Abstract
Plasmodium vivax is the most prevalent cause of human malaria in the world and can lead to severe disease with high potential for relapse. Its genetic and geographic diversities make it challenging to control. P. vivax is understudied and to achieve control of malaria in endemic areas, a rapid, accurate, and simple diagnostic tool is necessary. In this pilot study, we found that a colorimetric system using AuNPs and MSP10 DNA detection in urine can provide fast, easy, and inexpensive identification of P. vivax. The test exhibited promising sensitivity (84%), high specificity (97%), and only mild cross-reactivity with P. falciparum (21%). It is simple to use, with a visible color change that negates the need for a spectrometer, making it suitable for use in austere conditions. Using urine eliminates the need for finger-prick, increasing both the safety profile and patient acceptance of this model.
Author Summary
To control malaria, there is an urgent need for applying innovative diagnostics and new technologies. Nanoparticles can augment detection of malaria at lower parasite levels while providing fast and simple methodology. Novel use of MSP10 and gold nanoparticles to identify Plasmodium vivax’s DNA in urine can be utilized as screening tool with global application potentials. The proposed test could impact the control of the most common species of malaria in low resource settings as it could present a simple, fast, cheap and easy to interpret test. Furthermore, utilizing urine instead of blood eliminates the need for finger-prick which would increase safety profile and likely increase participation rate in mass screening programs.
Introduction
Malaria is the most common infectious disease in the tropics and subtropics [1]. Currently, P. vivax is endemic across Asia, the South Pacific, North Africa, Middle East, and South and Central America [2], and has recently reappeared in regions where it had previously been eradicated, including North America and Europe [3]. Currently, an estimated 2.9 billion people live at risk of P. vivax infection [4]. Research in malaria has been primarily focused on P. falciparum, the most fatal form of malaria. However, P. vivax can also cause severe illness with serious complications and costs, especially in children, in whom it has a major impact on growth [5–7].
Relying on microscopic identification of malaria species jeopardizes malaria control due to its limitations [8,9]. The WHO recognizes a need for rapid, accurate, and easy diagnostic tools in order to control malaria and need for such a test is mounting in developing countries [10]. Currently available rapid diagnostic tests (RDTs) are controversial due to their sensitivity and specificity, and differentiate poorly between plasmodium species [11–13].
With the expanding use of nanotechnology in the biomedical arena, nanoparticles can play a role in low cost, innovative diagnostics [14]. Gold nanoparticles aggregate and change their color from red to purple-blue upon exposure to single stranded DNA in aqueous solution while double stranded DNA stabilize them to preserve their red color and thus present an opportunity to develop a fast and easily interpreted diagnostic test [15–20].
Merozoite Surface Protein 10 (MSP10) is an immunogenic protein encoded by a single copy gene (in P. falciparum, GenBan Accession PF3D7 0620400; in P. vivax, PVX_114145) which is expressed in the asexual blood stages of Plasmodium falciparum and P. vivax [21]. One of at least 10 epidermal growth factor domain-containing proteins, the role of MSP10 in the biology of Plasmodium parasites has yet to be determined. Plasmodium MSP10 proteins have been identified as being subject to positive selection for amino acid-changing polymorphisms at the population genomic level [22,23]. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax [24].
Blood based tests can discourage screening both because of the pain associated with finger-prick and because of social and cultural beliefs about blood sampling. Less painful, more culturally sensitive, and safer tools for malaria diagnosis should encourage participation in mass screening programs and improve public health [25]. Urine contains circulating P. vivax DNA in detectable quantities [26–28] and can therefore serve as a less invasive and more acceptable sample for malaria screening and diagnosis. Also, urine contains less interfering proteins and inhibitors than blood which allows easier DNA extraction [29]. Furthermore, urine provides lower risks to healthcare personnel, with reliable amounts of malaria DNA found in urine despite being substantially lower than blood samples [30]. Additionally, urine color is not expected to obscure color change of gold nanoparticles.
In this pilot study, we tested the hypothesis that a colorimetric system using gold nanoparticles and MSP10 DNA detection in urine would be useful as a safe diagnostic and surveillance tool for P. vivax. Such a tool is needed for improving malaria control in the endemic setting.
Materials and Methods
Chemicals and Reagents
Citrate reduced gold 15nm nanoparticles, and KCl were purchased from Sigma Aldrich (St. Louis, MO, United States). PBS was obtained from Invitrogen (Grand Island, NY, United States). NaCl and NaOH were acquired from Merck Millipore (Kenilworth, NJ, United States).
MSP10 Oligonucleotides
The two MSP10 oligonucleotides utilized in this study were a generous gift from Professor Mirko Zimic (Universidad Peruana Cayetano Heredia, Lima, Peru) and designed by Dr. Joseph Vinetz (University of California San Diego, United States). Crafted to represent the C-Terminal segment of MSP10, the first oligonucleotide has a sequence of 5´CACCATGGAACAGTTTATCCTGAAGAC3'. The other oligonucleotide was used as a representative of the N-terminal segment of MSP10. It has a sequence of 5´AGCCATGGAACGTGCTAAGTGCAACA3’.
Urine Samples
Archived urine samples positive for P. vivax and P. falciparum were collected from Iquitos in Peru and Ghana, respectively. Negative control urine samples were collected from volunteers who were blood smear negative in Iquitos, Peru and in Ghana, as well as in Lima, Peru, which is a non-endemic site. All urine samples were collected by clean catch procedures. Ghana urine samples were pelleted in the field and shipped on dry ice, pH was adjusted and samples were refrozen at -80°C as described earlier [31]. Peru urine samples were stored initially at -20°C prior to freezing at -80°C (Table 1). Peru’s urine samples were stored for 8 months while all Ghana samples were stored for more than one year.
Table 1. Urine samples were collected from three different sites representing Africa and South America.
| Urine Sample | Origin | Quantity |
|---|---|---|
| Positive for P. vivax | Iquitos, Peru | 31 |
| Positive for P. falciparum | Ghana | 14 |
| South American Controls | Iquitos, Peru | 8 |
| South American Controls | Lima, Peru | 28 |
| African Controls | Ghana | 9 |
Blood Smears
During the epidemiological surveys on the communities, the field microscopist reports whether a slide is positive or negative, and identifies the species, P. vivax and P. falciparum. They read 300 microscopy fields before the slide is reported as negative.
In the laboratory, a second reader (an experienced microscopist working for research projects for more than 15 years) read the slide to report species and parasite density assuming a white blood cell count of 6,000/μl. The research microscopist read until 500 microscopy fields, before the slide is reported as negative.
For quality control, 10% randomly selected slides (positive and negative) were reexamined by two blinded, expert microscopists at a reference laboratory in Loreto from Peru’s Ministry of Health. From the quality control examinations, the level of concordance varies between 98–100% for species and parasite density.
Colorimetric System
Urine was thawed at 25°C. Once urine was at room temperature, dipsticks were carried out to determine urine pH and the presence of protein. Each urine sample was centrifuged at 15,000 rpm for 5 minutes to remove sediments and then filtered using a 0.2 mm membrane (Minisart, Bohemia, NY, United States) to remove possible confounding particulates. The urine samples were diluted 1:16 with PBS. Diluted samples’ pH was adjusted to reach ≈ 6.4 using pH meter, and HCl and NaOH solutions. 50 uL of each diluted urine sample was heated at 95°C for 30 seconds using a thermocycler. Samples were cooled at room temperature for 10 minutes and 10 uL of either C-Terminal or N-Terminal MSP10 oligonucleotides and 20 uL of 0.25 M NaCl were added. The sample was heated at 59°C for two minutes and allowed to cool to room temperature for ten minutes. Finally, 50 uL of citrate reduced AuNPs were added. Two minutes later, the system was read visually and by spectrophotometer.
Ethics Statement
Urine and blood were collected for previous studies that were approved by institutional review boards of Universidad Peruana Caytano Heredia and University of Ghana, respectively. Written informed consents were obtained prior to storing samples as anonymous and unidentified.
Results
Color Change and Detection of P. vivax
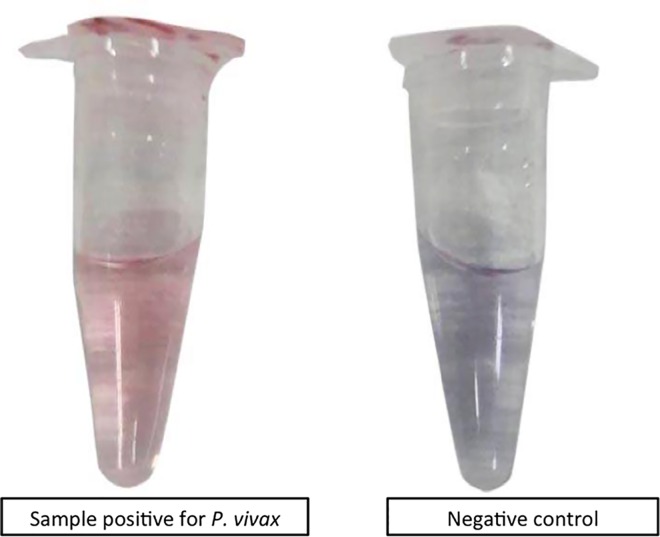
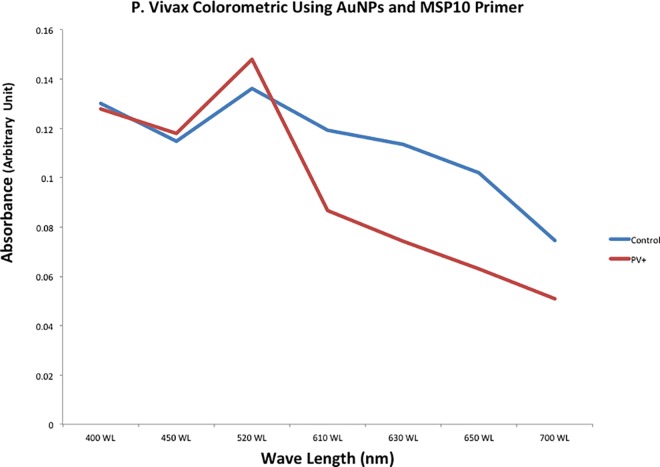
Up to 84% of P. vivax positive samples stabilized the gold nanoparticles and maintained a red color while 97% of negative controls induced aggregation and allowed color change to purple-blue. This color change was distinctly distinguished by naked eye (Fig 1). Additionally, the color difference was well defined by spectrophotometer, with positive samples at wavelengths of 520 and negative samples exhibiting wavelengths of 610–630 (Fig 2).
Fig 1. Urine from P. vivax negative volunteers turned AuNPs blue due to lack of targeted MSP10 DNA while positive P. vivax urine was able to form DS DNA and stabilized AuNPs to stay red in color.
Fig 2. The newly formed MSP10 double stranded DNA in urine was able to stabilize AuNPs to stay red in color.
P. vivax negative urine turned AuNPs’ color to blue which was detected by Spectrophotometer as color switches from red at 520 wavelength (P. vivax positive) into purple-blue at 610 wavelength (P. Vivax negative).
Sensitivity
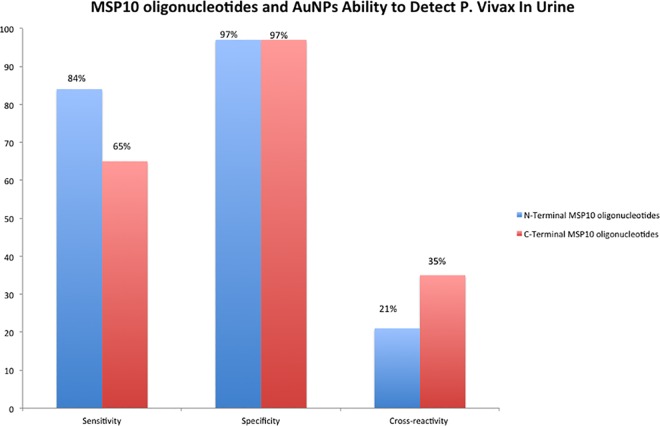
The colorimetric system was able to detect P. vivax with variable sensitivity. The sensitivity was dependent on which segment of MSP10 was used. The N-terminal segment distinguished 26 of 31 positive samples (84%) while the C-terminal segment distinguished only 20 of 31 samples (65%).
Specificity
Both the N-terminal and C-terminal segments of MSP10 had an overall specificity of 97% in urine, with only one false positive out of 45 control samples. The false positive was from a laboratory control in Lima (Fig 3).
Fig 3. N-Terminal MSP10 oligonucleotide has higher sensitivity, similar specificity and lower cross-reactivity in comparison to C-Terminal segment of MSP10 DNA in detecting P. vivax in urine using AuNPs.
Parasitemia Level
Parasitemia level was determined by blood smears collected at the same time as the urine samples. Data were shared after running the colorimetric system on urine to minimize investigator bias. There was no correlation between parasitemia level in blood and false negativity. The lowest parasitemia level observed in blood smear that also had a positive colorimetric test was 12 parasite/uL. However, there were two false negative samples with average parasitemia level in blood of 2510 parasite/uL (Table 2).
Table 2. Blood parasitemia levels showed poor correlation with both oligonucleotides in regard to their ability to detect P. vivax in urine using AuNPs.
| P. vivax Urine Sample | Age of study participant (in years) | N-Terminal Oligoneclutide | C-Terminal Oligoneclutide | Parasite Count by Microscope—Parasitos Asexuales (par/ul) |
|---|---|---|---|---|
| 1 | 56 | red | Red | 840 |
| 2 | 22 | red | Red | 24 |
| 3 | 24 | red | purple-blue | 36 |
| 4 | 37 | red | Red | 5310 |
| 5 | 51 | red | Red | 328 |
| 6 | 50 | red | Red | 360 |
| 7 | 40 | red | purple-blue | 36 |
| 8 | 31 | red | Red | 12 |
| 9 | 48 | red | Red | 720 |
| 10 | 18 | red | purple-blue | 1166 |
| 11 | 21 | red | purple-blue | 1080 |
| 12 | 68 | purple-blue | purple-blue | 420 |
| 13 | 24 | red | Red | 7620 |
| 14 | 42 | red | Red | 1680 |
| 15 | 55 | red | Red | 24 |
| 16 | 18 | red | purple-blue | 24 |
| 17 | 13 | red | purple-blue | 24 |
| 18 | 35 | purple-blue | purple-blue | 2350 |
| 19 | 40 | purple-blue | purple-blue | 2670 |
| 20 | 30 | red | red | 360 |
| 21 | 57 | red | red | 1098 |
| 22 | 54 | red | purple-blue | 600 |
| 23 | 26 | purple-blue | purple-blue | 24 |
| 24 | 68 | red | Red | 60 |
| 25 | 22 | red | Red | 108 |
| 26 | 28 | red | red | 12 |
| 28 | 50 | red | red | 2610 |
| 29 | 17 | purple-blue | red | 360 |
| 30 | 32 | red | Red | 5520 |
| 31 | 44 | red | red | 4470 |
Cross-Reactivity with P. falciparum
N-Terminal MSP10 oligonucleotide had a lower cross-reactivity (21%). C-Terminal MSP10 oligonucleotides showed high cross-reactivity with P. falciparum in urine utilizing colorimetric system (36%) (Table 3).
Table 3. MSP10 N-Terminal oligonucleotide and AuNPs were able to detect P. vivax in urine with higher sensitivity, specificity and lower cross-reactivity than C-Terminal oligonucleotide of MSP10.
| Origin | MSP10 N-Terminal oligonucleotide Detection | MSP10 C-Terminal oligonucleotideDetection | |
|---|---|---|---|
| Urine Samples Positive for P. vivax | Iquitos, Peru | 26 | 20 |
| (26/31: 84%) | (20/31: 65%) | ||
| Urine Samples Positive for P. falciparum | Ghana | 3 | 5 |
| (3/14: 21%) | (5/14: 36%) | ||
| South American Negative Urine Samples | Peru | 1 | 1 |
| (1/36: 3%) | (1:36: 3%) | ||
| African Negative Urine Samples | Ghana | 0 | 0 |
| (0/9: 0%) | (0/9: 0%) |
Duration and Cost
It took an average of 45 minutes from collecting urine to reading the test. Cost of the raw materials for each test is $0.20.
Discussion
Currently available RDTs suffer from limited sensitivity, specificity, genetic instability, the inability to differentiate Plasmodium species, expense, and limitation in sample types (blood only) [11–13]. Blood tests also cannot be performed in many field settings since expertise, temperature control, storage, and laboratory equipment are unavailable, [11]. Urine has been reported as an accessible and reliable source of malaria DNA with lower detection threshold than blood; however, it has lower levels of detectable antigen and current RDTs have a minimum detection level of 100 parasites per microliter [26,32]. However, urine contains smaller double stranded DNA of 150-to 250-nucleotide size that can interact with nanoparticles and still be utilized as a valid source of microorganisms’ DNA [28,33].
P. vivax presents unique diagnostic challenges because of its genetic and geographic diversities [34,35]. The emergence of sequencing technologies and malaria sequences is providing us with a greater understanding of conserved regions, which must be targeted for broad-applicability of diagnostic tests [36]. Merozoite Surface Protein 10 (MSP10) is one of the asexual stage proteins of P. vivax linked to erythrocyte invasion [37]. It has two prominent EGF-like domains at the C-terminus, which are highly conserved and carry close homogeneity among all Plasmodium species [38,39].
We used two segments in our study. The N terminal segment showed superior sensitivity and equal specificity when compared to the C terminal segment. It could be due to its higher adenine and guanine contents as they both have higher adsorption rate to AuNPs’ surface [40,41].
The sensitivity of MSP10 oligonucleotides in detecting DNA in urine samples is dependent on quality of urine. Stored urine is known to have less DNA than fresh urine and age could play an important role in determining sensitivity of MSP10 oligonucleotides [28].
MSP10 has little similarity among Plasmodium species apart from the two EGF-like domains [21], which could explain lower cross-reactivity of N-terminal segment with P. falciparum. To be able to lower cross-reactivity, optimization of MSP10 oligonucleotides is required in future work.
Currently, most commercially and widely used malaria RDTs employ monoclonal antibodies to identify histidine-rich protein two (HRPII) [32]. Although HRPII based RDTs reported variable sensitivity and specificity, HRPII was subject to genetic and geographical diversities along with gene polymorphism [42]. Additionally, monoclonal antibodies against HRPII might cross react with other proteins [42]. In Peru, approximately 30% of P. Falciparum had HRPII gene deletions, which might lead to false negative RDTs results [43]. Additionally, other RDTs employed malaria markers that have been subject to controversy. Aldolase, an isoenzyme widely used to diagnose P. falciparum and P. vivax, was criticized for its low sensitivity and genetic variability in diagnosing P. vivax [44]. P.vivax’s lactate dehydrogenase (pLDH), another common antigen used in malaria RDTs, was found to be affected by its gene polymorphism [45]. Furthermore, pLDH requires whole blood sampling and declines very fast, with clearance of asexual parasitemia [45]. This study provides evidence that MSP10 DNA can serve as a marker for malaria at a global scale, and may deliver innovative tools to aid in malaria control.
In developing other diagnostic tests, nanoparticles have been utilized both for concentrating and for detecting samples with low concentration of target molecules in austere settings [46–48]. Jeon et al in 2013 found that AuNPs could be used to recognize P. vivax DNA in diluted blood samples using pLDH with detection levels as low as 74 parasites/uL [46]. We were unable to determine our detection level due to lack of correlation between Plasmodium species’ blood and urine DNA levels, which has been previously reported [30]. Despite our ability to detect a positive urine sample in a patient with low blood parasitemia, we had false negative samples with very high blood parasitemia levels indicating poor correlation between our test and parasitemia levels in blood.
In spite of lack of correlation between blood and urine DNA, this test has many other advantages. With further simplification of the process, it has potential to represent a simple, rapid test that does not require extensive lab skills, which will make it suitable as point of care test for low resource settings [49].
Conclusion
To our knowledge, this test is the first RDT utilizing urine samples rather than blood and employing nanoparticles. The colorimetric assay using AuNPs and MSP10 oligonucleotides to detect P. vivax in urine holds potentials to provide a safe, simple, rapid, and cheap tool to diagnose one of the most common form of malaria. Innovative use of MSP10 as a marker for P. vivax has potential for global application in mass screening programs.
Supporting Information
(DOCX)
(DOCX)
Acknowledgments
The authors would thank Dr. Mirko Zimic and Dr. Patricia Sheen for their generous gifts and continuous support. They also extend gratitude to Dr. Sujatha Kannan and Dr. Elizabeth Nance for their technical support and guidance through study design.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by NIAID/ICEMR Amazonia (U19AI089702). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tangpukdee N., Duangdee C., Wilairatana P., and Krudsood S., “Malaria Diagnosis: A Brief Review,” Korean J. Parasitol., vol. 47, no. 2, p. 93, 2009. 10.3347/kjp.2009.47.2.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hay S. I., Guerra C. A., Tatem A. J., Noor A. M., and Snow R. W., “Europe PMC Funders Group The global distribution and population at risk of malaria: past, present, and future,” vol. 4, no. 6, pp. 327–336, 2011. 10.1016/S1473-3099(04)01043-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens P. and Hall L., “Malaria on the move: human population movement and malaria transmission. Malaria on the move: human population movement and malaria transmission.,” Emerg. Infect. Dis., vol. 103, no. April, pp. 1–7, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerra C. A., Howes R. E., Patil A. P., Gething P. W., van Boeckel T. P., Temperley W. H., Kabaria C. W., Tatem A. J., Manh B. H., Elyazar I. R. F., Baird J. K., Snow R. W., and Hay S. I., “The international limits and population at risk of Plasmodium vivax transmission in 2009,” PLoS Negl. Trop. Dis., vol. 4, no. 8, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice B. L., Acosta M. M., Pacheco M. A., and a Escalante A., “Merozoite surface protein-3 alpha as a genetic marker for epidemiologic studies in Plasmodium vivax: a cautionary note.,” Malar. J., vol. 12, p. 288, January 2013. 10.1186/1475-2875-12-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ketema T. and Bacha K., “Plasmodium vivax associated severe malaria complications among children in some malaria endemic areas of Ethiopia.,” BMC Public Health, vol. 13, no. 1, p. 637, January 2013. 10.1186/1471-2458-13-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee G., Yori P., Olortegui M. P., Pan W., Caulfield L., Gilman R. H., Sanders J. W., Delgado H. S., and Kosek M., “Comparative effects of vivax malaria, fever and diarrhoea on child growth,” Int. J. Epidemiol., vol. 41, no. 2, pp. 531–539, 2012. 10.1093/ije/dyr190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hänscheid T., Valadas E., and Grobusch M. P., “Pigment Detection,” Parasitol. Today, vol. 4758, no. 00, pp. 2000–2002, 2000. [DOI] [PubMed] [Google Scholar]
- 9.Erdman L. K. and Kain K. C., “Molecular diagnostic and surveillance tools for global malaria control.,” Travel Med. Infect. Dis., vol. 6, no. 1–2, pp. 82–99, 2008. 10.1016/j.tmaid.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Asiimwe C., Kyabayinze D. J., Kyalisiima Z., Nabakooza J., Bajabaite M., Counihan H., and Tibenderana J. K., “Early experiences on the feasibility, acceptability, and use of malaria rapid diagnostic tests at peripheral health centres in Uganda-insights into some barriers and facilitators.,” Implement. Sci., vol. 7, no. 1, p. 5, January 2012. 10.1186/1748-5908-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray C. K., a Gasser R., Magill A. J., and Miller R. S., “Update on rapid diagnostic testing for malaria.,” Clin. Microbiol. Rev., vol. 21, no. 1, pp. 97–110, January 2008. 10.1128/CMR.00035-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Demas, J. Oberstaller, J. Debarry, W. Lucchi, G. Srinivasamoorthy, D. Sumari, A. M. Kabanywanyi, L. Villegas, A. Ananias, S. P. Kachur, J. W. Barnwell, S. David, V. Udhayakumar, C. Jessica, N. W. Lucchi, A. A. Escalante, D. S. Peterson, and J. C. Kissinger, “Applied Genomics: Data Mining Reveals Species-Specific Malaria Diagnostic Targets More Sensitive than 18S rRNA Applied Genomics: Data Mining Reveals Species-Specific Malaria Diagnostic Targets More Sensitive than 18S rRNA,” 2011.
- 13.Makler M. T., Piper R. C., and Milhous W. K., “Lactate dehydrogenase and the diagnosis of malaria.,” Parasitol. Today, vol. 14, no. 9, pp. 376–7, September 1998. 10.1016/s0169-4758(98)01284-8 [DOI] [PubMed] [Google Scholar]
- 14.Burda C. and Doane T. L., “The unique role of nanoparticles in nanomedicine: imaging, drug delivery and therapy,” Chem. Soc. Rev., vol. 41, no. 7, pp. 2885–2911, 2012. 10.1039/c2cs15260f [DOI] [PubMed] [Google Scholar]
- 15.Jung Y. L., Jung C., Parab H., Li T., and Park H. G., “Direct colorimetric diagnosis of pathogen infections by utilizing thiol-labeled PCR primers and unmodified gold nanoparticles.,” Biosens. Bioelectron., vol. 25, no. 8, pp. 1941–6, April 2010. 10.1016/j.bios.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 16.Yang J., Lee Y., Deivaraj T. C., and Too H., “Single Stranded DNA Induced Assembly of Gold Nanoparticles,” Mol. Eng. Biol. Chem. Syst., 2003. [Google Scholar]
- 17.Sato K., Hosokawa K., and Maeda M., “Non-cross-linking gold nanoparticle aggregation as a detection method for single-base substitutions.,” Nucleic Acids Res., vol. 33, no. 1, p. e4, January 2005. 10.1093/nar/gni007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardenas M., Barauskas J., Schillen K., Brennan J. L., Brust M., and Nylander T., “Thiol-speciric and nonspecific interactions between DNA and gold nanoparticles,” Langmuir, vol. 22, no. 7, pp. 3294–3299, 2006. 10.1021/la0530438 [DOI] [PubMed] [Google Scholar]
- 19.Sandström P., Boncheva M., and Åkerman B., “Nonspecific and thiol-specific binding of DNA to gold nanoparticles,” Langmuir, vol. 19, no. 18, pp. 7537–7543, 2003. 10.1021/la034348u [DOI] [Google Scholar]
- 20.Padmavathy B., Vinoth Kumar R., and Jaffar Ali B. M., “A direct detection of Escherichia coli genomic DNA using gold nanoprobes.,” J. Nanobiotechnology, vol. 10, no. 1, p. 8, January 2012. 10.1186/1477-3155-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacheco M. A., Elango A. P., a Rahman A., Fisher D., Collins W. E., Barnwell J. W., and a Escalante A., “Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp.,” Infect. Genet. Evol., vol. 12, no. 5, pp. 978–86, July 2012. 10.1016/j.meegid.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres K. J., Castrillon C. E., Moss E. L., Saito M., Tenorio R., Molina D. M., Davies H., Neafsey D. E., Felgner P., Vinetz J. M., and Gamboa D., “Genome-level determination of plasmodium falciparum blood-stage targets of malarial clinical immunity in the peruvian amazon,” J. Infect. Dis., vol. 211, no. 8, pp. 1342–1351, 2015. 10.1093/infdis/jiu614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuquiyauri R., Molina D. M., Moss E. L., Wang R., Gardner M. J., Brouwer K. C., Torres S., Gilman R. H., Llanos-Cuentas A., Neafsey D. E., Felgner P., Liang X., and Vinetz J. M., “Genome-Scale Protein Microarray Comparison of Human Antibody Responses in Plasmodium vivax Relapse and Reinfection.,” Am. J. Trop. Med. Hyg., vol. 93, no. 4, pp. ajtmh.15–0232–, 2015. 10.4269/ajtmh.15-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neafsey D. E., Galinsky K., Jiang R. H. Y., Young L., Sykes S. M., Saif S., Gujja S., Goldberg J. M., Young S., Zeng Q., Chapman S. B., Dash A. P., Anvikar A. R., Sutton P. L., Birren B. W., a Escalante A., Barnwell J. W., and Carlton J. M., “The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum.,” Nat. Genet., vol. 44, no. 9, pp. 1046–50, 2012. 10.1038/ng.2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sin M. L. Y., Mach K. E., Wong P. K., and Liao J. C., “Advances and challenges in biosensor-based diagnosis of infectious diseases.,” Expert Rev. Mol. Diagn., vol. 14, no. 2, pp. 225–44, March 2014. 10.1586/14737159.2014.888313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nwakanma D. C., Gomez-Escobar N., Walther M., Crozier S., Dubovsky F., Malkin E., Locke E., and Conway D. J., “Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine.,” J. Infect. Dis., vol. 199, no. 11, pp. 1567–74, June 2009. 10.1086/598856 [DOI] [PubMed] [Google Scholar]
- 27.Buppan P., Putaporntip C., Pattanawong U., and Seethamchai S., “Comparative detection of Plasmodium vivax and Plasmodium falciparum DNA in saliva and urine samples from symptomatic malaria patients in a low endemic area,” Malar. J., vol. 9, no. 72, pp. 1–7, 2010. 10.1186/1475-2875-9-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Y.-H., Wang M., Brenner D. E., Ng A., Melkonyan H., Umansky S., Syngal S., and Block T. M., “Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer.,” J. Mol. Diagn., vol. 6, no. 2, pp. 101–7, 2004. 10.1016/s1525-1578(10)60497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botezatu I., Serdyuk O., Potapova G., Shelepov V., Alechina R., Molyaka Y., Anan'ev V., Bazin I., Garin A., Narimanov M., Knysh V., Melkonyan H., Umansky S., and Lichtenstein A., “Genetic analysis of DNA excreted in urine: A new approach for detecting specific genomic DNA sequences from cells dying in an organism,” Clin. Chem., vol. 46, no. 8 I, pp. 1078–1084, 2000. [PubMed] [Google Scholar]
- 30.Putaporntip C., Buppan P., and Jongwutiwes S., “Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays,” Clin. Microbiol. Infect., vol. 17, no. 10, pp. 1484–1491, 2011. 10.1111/j.1469-0691.2011.03507.x [DOI] [PubMed] [Google Scholar]
- 31.Howard C. T., McKakpo U. S., Quakyi I. A., Bosompem K. M., Addison E. A., Sun K., Sullivan D., and Semba R. D., “Relationship of hepcidin with parasitemia and anemia among patients with uncomplicated Plasmodium falciparum malaria in Ghana,” Am. J. Trop. Med. Hyg., vol. 77, no. 4, pp. 623–626, 2007. [PubMed] [Google Scholar]
- 32.Moody A., “Rapid Diagnostic Tests for Malaria Parasites Rapid Diagnostic Tests for Malaria Parasites,” Clin. Microbiol. Rev., vol. 15, no. 1, pp. 66–77, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilhorst M., Theunissen R., van Rie H., van Paassen P., and Tervaert J. W., “DNA extraction from long-term stored urine.,” BMC Nephrol., vol. 14, no. 1, p. 238, 2013. 10.1186/1471-2369-14-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Collins W. E., a Wirtz R., Rathore D., Lal a, and McCutchan T. F., “Geographic subdivision of the range of the malaria parasite Plasmodium vivax.,” Emerg. Infect. Dis., vol. 7, no. 1, pp. 35–42, 2001. 10.3201/eid0701.700035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonilla J. A., Validum L., Cummings R., and Palmer C. J., “Genetic diversity of Plasmodium vivax Pvcsp and Pvmsp1 in Guyana, South America.,” Am. J. Trop. Med. Hyg., vol. 75, no. 5, pp. 830–5, November 2006. [PubMed] [Google Scholar]
- 36.Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., Caler E., Crabtree J., V Angiuoli S., Merino E. F., Amedeo P., Cheng Q., Coulson R. M. R., Crabb B. S., a Del Portillo H., Essien K., V Feldblyum T., Fernandez-Becerra C., Gilson P. R., Gueye A. H., Guo X., Kang’a S., a Kooij T. W., Korsinczky M., Meyer E. V.-S., Nene V., Paulsen I., White O., a Ralph S., Ren Q., Sargeant T. J., Salzberg S. L., Stoeckert C. J., a Sullivan S., Yamamoto M. M., Hoffman S. L., Wortman J. R., Gardner M. J., Galinski M. R., Barnwell J. W., and Fraser-Liggett C. M., “Comparative genomics of the neglected human malaria parasite Plasmodium vivax.,” Nature, vol. 455, no. 7214, pp. 757–63, October 2008. 10.1038/nature07327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders P. R., Gilson P. R., Cantin G. T., Greenbaum D. C., Nebl T., Carucci D. J., McConville M. J., Schofield L., Hodder A. N., Yates J. R., and Crabb B. S., “Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum.,” J. Biol. Chem., vol. 280, no. 48, pp. 40169–76, December 2005. 10.1074/jbc.M509631200 [DOI] [PubMed] [Google Scholar]
- 38.Perez-Leal O., Sierra A. Y., a Barrero C., Moncada C., Martinez P., Cortes J., Lopez Y., Torres E., Salazar L. M., and a Patarroyo M., “Plasmodium vivax merozoite surface protein 8 cloning, expression, and characterisation.,” Biochem. Biophys. Res. Commun., vol. 324, no. 4, pp. 1393–9, November 2004. 10.1016/j.bbrc.2004.09.202 [DOI] [PubMed] [Google Scholar]
- 39.Black C. G., Wang L., Wu T., and Coppel R. L., “Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum,” Mol. Biochem. Parasitol., vol. 127, no. 1, pp. 59–68, March 2003. 10.1016/s0166-6851(02)00308-0 [DOI] [PubMed] [Google Scholar]
- 40.Koo K. M., Sina A. a. I., Carrascosa L. G., Shiddiky M. J. a., and Trau M, “DNA–bare gold affinity interactions: mechanism and applications in biosensing,” Anal. Methods, vol. 7, no. 17, pp. 7042–7054, 2015. [Google Scholar]
- 41.Zhang X., Sun C. Q., and Hirao H., “Guanine binding to gold nanoparticles through nonbonding interactions.,” Phys. Chem. Chem. Phys., vol. 15, no. 44, pp. 19284–92, 2013. 10.1039/c3cp52149d [DOI] [PubMed] [Google Scholar]
- 42.Lee N., Baker J., Andrews K. T., Gatton M. L., Bell D., Cheng Q., and McCarthy J., “Effect of sequence variation in Plasmodium falciparum histidine- rich protein 2 on binding of specific monoclonal antibodies: Implications for rapid diagnostic tests for malaria.,” J. Clin. Microbiol., vol. 44, no. 8, pp. 2773–2778, 2006. 10.1128/JCM.02557-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akinyi S., Hayden T., Gamboa D., Torres K., Bendezu J., Abdallah J. F., Griffing S. M., Quezada W. M., Arrospide N., De Oliveira A. M., Lucas C., Magill A. J., Bacon D. J., Barnwell J. W., and Udhayakumar V., “Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru.,” Sci. Rep., vol. 3, p. 2797, 2013. 10.1038/srep02797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J.-Y., Kim H.-H., Shin H., Sohn Y., Kim H., Lee S.-W., Lee W.-J., and Lee H.-W., “Genetic variation of aldolase from Korean isolates of Plasmodium vivax and its usefulness in serodiagnosis,” Malar. J., vol. 11, no. 1, p. 159, 2012. 10.1186/1475-2875-11-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin H.-I., Kim J.-Y., Lee W.-J., Sohn Y., Lee S.-W., Kang Y.-J., and Lee H.-W., “Polymorphism of the parasite lactate dehydrogenase gene from Plasmodium vivax Korean isolates.,” Malar. J., vol. 12, no. 1, p. 166, 2013. 10.1186/1475-2875-12-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon W., Lee S., Manjunatha D. H., and Ban C., “A colorimetric aptasensor for the diagnosis of malaria based on cationic polymers and gold nanoparticles.,” Anal. Biochem., vol. 439, no. 1, pp. 11–6, August 2013. 10.1016/j.ab.2013.03.032 [DOI] [PubMed] [Google Scholar]
- 47.Yuen C. and Liu Q., “Magnetic field enriched surface enhanced resonance Raman spectroscopy for early malaria diagnosis.,” J. Biomed. Opt., vol. 17, no. 1, p. 017005, January 2012. 10.1117/1.JBO.17.1.017005 [DOI] [PubMed] [Google Scholar]
- 48.Xia F., Zuo X., Yang R., Xiao Y., Kang D., Vallée-Bélisle A., Gong X., Yuen J. D., Hsu B. B. Y., Heeger A. J., and Plaxco K. W., “Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes.,” Proc. Natl. Acad. Sci. U. S. A., vol. 107, no. 24, pp. 10837–41, June 2010. 10.1073/pnas.1005632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pai N. P., Vadnais C., Denkinger C., Engel N., and Pai M., “Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries.,” PLoS Med., vol. 9, no. 9, p. e1001306, January 2012. 10.1371/journal.pmed.1001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.