assays of more than 1000 cancer-associated proteins were generated and then used to determine the detectability of those proteins in human plasma and urine [110]. In depleted human plasma, 182 proteins were detected, spanning five orders of magnitude in abundance and reaching below a concentration of 10 ng/mL, whereas in human urine 408 proteins were confidently detected due to the narrower concentration range of proteins in urine.
Table 2.
Preclinical verification of candidate protein biomarkers by targeted proteomics assays in 2012–2015
| Candidate protein biomarker | Disease type | Specimen | Assay type | Ref. |
|---|---|---|---|---|
| AGR2 | Prostate | Urine | PRISM-SRM | [59] |
| TMPRSS2-ERG fusion protein products | Prostate | Tissue | PRISM-SRM | [60,61] |
| CD44 antigen, immunoglobulin γ-2 heavy chain, ITIH2, cadherin-13 | Prostate | Serum | SPEG-LC-PRMa) | [86] |
| zyxin | Non-small-cell Lung | Plasma | LC-SRM | [87] |
| SAA1, SAA2 | Lung | Plasma | LC-PRM | [88] |
| SAA1, SAA2 | Lung | Serum | LC-SRM | [89] |
| TFPI, MDK, OPN, MMP2, TIMP1, CEA, CYFRA 21–1, SCC | Lung | Tissue, serum | LC-SRM | [90] |
| LRP1, BGH3, COIA1, TETN, TSP1, ALDOA, GRP78, ISLR, FRIL, LG3BP, PRDX1, FIBA, GSLG1 | Lung | Plasma | IgY14-LC-SRM | [91] |
| ALCAM, CDH1, MUC1, SPINT1, THBS4, SVEP1 | Lung | Plasma | MARS6-LC-SRM | [92] |
| ITGA5, GPRC5A, PDGFRB, TFRCC, 8orf55 | Colorectal | Tissue | LC-SRM | [93] |
| CP, TIMP1, LRG1, PON1, SERPINA3 | Colorectal | Plasma | Glyco-LC-SRMb) | [94] |
| HLA-A, CFH, CD44, PTPRJ, HP, CDH5 | Colorectal | Plasma | Glyco-LC-SRMb) | [95] |
| LDH-B, CKMB, myoglobin, troponin I | Cardiovascular | Serum | MARS6-LC-SRM | [84] |
| PRNP, HSPG2, MEGF8, NCAM1 | Parkinson | Plasma | LC-SRM | [98] |
| SPP1, LPR1, CSF1R. EPHA4, TIMP1 | Parkinson | CSF | LC-SRM | [99] |
| GSN, MSN, LSP1, SEPT6, TALDO1, TWF2, VIM | Parkinson | T-lymphocyte | LC-SRM | [100] |
| Ubiquitin | Neurodegenerative | CSF | LC-SRM | [101] |
| CEL, CPA1, MUC13, CLCA1, MUC5AC, HAPLN1 | Down syndrome | Amniotic fluid | LC-SRM | [102] |
| CEL, MUC13, CPA1, DPP4, MMP2 | Down syndrome | Amniotic fluid | LC-SRM | [103] |
| SAP, C1-inhibitor | Down syndrome | Plasma | LC-SRM | [104] |
| 244 NLF proteins | Upper airway | NLFc) | LC-SRM | [200] |
| C3, CD3E, DPT, MCM4, PMEL, S100A8, S100A13, S100B, TAGLN2 | Malignant melanoma | Tissue | LC-SRM | [201] |
| Proline-hydroxylated α-fibrinogen | Pancreatic | Plasma | LC-SRM | [96] |
| SFN, GSN, LUM, TIMP1 | Pancreatic | Plasma | IgG-LC-SRM | [97] |
| A1BG, GFH, IGFALS, PROC, RBP4 | Hepatic fibrosis | Serum | IgY14-LC-SRM | [105] |
| ORM1, PFN1, H4, H2AFX, MPO, M2BP, C4BP, CRP, S100A9, MMP3, DEFA1, CD5L | Psoriatic arthritis | Synovial fluid | LC-SRM | [106] |
| A4, APOE, KLK6, MY15B, PEDF, 1433F, 1433G, AACT, CAD13, TAU, NFH, NFL, NFM, NRCAM, OSTP, SAMP | Multiple sclerosis | CSF | LC-SRM | [107] |
| ICAM1, HSPG2, ANTXR1, PON1, HYOU1, THBS1, MSLN | Malignant pleural mesothelioma | Serum | SPEG-LC-SRMa) | [108] |
| PPBP, SERPING1 | Type 1 diabetes | Serum | LC-SRM | [8] |
| A1AG1, AACT, A1AT, CERU | Liver | Plasma | Lectin-LC-SRM | [109] |
SPEG: solid-phase extraction of N-linked glycopeptides.
Glycoprotein enrichment.
NLF: nasal lavage fluid.
Due to the lack of specificity to recognize individual protein isoforms, antibody-based assays can only be used to differentially measure certain protein isoforms [17]. SRM assays have been demonstrated to be well suited for site-specific quantification protein isoforms or PTMs. Isoform-specific SRM assays were recently developed for reliably quantifying protein isoforms in many studies (e.g., 14 UGT1As and UGT2Bs in liver matrices [114], three polyphenol oxidase isoforms in loquat fruits [115], 3 Apolipoprotein E (ApoE) isoforms that differ in only one or two amino acids in cerebrospinal fluid and plasma samples [116], and TMPRSS2-ERG fusion protein products in prostate cancer [60]). Two SAA isoforms, SAA1 and SAA2 that have high similarity (92%) amino acid sequences can be accurately measured by SRM assays for the first time [89]. Verification of SAA1 and SAA2 in clinical crude serum samples indicated that SAA2 could be a good biomarker for the detection of lung cancer. Another two elegant studies are to design SRM assays for identification of a novel PSA isoform coded by SNP-L132I (rs2003783) in clinical prostate samples [117], and to use de novo sequencing for unambiguous characterization of two VTG isoforms, Dc-VTG1 and Dc-VTG2, followed by SRM quantification of the two isoforms [118]. Plasma levels of Dc-VTG1 and Dc-VTG2 were found to be decreased as the nesting season proceeded, and were closely related to the increased levels of reproductive effort. In addition, several studies were reported for using PTM SRM assays for site-specific quantification of PTMs, such as selective measurement of free monoubiquitin (a potential neurodegenerative disease biomarker) in cerebrospinal fluid [101], and evaluation of a large set of candidate serum biomarker glycopeptides for malignant pleural mesothelioma (MPM), resulting in a seven glycopeptide signature with diagnostic potential for MPM [108].
2.3.2 Systems biology
Besides application in biomarker studies, SRM has also been used for accurate quantification of the dynamics of signaling pathways or networks in response to a given perturbation to improve our understanding of molecular mechanisms of signal transduction [7, 119–129]. Using the combination of affinity purification (AP) and quantitative SRM temporal regulation of EGF signaling networks by the scaffold protein Shc1 was investigated in detail, resulting in a significant finding that the Shc1 directs the temporal flow of signaling information after EGF stimulation [7]. AP-SRM has been widely used for measuring the concentrations of protein components including PTMs in target protein complexes. For example, it was used to quantify the dynamics of protein interaction networks in pre-60S particles after nuclear export [130]. From the dynamic data assembly factors were found to travel with pre-60S particles to the cytoplasm and a novel shuttling factor that facilitates nuclear export of pre-60S particles was identified. Other types of affinity enrichment combined with SRM were employed for comprehensively monitoring signal transduction pathway dynamics. For example, IMAC-SRM was used to measure phosphorylation dynamics of the PI3K-mTOR/MAPK signaling networks in oncogene-induced senescence [120].
Besides using SRM to measure the concentrations of a set of enriched proteins SRM is more broadly used for accurate quantification of the dynamics of specifically selected proteins (e.g., 76 proteins in the energy metabolic pathways and more than 80 cancer signaling proteins [119]) involved in the specific signal transduction pathways. 144 proteins involved in the insulin-signaling pathway and central metabolism in mouse liver homogenates were selected and measured by SRM across 36 samples (12 conditions and three replicates for each condition) [119]. The nearly complete quantitative pathway data revealed strain-specific changes in the mouse insulin and central metabolic pathways after a sustained high-fat diet. Very recently, SRM was combined with RNA-seq and rule-based pathway modeling to quantitatively explore the chemotaxis signaling pathway mediated by sphingosine-1-phosphate [126]. Using absolute protein concentrations measured by SRM as input parameters of a mathematic model, the resulting in silico pathway behavior matched experimental measurements. This finding demonstrates the feasibility and value of combining SRM with pathway modeling for advancing biological insight.
In addition, SRM was also applied in other systems biology studies, such as systematic measurement of transcription factor-DNA interactions [131], global kinetic analysis of proteolysis [132], and a sentinel protein assay for simultaneously quantifying cellular processes [133]. SRM assays of sentinel proteins (i.e., biological markers) were demonstrated for enabling simultaneously probed 188 biological processes in Saccharomyces cerevisiae under a set of environmental perturbations. Another elegant study is global analysis of protein structural changes in complex proteomes by an approach of coupling limited proteolysis with SRM [134]. This approach potentially enables probing both subtle and pronounced structural changes of proteins on a large scale.
3 New developments in other types of targeted MS/MS quantification and their applications
3.1 PRM
With recent introduction of fast data acquisition HR/AM MS (e.g., Q Exactive quadrupole Orbitrap (QqOrbi) [22, 23], and TripleTOF quadrupole TOF (QqTOF) [11, 25]), the third quadrupole is replaced by a HR/AM Orbitrap or TOF mass analyzer (Fig. 1A). Targeted MS/MS, when operated in PRM mode [11, 22, 23, 25], can be used for reliable quantification of target proteins in complex matrices. PRM was demonstrated to have similar analytical performance as SRM in sensitivity, reproducibility, accuracy, and dynamic range [22, 23, 135–137]. In the urine samples with 35 peptides spiked-in, PRM has shown to provide lower LOQs for 61% of 175 transitions than SRM and 20% of the transitions have similar LOQs between PRM and SRM [135]. For synthetic peptides spiked in yeast cell lysates PRM yielded quantitative data over a wider dynamic range than SRM [22]. Compared to SRM the analytical workflow of PRM is simplified without preselecting/optimizing target peptide transitions because it allows for detection of all product ions simultaneously [22,23]. Another advantage of PRM over SRM is the improved selectivity (or specificity) from the HR/AM quantification by discriminating target peptides more effectively from background interferences, which directly translates into the increased assay and data quality [22, 23]. With the use of trapping device in QqOrbi MS or longer accumulation time in QqTOF MS, PRM allows improving the S/N ratio (i.e., sensitivity) and thus lowering the LOQ values for detecting low-abundance target peptides [23, 27] but at the expense of duty cycle (i.e., multiplexing). For example, Majovsky et al. developed PRM assays on lower scan speed LTQ-Orbitrap Velos MS for quantification of protein degradation in plant signaling and concluded that the instrument duty cycle was a critical parameter limiting sensitivity and thus only a small number of proteins can be reliably quantified [138]. In addition, comparisons of PRM in QqTOF with SRM using six model peptides indicated that increasing the accumulation time in PRM was required to match the performance of SRM [139].
3.1.1 Multiplexing
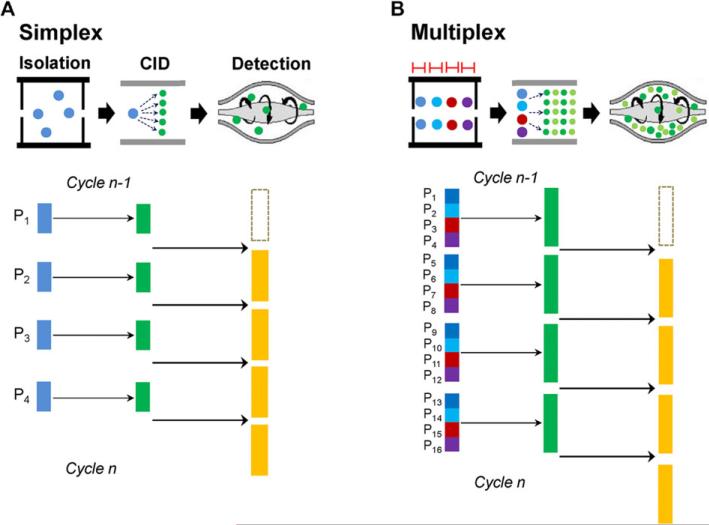
For measuring limited peptide sets (typically 50 peptides or less) in complex matrices PRM was demonstrated to have high assay performance in terms of sensitivity and reproducibility [23,27]. However, similar to SRM it is still challenging for applying PRM to measure a large number of peptides without sacrificing the sensitivity and reproducibility [23,27]. The different PRM acquisition methods in QqOrbi MS (simplex, broadband, and multiplex) [23, 28] have been discussed in details (Fig. 3). In principle, the number of peptides that can be analyzed in one PRM run is determined by the cycle time, the transient time (the MS resolution), and the degree of multiplexing [23,28,140]. The cycle time is determined by the peak width of peptide LC elution (e.g., 8–10 data points across peptide peak for achieving precise quantification). The transient time (i.e., the resolving power) is directly correlated to the number of peptides to be monitored. The defined parameters determine the number of peptides measured in one cycle (i.e., the number of peptides = [cycle time × multiplexing degree]/transient time) [23, 28]. The PRM sensitivity is affected by the length of fill time [23,28,140]. The maximum fill time is generally adjusted according to the transient time and the multiplexing degree to ensure isolation and fragmentation of a given peptide precursor ion concomitantly in the Orbitrap analysis (i.e., maximum fill time = [transient time / multiplexing degree]) [23, 28]. Thus, when the same fill time is used for both simplex and multiplex PRM the multiplex PRM could have similar sensitivity to the simplex PRM. By using multiplexing degree of 4, a resolving power of 35 000 at m/z 200 operated on the Q Exactive or Q Exactive Plus platform (i.e., 128 ms of transient time), and 2 s of cycle time, up to 64 peptides can be analyzed in one cycle but with compromising the fill time of 30 ms for each peptide precursor ion [28]. Thus, 4-plex PRM could have the similar multiplexing power as SRM, in which ~66 peptides can be simultaneously monitored in one 2 s cycle (i.e., the number of peptides ≈ [2000 ms / 30 ms]) when three transitions per peptide and the standard dwell time (10 ms per transition) are used. However, 4-plex PRM provides full MS/MS spectra with all the fragment ions while SRM only monitors the selected three fragment ions for the precursor ion. With the implementation of the new generation Q Exactive HF platform, which has nearly two times faster in scan speed [141], up to 128 peptides can be measured in one cycle under the same condition.
Figure 3.
Data acquisition methods in PRM modified from Gallien et al. [23, 28] with permission. (A) In the simplex mode, the co-eluting peptides are sequentially analyzed (i.e., isolating one precursor of interest at a time for MS/MS and then measuring its product ions). (B) In the multiplex mode the precursor ions of subsets of co-eluting peptides are isolated sequentially, and then sequentially fragmented in the HCD cell, where all products are trapped and accumulated in the C-trap. When the last precursor ion is fragmented, the mixture of product ions is transferred into the Orbitrap mass analyzer for detection, where a single combined MS/MS spectrum is acquired.
For a given number of peptides different combinations of parameters could lead to a trade-off between specificity (i.e., the transient time and quadrupole isolation window) and sensitivity (i.e., the fill time) [28]. Thus, large-scale quantification with PRM requires the adjustment of MS acquisition parameters, which dramatically affects the assay and data quality (e.g., sensitivity, accuracy, and reproducibility). Very recently, a new data acquisition method, internal standard triggered-PRM (i.e., IS-PRM) [27] was designed to maximize the efficient use of the instrument time for sensitive quantification of endogenous peptides. The data acquisition scheme switches between two PRM modes: a fast low-resolution “watch mode” for scanning heavy internal standards (e.g., the maximum fill time of 60 ms) and a slow high-resolution “quantitative mode” for scanning endogenous peptides (e.g., the maximum fill time of 360 ms). IS-PRM was demonstrated for achieving both the number of endogenous peptides and the quantification sensitivity. Application of IS-PRM to the analysis of large peptide sets (up to 600) in complex samples has shown an unprecedented combination of scale and analytical performance, with the LOQs at the low amol range [27]. However, this method involves the front truncation of the PRM signal and sophisticated MS software controls to trigger endogenous peptide measurement, which could affect the quantification accuracy and reliability. Thus, at present the broad utility of IS-PRM is not clear in the targeted proteomics field.
3.1.2 Application
Unlike SRM, the application of PRM in targeted quantification is still in its infancy, though PRM method development has been well documented in several reviews [26, 28, 140, 142, 143] and the data analysis of PRM is very similar to that of SRM [30, 72, 73]. In the past three years there are only a handful of reports of applying PRM for target protein quantification. Multiplexed PRM assays were recently developed for isotype-specific SAA1 (1α, 1β, 1γ) and SAA2 (2α, 2β) variants in human plasma and successfully applied to analyze plasma samples from lung cancer patients [88]. The high data quality from PRM assays allows interference-free quantification of all variants in large clinical cohorts of lung cancer patient plasma. The combination of immunoaffinity purification and PRM (i.e., immuno-PRM) allows fast screening driver mutations in tissue and tumor markers in human plasma [144]. Very recently, Thomas et al. developed multiplexed PRM assays for quantification of 43 N-linked glycosite-containing peptides in prostate patient sera [86]. A total 41 N-linked glycosite-containing peptides (corresponding to 37 proteins) were reproducibly quantified with four proteins showing differential significance between nonaggressive and aggressive prostate cancer patient sera. Kim et al. developed and implemented multiplexed PRM assays for quantitative profiling 83 protein tyrosine kinases in human cancer cell lines [145]. The PRM assays enabled to detect 308 proteotypic peptides from 54 receptor tyrosine kinases and 29 nonreceptor tyrosine kinases in a single run. In addition, PRM was also used for sensitive quantification of protein modifications (e.g., histone lysine acetylation and methylation [146, 147], and polyubiquitin chains [148]).
3.2 DIA for proteome-wide quantification
Currently the top choice for protein targeted quantification is still SRM primarily because it has matured for the past two decades [21]. However, SRM quantification involves front-end assay development (e.g., peptide selection, transition and collision energy optimization, and interference-free transition selection, etc.) [19, 21]. The recently introduced PRM has partially alleviated this issue with more simplified analytical workflow. However, the scale of SRM/PRM quantification is limited, typically to several hundred of peptides per a single run for reliable quantification [21, 23]. Increasing the number of target peptides results in a significant trade-off in quantification sensitivity [27]. Often times, additional experiments are needed for achieving both the scale of quantification and sensitivity.
3.2.1 DIA method development
Data-independent acquisition (DIA) is an emerging MS technique to combine advantages of shotgun (i.e., large-scale based on DDA) and targeted proteomics (i.e., high reproducibility and accuracy) [11, 149, 150]. The data creation of DIA is more flexible and simpler compared to DDA or SRM/PRM experiments. DIA collects all MS/MS scans irrespective of precursor ion selections from a survey scan or full MS scan, in which DDA necessitates. The predefinition of target lists, which SRM/PRM requires, is unnecessary for DIA experiment. A broad range of precursors and corresponding transitions can be extracted after the data procurement. Thus, in targeted proteomics, DIA aims at inclusive proteome-wide quantification using targeted data extraction strategy. However, when compared to SRM/PRM DIA-based targeted method in general provides lower sensitivity, specificity, and reproducibility as well as smaller dynamic range in protein quantification [11].
The term DIA was introduced by Venable et al. using LTQ-linear ion trap (LIT) mass spectrometer [151]. They used wide precursor isolation window (10 m/z) to perform sequential isolation and fragmentation of a predefined m/z range. Compared to the MS level quantification, the S/N ratio was greatly improved (higher than 350%) with a good linear dynamic range. They also demonstrated the applicability of ion extractions in MS/MS level of DIA quantification. However, such low resolution DIA MS/MS with the wide precursor isolation window decreased the mass accuracy and the confidence in peptide identification, which potentially resulted in increasing the false positive discovery rate. Since then, other modified DIA have been introduced. In 2005, MSE was introduced by Silva et al. and operated on QqTOF instrument [152]. Precursor acquisition independent from ion count (PAcIFIC) [153,154] was a modified DIA version, and initially performed on LIT MS [153] and later on a faster LTQ Velos MS [153]. The use of smaller isolation windows (2.5u) led to the improvement of protein identification though the overall data acquisition covering the whole target mass range required multiple injections (67 injections for 5 days). A much faster scanning ion trap MS (e.g., LTQ Orbitrap Velos MS) reduced the whole data acquisition time to ~2 days. In 2010, with an introduction of a bench top Exactive MS, Geiger et al. demonstrated the application of all-ion fragmentation (AIF) [155], in which peptides were injected to HCD collision cell for fragmentation without precursor selection and the fragments were returned back to C-trap and analyzed through Orbitrap mass analyzer. The assignment of fragment ions to co-eluting precursor ions was facilitated by high resolution (100 000 at m/z 200) and high mass accuracy. This concept significantly decreases the duty cycle time, but introduces more interferences from AIF at a certain retention time. In the same year, another DIA approach was introduced by Carvalho et al., namely extended data-independent acquisition (XDIA) [156], which was performed on a different type of Orbitrap MS with the capability of electron transfer dissociation (ETD). When compared to DDA-based ETD analysis, the DIA-based ETD approach significantly increased the number of identified spectra (~250%) and the number of unique peptides (~30%), which could potentially facilitate the low-abundance PTM study. However, it is worth noting that ETD was performed on a low-resolution ion trap, and thus there are some challenges in differentiating isobaric peptides. FT-all reaction monitoring (FT-ARM) developed by Weisbrod et al. is to fragment all ions within a broad m/z window (100 m/z) [157]. Different types of fragmentation were applied (e.g., CID or infrared multiphoton dissociation).
Significant improvement in DIA has been achieved along with development of fast scanning HR/AM instruments for the past 3 years. Gillet et al. introduced a variation of DIA using QqTOF MS, termed SWATH, to conceptually refer the utilization of a broad isolation window (typically 25 m/z) consisting of multiplexed spectra [11]. One key feature of using QqTOF MS is the fastest data acquisition rate. Another DIA strategy was introduced by Egertson et al. with the use of QqOrbi MS, in which a novel acquisition method, namely MSX [158], was incorporated to improve the instrumental speed, selectivity, and sensitivity. The isolation window was set to 4 m/z covering a mass range from 500 to 900 m/z. Among those 100 windows (400 m/z divided by 4 m/z), five random windows were selected, stored in C-trap, and transferred to Orbitrap analyzer. The resultant spectra are multiplexed MS/MS of the selected windows [158]. However, more intelligent data processing was required to de-multiplex spectra for reliable identification and quantification of peptides. Two more recent improvements in DIA data acquisition and processing were aided by newer version of Orbitrap MS instruments. The first one termed pSMART, which was implemented on Q Exactive MS, utilized asymmetric isolation windows over the mass range: 5 Da window covering 400~800 m/z, 10 Da window covering 800–1000 m/z, and 20 Da window covering 1000–1200 m/z [159]. In another DIA method termed wide selected-ion monitoring DIA (wiSIM-DIA) [160], which was implemented on the new hybrid Q-HCD-Orbitrap-LIT MS (i.e., Orbitrap Fusion and Lumos), three HR/AM SIM scans with 200 Da isolation windows were used to cover all precursor ions of 400–1000 m/z. In parallel with each SIM scan, 17 sequential ion-trap MS/MS with 12-Da isolation windows were acquired to cover the associated 200-Da SIM mass range. Different from the standard wide-window MS/MS-only DIA methods, for pSMART and wiSIM-DIA the MS1 data (i.e., HR/AM precursor data with a longer fill time and enough precursor ion data points across the LC elution profile) is used for sensitive detection and quantification, while the MS/MS data from the fast ion trap MS/MS scan is used only for peptide identification/confirmation. Compared to the standard DIA methods with a complete recording of all fragment ions of the detectable peptide precursors but with sophisticated data analysis, pSMART and wiSIM-DIA can only provide a smaller number of MS/MS spectra for the detectable precursors with relatively easier data analysis because the majority of the duty cycle time is used for generating the high-quality MS1 data [159, 160]. Thus, their sensitivity and precision are higher than those provided by the standard DIA methods, while their quantification accuracy (i.e., specificity or selectivity) could be somewhat lower than that from the standard DIA methods due to the increased chances for having co-eluting interference from MS1 than MS/MS. Very recently, Bruderer et al. introduced a novel DIA method termed hyper reaction monitoring (HRM) [161]. This consists of comprehensive DIA acquisition on a Q Exactive MS platform and target data analysis with retention-time-normalized (iRT) spectral libraries. HRM was demonstrated to outperform shotgun proteomics both in the number of consistently identified peptides and in reliable quantification of different abundant proteins across multiple measurements.
3.2.2 Bioinformatics tools for DIA data analysis
DIA spectra are highly multiplexed and thus more elaborate data interpretation algorithm is needed compared to DDA or SRM/PRM (reviewed in [162]). Currently, there are three approaches that are used to decipher DIA spectra. The first one is to construct pseudo-DDA spectra from DIA spectra, such as Demux [163], MaxQuant [155], XDIA processor [156, 164, 165], and Complementary Finder [166]. Those reconstructed pseudo-DDA spectra are then processed through conventional search engine tools such as MSGF+ [167], MaxQuant [168], MASCOT [169], or other spectra libraries [170, 171]. Some of the schemes implemented the use of chromatographic elution profiles to improve identification of peptides [155, 165, 172]. The recent publication by Tsou et al. described the development of a computational approach, termed DIA-Umpire [172]. DIA-Umpire starts with two dimensional (m/z and retention time) feature-detection algorithm to discover all possible precursor and fragment ion signals in MS and MS/MS data. Fragment ions are grouped with a precursor ion that has a correlation of LC elution peak and retention time at a peak apex. Generated pseudo-MS/MS spectra for each precursor-fragment group are then processed with conventional database search engine including the abovementioned tools. The other approach is to match multiplexed MS/MS to theoretical spectra of peptides (e.g., ProbIDtree [173], Ion Accounting [174], M-SPLIT [175], MixDB [176], and FT-ARM [157]). The scoring algorithms are directly based on how many theoretical fragment ions of a peptide from sequence databases or spectral libraries are found on multiplexed spectra with a high mass accuracy. The first two approaches have greatly tackled identification of peptides from DIA spectra prior to further quantification. Freely-available automated or semi-automated tools such as Skyline [30,177], mProphet [178], OpenSWATH [29], and DI-ANA [179] and two commercial software, PeakView® from AB/Sciex and Spectronaut™ from Biognosys, have been employed to process quantitative targeted DIA data using the targeted data extraction strategy [11, 29, 158, 177]. However, more powerful computational tools are still needed for more effective analysis of DIA data.
3.2.3 Recent applications of DIA-based targeted quantification
DIA-based targeted proteomics provides SRM-like reproducibility, linearity, and accuracy while the sensitivity is still less than SRM [133,149,180]. Gillet et al. showed an excellent linear correlation of fold changes between SRM and SWATH with R2 ≥ 0.95, and a linear dynamic range of 4 orders of magnitude for SWATH [11]. Compared to DDA the quantification sensitivity was increased by 2–8 fold but still ~10 fold lower than SRM/PRM. Egertson et al. applied MSX strategy to increase the selectivity of precursors, showing LOD of 0.41 fmol with R2 ≥ 0.95 [158]. Compared to SRM/PRM the major advantage of DIA-based targeted quantification is the proteome-scale of quantification and thousands of proteins can be quantified simultaneously [149, 150]. Another advantage is that one-time DIA data procurement can serve as both protein identification and quantitation, which should accelerate the biomarker discovery process [149,150].
Recently targeted DIA approaches were applied for large-scale protein quantification. When combined with pressure cycling technology SWATH-MS allows detection and quantification of more than 2000 proteins with a high degree of reproducibility across 18 small biopsy samples from 9 patients [181]. Selevsek et al. demonstrated the reproducibility and consistency for quantification of more than 15 000 peptides and 2500 proteins of Saccharomyces cerevisiae using SWATH-MS [182]. They completed measuring proteins for 18 osmotic shock time course samples in less than 3 days. With the use of affinity purification for enriching protein complexes, targeted DIA has been applied for efficient quantification of large-scale proteome dynamic changes (e.g., interactomes of the 14-3-3 system [183] and the HSP90 inhibitor NVP-AUY922 or melanoma-associated mutations in the human kinase CDK4 [184]). In combination with solid phase extraction of glycopeptides (SPEG) Liu et al. applied SWATH MS for glycoproteomic analysis of prostate cancer tissues [185]. This approach was validated in human plasma with the LOQ of 5 ng/mL and a dynamic range of 4 orders of magnitude [180]. Quantitative glycoproteomics analysis showed that 220 glycoproteins appeared to be significant associated with prostate cancer aggressiveness and metastasis. Another example is the application of targeted DIA to investigate the effects of the histone deacetylase inhibitor SAHA (suberoylanilide hydroxamic acid) on the global histone PTM state of MCF7 cells [186]. A total of 62 unique histone PTMs were quantified accounting for less than 0.01% of a total peptides, which was also supported by another histone PTM study with SWATH MS (i.e., remarkably low abundance of hi-stone PTMs (H3K9me2S10ph, relative abundance <0.02%) in mouse trophoblast stem cells [187]).
4 Future perspectives
Targeted proteomics has become an indispensable tool for cost-effective and reliable measurement of hundreds of candidate protein biomarkers or signaling pathway proteins across many samples in term of specificity, reproducibility, multiplexing capability, and being independent of affinity reagents [17,19,149,188]. However, the major constraint of current target proteomics approaches is the lack of sufficient sensitivity for measuring low-abundance proteins in complex biological matrices. Enhancing the sensitivity is often accomplished by the front-end sample enrichment or fractionation followed by subsequent LC separation (a typical 60 min run time) to increase analyte concentration and reduce sample complexity but with significantly sacrificing sample throughput (see [17]). Thus, it is a formidable challenge in time and cost for validation of multiple low-abundance protein biomarkers in large clinical cohorts (e.g., only two publications were found to use SRM to measure more than 1000 patient samples [189,190]) with the current targeted proteomics platforms.
Recent advances in coupling SISCAPA enrichment to RapidFire SPE-SRM [191] or MALDI-TOF MS [192, 193] significantly improve sample throughput with an average ~7 s per sample for RapidFire SPE-SRM and ~30 s per sample for MALDI-TOF MS. It could be practical for measuring ~1000 clinical samples per day if all components including sample processing (e.g., trypsin digestion and SISCAPA enrichment), MS measurement, and data analysis are effectively co-ordinated. However, quantification accuracy and assay reproducibility from the two SISCAPA-based methods primarily rely on peptide antibody quality (i.e., the purity of enriched target peptides) because there are no additional separation steps that are used to further remove co-eluting interferences. This one-step SISCAPA enrichment without additional separation also limits the multiplexing power (e.g., less than ten target peptides with monitoring only one transition per peptide [191]) and lowers the detection sensitivity (e.g., 100-fold less sensitive for SISCAPA-MALDI-TOF MS when compared to SISCAPA-LC-SRM [191]). Furthermore, SISCAPA-based targeted quantification shares the similar shortcomings as the antibody-based immunoassays with the difficulty of generating high-quality antibodies and a high failure rate.
To achieve high-throughput sensitive measurement of hundreds of target proteins we believe that advancing MS instrumentation is probably the most effective way to improve both sample throughput and MS sensitivity because any additional front-end sample handling will enhance MS sensitivity but at the expense of overall sample throughput [17]. This can be achieved by implementing advanced MS interface technologies (e.g., gas-phase ion mobility separations in long-path structures for lossless ion manipulation separation, termed SLIM [194–196] and the use of cutting-edge ion sources [197]), as well as increasing the resolving power of mass analyzer [23, 137]. For example, in the PRM mode HR/AM mass analyzer was demonstrated to provide high specificity and sensitivity in targeted quantification [23, 137]. A combination of high-resolution LC and gas-phase SLIM IMS separations could lead to achieve extremely high peak capacity because the two separation methods are fully orthogonal and each separation method should have at least 100 s of peak capacity in a short separation time (e.g., ~150 of peak capacities in less than 10 min LC separation [198]). Thus, LC×SLIM IMS may have great potential to achieve full-baseline separation of target peptides from complex biological matrices in a high-throughput manner without the development of specific antibodies for target proteins or peptides. It is very optimistic that in the near future ~1000 clinical samples with 100 s of target proteins can be analyzed by advanced targeted MS within a week. We expect that the new generation MS will revolutionize current targeted MS quantification and be more broadly and readily adopted in many biomedical and clinical laboratories.
5 Concluding remarks
Targeted proteomics has emerged as a powerful protein quantification tool for reliably measuring target protein concentrations across many biological or clinical samples in a consistent, reproducible, and accurate manner. SRM is a classic targeted proteomics approach and has matured over the past two decades. It has been widely applied for accurate quantification of cellular signaling networks or pathways and pre-clinical verification of candidate protein biomarkers. With recent advances in enhancing SRM sensitivity (e.g., PRISM-SRM [18, 58]), SRM enables quantification of target proteins in human plasma/serum at 50–100 pg/mL levels [18] and in human cells at ≤100 copies per cell [199]. Such sensitivity is comparable to or even better than that provided by many analytically validated ELISA. This could bring immediate impact on systems biology and biomedical research because the new informative protein biomarkers or feedback regulatory proteins in signaling pathways are generally less abundant and antibodies are available only for several protein biomarkers or regulatory proteins. However, SRM still has limitations in selectivity and requires more assay development/validation for achieving high quality quantification assays because it is operated on a low-resolution QqQ MS.
The recent introduction of fast HR/AM MS (e.g., QqOrbi MS and QqTOF MS) resulted in two new types of targeted MS quantification, PRM and DIA. They offer new avenues for HR/AM quantification of target proteins and overcome some SRM limitations. Compared to SRM the analytical workflow of PRM is simplified without the need for transition selection and optimization. For a smaller number of target peptides PRM was demonstrated to have similar performance as SRM in terms of sensitivity, multiplexing, reproducibility, and linear dynamic range and better performance than SRM in specificity (selectivity). Similar to SRM the daunting challenge for PRM is the scale of quantification because increasing the degree of multiplexing leads to significantly lowering sensitivity. Thus, further method development of PRM is required for enabling effective combination of scale and sensitivity for large-scale protein quantification. In contrast to SRM or PRM, DIA requires minimal assay development and offers proteome-scale targeted quantification using the targeted data extraction strategy. However, DIA has lower sensitivity and specificity, poorer reproducibility, smaller dynamic range, and much less matured informatics tools than SRM or PRM. Thus, at present DIA is suited for initial screening of a large number of high- to moderate-abundance proteins, and SRM or PRM is then used for sensitive accurate quantification of the remaining set of low-abundance proteins. It has been recognized by the proteomics community that a paradigm has already progressively shifted from un-targeted global proteomics to targeted proteomics as well as from small-scale to proteome-wide targeted quantification. With further advances in instrumentation and bioinformatics tools, we anticipate that targeted proteomics quantification will become a routinely accessible protein measurement tool for enabling cost-effective, rapid, robust quantification of target proteins including low-abundance proteins, isoforms and PTMs at a proteome scale in a high throughput manner. This could lead to significantly improving our understanding of aberrant signal transduction networks underlying human diseases for identification of rational therapeutic targets as well as accelerating biomarker verification for timely translating promising biomarkers into clinical use.
Acknowledgments
Portions of the research were supported by NIH Grants P41GM103493, Y01CN0501329, U24CA160019, and UC4DK104167. The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the DOE under Contract DE-AC05-76RL0 1830.
Abbreviations
- AP
affinity purification
- DDA
data-dependent acquisition
- DIA
data-independent acquisition
- HILIC
hydrophilic interaction liquid chromatography
- HR/AM
high-resolution accurate-mass
- LG
long gradient
- MRM
multiple reaction monitoring
- MSIA
mass spectrometric immunoassay
- PRISM
high-pressure, high-resolution separations with intelligent selection and multiplexing
- PRM
parallel reaction monitoring
- QqOrbi
quadrupole-Orbitrap
- QqQ
triple quadrupole mass spectrometer
- QqTOF
quadrupole-TOF
- RP
reversed-phase
- SIM
selectedion monitoring
- SISCAPA
stable isotope standard and capture by the anti-peptide antibody
- SRM
selected reaction monitoring
- SWATH
sequential windowed acquisition of all theoretical fragment ion mass spectra
Footnotes
The authors have declared no conflict of interest.
References
- 1.Zhou F, Lu Y, Ficarro SB, Adelmant G, et al. Genome-scale proteome quantification by DEEP SEQ mass spec-trometry. Nat. Commun. 2013;4:2171. doi: 10.1038/ncomms3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MS, Pinto SM, Getnet D, Nirujogi RS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 4.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphrey SJ, Azimifar SB, Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 6.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nature Rev. Genetics. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Y, Zhang C, Croucher DR, Soliman MA, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Fillmore TL, Schepmoes AA, Clauss TR, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J. Exp. Med. 2013;210:191–203. doi: 10.1084/jem.20111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keshishian H, Burgess MW, Gillette MA, Mertins P, et al. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol. Cell. Proteomics. 2015;14:2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 11.Gillet LC, Navarro P, Tate S, Rost H, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomark Insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BT, Liew L, Lim J, Tan JK, et al. Candidate List of yoUr Biomarker (CLUB): A Web-based Platform to Aid Cancer Biomarker Research. Biomark Insights. 2008;3:65–71. doi: 10.4137/bmi.s467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin. Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 16.Anderson L. Six decades searching for meaning in the proteome. J. Proteomics. 2014;107:24–30. doi: 10.1016/j.jprot.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Shi T, Su D, Liu T, Tang K, et al. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12:1074–1092. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi T, Fillmore TL, Sun X, Zhao R, et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 20.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson AC, Russell JD, Bailey DJ, Westphall MS, et al. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallien S, Duriez E, Crone C, Kellmann M, et al. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteomics. 2012;11:1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: time to turn the tables? Mol. Cell. Proteomics. 2013;12:2381–2382. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schilling B, MacLean B, Held JM, Sahu AK, et al. Multiplexed, scheduled, high-resolution parallel reaction monitoring on a full scan qqtof instrument with integrated data-dependent and targeted mass spectrometric work-flows. Anal. Chem. 2015;87:10222–10229. doi: 10.1021/acs.analchem.5b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domon B, Gallien S. Recent advances in targeted proteomics for clinical applications. Proteomics Clin. Appl. 2015;9:423–431. doi: 10.1002/prca.201400136. [DOI] [PubMed] [Google Scholar]
- 27.Gallien S, Kim SY, Domon B. Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM). Mol. Cell. Proteomics. 2015;14:1630–1644. doi: 10.1074/mcp.O114.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallien S, Bourmaud A, Kim SY, Domon B. Technical considerations for large-scale parallel reaction monitoring analysis. J. Proteomics. 2014;100:147–159. doi: 10.1016/j.jprot.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 29.Rost HL, Rosenberger G, Navarro P, Gillet L, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 30.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addona TA, Abbatiello SE, Schilling B, Skates SJ, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol. Cell. Proteomics. 2012;11:M111 013854. doi: 10.1074/mcp.M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy JJ, Abbatiello SE, Kim K, Yan P, et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat. Methods. 2014;11:149–155. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbatiello SE, Mani DR, Schilling B, Maclean B, et al. Design, implementation and multisite evaluation of a system suitability protocol for the quantitative assessment of instrument performance in liquid chromatography-multiple reaction monitoring-MS (LC-MRM-MS). Mol. Cell. Proteomics. 2013;12:2623–2639. doi: 10.1074/mcp.M112.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruppen-Canas I, Lopez-Casas PP, Garcia F, Ximenez-Embun P, et al. An improved quantitative mass spectrometry analysis of tumor specific mutant proteins at high sensitivity. Proteomics. 2012;12:1319–1327. doi: 10.1002/pmic.201100611. [DOI] [PubMed] [Google Scholar]
- 36.Katafuchi T, Esterhazy D, Lemoff A, Ding X, et al. Detection of FGF15 in plasma by stable isotope standards and capture by anti-peptide antibodies and targeted mass spec-trometry. Cell metabolism. 2015;21:898–904. doi: 10.1016/j.cmet.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteaker JR, Zhao L, Lin C, Yan P, et al. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol. Cell. Proteomics. 2012;11:M111 015347. doi: 10.1074/mcp.M111.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krastins B, Prakash A, Sarracino DA, Nedelkov D, et al. Rapid development of sensitive, high-throughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clin. Biochem. 2013;46:399–410. doi: 10.1016/j.clinbiochem.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiteaker JR, Zhao L, Frisch C, Ylera F, et al. High-affinity recombinant antibody fragments (Fabs) can be applied in peptide enrichment immuno-MRM assays. J. Proteome Res. 2014;13:2187–2196. doi: 10.1021/pr4009404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteaker JR, Zhao L, Yan P, Ivey RG, et al. Peptide immunoaffinity enrichment and targeted mass spectrometry enables multiplex, quantitative pharmacodynamic studies of phospho-signaling. Mol. Cell. Proteomics. 2015;14:2261–2273. doi: 10.1074/mcp.O115.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neubert H, Muirhead D, Kabir M, Grace C, et al. Sequential protein and peptide immunoaffinity capture for mass spectrometry-based quantification of total human beta-nerve growth factor. Anal. Chem. 2013;85:1719–1726. doi: 10.1021/ac303031q. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Trudeau B, Xie H, Prostko J, et al. Epitope mapping and targeted quantitation of the cardiac biomarker troponin by SID-MRM mass spectrometry. Proteomics. 2014;14:1311–1321. doi: 10.1002/pmic.201300150. [DOI] [PubMed] [Google Scholar]
- 43.Xu Q, Zhu M, Yang T, Xu F, et al. Quantitative assessment of human serum transferrin receptor in breast cancer patients pre- and post-chemotherapy using peptide immunoaffinity enrichment coupled with targeted proteomics. Clin. Chim. Acta. 2015;448:118–123. doi: 10.1016/j.cca.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Gauthier MS, Perusse JR, Awan Z, Bouchard A, et al. A semi-automated mass spectrometric immunoassay coupled to selected reaction monitoring (MSIA-SRM) reveals novel relationships between circulating PCSK9 and metabolic phenotypes in patient cohorts. Methods. 2015;81:66–73. doi: 10.1016/j.ymeth.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Gold L, Ayers D, Bertino J, Bock C, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostroff RM, Bigbee WL, Franklin W, Gold L, et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS One. 2010;5:e15003. doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraemer S, Vaught JD, Bock C, Gold L, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostroff RM, Mehan MR, Stewart A, Ayers D, et al. Early detection of malignant pleural mesothelioma in asbestos-exposed individuals with a noninvasive proteomics-based surveillance tool. PLoS One. 2012;7:e46091. doi: 10.1371/journal.pone.0046091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Groote MA, Nahid P, Jarlsberg L, Johnson JL, et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One. 2013;8:e61002. doi: 10.1371/journal.pone.0061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiddle SJ, Sattlecker M, Proitsi P, Simmons A, et al. Candidate blood proteome markers of Alzheimer's disease onset and progression: a systematic review and replication study. J. Alzheimers Dis. 2014;38:515–531. doi: 10.3233/JAD-130380. [DOI] [PubMed] [Google Scholar]
- 51.Hathout Y, Brody E, Clemens PR, Cripe L, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7153–7158. doi: 10.1073/pnas.1507719112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehan MR, Williams SA, Siegfried JM, Bigbee WL, et al. Validation of a blood protein signature for non-small cell lung cancer. Clinical proteomics. 2014;11:32. doi: 10.1186/1559-0275-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Widen SG, Jamaluddin M, Tian B, et al. Quantification of activated NF-kappaB/RelA complexes using ss-DNA aptamer affinity-stable isotope dilution-selected reaction monitoring-mass spectrometry. Mol. Cell. Proteomics. 2011;10:M111 008771. doi: 10.1074/mcp.M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simicevic J, Schmid AW, Gilardoni PA, Zoller B, et al. Absolute quantification of transcription factors during cellular differentiation using multiplexed targeted proteomics. Nature Methods. 2013;10:570–576. doi: 10.1038/nmeth.2441. [DOI] [PubMed] [Google Scholar]
- 55.Such-Sanmartin G, Bache N, Callesen AK, Rogowska-Wrzesinska A, et al. Targeted mass spectrometry analysis of the proteins IGF1, IGF2, IBP2, IBP3 and A2GL by blood protein precipitation. J. Proteomics. 2015;113:29–37. doi: 10.1016/j.jprot.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 56.Halvey PJ, Ferrone CR, Liebler DC. GeLC-MRM quantitation of mutant KRAS oncoprotein in complex biological samples. J. Proteome Res. 2012;11:3908–3913. doi: 10.1021/pr300161j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sano S, Tagami S, Hashimoto Y, Yoshizawa-Kumagaye K, et al. Absolute quantitation of low abundance plasma apl1 beta peptides at sub-fmol/ml level by srm/mrm without immunoaffinity enrichment. J. Proteome Res. 2014;13:1012–1020. doi: 10.1021/pr4010103. [DOI] [PubMed] [Google Scholar]
- 58.Shi T, Sun X, Gao Y, Fillmore TL, et al. Targeted quantification of low ng/mL level proteins in human serum without immunoaffinity depletion. J. Proteome Res. 2013;12:3353–3361. doi: 10.1021/pr400178v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi T, Gao Y, Quek SI, Fillmore TL, et al. A Highly sensitive targeted mass spectrometric assay for quantification of agr2 protein in human urine and serum. J. Proteome Res. 2014;13:875–882. doi: 10.1021/pr400912c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He J, Sun X, Shi T, Schepmoes AA, et al. Antibody-independent targeted quantification of TMPRSS2-ERG fusion protein products in prostate cancer. Molecular Oncology. 2014;8:1169–1180. doi: 10.1016/j.molonc.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He J, Schepmoes AA, Shi T, Wu C, et al. Analytical platform evaluation for quantification of ERG in prostate cancer using protein and mRNA detection methods. J. Transl. Med. 2015;13:54. doi: 10.1186/s12967-015-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi TJ, Gao YQ, Gaffrey MJ, Nicora CD, et al. Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal Chem. 2015;87:1103–1110. doi: 10.1021/ac503797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim JS, Ahn HS, Cho SM, Lee JE, et al. Detection and quantification of plasma amyloid-beta by selected reaction monitoring mass spectrometry. Anal. Chim. Acta. 2014;840:1–9. doi: 10.1016/j.aca.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 64.Percy AJ, Simon R, Chambers AG, Borchers CH. Enhanced sensitivity and multiplexing with 2D LC/MRM-MS and labeled standards for deeper and more comprehensive protein quantitation. J. Proteomics. 2014;106:113–124. doi: 10.1016/j.jprot.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 65.Keshishian H, Addona T, Burgess M, Kuhn E, et al. multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J. Sep. Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Yang F, Gritsenko MA, Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon R, Passeron S, Lemoine J, Salvador A. Hydrophilic interaction liquid chromatography as second dimension in multidimensional chromatography with an anionic trapping strategy: application to prostate-specific antigen quantification. J. Chromatogr A. 2014;1354:75–84. doi: 10.1016/j.chroma.2014.05.063. [DOI] [PubMed] [Google Scholar]
- 69.Shi T, Fillmore TL, Gao Y, Zhao R, et al. Long-gradient separations coupled with selected reaction monitoring for highly sensitive, large scale targeted protein quantification in a single analysis. Anal. Chem. 2013;85:9196–9203. doi: 10.1021/ac402105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burgess MW, Keshishian H, Mani DR, Gillette MA, et al. Simplified and efficient quantification of low-abundance proteins at very high multiplex via targeted mass spectrometry. Mol. Cell. Proteomics. 2014;13:1137–1149. doi: 10.1074/mcp.M113.034660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi T, Zhou JY, Gritsenko MA, Hossain M, et al. IgY14 and SuperMix immunoaffinity separations coupled with liquid chromatography-mass spectrometry for human plasma proteomics biomarker discovery. Methods. 2012;56:246–253. doi: 10.1016/j.ymeth.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mani DR, Abbatiello SE, Carr SA. Statistical characterization of multiple-reaction monitoring mass spectrometry (MRM-MS) assays for quantitative proteomics. BMC Bioinformatics. 2012;13(Suppl 16):S9. doi: 10.1186/1471-2105-13-S16-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohammed Y, Percy AJ, Chambers AG, Borchers CH. Qualis-SIS: automated standard curve generation and quality assessment for multiplexed targeted quantitative proteomic experiments with labeled standards. J. Proteome Res. 2015;14:1137–1146. doi: 10.1021/pr5010955. [DOI] [PubMed] [Google Scholar]
- 74.Percy AJ, Tamura-Wells J, Albar JP, Aloria K, et al. Inter-laboratory evaluation of instrument platforms and experimental workflows for quantitative accuracy and reproducibility assessment. EuPA Open Proteomics. 2015;8:6–15. [Google Scholar]
- 75.Abbatiello SE, Schilling B, Mani DR, Zimmerman LJ, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol. Cell. Proteomics. 2015;14:2357–2374. doi: 10.1074/mcp.M114.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Method of the Year 2012. Nat. Methods. 2013;10:1. doi: 10.1038/nmeth.2329. [DOI] [PubMed] [Google Scholar]
- 78.Ebhardt HA, Root A, Sander C, Aebersold R. Applications of targeted proteomics in systems biology and translational medicine. Proteomics. 2015;15:3193–3208. doi: 10.1002/pmic.201500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Picotti P, Bodenmiller B, Aebersold R. Proteomics meets the scientific method. Nat. Methods. 2013;10:24–27. doi: 10.1038/nmeth.2291. [DOI] [PubMed] [Google Scholar]
- 80.Shi T, Qian WJ. Antibody-free PRISM-SRM for multiplexed protein quantification: is this the new competition for immunoassays in bioanalysis? Bioanalysis. 2013;5:267–269. doi: 10.4155/bio.12.336. [DOI] [PubMed] [Google Scholar]
- 81.Diamandis EP. Towards identification of true cancer biomarkers. BMC medicine. 2014;12:156. doi: 10.1186/s12916-014-0156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodland KD. As if biomarker discovery isn't hard enough: the consequences of poorly characterized reagents. Clin. Chem. 2014;60:290–291. doi: 10.1373/clinchem.2013.216382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu T, Hossain M, Schepmoes AA, Fillmore TL, et al. Analysis of serum total and free PSA using immunoaffinity depletion coupled to SRM: correlation with clinical immunoassay tests. J. Proteomics. 2012;75:4747–4757. doi: 10.1016/j.jprot.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huillet C, Adrait A, Lebert D, Picard G, et al. Accurate quantification of cardiovascular biomarkers in serum using Protein Standard Absolute Quantification (PSAQ) and selected reaction monitoring. Mol. Cell. Proteomics. 2012;11:M111 008235. doi: 10.1074/mcp.M111.008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin D, Alborn WE, Slebos RJ, Liebler DC. Comparison of protein immunoprecipitation-multiple reaction monitoring with ELISA for assay of biomarker candidates in plasma. J. Proteome Res. 2013;12:5996–6003. doi: 10.1021/pr400877e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomas SN, Harlan R, Chen J, Aiyetan P, et al. Multiplexed targeted mass spectrometry-based assays for the quantification of n-linked glycosite-containing peptides in serum. Anal. Chem. 2015;87:10830–10838. doi: 10.1021/acs.analchem.5b02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim YJ, Sertamo K, Pierrard MA, Mesmin C, et al. Verification of the biomarker candidates for non-small-cell lung cancer using a targeted proteomics approach. J. Proteome Res. 2015;14:1412–1419. doi: 10.1021/pr5010828. [DOI] [PubMed] [Google Scholar]
- 88.Kim YJ, Gallien S, El-Khoury V, Goswami P, et al. Quantification of SAA1 and SAA2 in lung cancer plasma using the isotype-specific PRM assays. Proteomics. 2015;15:3116–3125. doi: 10.1002/pmic.201400382. [DOI] [PubMed] [Google Scholar]
- 89.Sung HJ, Jeon SA, Ahn JM, Seul KJ, et al. Large-scale isotype-specific quantification of Serum amyloid A 1/2 by multiple reaction monitoring in crude sera. J. Proteomics. 2012;75:2170–2180. doi: 10.1016/j.jprot.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 90.Birse CE, Lagier RJ, FitzHugh W, Pass HI, et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clinical Proteomics. 2015;12:18. doi: 10.1186/s12014-015-9090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li XJ, Hayward C, Fong PY, Dominguez M, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci. Transl. Med. 2013;5:207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen CD, Wang CL, Yu CJ, Chien KY, et al. Targeted proteomics pipeline reveals potential biomarkers for the diagnosis of metastatic lung cancer in pleural effusion. J. Proteome Res. 2014;13:2818–2829. doi: 10.1021/pr4012377. [DOI] [PubMed] [Google Scholar]
- 93.Kume H, Muraoka S, Kuga T, Adachi J, et al. Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using selected reaction monitoring (SRM) and tissue microarray (TMA) analysis. Mol. Cell. Proteomics. 2014;13:1471–1484. doi: 10.1074/mcp.M113.037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Surinova S, Choi M, Tao S, Schuffler PJ, et al. Prediction of colorectal cancer diagnosis based on circulating plasma proteins. Embo Mol. Med. 2015;7:1166–1178. doi: 10.15252/emmm.201404873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Surinova S, Radova L, Choi M, Srovnal J, et al. Noninvasive prognostic protein biomarker signatures associated with colorectal cancer. Embo Mol. Med. 2015;7:1153–1165. doi: 10.15252/emmm.201404874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoneyama T, Ohtsuki S, Ono M, Ohmine K, et al. Quantitative targeted absolute proteomics-based large-scale quantification of proline-hydroxylated alpha-fibrinogen in plasma for pancreatic cancer diagnosis. J. Proteome Res. 2013;12:753–762. doi: 10.1021/pr3008144. [DOI] [PubMed] [Google Scholar]
- 97.Pan S, Chen R, Brand RE, Hawley S, et al. Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J. Proteome Res. 2012;11:1937–1948. doi: 10.1021/pr201117w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan C, Zhou Y, Dator R, Ginghina C, et al. Targeted discovery and validation of plasma biomarkers of Parkinson's disease. J. Proteome Res. 2014;13:4535–4545. doi: 10.1021/pr500421v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi M, Movius J, Dator R, Aro P, et al. Cerebrospinal fluid peptides as potential parkinson disease biomarkers: a staged pipeline for discovery and validation. Mol. Cell. Proteomics. 2015;14:544–555. doi: 10.1074/mcp.M114.040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alberio T, McMahon K, Cuccurullo M, Gethings LA, et al. Verification of a Parkinson's disease protein signature in T-lymphocytes by multiple reaction monitoring. J. Proteome Res. 2014;13:3554–3561. doi: 10.1021/pr401142p. [DOI] [PubMed] [Google Scholar]
- 101.Oeckl P, Steinacker P, von Arnim CAF, Straub S, et al. Intact protein analysis of ubiquitin in cerebrospinal fluid by multiple reaction monitoring reveals differences in alzheimer's disease and frontotemporal lobar degeneration. J. Proteome Res. 2014;13:4518–4525. doi: 10.1021/pr5006058. [DOI] [PubMed] [Google Scholar]
- 102.Martinez-Morillo E, Cho CKJ, Drabovich AP, Shaw JLV, et al. Development of a multiplex selected reaction monitoring assay for quantification of biochemical markers of down syndrome in amniotic fluid samples. J. Proteome Res. 2012;11:3880–3887. doi: 10.1021/pr300355a. [DOI] [PubMed] [Google Scholar]
- 103.Cho CK, Drabovich AP, Batruch I, Diamandis EP. Verification of a biomarker discovery approach for detection of Down syndrome in amniotic fluid via multiplex selected reaction monitoring (SRM) assay. J. Proteomics. 2011;74:2052–2059. doi: 10.1016/j.jprot.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 104.Heywood W, Wang D, Madgett TE, Avent ND, et al. The development of a peptide SRM-based tandem mass spectrometry assay for prenatal screening of Down syndrome. J. Proteomics. 2012;75:3248–3257. doi: 10.1016/j.jprot.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 105.Qin S, Zhou Y, Lok AS, Tsodikov A, et al. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics. 2012;12:1244–1252. doi: 10.1002/pmic.201100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cretu D, Prassas I, Saraon P, Batruch I, et al. Identification of psoriatic arthritis mediators in synovial fluid by quantitative mass spectrometry. Clinical proteomics. 2014;11:27. doi: 10.1186/1559-0275-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia Y, Wu T, Jelinek CA, Bielekova B, et al. Development of protein biomarkers in cerebrospinal fluid for secondary progressive multiple sclerosis using selected reaction monitoring mass spectrometry (SRM-MS). Clinical Proteomics. 2012;9:9. doi: 10.1186/1559-0275-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cerciello F, Choi M, Nicastri A, Bausch-Fluck D, et al. Identification of a seven glycopeptide signature for malignant pleural mesothelioma in human serum by selected reaction monitoring. Clin. Proteomics. 2013;10:16. doi: 10.1186/1559-0275-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ahn YH, Shin PM, Oh NR, Park GW, et al. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J. Proteomics. 2012;75:5507–5515. doi: 10.1016/j.jprot.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 110.Huttenhain R, Soste M, Selevsek N, Rost H, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci. Transl. Med. 2012;4:142ra194. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Percy AJ, Chambers AG, Yang J, Borchers CH. Multiplexed MRM-based quantitation of candidate cancer biomarker proteins in undepleted and non-enriched human plasma. Proteomics. 2013;13:2202–2215. doi: 10.1002/pmic.201200316. [DOI] [PubMed] [Google Scholar]
- 112.Vegvari A, Sjodin K, Rezeli M, Marko-Varga G. Quantification of human kallikrein-2 in clinical samples by selected reaction monitoring. J. Proteome Res. 2013;12:4612–4616. doi: 10.1021/pr400423k. [DOI] [PubMed] [Google Scholar]
- 113.Percy AJ, Yang J, Chambers AG, Simon R, et al. Multiplexed MRM with internal standards for cerebrospinal fluid candidate protein biomarker quantitation. J. Proteome Res. 2014;13:3733–3747. doi: 10.1021/pr500317d. [DOI] [PubMed] [Google Scholar]
- 114.Fallon JK, Neubert H, Hyland R, Goosen TC, et al. Targeted quantitative proteomics for the analysis of 14 UGT1As and -2Bs in human liver using NanoUPLC-MS/MS with selected reaction monitoring. J. Proteome Res. 2013;12:4402–4413. doi: 10.1021/pr4004213. [DOI] [PubMed] [Google Scholar]
- 115.Martinez-Marquez A, Morante-Carriel J, Selles-Marchart S, Martinez-Esteso MJ, et al. Development and validation of MRM methods to quantify protein isoforms of polyphenol oxidase in loquat fruits. J. Proteome Res. 2013;12:5709–5722. doi: 10.1021/pr4006712. [DOI] [PubMed] [Google Scholar]
- 116.Martinez-Morillo E, Nielsen HM, Batruch I, Drabovich AP, et al. Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein e iso-forms. J. Proteome Res. 2014;13:1077–1087. doi: 10.1021/pr401060x. [DOI] [PubMed] [Google Scholar]
- 117.Vegvari A, Sjodin K, Rezeli M, Malm J, et al. Identification of a novel proteoform of prostate specific antigen (SNP-L132I) in clinical samples by multiple reaction monitoring. Mol. Cell. Proteomics. 2013;12:2761–2773. doi: 10.1074/mcp.M113.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plumel MI, Wasselin T, Plot V, Strub JM, et al. Mass spectrometry-based sequencing and srm-based quantitation of two novel vitellogenin isoforms in the leatherback sea turtle (Dermochelys coriacea). J. Proteome Res. 2013;12:4122–4135. doi: 10.1021/pr400444m. [DOI] [PubMed] [Google Scholar]
- 119.Sabido E, Wu YB, Bautista L, Porstmann T, et al. Targeted proteomics reveals strain-specific changes in the mouse insulin and central metabolic pathways after a sustained high-fat diet. Mol. Syst. Biol. 2013;9:681. doi: 10.1038/msb.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Graaf EL, Kaplon J, Mohammed S, Vereijken LAM, et al. Signal transduction reaction monitoring deciphers site-specific pi3k-mtor/mapk pathway dynamics in onaigene-induced senescence. J. Proteome Res. 2015;14:2906–2914. doi: 10.1021/acs.jproteome.5b00236. [DOI] [PubMed] [Google Scholar]
- 121.Drabovich AP, Pavlou MP, Dimitromanolakis A, Diamandis EP. Quantitative analysis of energy metabolic pathways in mcf-7 breast cancer cells by selected reaction monitoring assay. Mol. Cell. Proteomics. 2012;11:422–434. doi: 10.1074/mcp.M111.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Krieger JR, Taylor P, Gajadhar AS, Guha A, et al. Identification and selected reaction monitoring (srm) quantification of endocytosis factors associated with numb. Mol. Cell. Proteomics. 2013;12:499–514. doi: 10.1074/mcp.M112.020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao YX, Tian B, Edeh CB, Brasier AR. Quantitation of the dynamic profiles of the innate immune response using multiplex selected reaction monitoring-mass spec-trometry. Mol. Cell. Proteomics. 2013;12:1513–1529. doi: 10.1074/mcp.M112.023465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tong JF, Taylor P, Moran MF. Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol. Cell. Proteomics. 2014;13:1644–1658. doi: 10.1074/mcp.M114.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rebecca VW, Wood E, Fedorenko IV, Paraiso KH, et al. Evaluating melanoma drug response and therapeutic escape with quantitative proteomics. Mol. Cell. Proteomics. 2014;13:1844–1854. doi: 10.1074/mcp.M113.037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Manes NP, Angermann BR, Koppenol-Raab M, An E, et al. Targeted proteomics-driven computational modeling of macrophage S1P chemosensing. Mol. Cell. Proteomics. 2015;14:2661–2681. doi: 10.1074/mcp.M115.048918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kiel C, Ebhardt HA, Burnier J, Portugal C, et al. Quantification of ErbB network proteins in three cell types using complementary approaches identifies cell-general and cell-type-specific signaling proteins. J. Proteome Res. 2014;13:300–313. doi: 10.1021/pr400878x. [DOI] [PubMed] [Google Scholar]
- 128.Sangar V, Funk CC, Kusebauch U, Campbell DS, et al. Quantitative proteomic analysis reveals effects of epidermal growth factor receptor (EGFR) on invasion-promoting proteins secreted by glioblastoma cells. Mol. Cell. Proteomics. 2014;13:2618–2631. doi: 10.1074/mcp.M114.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tian R, Jin J, Taylor L, Larsen B, et al. Rapid and sensitive MRM-based mass spectrometry approach for systematically exploring ganglioside-protein interactions. Proteomics. 2013;13:1334–1338. doi: 10.1002/pmic.201200410. [DOI] [PubMed] [Google Scholar]
- 130.Altvater M, Chang YM, Melnik A, Occhipinti L, et al. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Molecular Systems Biology. 2012;8:628. doi: 10.1038/msb.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mirzaei H, Knijnenburg TA, Kim B, Robinson M, et al. Systematic measurement of transcription factor-DNA interactions by targeted mass spectrometry identifies candidate gene regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3645–3650. doi: 10.1073/pnas.1216918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Agard NJ, Mahrus S, Trinidad JC, Lynn A, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Soste M, Hrabakova R, Wanka S, Melnik A, et al. A sentinel protein assay for simultaneously quantifying cellular processes. Nat. Methods. 2014;11:1045–1048. doi: 10.1038/nmeth.3101. [DOI] [PubMed] [Google Scholar]
- 134.Feng Y, De Franceschi G, Kahraman A, Soste M, et al. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 135.Gallien S, Duriez E, Demeure K, Domon B. Selectivity of LC-MS/MS analysis: implication for proteomics experiments. J. Proteomics. 2013;81:148–158. doi: 10.1016/j.jprot.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 136.Schiffmann C, Hansen R, Baumann S, Kublik A, et al. Comparison of targeted peptide quantification assays for reductive dehalogenases by selective reaction monitoring (SRM) and precursor reaction monitoring (PRM). Anal. Bioanal. Chem. 2014;406:283–291. doi: 10.1007/s00216-013-7451-7. [DOI] [PubMed] [Google Scholar]
- 137.Ronsein GE, Pamir N, von Haller PD, Kim DS, et al. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics. 2015;113:388–399. doi: 10.1016/j.jprot.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Majovsky P, Naumann C, Lee CW, Lassowskat I, et al. Targeted proteomics analysis of protein degradation in plant signaling on an LTQ-Orbitrap mass spectrometer. J. Proteome Res. 2014;13:4246–4258. doi: 10.1021/pr500164j. [DOI] [PubMed] [Google Scholar]
- 139.Morin LP, Mess JN, Garofolo F. Large-molecule quantification: sensitivity and selectivity head-to-head comparison of triple quadrupole with Q-TOF. Bioanalysis. 2013;5:1181–1193. doi: 10.4155/bio.13.87. [DOI] [PubMed] [Google Scholar]
- 140.Gallien S, Domon B. Detection and quantification of proteins in clinical samples using high resolution mass spec-trometry. Methods. 2015;81:15–23. doi: 10.1016/j.ymeth.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 141.Scheltema RA, Hauschild JP, Lange O, Hornburg D, et al. The Q Exactive HF, a benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field orbitrap analyzer. Mol. Cell. Proteomics. 2014;13:3698–3708. doi: 10.1074/mcp.M114.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gallien S, Domon B. Quantitative proteomics using the high resolution accurate mass capabilities of the quadrupole-orbitrap mass spectrometer. Bioanalysis. 2014;6:2159–2170. doi: 10.4155/bio.14.115. [DOI] [PubMed] [Google Scholar]
- 143.Lesur A, Domon B. Advances in high-resolution accurate mass spectrometry application to targeted proteomics. Proteomics. 2015;15:880–890. doi: 10.1002/pmic.201400450. [DOI] [PubMed] [Google Scholar]
- 144.Lesur A, Ancheva L, Kim YJ, Berchem G, et al. Screening protein isoforms predictive for cancer using immunoaffinity capture and fast LC-MS in PRM mode. Proteomics Clin. Appl. 2015;9:695–705. doi: 10.1002/prca.201400158. [DOI] [PubMed] [Google Scholar]
- 145.Kim HJ, Lin D, Li M, Liebler DC. Quantitative profiling of protein tyrosine kinases in human cancer cell lines by multiplexed parallel reaction monitoring assays. Mol. Cell. Proteomics. 2016;15:682–691. doi: 10.1074/mcp.O115.056713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang H, Fang H, Yin E, Brasier AR, et al. Multiplexed parallel reaction monitoring targeting histone modifications on the QExactive mass spectrometer. Anal. Chem. 2014;86:5526–5534. doi: 10.1021/ac500972x. [DOI] [PubMed] [Google Scholar]
- 147.Sowers JL, Mirfattah B, Xu P, Tang H, et al. Quantification of Histone Modifications by Parallel-Reaction Monitoring: A Method Validation. Anal. Chem. 2015;87:10006–10014. doi: 10.1021/acs.analchem.5b02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tsuchiya H, Tanaka K, Saeki Y. The parallel reaction monitoring method contributes to a highly sensitive polyubiquitin chain quantification. Biochem. Biophys. Res. Commun. 2013;436:223–229. doi: 10.1016/j.bbrc.2013.05.080. [DOI] [PubMed] [Google Scholar]
- 149.Liu Y, Huttenhain R, Collins B, Aebersold R. Mass spec-trometric protein maps for biomarker discovery and clinical research. Exp. Rev. Mol. Diagn. 2013;13:811–825. doi: 10.1586/14737159.2013.845089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sajic T, Liu YS, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: Perspectives and clinical applications. Proteom Clin. Appl. 2015;9:307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 151.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 152.Silva JC, Denny R, Dorschel CA, Gorenstein M, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 153.Panchaud A, Scherl A, Shaffer SA, von Haller PD, et al. Precursor acquisition independent from ion count: how to dive deeper into the proteomics Ocean. Anal. Chem. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Panchaud A, Jung S, Shaffer SA, Aitchison JD, Goodlett DR. Faster, quantitative, and accurate precursor acquisition independent from ion count. Anal. Chem. 2011;83:2250–2257. doi: 10.1021/ac103079q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Geiger T, Cox J, Mann M. Proteomics on an orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics. 2010;9:2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Carvalho PC, Han XM, Xu T, Cociorva D, et al. XDIA: improving on the label-free data-independent analysis. Bioinformatics. 2010;26:847–848. doi: 10.1093/bioinformatics/btq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weisbrod CR, Eng JK, Hoopmann MR, Baker T, et al. Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 2012;11:1621–1632. doi: 10.1021/pr2008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Egertson JD, Kuehn A, Merrihew GE, Bateman NW, et al. Multiplexed MS/MS for improved data-independent acquisition. Nature Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Prakash A, Peterman S, Ahmad S, Sarracino D, et al. Hybrid data acquisition and processing strategies with increased throughput and selectivity: pSMART analysis for global qualitative and quantitative analysis. J. Proteome Res. 2014;13:5415–5430. doi: 10.1021/pr5003017. [DOI] [PubMed] [Google Scholar]
- 160.Kiyonami Reiko, Patel Bhavin, Senko Michael, Zabrouskov Vlad, et al. Large scale targeted protein quantification using WiSIM-DIA workflow on a orbitrap fusion tribrid mass spectrometer. Thermo Scientific Application Note 600. 2014 [Google Scholar]
- 161.Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtis-sues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bilbao A, Varesio E, Luban J, Strambio-De-Castillia C, et al. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics. 2015;15:964–980. doi: 10.1002/pmic.201400323. [DOI] [PubMed] [Google Scholar]
- 163.Bern M, Finney G, Hoopmann MR, Merrihew G, et al. Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal. Chem. 2010;82:833–841. doi: 10.1021/ac901801b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ledvina AR, Savitski MM, Zubarev AR, Good DM, et al. Increased throughput of proteomics analysis by multiplexing high-resolution tandem mass spectra. Anal. Chem. 2011;83:7651–7656. doi: 10.1021/ac201843e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pak H, Nikitin F, Gluck F, Lisacek F, et al. Clustering and filtering tandem mass spectra acquired in data-independent mode. J. Am. Soc. Mass Spectrom. 2013;24:1862–1871. doi: 10.1007/s13361-013-0720-z. [DOI] [PubMed] [Google Scholar]
- 166.Kryuchkov F, Verano-Braga T, Hansen TA, Sprenger RR, et al. Deconvolution of mixture spectra and increased throughput of peptide identification by utilization of intensified complementary ions formed in tandem mass spec-trometry. J. Proteome Res. 2013;12:3362–3371. doi: 10.1021/pr400210m. [DOI] [PubMed] [Google Scholar]
- 167.Kim S, Mischerikow N, Bandeira N, Navarro JD, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 169.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 170.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 171.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 172.Tsou CC, Avtonomov D, Larsen B, Tucholska M, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nature Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang N, Li XJ, Ye M, Pan S, et al. ProbIDtree: an automated software program capable of identifying multiple peptides from a single collision-induced dissociation spectrum collected by a tandem mass spectrometer. Proteomics. 2005;5:4096–4106. doi: 10.1002/pmic.200401260. [DOI] [PubMed] [Google Scholar]
- 174.Li GZ, Vissers JP, Silva JC, Golick D, et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 175.Wang J, Perez-Santiago J, Katz JE, Mallick P, Bandeira N. Peptide identification from mixture tandem mass spectra. Mol. Cell. Proteomics. 2010;9:1476–1485. doi: 10.1074/mcp.M000136-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang J, Bourne PE, Bandeira N. Peptide identification by database search of mixture tandem mass spectra. Mol. Cell. Proteomics. 2011;10:M111 010017. doi: 10.1074/mcp.M111.010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Egertson JD, MacLean B, Johnson R, Xuan Y, et al. Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat. Protoc. 2015;10:887–903. doi: 10.1038/nprot.2015.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reiter L, Rinner O, Picotti P, Huttenhain R, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 179.Teleman J, Rost HL, Rosenberger G, Schmitt U, et al. DIANA-algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2015;31:555–562. doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- 180.Liu Y, Huttenhain R, Surinova S, Gillet LC, et al. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 181.Guo T, Kouvonen P, Koh CC, Gillet LC, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nature Medicine. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Selevsek N, Chang CY, Gillet LC, Navarro P, et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae Proteome by SWATH-mass spectrometry. Mol. Cell. Proteomics. 2015;14:739–749. doi: 10.1074/mcp.M113.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Collins BC, Gillet LC, Rosenberger G, Rost HL, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 184.Lambert JP, Ivosev G, Couzens AL, Larsen B, et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nature Methods. 2013;10:1239–1245. doi: 10.1038/nmeth.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu YS, Chen J, Sethi A, Li QK, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers n-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol. Cell. Proteomics. 2014;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Krautkramer KA, Reiter L, Denu JM, Dowell JA. Quantification of SAHA-dependent changes in his-tone modifications using data-independent acquisition mass spectrometry. J. Proteome Res. 2015;14:3252–3262. doi: 10.1021/acs.jproteome.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sidoli S, Lin S, Xiong L, Bhanu NV, et al. Sequential window acquisition of all theoretical mass spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol. Cell. Proteomics. 2015;14:2420–2428. doi: 10.1074/mcp.O114.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gallien S, Domon B. Advances in high-resolution quantitative proteomics: implications for clinical applications. Exp. Rev. Proteomics. 2015;12:489–498. doi: 10.1586/14789450.2015.1069188. [DOI] [PubMed] [Google Scholar]
- 189.Garcia-Bailo B, Brenner DR, Nielsen D, Lee HJ, et al. Dietary patterns and ethnicity are associated with distinct plasma proteomic groups. Am. J. Clin. Nutr. 2012;95:352–361. doi: 10.3945/ajcn.111.022657. [DOI] [PubMed] [Google Scholar]
- 190.Fernandez Ocana M, James IT, Kabir M, Grace C, et al. Clinical pharmacokinetic assessment of an anti-MAdCAM monoclonal antibody therapeutic by LC-MS/MS. Anal. Chem. 2012;84:5959–5967. doi: 10.1021/ac300600f. [DOI] [PubMed] [Google Scholar]
- 191.Razavi M, Frick LE, LaMarr WA, Pope ME, et al. High-throughput SISCAPA quantitation of peptides from human plasma digests by ultrafast, liquid chromatography-free mass spectrometry. J. Proteome Res. 2012;11:5642–5649. doi: 10.1021/pr300652v. [DOI] [PubMed] [Google Scholar]
- 192.Anderson NL, Razavi M, Pearson TW, Kruppa G, et al. Precision of heavy-light peptide ratios measured by malditof mass spectrometry. J. Proteome Res. 2012;11:1868–1878. doi: 10.1021/pr201092v. [DOI] [PubMed] [Google Scholar]
- 193.Razavi M, Johnson LD, Lum JJ, Kruppa G, et al. Quantification of a proteotypic peptide from protein C inhibitor by liquid chromatography-free SISCAPA-MALDI mass spectrometry: application to identification of recurrence of prostate cancer. Clin. Chem. 2013;59:1514–1522. doi: 10.1373/clinchem.2012.199786. [DOI] [PubMed] [Google Scholar]
- 194.Webb IK, Garimella SV, Tolmachev AV, Chen TC, et al. Experimental evaluation and optimization of structures for lossless ion manipulations for ion mobility spectrometry with time-of-flight mass spectrometry. Anal. Chem. 2014;86:9169–9176. doi: 10.1021/ac502055e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Webb IK, Garimella SV, Tolmachev AV, Chen TC, et al. Mobility-resolved ion selection in uniform drift field ion mobility spectrometry/mass spectrometry: dynamic switching in structures for lossless ion manipulations. Anal. Chem. 2014;86:9632–9637. doi: 10.1021/ac502139e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang X, Garimella SV, Prost SA, Webb IK, et al. Ion trapping, storage, and ejection in structures for lossless ion manipulations. Anal. Chem. 2015;87:6010–6016. doi: 10.1021/acs.analchem.5b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Page JS, Tang K, Kelly RT, Smith RD. Subambient pressure ionization with nanoelectrospray source and interface for improved sensitivity in mass spectrometry. Anal. Chem. 2008;80:1800–1805. doi: 10.1021/ac702354b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shen Y, Smith RD, Unger KK, Kumar D, et al. Ultrahigh-throughput proteomics using fast RPLC separations with ESI-MS/MS. Anal. Chem. 2005;77:6692–6701. doi: 10.1021/ac050876u. [DOI] [PubMed] [Google Scholar]
- 199.Shi T, Niepel M, McDermott JE, Gao Y, et al. Conserved protein abundance pattern of the EGFR-MAPK pathway reveals its regulatory architecture. Science Signaling. 2016 doi: 10.1126/scisignal.aaf0891. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mortstedt H, Karedal MH, Jonsson BAG, Lindh CH. Screening method using selected reaction monitoring for targeted proteomics studies of nasal lavage fluid. J. Proteome Res. 2013;12:234–247. doi: 10.1021/pr300802g. [DOI] [PubMed] [Google Scholar]
- 201.Welinder C, Jonsson G, Ingvar C, Lundgren L, et al. Feasibility study on measuring selected proteins in malignant melanoma tissue by SRM quantification. J. Proteome Res. 2014;13:1315–1326. doi: 10.1021/pr400876p. [DOI] [PubMed] [Google Scholar]