Abstract
Nucleosomes containing the CenH3 (CENPA or CENP-A) histone variant replace H3 nucleosomes at centromeres to provide a foundation for kinetochore assembly. CENPA nucleosomes are part of the constitutive centromere associated network (CCAN) that forms the inner kinetochore on which outer kinetochore proteins assemble. Two components of the CCAN, CENPC and the histone-fold protein CENPT, provide independent connections from the ∼171-bp centromeric α-satellite repeat units to the outer kinetochore. However, the spatial relationship between CENPA nucleosomes and these two branches remains unclear. To address this issue, we use a base-pair resolution genomic readout of protein–protein interactions, comparative chromatin immunoprecipitation (ChIP) with sequencing, together with sequential ChIP, to infer the in vivo molecular architecture of the human CCAN. In contrast to the currently accepted model in which CENPT associates with H3 nucleosomes, we find that CENPT is centered over the CENPB box between two well-positioned CENPA nucleosomes on the most abundant centromeric young α-satellite dimers and interacts with the CENPB/CENPC complex. Upon cross-linking, the entire CENPA/CENPB/CENPC/CENPT complex is nuclease-protected over an α-satellite dimer that comprises the fundamental unit of centromeric chromatin. We conclude that CENPA/CENPC and CENPT pathways for kinetochore assembly are physically integrated over young α-satellite dimers.
Centromeres are unique structures located at the primary constriction of chromosomes that mediate faithful chromosome segregation. Although centromeric DNA sequences evolve rapidly, components of the proteinaceous kinetochore that connect centromeres to spindle microtubules at mitosis and meiosis are highly conserved (Henikoff et al. 2001; Cheeseman and Desai 2008). In most eukaryotes, canonical histone H3 is replaced by the CenH3 variant, called CENPA in mammals, to form specialized chromatin that acts as the foundation for kinetochore assembly (Quénet and Dalal 2012). Defining the molecular architecture of centromeric chromatin is essential to understand the mechanism by which the kinetochore assembles on centromeres to connect with spindle microtubules.
At human centromeres, blocks of CENPA nucleosomes have been shown to alternate with blocks of H3 nucleosomes, each occupying ∼15–40 kb of α-satellite DNA consisting of ∼171-bp repeat units (Blower et al. 2002). CENPA containing centromeric chromatin is associated with a set of proteins that form the Constitutive Centromere-Associated Network (CCAN) (Foltz et al. 2006). The CCAN is essential for kinetochore assembly, and loss of CCAN components leads to errors in chromosome segregation (Hori et al. 2008; Amano et al. 2009; Gascoigne et al. 2011; Nishino et al. 2013; Rago et al. 2015). Despite its fundamental role in kinetochore assembly, the molecular architecture of the CCAN in vivo remains unclear. CENPB, the only CCAN component that binds to centromeres in a sequence-dependent manner, is present on both active and inactive centromeres of human dicentric chromosomes (Kipling and Warburton 1997). The CENPB box, a consensus binding site for CENPB, is required for establishment of centromeres on human artificial chromosomes (Ohzeki et al. 2002; Okada et al. 2007). The histone fold-containing CENPT protein and the widely conserved CENPC protein are two major components of the CCAN that play fundamental roles in kinetochore structure and function by providing two alternative attachments to outer kinetochore proteins (Hori et al. 2008; Screpanti et al. 2011; Gascoigne and Cheeseman 2012; Kato et al. 2013; Rago et al. 2015; Wood et al. 2016). Both CENPC and CENPT-mediated pathways are involved in the recruitment of Ndc80, an outer kinetochore complex that interacts directly with spindle microtubules (Nishino et al. 2013; Rago et al. 2015). CENPC physically associates with CENPA nucleosomes and extends to the outer kinetochore through the KNL1/Mis12 complex/Ndc80 complex (KMN) network (Carroll et al. 2010; Kato et al. 2013; Rago et al. 2015). Independently, CENPT makes a direct connection with the Ndc80 complex (Gascoigne et al. 2011; Nishino et al. 2013; Rago et al. 2015). CENPT, together with three other histone fold-containing proteins, CENPW, APITD1 (also known as CENPS), and CENPX, forms a heterotetrameric nucleosome-like structure, which can induce positive supercoils in DNA in vitro (Nishino et al. 2012; Takeuchi et al. 2014). The DNA binding activity of CENPT/CENPW is essential for kinetochore formation, as the localization of Ndc80 is abolished in mutants defective in DNA binding activity of CENPW (Nishino et al. 2012). Although CENPT and CENPW localize exclusively to centromeres, CENPS and CENPX localize to both centromeres and euchromatic arms (Huang et al. 2010; Singh et al. 2010). CENPS and CENPX have been shown to play an important role in the assembly of the outer kinetochore, but depletion of CENPS does not affect localization of CCAN components (Amano et al. 2009).
The spatial relationship between CENPC, CENPT, and CENPA has been the subject of controversy. CENPA nucleosomal arrays were found to interact with various components of the CCAN, including CENPC and CENPT in human cells (Foltz et al. 2006) and in fission yeast (Thakur et al. 2015). Consistent with a direct physical interaction with CENPA nucleosomes, centromere association of both CENPC and CENPT is compromised in cells containing reduced levels of CENPA (Gascoigne et al. 2011). However, under conditions optimized to isolate protein subcomplexes at nucleosomal levels in DT40 cells, both CENPT and CENPC were found to coimmunoprecipitate with H3 and not with CENPA (Hori et al. 2008). These latter results led to the currently accepted model in which CENPT/CENPW/CENPS/CENPX forms a nucleosome-like structure within H3 nucleosome arrays and are physically separated from CENPA nucleosomes at centromeres (Hori et al. 2008; Fukagawa and Earnshaw 2014; McKinley et al. 2015).
Mapping of centromeric proteins at human centromeres is difficult due to lack of a complete genomic assembly of highly repetitive centromeric sequences. Only the higher order repeats (HORs) proximal to the centromere edges have been assembled, leaving behind unmappable megabase-size gaps (Schueler et al. 2001; Hayden et al. 2013). Recently, using high-resolution native Chromatin Immunoprecipitation (ChIP) of CENPA combined with a bottom-up approach of clustering of enriched sequences, we discovered that CENPA nucleosomes are most abundantly enriched at two homogeneous α-satellite arrays consisting of distinct dimers of 340-bp (Cen1-like) and 342-bp (Cen13-like) α-satellites (Henikoff et al. 2015). In that study, we found that homogeneous α-satellite arrays were highly enriched for CENPA relative to H3 nucleosomes, in contrast to pericentric α-satellite arrays. We also showed that both dimeric units precisely position two CENPA nucleosomes, each protecting 100 bp DNA.
To resolve the inconsistencies regarding the physical relationship between the two essential kinetochore assembly pathways and to elucidate the molecular architecture of the DNA-binding core of the CCAN complex in vivo, we apply a comprehensive genomics-based approach for high-resolution mapping of protein–protein interactions. We develop a general framework for Comparative ChIP-seq by using Micrococcal Nuclease (MNase)-based native ChIP (N-ChIP), cross-linking ChIP (X-ChIP), and sequential ChIP with sequencing. Our analysis of the key CCAN components to CENPA-enriched α-satellite dimers defines the fundamental units of human centromeres.
Results
Comparative ChIP-seq reveals that CENPT particles are highly sensitive to MNase digestion
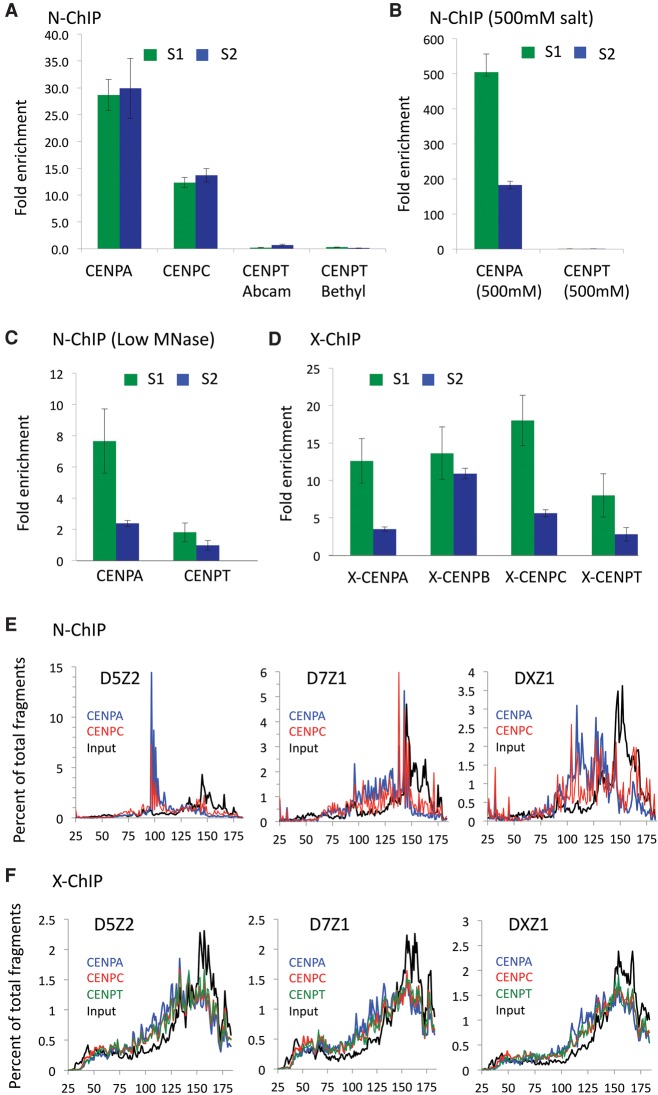
In light of previous studies, which have led to contradictory conclusions as to the relationship between core CCAN components CENPT and CENPC, we developed a MNase-based comparative ChIP method (Fig. 1) to precisely map CENPT to centromeric α-satellites. MNase preferentially cuts linker DNA, leaving behind DNA that is protected by nucleosomes, nonhistone chromatin proteins and protein complexes. We first performed N-ChIP in the HuRef lymphoblastoid cell line using antibodies against centromeric proteins and assayed by qPCR to quantify enrichment of α-satellites in the DNA associated with CENPA, CENPC, and CENPT N-ChIP. At α-satellite sequences derived from Chromosomes 1, 5, and 19 and from Chromosomes 13 and 21, CENPA and CENPC N-ChIP showed ∼30-fold and ∼10-fold enrichment, respectively, but CENPT enrichment was comparable to background (Fig. 2A).
Figure 1.
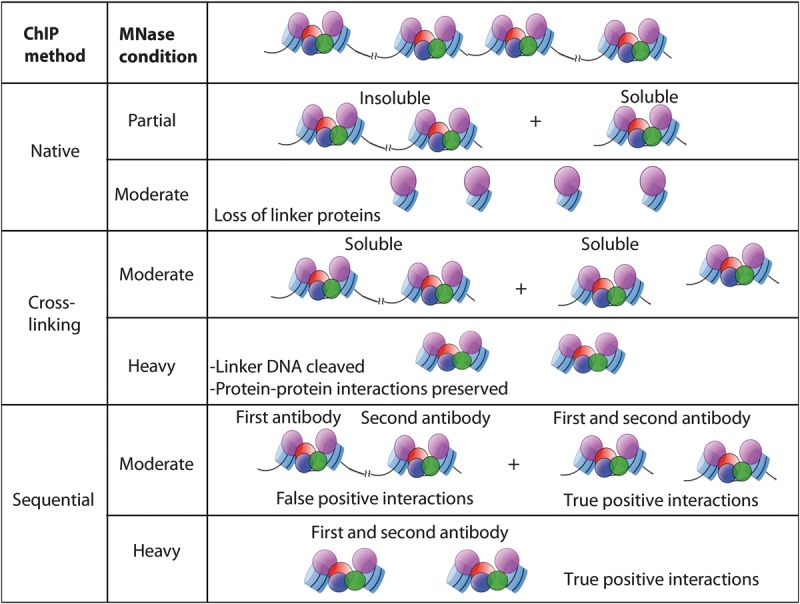
Schematic representation of comparative ChIP methods. Under native conditions, partial MNase digestion produces insoluble chromatin arrays as the major population. Upon moderate MNase digestion under similar conditions, only nucleosomes or similar structures that wrap DNA are protected; and therefore, proteins associated with the linker DNA are lost from the chromatin. In X-ChIP and sequential ChIP, however, protein–protein interactions are stabilized by cross-linking, and the solubility of chromatin is enhanced by detergents, resulting in ChIP signals after both moderate and heavy MNase digestion. In sequential ChIP, false positives can arise from there being multiple complexes pulled down from longer arrays, in which the first antibody pulls down one complex and the second antibody pulls down a different complex: (DNA) black line; (nucleosomes) blue barrels; (DNA-binding proteins) colored balls.
Figure 2.
CENPT is not retained at α-satellites by N-ChIP but is retained by X-ChIP. (A) Real-time PCR analysis of DNA obtained by native ChIP of CENPA, CENPC, and CENPT (using two different antibodies). The S1 primer pair was designed from Chromosome 21 alphoid arrays but is also present on Chromosome 13. Primer pair S2 was designed from the Cen1-like (SF1 family) sequences and is also present on Chromosomes 5 and 19 (Supplemental Table 1). Fold-enrichment over input was calculated relative to a control noncentromeric sequence from the 5S rDNA locus: Centromere (ChIP/input) ÷ 5S rDNA (ChIP/input). (B) CENPA and CENPT enrichment from native ChIP assays performed under 500 mM salt conditions. (C) CENPA and CENPT enrichment on DNA obtained from native ChIP assays performed on partially digested chromatin. (D) Enrichment of centromeric sequences in CENPA, CENPC, and CENPT cross-linking ChIP (X-ChIP). (E,F) Fragment length analysis of merged pairs obtained from (E) native or (F) X-ChIP data sets on D5Z2 (left), D7Z1 (middle), and DXZ1 (right) sequences. The sharp reduction in the size distribution above ∼160 bp and truncation at 185 bp is attributable to mapping of only paired-end reads in which the two 100-bp reads in a pair overlapped by 15 bp.
It seemed possible that the large size of the kinetochore complex (Cheeseman and Desai 2008), which is highly insoluble under N-ChIP conditions (Krassovsky et al. 2012; Steiner and Henikoff 2014), is responsible for the differential recovery of centromere proteins in our ChIP assay. To increase kinetochore solubility, we extracted MNase-digested chromatin with 500 mM NaCl and observed dramatic enrichment of CENPA on centromeric α-satellite DNA by ChIP qPCR (Fig. 2B). However, even under these conditions, we observed only background levels of CENPT at centromeric α-satellite DNA.
To determine whether the lack of CENPT N-ChIP is general for all centromeric α-satellite DNA, we performed Illumina sequencing on CENPT N-ChIP DNA fragments and mapped the paired-end reads to dimeric units derived from a variety of α-satellite classes. Human centromeres are characterized by two major families of homogeneous α-satellite dimeric arrays—Suprachromosomal Family 1 (SF1, in nine centromeres) and Suprachromosomal Family 2 (SF2, in 11 centromeres) (Alexandrov et al. 2001)—and by higher-order repeats (HORs) that have been mapped to centromere edges (Rudd et al. 2003). We previously showed that SF1, represented by a Cen1-like consensus α-satellite dimer, and to a lesser extent SF2, represented by a Cen13-like consensus dimer, likely account for most of the CENPA and CENPC N-ChIP signals at annotated α-satellites (Henikoff et al. 2015). These dimeric units showed striking CENPA and CENPC ChIP patterns, with each monomeric unit of the dimer protected from MNase digestion by a precisely positioned ∼100-bp particle, separated by an ∼60-bp linker with a CENPB box. We quantified the total CENPA ChIP signals from these 340-bp dimer consensus sequences and from representative HOR dimers of the same length. As shown previously (Henikoff et al. 2015), CENPA and CENPC were enriched and H3 depleted relative to input on Cen1-like and Cen13-like dimer consensus sequences and on D5Z2, D7Z1, and DXZ1 HORs, but not on noncentromeric D19Z1, X-monomeric (Xmono) and D5Z1 α-satellite arrays (Supplemental Fig. S1, top). In contrast, the CENPT N-ChIP profiles of the same α-satellite dimers showed ∼100-fold less enrichment relative to noncentromeric sequences (Supplemental Fig. S1, bottom). For example, the CENPA N-ChIP enrichment over D5Z2 was >3000-fold higher than that over the noncentromeric D5Z1, despite showing fluorescence in situ hybridization signals of similar magnitude (Finelli et al. 1996; Slee et al. 2012), and yet CENPT N-ChIP enrichment was only 20-fold higher for D5Z2 relative to D5Z1. Therefore, CENPT is only marginally enriched over CENPA- and CENPC-containing centromeric α-satellites when compared to noncentromeric α-satellites.
CenH3 nucleosomes in some organisms are hypersensitive to MNase digestion (Dimitriadis et al. 2010; Steiner and Henikoff 2014), and so we wondered whether the lack of CENPT enrichment at α-satellites might have been caused by hypersensitivity to MNase. Consistent with this possibility, N-ChIP after light MNase digestion resulted in enrichment of CENPA and CENPT (Fig. 2C). The fold-enrichment of CENPA was lower than that obtained using standard MNase digestion conditions, presumably due to inefficient immunoprecipitation of larger chromatin arrays. Our results suggest that depletion of CENPT occurred during MNase fragmentation used to prepare chromatin for N-ChIP.
Next, we applied MNase X-ChIP (cross-linking ChIP), which provides nearly base-pair resolution by combining MNase digestion of cross-linked chromatin with light sonication to dissolve otherwise insoluble protein complexes (Skene et al. 2014; Skene and Henikoff 2015). Because CENPT is part of the CCAN network, we reasoned that its loss during N-ChIP could be prevented by formaldehyde cross-linking, which stabilizes protein–protein interactions (Zentner and Henikoff 2014). We obtained robust ChIP qPCR enrichment for all CENPs: CENPA, CENPC, and CENPT (Fig. 2D), confirming that the relative lack of CENPT in N-ChIP results from its preferential loss during chromatin preparation.
CENPA, CENPC, and CENPT cross-link to the same α-satellite sequences
As we successfully recovered centromeric α-satellites in CENPT, CENPA, and CENPC X-ChIP assays, we subjected the ChIP DNA to Illumina sequencing and mapped paired-end 2 × 100-bp reads to α-satellite dimer sequences, as previously described (Henikoff et al. 2015). In sharp contrast to N-ChIP, in which CENPA and CENPC showed size distributions of immunoprecipitated DNA fragments that differed between various dimeric units (Fig. 2E), CENPA, CENPC, and CENPT X-ChIP profiles showed a similar broad distribution of lengths for all dimers. Thus, all three proteins were cross-linked to protect complexes of similar size from MNase digestion (Fig. 2F).
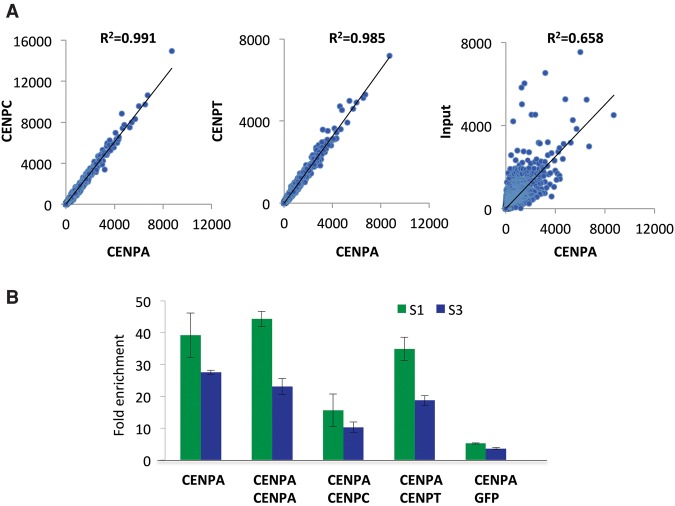
The similar size distribution of protected fragments over all dimeric units from CENPA, CENPC, and CENPT ChIP-seq suggests that the same cross-linked particles were being immunoprecipitated. To test this possibility, we clustered X-ChIP fragments based on sequence identity and compared the frequency distributions of fragments within clusters between data sets. We found remarkably strong correlations between distributions (Fig. 3A) (R2 = 0.991 for CENPA and CENPC and R2 = 0.985 for CENPA and CENPT). Together with the similar fragment size distributions, these highly concordant cluster frequency distributions suggest that CENPA, CENPC, and CENPT cross-link in the same α-satellite-containing chromatin complex.
Figure 3.
CENPA, CENPC, and CENPT cross-link in a single chromatin complex. (A) Close correspondence between X-ChIP of CENPA, CENPC, and CENPT data sets. X-ChIP fragments were clustered based on sequence identity, and frequency distributions of fragments within clusters between data sets were compared. Scatter plots show the regression of the number of distinct mapped merged pairs for the CENPC, CENPT, and input X-ChIP data sets on the CENPA X-ChIP data set. (B) Real-time PCR analysis of DNA obtained by sequential ChIP with indicated pairs of antibodies. The S1 primer pair was designed as described in the legend to Figure 2, and the S3 primer pair was designed from the Cen1-like (SF1 family) sequences and is also present on Chromosomes 1, 5, and 19 (Supplemental Table 1). Fold enrichment over input was calculated relative to a control noncentromeric sequence from the 5S rDNA locus.
CENPA, CENPC, and CENPT cross-link in a single chromatin complex
The extremely high correlations between CENPA-, CENPC-, and CENPT-associated α-satellite sequence and the similar size distributions strongly suggest that all are part of the same complex. However, because of the high repetitiveness of α-satellite DNA, it is possible that the three different proteins reside on identical or highly similar α-satellite sequences. Sequential ChIP assays can potentially distinguish these alternative possibilities, because a sequence will be recovered in the second ChIP above background levels only if both proteins are present in the same complex. However, an efficient first ChIP will recover only a very small fraction of the original chromatin, so there will be very little input for the second ChIP. Because there are only roughly 400 CENPA molecules per human chromosome (Bodor et al. 2014), sequential ChIP of centromeric chromatin is very challenging. Moreover, failure to completely remove the first antibody in the sequential precipitation can lead to false positive signals. To minimize these issues, we first created a cell line expressing a CENPA-FLAG fusion protein and performed X-ChIP using anti-FLAG magnetic beads. To increase the recovery of protein complexes from FLAG magnetic beads and avoid contamination of the first antibody in the subsequent immunoprecipitation, we took advantage of highly efficient competitive elution of protein complexes from the FLAG magnetic beads by FLAG peptide under mild buffer conditions. Moreover, to control for false positive signals, we also performed sequential ChIP with a green fluorescent protein (GFP) antibody in parallel. To exclude the possibility that a sequential ChIP signal was caused by adjacent particles, we subjected the cross-linked input chromatin to heavy MNase digestion. Sequential CENPA/CENPA ChIP resulted in 45-fold enrichment of centromeric α-satellite dimers relative to noncentromeric sequences (Fig. 3B). In contrast, sequential CENPA/GFP ChIP resulted in only background levels of enrichment of centromeric sequences relative to noncentromeric sequences, suggesting that our sequential ChIP method is highly specific and efficient. Importantly, sequential CENPA/CENPC and CENPA/CENPT ChIP resulted in approximately 10- to 35-fold enrichment of all centromeric α-satellite dimers relative to noncentromeric sequences (Fig. 3B). The strong enrichment of CENPC and CENPT in the CENPA ChIP implies that all three CCAN proteins are part of the same multisubunit chromatin complex.
To comprehensively confirm the qPCR results, we subjected the DNA from all the sequential ChIP experiments to Illumina sequencing. After mapping the reads to previously characterized α-satellite arrays (Hayden et al. 2013; Henikoff et al. 2015), we calculated the number of reads mapped to an array as a fraction of the total number of mapped reads (Fig. 4A). As was the case for qPCR, we observed a high fraction of sequential CENPA/CENPC, CENPA/CENPB, and CENPA/CENPT ChIP signals over centromeric α-satellites (∼4%–8% of both Cen1-like and D5Z2) relative to CENPA/GFP and input data sets. In contrast, we observed only background enrichment of noncentromeric α-satellites (∼0.002%–0.012% of both Xmono and D19Z1). Sequential ChIP fragment distributions were well correlated between CENPA and CENPA/CENPC and between CENPA and CENPA/CENPT but were essentially uncorrelated between CENPA and CENPA/GFP (Fig. 4B), consistent with CENPA, CENPC, and CENPT being present in the same cross-linked particle at sequences where they are enriched. We conclude that CENPA, CENPB, CENPC, and CENPT are part of the same complex at human centromeres.
Figure 4.
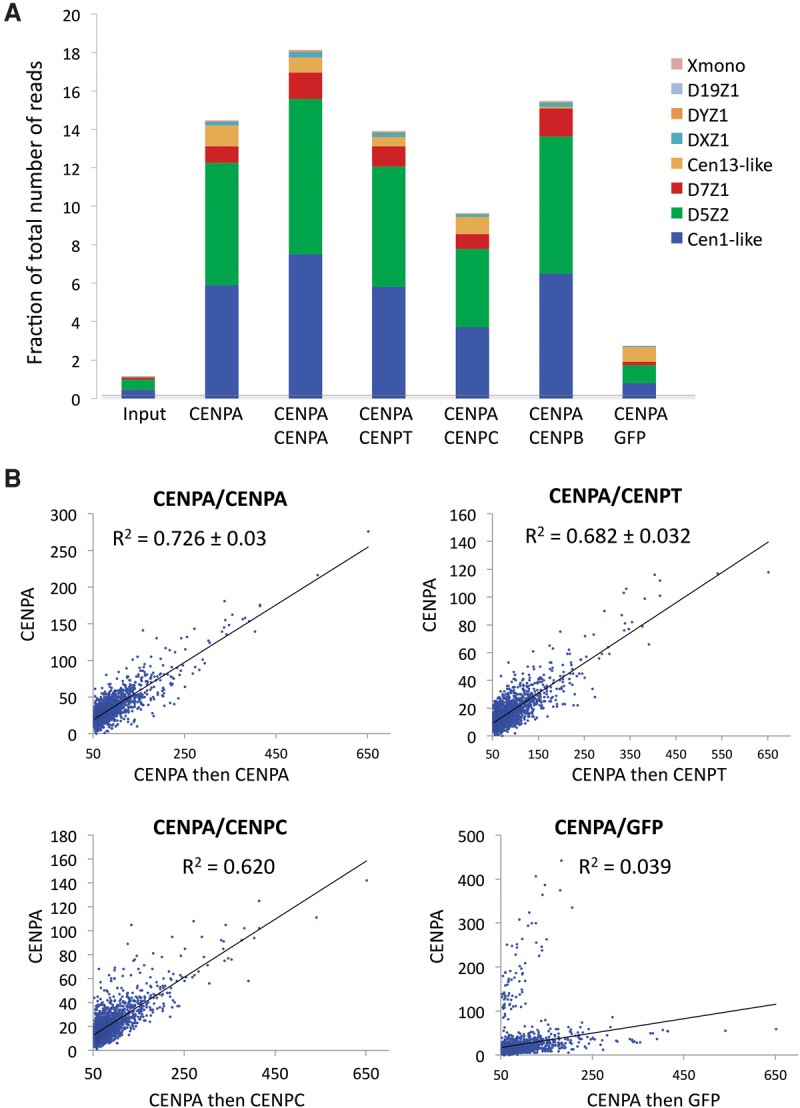
Sequential ChIP-seq data sets of CCAN components are enriched for centromeric α-satellites and are highly correlated. (A) Sequential ChIP-seq data sets show that abundant centromeric α-satellite arrays (D5Z2 and Cen1-like) are enriched for CENPA, CENPT, CENPC, and CENPB, whereas representative noncentromeric satellites (Xmono and D19Z1) are not. The reads were mapped to previously characterized α-satellite arrays (Hayden et al. 2013; Henikoff et al. 2015), and the number of reads mapped to an array were calculated (using Trim Galore!) as a fraction of the total number of mapped reads processed. (B) Close correspondence between sequential ChIP-seq data sets: CENPA/CENPA, CENPA/CENPC, and CENPA/CENPT. Scatter plots show the regression of the number of mapped reads for the CENPA/CENPA, CENPA/CENPC, CENPA/CENPT, and CENPA/GFP sequential ChIP data sets on the CENPA X-ChIP data set.
The CENPT particle bridges the gap between two precisely positioned CENPA nucleosomes at centromeres
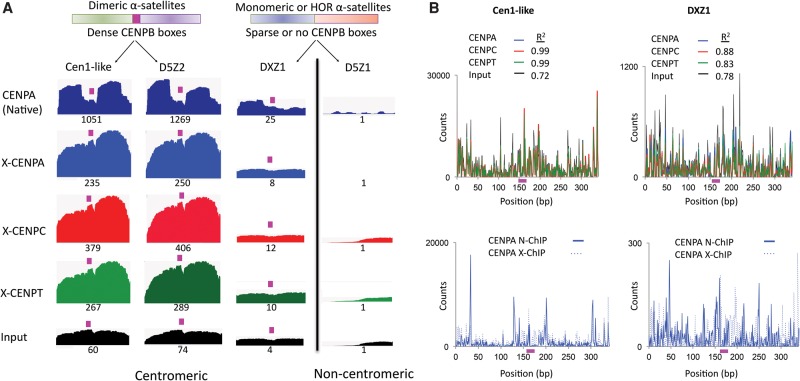
To determine whether the retention of CENPT affects the distinctive chromatin profiles seen with N-ChIP, we mapped total MNase X-ChIP paired-end reads to BACs containing the same α-satellite dimers on which N-ChIP paired-end reads were mapped. Interestingly, instead of two 100-bp “pillars” seen with N-ChIP for the most abundant α-satellite dimers, we observed a single broad peak for CENPA, CENPC, and CENPT (Fig. 5A; Supplemental Fig. S2). This filling of the gap between the two CENPA nucleosomes implies that cross-linking stabilizes CENPT, which helps to protect the ∼60-bp linker containing the CENPB box from MNase digestion. Mapping of CENPT to the linker also implies that it interacts with adjacent CENPA nucleosomes and with CENPC within the dimeric unit. This interaction leads to an increase in fragment length under the MNase digestion conditions that are used for X-ChIP (Fig. 2B). The DXZ1 dimer also showed similar X-ChIP profiles for CENPA, CENPC, and CENPT, as did all other centromeric α-satellites tested (Fig. 5A), confirming the inference from the highly concordant cluster frequency distributions and coherent size distribution of X-ChIP fragments that these three proteins are part of a single complex for HORs.
Figure 5.
CENPT and CENPC bridge the gap between adjacent CENPA nucleosomes over the CENPB box on young α-satellite dimers. (A) Mapping of X-ChIP merged pairs to previously characterized α-satellite arrays (Hayden et al. 2013; Henikoff et al. 2015). The profiles display normalized counts as described in Methods, in which entire fragments are “stacked,” and for each base-pair position the total number of counts is measured on the y-axis. A single α-satellite dimer from each array is shown. The CENPB box region is indicated by the magenta box. Cen1-like and D5Z2 arrays contain CENPB boxes in every α-satellite dimer (dense) (Henikoff et al. 2015). The DXZ1 tandem array, which is a 12-copy HOR, contains CENPB boxes only in a subset of dimeric units of the array (sparse), and a single CENPB box-containing dimer, which is embedded in α-satellite units that lack a CENPB box, is shown. The D5Z1 array does not contain CENPB boxes. The relative scale is the area of the indicated profile divided by the area of the D5Z1 profile, setting the D5Z1 value to 1, in which the numbers reflect the product of the total sequence abundance and enrichment. For example, Cen1-like is 60-fold enriched in the X-ChIP input, reflecting higher sequence abundance, and 235-fold enriched in CENPA ChIP, which implies that ChIP enrichment per copy is 235/60 or approximately fourfold. Biological replicates using different CENPT antibodies gave nearly identical patterns and relative scale values (Supplemental Fig. S2). (B) Fragment ends from native and X-ChIP data sets were mapped to the Cen1-like consensus and DXZ1 sequences. The CENPB box region is indicated by the magenta box. The top panel shows ends from CENPA, CENPC, CENPT X-ChIP, and input. The bottom panel compares ends of CENPA reads in N-ChIP (solid line) and X-ChIP (dotted line).
To detect possible chromatin particle substructures, we mapped MNase cleavage sites over dimeric units (Ramachandran et al. 2015). For all X-ChIP data sets (Fig. 5B, top; Supplemental Fig. S3), cleavage peaks were dispersed over the full 340-bp unit, with noticeable enrichment in the middle. In contrast, N-ChIP CENPA data sets (Fig. 5B, bottom; Supplemental Fig. S3) showed specific cleavages that represent the two precisely positioned 100-bp nucleosomes on the Cen1-like α-satellite dimer (Henikoff et al. 2015). Despite the lack of a distinct substructure in X-ChIP, we observed a striking overlap of cleavage peaks for CENPA, CENPC, and CENPT, with very high correlations (R2 = 0.99, Fig. 5B), further confirming that all three proteins reside within the same cross-linked complex. Strong overlap of cleavage patterns was also observed for X-ChIP of DXZ1.
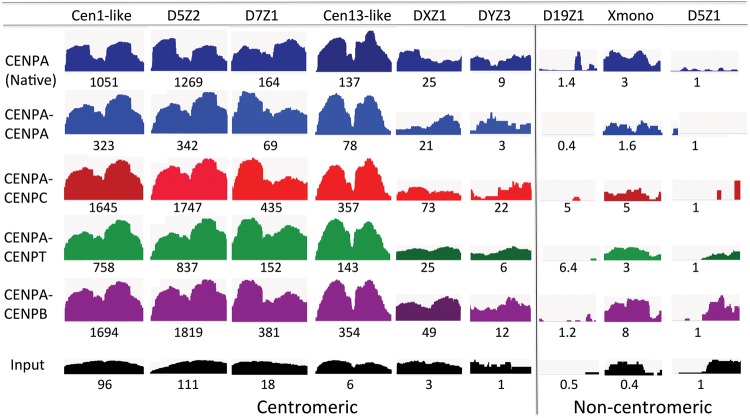
We also mapped sequential ChIP-seq reads to both centromeric and noncentromeric α-satellite arrays. The fragment length distribution of reads mapped to centromeric α-satellites revealed a sharp peak at ∼100 bp, resulting from enhanced cleavage between the two dimeric units despite protection by cross-linking (Supplemental Fig. S4). This also implies that we have successfully removed arrays from our sequential ChIP by heavy MNase digestion of cross-linked chromatin, and CENPC and CENPT are pulled down via their protein–protein interactions with CENPA nucleosomes. We observed similar profiles of CENPA, CENPB, CENPC, and CENPT on various α-satellite sequences in the sequential ChIP experiment (Fig. 6), further demonstrating their presence in a single complex.
Figure 6.
Sequential ChIP-seq profiles of CCAN components are nearly identical. Mapping of sequential ChIP-seq reads to α-satellite arrays (Hayden et al. 2013; Henikoff et al. 2015). A single α-satellite dimer from each array is shown, and the relative scale is the area of the indicated profile divided by the area of the D5Z1 profile, in which the numbers reflect the product of the total sequence abundance and enrichment. Because DXZ1, DYZ3, D19Z1, Xmono, and D5Z1 are not dimeric units, we chose pairs of tandem monomers as representatives.
Taken together with the retention of CENPT with cross-linking, the reduced MNase sensitivity, and the filling in of the ∼60-bp gap between CENPA nucleosomes, sequential ChIP of CENPT, CENPA, CENPB, and CENPC from CENPA nucleosomes confirms that these proteins reside in a single inner kinetochore complex.
Discussion
Mapping of human centromeric proteins has been impeded by the intractability of assembling homogeneous α-satellite sequences that span centromeric regions. Only HORs at centromere edges have been fully assembled, leaving unmapped multi-megabase gaps (Schueler et al. 2001; Hayden et al. 2013). Previously, using high-resolution N-ChIP-seq of CENPA and clustering of sequencing reads, we discovered that CENPA- and CENPC-enriched α-satellites are dominated by two distinct dimeric units, Cen1-like (340 bp) and Cen13-like (342 bp) (Henikoff et al. 2015), respectively derived from SF1 and SF2, which have been mapped to 20 human chromosomes (Alexandrov et al. 2001). In the present study, we used a genomic readout of protein–protein interactions to infer CCAN molecular architecture. We adopted a comparative ChIP approach to map the key components of the DNA-binding core CCAN complex on abundant α-satellite dimers and HORs. Our results revealed that the CENPT particle is highly sensitive to MNase digestion in vivo. We found that cross-linking retains CENPT, which is otherwise lost during chromatin preparation for N-ChIP. Our findings suggest that CENPT resides in the CENPB-box-containing linker between the two ∼100-bp CENPA particles and partially protects the larger complex from internal MNase cleavage.
The CENPC- and CENPT-mediated pathways for outer kinetochore assembly genetically interact with each other. For example, human CENPC requires CENPA for assembly, but CENPT requires both CENPA and CENPC (Fachinetti et al. 2013; Klare et al. 2015; Tachiwana et al. 2015). However, other evidence has suggested that CENPC and CENPT are spatially separated. For example, CENPA and CENPT did not appear to colocalize on a stretched chromosome fiber in DT40 cells (Ribeiro et al. 2010). FRET studies in human cells also showed that CENPT interacts with H3 but not with CENPA (Dornblut et al. 2014). These observations suggested that CENPT, together with its CENPW, CENPS, and CENPX histone-fold protein partners, is embedded within H3 nucleosome arrays at physically distinct locations from CENPA nucleosomes and forms an independent connection to the outer kinetochore. By mapping CENPT to the same young dimeric and pericentric HORs that are enriched in CENPA and depleted of H3, we can account for the genetic interactions between the two kinetochore assembly pathways. It is possible that the enormous abundance of H3 relative to CENPA complicates studies that do not physically separate CENPA chromatin from bulk chromatin. Because there are only approximately 400 CENPA molecules per human centromere (0.5–5 Mb), we infer that only the highly homogenous α-satellite dimers at the center of each centromere array are occupied by CENPA. In support of this interpretation, analysis of flow-sorted Chromosome 1 sequence libraries revealed that only a fraction of α-satellites on Chromosome 1 map with high identity to Cen1-like 340-bp dimeric consensus sequence (Supplemental Fig. S5A; S Kasinathan, pers. comm.). Both sequence similarity and CENPA enrichment decrease as the distance from the center increases, and H3 nucleosomes dominate the rest of the array (Henikoff et al. 2015). Because we cannot directly map larger arrays, it remains unclear whether homogeneous CENPA arrays at the center are continuous or interspersed with arrays of H3 nucleosomes as seen previously by fiber-FISH (Supplemental Fig. S5B; Blower et al. 2002).
Human CENPC binds to CENPA nucleosomes and dimerizes through its C terminus in vitro (Carroll et al. 2010; Kato et al. 2013). This implies that CENPC associates with CENPA nucleosomes, and two such CENPC molecules dimerize near the CENPB box to span adjacent CENPA nucleosomes over each young α-satellite dimer. A previous suggestion of CENPA and CENPT forming separate particles was based on complete MNase digestion to monomeric nucleosomes, which caused separation of the two proteins (Hori et al. 2008). Because protection of DNA is not assayed in such immunoprecipitation experiments, this result might be explained by the high sensitivity of CENPT chromatin to MNase digestion that we have observed. Therefore, our finding that both CENPT and CENPC are instead part of a single complex with CENPA resolves the question of how the two distinct pathways for assembling the inner kinetochore can be interdependent.
Our results suggest a model for centromeric chromatin structure at abundant homogeneous α-satellite dimers, in which each of the two 100-bp CENPA nucleosomes binds to a CENPC molecule (Fig. 7, left). The two CENPC molecules dimerize over the CENPB box. A CENPT/CENPW/CENPS/CENPX tetramer wraps the ∼60-bp CENPB-containing linker DNA and thus makes contacts with CENPC (Fig. 7, middle). This model is supported by the observation that CENPT/CENPW/CENPS/CENPX nucleosome-like particles assemble on the linker regions of reconstituted dimeric nucleosomes (Takeuchi et al. 2014).
Figure 7.
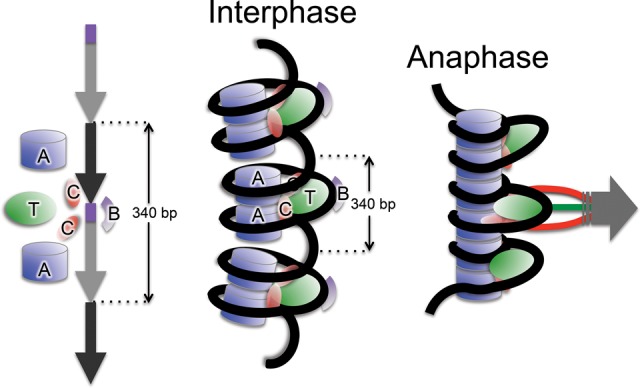
A model for the CENPA/CENPB/CENPC/CENPT chromatin complex. (Left) Hypothesized arrangement of CENPA, B, C, and T along a single 340-bp Cen1-like dimeric unit. (Middle) During interphase, the CENPT complex occupies the CENPB box-containing linker on young 340-bp α-satellite dimers between tandem CENPA nucleosomes. Each CENPA particle binds a CENPC on one side and wraps ∼100-bp DNA with right-handed chirality. (Right) At anaphase, tension causes the positively supercoiled DNA to overwind and tighten up around the complex, favoring stacking of tandem dimers to form a stiff platform that spreads pulling forces.
Our model can account for the recent observation that octameric CENPA nucleosomes reconstituted with 145 bp DNA from a single unit of the 12-unit DXZ1 HOR (with flanking 25-bp linkers) are stabilized by CENPC binding (Falk et al. 2015). The DXZ1 dimeric cross-linked particle resembles the Cen1-like particle (which is ∼50× more abundant than DXZ1) (Fig. 5A) in both composition and MNase protection in X-ChIP. We attribute the relative instability of the reconstituted particle that lacks CENPC both to the insufficiency of hybrid 195-bp monomeric α-satellite DNA to wrap the full in vivo complex detected by X-ChIP and to the lack of CENPT/CENPW/CENPS/CENPX in the reconstitution. Addition of CENPC would both stabilize the reconstituted particle and increase the fraction of the 100-bp MNase-protected particle, the most prominent size class seen by N-ChIP in vivo (Hasson et al. 2013; Henikoff et al. 2015).
The reconstituted CENPT/CENPW/CENPS/CENPX complex has been shown to induce positive DNA supercoils, which implies a right-handed wrap, opposite the wrap of octameric nucleosomes (Takeuchi et al. 2014). Insofar as budding yeast CENPA nucleosomes also wrap DNA with right-handed chirality in vivo (Furuyama and Henikoff 2009; Huang et al. 2011; Díaz-Ingelmo et al. 2015), it is tempting to speculate that CENPA and CENPT subcomplexes coordinate to wrap a right-handed inner kinetochore chromatin complex. As DNA overwinds when stretched (Gore et al. 2006), a right-handed wrap would be expected to tighten up under anaphase tension to provide a solid foundation for chromosome segregation (Fig. 7, right).
Methods
Cell culture
The HuRef lymphoblastoid cell line (Levy et al. 2007) was grown in RPMI media supplemented with 15% FBS and 1 mM pyruvate using standard protocols. The HT1080 and HT1080-1b cell lines were grown in DMEM media supplemented with 15% FBS. The HT1080-1b monoclonal cell line was generated by expressing CENPA-FLAG in the HT1080 background. Based on the difference between the ChIP signals for anti-CENPA in the untagged cell line and anti-FLAG in the HT1080-1b cell line, we estimate that there might have been a threefold overexpression of the CENPA-FLAG. We did not observe enrichment over the 5S control locus, which implies that this level of overexpression did not result in mislocalization.
Chromatin procedures
N-ChIP and X-ChIP assays were performed as described (Henikoff et al. 2015; Skene and Henikoff 2015). Illumina DNA sequencing libraries were produced as previously described (Henikoff et al. 2015). 2 × 25-bp paired-end sequencing was performed for CENPB and CENPT N-ChIP, 2 × 100-bp paired-end sequencing was performed for X-ChIP, and 250-bp single-end sequencing was performed for sequential ChIP. qPCR was performed using an Applied Biosystems Step One system following the manufacturer's instructions. The following antibodies were used: anti-CENPA (Abcam Ab13939), anti-CENPC (Abcam, Ab33034), and anti-CENPT (Abcam Ab114120 and Bethyl A302-313A).
For sequential ChIP, primary X-ChIP was performed as described previously (Skene et al. 2014) with modifications as follows. Briefly, 200 million cells per IP were cross-linked with 1% formaldehyde, and MNase digestion was carried out at a concentration of ∼0.5–1 unit per million cells for 45 min. MNase digested chromatin was lightly sonicated to solubilize chromatin as previously described (Skene et al. 2014), and solubilized chromatin was precleared using Protein A Sepharose beads for 30 min at 4°C. The precleared chromatin was incubated with Anti-FLAG-M2 magnetic beads (Sigma catalog #M8823) for 4 h at 4°C. The FLAG-M2 magnetic beads were washed once with buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl), four times with buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 500 mM NaCl), two times with buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.1]), and once with 1× TE. Protein complexes were eluted from magnetic beads using FLAG peptide in HE buffer (10 mM HEPES [pH 7.4], 0.25 mM EDTA, 0.5 mM phenylmethanesulfonyl fluoride). The eluent was subjected to subsequent ChIP reaction with anti-CENPA, anti-CENPB (Abcam catalog #Ab25734), anti-CENPC, anti-CENPT, or anti-GFP (Abcam catalog #Ab290) antibodies.
Sequence analysis
Paired-end 2 × 25-bp reads sequenced from N-ChIP libraries were mapped to previously described dimeric α-satellite units (GEO accession number GSE60951) using BWA (Version 0.7.10; http://bio-bwa.sourceforge.net/bwa.shtml; parameters aln and sampe -n 10) (Henikoff et al. 2015). BWA requires a maximum number of read matches to a reference sequence that it will save, and we chose 10 as a number that is much greater than the number of times any DNA fragment can plausibly match our short reference sequences using stringent criteria. This way, we would recover all of the best alignments over a tandemly repeated reference sequence of fixed length. Mapped fragments >185 bp in length were discarded. Paired-end 2 × 100-bp reads sequenced from X-ChIP libraries were merged using SeqPrep (https://github.com/jstjohn/SeqPrep; parameters -q 30 -L 25) to yield merged pairs ranging from 25 to 185 bp in length. Single-end 250-bp reads sequenced from X-ChIP libraries were trimmed with Trim Galore! version 0.3.7 using parameters –quality 20 –adapter AGATCGGAAGAGC –stringency 3 –phred33 –length 25 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). BWA (Li and Durbin 2010) was then used to map the merged pairs to 1-kb segments of BACs characterized by Hayden et al. (2013), saving up to 10 alignments per merged pair (Version 0.7.10, using the aln defaults and samse -n 10). The BACs were identified from GenBank for each of the ∼340-bp sequences, and one of the sequences with the highest BLAST score was extracted with its flanking dimeric units to give a 1-kb reference sequence of three tandem dimers. For tracks from all types of alignments, we counted the number of fragments aligned over each base pair and normalized base-pair counts by dividing by the number of paired reads (2 × 25-bp), merged pairs (2 × 100-bp), or trimmed reads (1 × 250-bp) presented to the alignment program. The number of merged pairs or trimmed reads mapped to a reference sequence was counted for regression analysis (Figs. 3A, 4B). For display in Figures 5 and 6, representative tandem dimers were cut out from the 1-kb reference sequences. For fragment end analysis (Fig. 5B), the reference sequence consisted of the most abundant α-satellite dimer flanked by its neighboring dimers to capture fragments spanning the edges of the reference sequence, and left and right mapped ends were combined for display.
Data access
Sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE69839.
Supplementary Material
Acknowledgments
We thank Jorja Henikoff for computational support and data analysis, Christine Codomo for preparing DNA sequencing libraries, the Fred Hutch Genomics Shared Resource for Illumina sequencing, Kayoko Maehara for providing the CENPA-FLAG construct, Peter Skene for sharing MNase X-ChIP protocols prior to publication, and Srinivas Ramachandran for thoughtful discussions. We also thank Siva Kasinathan for communicating unpublished results on the centromere-specific abundances of the Cen1-like dimer, and Paul Talbert, Srinivas Ramachandran, Sue Biggins, Siva Kasinathan, and Kami Ahmad for comments on the manuscript. This work was supported by the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.204784.116.
Freely available online through the Genome Research Open Access option.
References
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. 2001. α-Satellite DNA of primates: old and new families. Chromosoma 110: 253–266. [DOI] [PubMed] [Google Scholar]
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 186: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. 2014. The quantitative architecture of centromeric chromatin. eLife 3: e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. 2008. Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol 9: 33–46. [DOI] [PubMed] [Google Scholar]
- Díaz-Ingelmo O, Martínez-García B, Segura J, Valdés A, Roca J. 2015. DNA topology and global architecture of point centromeres. Cell Rep 13: 667–677. [DOI] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. 2010. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci 107: 20317–20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornblut C, Quinn N, Monajambashi S, Prendergast L, van Vuuren C, Münch S, Deng W, Leonhardt H, Cardoso MC, Hoischen C, et al. 2014. A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle. Open Biol 4: 130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 15: 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LE, Van Duyne GD, Vinogradov SA, et al. 2015. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348: 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli P, Antonacci R, Marzella R, Lonoce A, Archidiacono N, Rocchi M. 1996. Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics 38: 325–330. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. 2009. Centromeric nucleosomes induce positive DNA supercoils. Cell 138: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. 2012. T time for point centromeres. Nat Cell Biol 14: 559–561. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Bryant Z, Nöllmann M, Le MU, Cozzarelli NR, Bustamante C. 2006. DNA overwinds when stretched. Nature 442: 836–839. [DOI] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol 20: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. 2013. Sequences associated with centromere competency in the human genome. Mol Cell Biol 33: 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Thakur J, Kasinathan S, Henikoff S. 2015. A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci Adv 1: e14000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052. [DOI] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D'Andrea AD. 2010. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell 39: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Chang KM, Cui H, Jayaram M. 2011. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc Natl Acad Sci 108: 13671–13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Warburton PE. 1997. Centromeres, CENP-B and Tigger too. Trends Genet 13: 141–145. [DOI] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. 2015. CENP-C is a blueprint for constitutive centromere–associated network assembly within human kinetochores. J Cell Biol 210: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K, Henikoff JG, Henikoff S. 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci 109: 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. 2007. The diploid genome sequence of an individual human. PLoS Biol 5: e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM. 2015. The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol Cell 60: 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300. [DOI] [PubMed] [Google Scholar]
- Quénet D, Dalal Y. 2012. The CENP-A nucleosome: a dynamic structure and role at the centromere. Chromosome Res 20: 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago F, Gascoigne KE, Cheeseman IM. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol 25: 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Zentner GE, Henikoff S. 2015. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res 25: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. 2010. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci 107: 10484–10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF. 2003. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol 68: 141–149. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. 2010. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 37: 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2015. A simple method for generating high-resolution maps of genome-wide protein binding. eLife 4: e09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Hernandez AE, Groudine M, Henikoff S. 2014. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife 3: e02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP, Schindelhauer D, et al. 2012. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene 31: 3244–3253. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S. 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 3: e02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Müller S, Blümer J, Klare K, Musacchio A, Almouzni G. 2015. HJURP involvement in de novo CenH3CENP-A and CENP-C recruitment. Cell Rep 11: 22–32. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Nishino T, Mayanagi K, Horikoshi N, Osakabe A, Tachiwana H, Hori T, Kurumizaka H, Fukagawa T. 2014. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res 42: 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Talbert PB, Henikoff S. 2015. Inner kinetochore protein interactions with regional centromeres of fission yeast. Genetics 201: 543–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L, Booth DG, Vargiu G, Ohta S, deLima Alves F, Samejima K, Fukagawa T, Rappsilber J, Earnshaw WC. 2016. Auxin/AID versus conventional knockouts: distinguishing the roles of CENP-T/W in mitotic kinetochore assembly and stability. Open Biol 6: 150230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. 2014. High-resolution digital profiling of the epigenome. Nat Rev Genet 15: 814–827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.