Abstract
Alpha satellite is a tandemly organized type of repetitive DNA that comprises 5% of the genome and is found at all human centromeres. A defined number of 171-bp monomers are organized into chromosome-specific higher-order repeats (HORs) that are reiterated thousands of times. At least half of all human chromosomes have two or more distinct HOR alpha satellite arrays within their centromere regions. We previously showed that the two alpha satellite arrays of Homo sapiens Chromosome 17 (HSA17), D17Z1 and D17Z1-B, behave as centromeric epialleles, that is, the centromere, defined by chromatin containing the centromeric histone variant CENPA and recruitment of other centromere proteins, can form at either D17Z1 or D17Z1-B. Some individuals in the human population are functional heterozygotes in that D17Z1 is the active centromere on one homolog and D17Z1-B is active on the other. In this study, we aimed to understand the molecular basis for how centromere location is determined on HSA17. Specifically, we focused on D17Z1 genomic variation as a driver of epiallele formation. We found that D17Z1 arrays that are predominantly composed of HOR size and sequence variants were functionally less competent. They either recruited decreased amounts of the centromere-specific histone variant CENPA and the HSA17 was mitotically unstable, or alternatively, the centromere was assembled at D17Z1-B and the HSA17 was stable. Our study demonstrates that genomic variation within highly repetitive, noncoding DNA of human centromere regions has a pronounced impact on genome stability and basic chromosomal function.
Genomic variation, in the form of single-nucleotide polymorphisms (SNPs) and insertion-deletions (indels) within coding and regulatory regions and copy number variation (CNV) within both coding and noncoding regions, is often linked to alterations in gene expression and function (Hamilton 2002; Haraksingh and Snyder 2013). Most studies of genome variation have focused on gene expression, so little is known about how variation within highly repetitive sequences might be linked to broader chromosomal function. Alpha satellite DNA, a repetitive DNA that is present at all human centromeres, is a good example of this gap between difficult genomic regions and functional consequences of variation. Alpha satellite is defined by 171-bp monomers that are 50%–70% identical in sequence (Willard 1985). Monomers are typically tandemly arranged, so that a defined number of monomers creates a higher-order repeat (HOR) array. The size of the HOR (i.e., the number of monomers) gives rise to chromosome specificity. For instance, the HOR of Homo sapiens Chromosome X (HSAX) is defined by a 12-monomer HOR, while the HOR of HSA8 is defined by six monomers. Monomers can be grouped into suprachromosomal families, based on the organization of specific monomers into groups on different chromosomes (for detailed information on alpha satellite organization, the reader is referred to Willard 1985; Willard and Waye 1987; Alexandrov et al. 1988, 1993, 2001; Shepelev et al. 2015). HORs that are 97%–100% identical are reiterated hundreds to thousands of times, creating highly homogeneous alpha satellite arrays that stymie standard genome assembly (for review, see Miga 2015). As a result, centromeric gaps in genome assemblies have precluded cataloguing the amount and extent of variation within this type of DNA. Capturing variation in highly repetitive sequences even in a single individual like HuRef is difficult, although a few recent studies have highlighted the possibility of assessing variation within complex satellite DNA (Miga et al. 2014, 2015; Miga 2015).
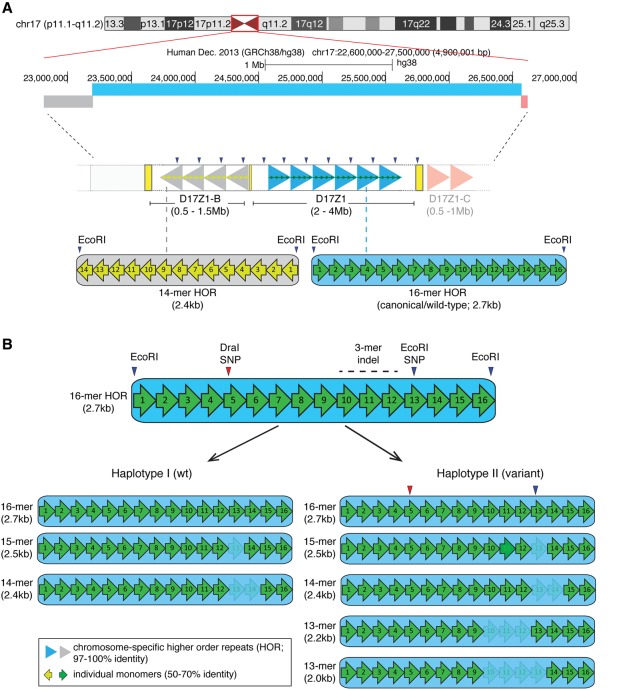
HSA17 has a complex centromere region. It contains three distinct alpha satellite arrays, D17Z1, D17Z1-B, and D17Z1-C (Fig. 1; Waye and Willard 1986b; Willard and Waye 1987; Rudd and Willard 2004; Shepelev et al. 2009). D17Z1 is the larger array, spanning 1–4 megabases (Mb) in various individuals (Wevrick and Willard 1989; Warburton and Willard 1990). D17Z1-B, oriented toward the short arm side of the D17Z1, and D17Z1-C, located on the long arm side of D17Z1, are smaller arrays, each estimated to be <1 Mb in size (Rudd et al. 2006; Shepelev et al. 2009; K Chew and BA Sullivan, unpubl.). There is little contiguous sequence information between D17Z1-B/D17Z1 and D17Z1/D17Z1-C; however, BAC end sequencing supports that the arrays are essentially adjacent (Rudd et al. 2006). In previous studies, we demonstrated that the functional centromere on HSA17 is assembled at either D17Z1 or D17Z1-B (Maloney et al. 2012). We showed that in ∼70% of individuals studied, the centromere is assembled at D17Z1, while in 30% of individuals, the centromere is assembled at D17Z1 on one homolog and at D17Z1-B on the other HSA17 homolog. To date, no individuals have been identified that exhibit centromere assembly at D17Z1-B on both HSA17 homologs. D17Z1-B can robustly support centromere assembly in human artificial chromosome (HAC) assays (Maloney et al. 2012; Hayden et al. 2013); therefore, it is possible that D17Z1-B/D17Z1-B homozygous centromeres represent rare alleles that may only be identified by deeper screening of the population. The molecular basis of these HSA17 centromeric epialleles and (epi)genomic features that direct their assembly are unknown. We hypothesized that genomic variation within alpha satellite DNA could be one factor that influences epiallele choice on HSA17.
Figure 1.
Homo sapiens Chromosome 17 (HSA17) centromeric satellite organization. (A) HSA17, illustrated by the UCSC Genome Browser chromosome ideogram, has three distinct alpha satellite arrays with different monomer organizations of higher order repeats (HORs). D17Z1 (blue bar) is comprised of canonical/wild-type 16-monomer (16-mer) HORs (large blue arrowheads) that are operationally defined by EcoRI restriction sites. D17Z1-B (gray bar), located toward the short arm side of the centromere region, is based on a 14-mer HOR (large gray arrowheads) that is also defined by EcoRI sites. D17Z1-C, a third array oriented toward the long arm side of the centromere (light red bar) is also defined by a 14-mer HOR (light red arrowheads) but is less well characterized and not a focus of this manuscript. Individual monomers (green or yellow arrows) in the HORs are <70% identical to each other but are arranged nonrandomly in the same order (i.e., monomer 1 through monomer 16). The HORs are repeated hundreds to thousands of times to create highly homogenous arrays that span multiple megabases. This high degree of homogeneity has confounded standard genomic assemblies of centromere regions. (B) D17Z1 is an extremely polymorphic alpha satellite array; D17Z1-B exists exclusively as a 14-mer HOR array. In D17Z1, single and multiple monomer deletions caused by repeated rounds of unequal crossing over and/or gene conversion produce HOR variants that differ in length by an integral number of monomers. In the general population, HOR variants range from 15-mers to 12-mers, with rare 11-mers (not shown) occurring in isolated populations or individuals (Warburton and Willard 1995). Two major D17Z1 haplotypes (I, II) exist in the population and are primarily distinguished by the presence or absence of a 13-mer, created by a three-monomer deletion (3-mer indel). Additional restriction enzyme polymorphisms (DraI in monomer 5 and a second EcoRI SNP in monomer 13) are in linkage disequilibrium with the 13-mer HOR (Warburton et al. 1991). Individual monomers are denoted by numbered arrows. Shading/lightness indicates monomers that are deleted in specific HOR variants.
Alpha satellite arrays vary in several ways. First, the different monomer organizations of the HOR distinguish distinct alpha satellite arrays. The overall size of an array can vary based on the number of times an HOR is repeated, such that between homologs in the same individual or between individuals, the same alpha satellite array can range from 1–4 Mb. These are important features that allow arrays on the same chromosome to be molecularly distinguished. On HSA17, D17Z1-B and D17Z1-C appear to exist only as 14-mer HORs; the amount of sequence variation within the HORs is not known. Conversely, D17Z1 exhibits a high degree of polymorphism, including overall array size, HOR size, and specific sequence features (Fig. 1B). The enzyme EcoRI operationally defines the D17Z1 HOR, designating monomer 1 of the HOR and demarcating the boundary between individual HORs (Waye and Willard 1986b). A common SNP that introduces an EcoRI site in monomer 13 is located in a subset of HORs. A second major polymorphism is the deletion of three monomers (monomers 10–12), resulting in HORs that consist of only 13 monomers (13-mer) (Fig. 1B; Waye and Willard 1986b). D17Z1 exhibits additional HOR variants that depart from the canonical or wild-type (16-mer) size. Single and multiple monomeric deletions caused by unequal crossing over produce other HORs that range from 15-mers to 11-mers (Fig. 1B; Waye and Willard 1986a; Warburton and Willard 1995). Arrays containing the EcoRI SNP and these monomeric deletions exist in distinct haplotypes in the population (Warburton and Willard 1995). The “wild-type” haplotype (Haplotype I) consists of 16-, 15-, and 14-mers and occurs in ∼65% of the population. Individuals in Haplotype I have D17Z1 arrays consisting of HORs that are pure 16-mers or a mixture of 16-, 15-, and 14-mers. D17Z1 arrays in individuals with Haplotype II (∼35% of population) contain 16-, 15-, and 14-mers, as well as additional 13- and 12-mers. The polymorphic EcoRI site exists in moderate linkage disequilibrium with the 13-mer variant (Haplotype II). Finally, D17Z1 exhibits array size variation that reflects the total number of HORs. D17Z1 arrays vary in size from 2–4 Mb (Wevrick and Willard 1989).
The frequency of individuals that have HSA17s in which the centromere assembles at D17Z1-B and the proportion of HSA17s that contain variant HORS (indel and SNP) are remarkably similar (∼30%) (Warburton and Willard 1995; Maloney et al. 2012). We hypothesized based on our previous studies that D17Z1 variation might influence centromere location and function. In this work, we investigated the role of sequence (SNP, indel) and structural variation (SV; satellite copy number) within alpha satellite DNA in the establishment of centromeric epialleles on HSA17. We find that extensive sequence and structural variation negatively correlates with the location of centromere assembly and/or impairs centromere function and HSA17 chromosome stability.
Results
D17Z1 HOR sequence variation is associated with metastable centromeric epialleles
Centromere location on HSA17s in the current data set had either been determined previously or had to be established in this study (Table 1). We used CENPA immunostaining and CENPA ChIP-PCR to assign the location of centromere assembly, so that each HSA17 in our data set was functionally characterized before proceeding to the analysis of molecular variation (Table 1; Supplemental Fig. S1; Maloney et al. 2012). Our data from previously published studies suggested that variation within the HORs of D17Z1 arrays could be linked to centromere location on HSA17 (Maloney et al. 2012). Comparison of the ratio of 16-mers and 13-mers had revealed that D17Z1 arrays with a greater proportion of 13-mers were less likely to be the site of centromere assembly (Maloney et al. 2012). However, our earlier approach did not take into account any other HOR variants (i.e., 15-, 14-, 12-, 11-mers) or sequence variants, such as the EcoRI SNP, that are found within a substantial subset of D17Z1 arrays (Willard et al. 1987; Warburton and Willard 1995). The EcoRI SNP found within monomer 13 of D17Z1 is easily identified by limited PCR amplification of the D17Z1 array (monomers 6–16), followed by digestion with EcoRI. In this approach, the entire spectrum of HOR size variants are revealed, from 16-mer to 11-mer, as well as the EcoRI SNP that yields the 9-/4-mer combination found in polymorphic 13-mers (3-mer indel). The PCR-restriction digestion assay also reveals rare 16-mer HORs that contain the EcoRI SNP in monomer 13. Furthermore, the assay is quantitative, in that band intensities are representative of HOR variant abundance within a multi-megabase-sized array.
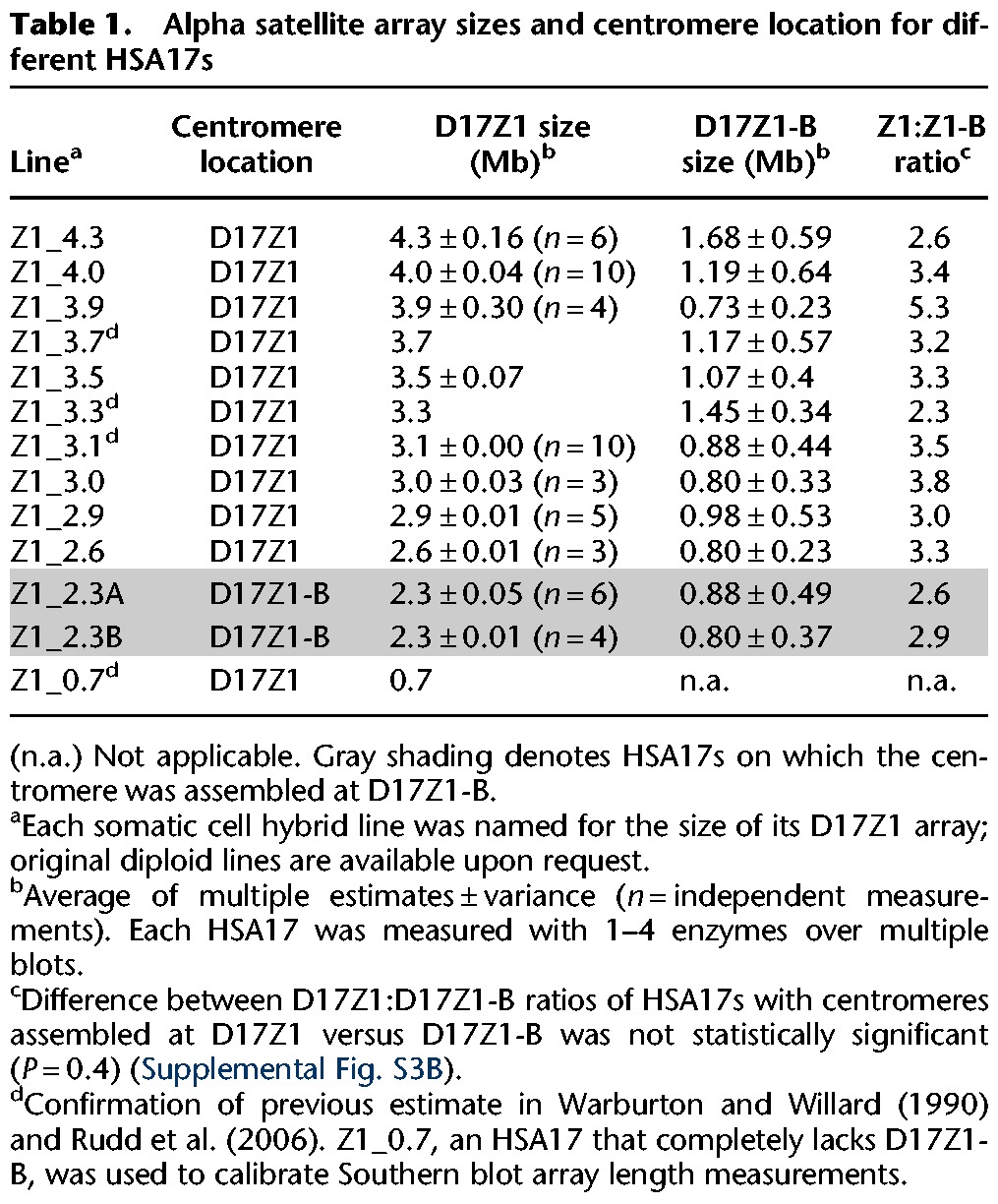
Table 1.
Alpha satellite array sizes and centromere location for different HSA17s
A second approach that we used to measure HOR level variation was EcoRI digestion of genomic DNA, followed by conventional gel electrophoresis and quantitative Southern blotting. This was the classical approach taken decades ago to define and characterize HOR variation within alpha satellite arrays. An advantage of the PCR assay is that it can be performed in half the time required for the Southern blotting approach. Moreover, it is as quantitative as Southern blotting, and in fact, we found no statistically significant differences in the amount of variation measured in multiple diploid and single HSA17 lines when comparing the PCR assay and Southern blotting approach (Supplemental Fig. S2B). Going forward, this PCR approach will be advantageous for screening a larger section of the population in order to identify additional individuals who have HSA17 epialleles.
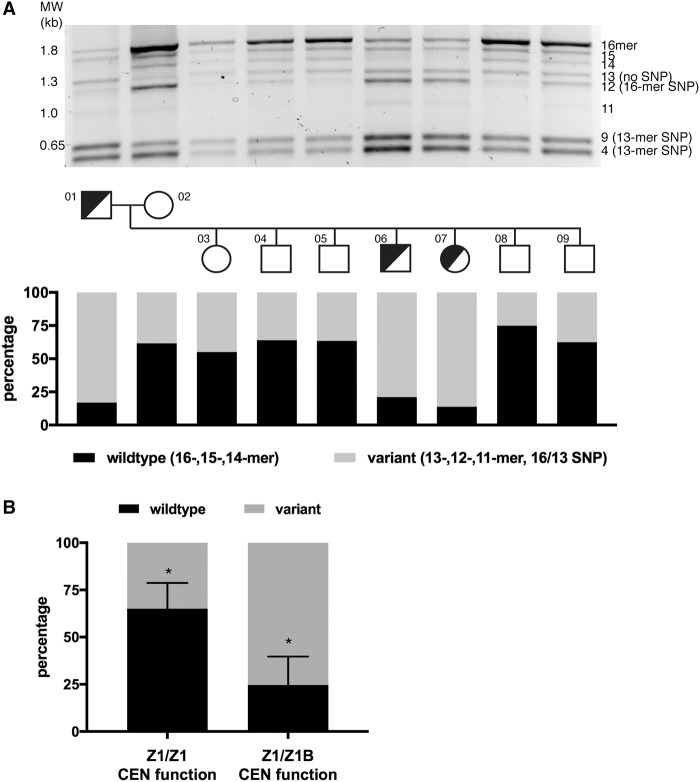
We applied these two (semi-)quantitative assays to measure variation in a multigenerational family (CEPH 1345) that segregates HSA17 epialleles (Fig. 2A; Supplemental Fig. S2A; Maloney et al. 2012). The proportion of wild-type versus variant HORs was calculated by digitally quantitating band intensities from PCR agarose gels and/or Southern blots. Both PCR and Southern blotting were quantitatively similar (Supplemental Fig. S2B). The wild-type designation included 16-mer HORs, as well as 15- and 14-mer HORs, since they carry the major alleles at the EcoRI SNP and 3-mer indel (Fig. 2; Supplemental Fig. S2A). Variant HORs included the indel HOR (13-mer) as well as 12-, 11-, 9-, and 4-mers. The 12-/4-mer bands included rare 16-mer HORs containing the EcoRI SNP, while the 9-/4-mer combination represented indel/13-mers containing the SNP.
Figure 2.
Extensive D17Z1 variation is associated with centromeric epialleles. (A) D17Z1 variation was detected using PCR followed by restriction digestion to reveal HOR size variation as well as identify HORs containing the EcoRI SNP that segregates with the 13-mer/indel HOR. D17Z1 variation within two generations of the three generation CEPH/Utah 1345 family is shown (for data on the third generation, see Supplemental Fig. S2). This family has individuals that are centromeric functional heterozygotes (half-shaded circles or squares): One homolog assembles the centromere at D17Z1 (Z1) and the other homolog assembles the centromere at D17Z1-B (Z1-B). Squares represent males; circles represent females. Each family member is numbered according to the original classification of the pedigrees (Dausset et al. 1990). Agarose gels were imaged as white bands on black background; the images were inverted for presentation purpose only. Quantitation of the amount of wild-type HORs (16-, 15-, 14-mers, Haplotype I) versus variant HORs (13-, 12-, 11-mers, and 13-mer SNP represented by 9 + 4 bands, Haplotype II) was measured. Individuals with HSA17 centromere epialleles (half-shaded squares and circles) had D17Z1 arrays with >80% variation. (B) In CEPH family 1345, the correlation between all individuals with D17Z1 variation and centromeric epialleles (D17Z1-B CEN function on one homolog only) compared to individuals lacking epialleles (D17Z1/D17Z1 CEN function) was statistically significant (asterisks indicate P < 0.001).
Quantification of HOR variation within CEPH 1345 (diploid) individuals revealed that some arrays were composed entirely of wild-type HORs, while others contained few wild-type HORs (Fig. 2B; Supplemental Fig. S2C). The presence of the indel 13-mer HOR was not significantly associated with active or inactive D17Z1 arrays (Supplemental Fig. S2C,D). However, a high proportion of HORs containing the EcoRI SNP was significantly associated with inactive D17Z1 (Fig. 2B,C; Supplemental Fig. S2D). Overall, the total variant composition (indel plus EcoRI SNP) between active versus inactive D17Z1 arrays was statistically significant (Fig. 2C). Individuals with D17Z1 arrays that contained >50% wild-type HORs were associated with active D17Z1 arrays. Alternatively, individuals in which D17Z1 exhibits >80% variation carried HSA17 epialleles (i.e., centromere assembly at D17Z1-B on one homolog) (Fig. 2B). To determine if these genomic predictions extended beyond the CEPH 1345 family, we analyzed additional diploid lines in which HSA17 had been functionally characterized. Again, highly variant D17Z1 arrays were associated with centromere assembly at D17Z1-B (Supplemental Fig. S2E). Collectively, these results suggest that centromere assembly on HSA17 is negatively associated with highly variant arrays.
Large homogeneous arrays of D17Z1 are the sites of centromere assembly on HSA17
We predicted that centromere assembly would occur at D17Z1 on invariant arrays, while assembly at D17Z1-B would occur on HSA17s containing highly variant D17Z1 arrays. A caveat of the previous set of experiments (i.e., CEPH 1345 family) was that the diploid individuals/cell lines were analyzed. This made it difficult to definitively determine the amount of variant HORs within the D17Z1 array of a single HSA17 homolog. In addition, it is difficult to determine the size of each alpha satellite array in diploid cells. This is relevant because the total size of an alpha satellite array might predict where the centromere assembles, either independently or dependent on proportion of variation.
Therefore, we studied individual HSA17s that had been isolated from diploid human cell lines and transferred into human-mouse somatic cell hybrid lines. Our samples included those from our previous study (Maloney et al. 2012) as well as several new HSA17s (n = 12 total) (Table 1). This sample size allowed us to achieve sufficient statistical power (90% power, 0.01 CI) to test the hypothesis that large D17Z1 arrays are more likely to be the site of centromere assembly on D17Z1. Total D17Z1 alpha satellite array sizes were estimated using restriction digestion of high molecular weight DNA that releases the D17Z1 as one or a few large fragments that can be resolved by pulsed field gel electrophoresis (Fig. 3A; Mahtani and Willard 1990; Sullivan et al. 2011). Although some of the D17Z1 array sizes were previously known, D17Z1-B array sizes have not been previously reported. Therefore, we estimated the size of D17Z1-B arrays for every HSA17 in our entire data set using DNA fiber FISH or interphase FISH with probes specific to D17Z1 and D17Z1-B (Table 1; Fig. 3B; Supplemental Fig. S4A). Since each D17Z1 array size was known (from the present study or Maloney et al. 2012), the D17Z1 fluorescent signal was measured in microns and assigned a value in megabases. The D17Z1-B probe was also measured, and the D17Z1-B array size was calculated based on the D17Z1 fluorescent probe/array size. D17Z1 sizes ranged in size from 2.3–4.3 Mb (Supplemental Fig. S3), and D17Z1-B arrays ranged from 0.5–1.6 Mb (Table 1; Fig. 3B; Supplemental Fig. S4A). We found that HSA17s in which D17Z1-B was the active centromere tended to have the smallest D17Z1 arrays (Supplemental Fig. S3). This raised the possibility that the ratio of D17Z1:D17Z1-B size might be related to centromere location, such that D17Z1 and D17Z1-B arrays of similar size may compete for centromere function. However, when we compared D17Z1:D17Z1-B ratios for HSA17s in which the centromere formed at Z1 versus Z1-B, there was not a statistically significant difference (Supplemental Fig. S4B).
Figure 3.
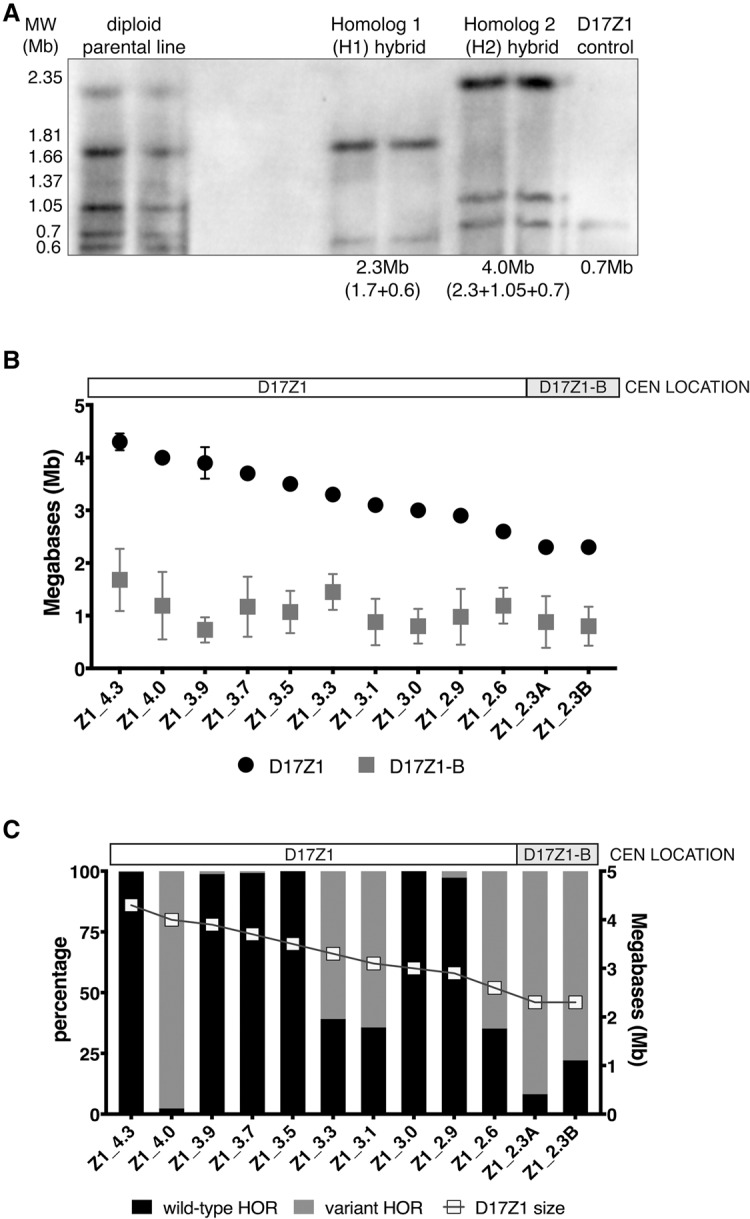
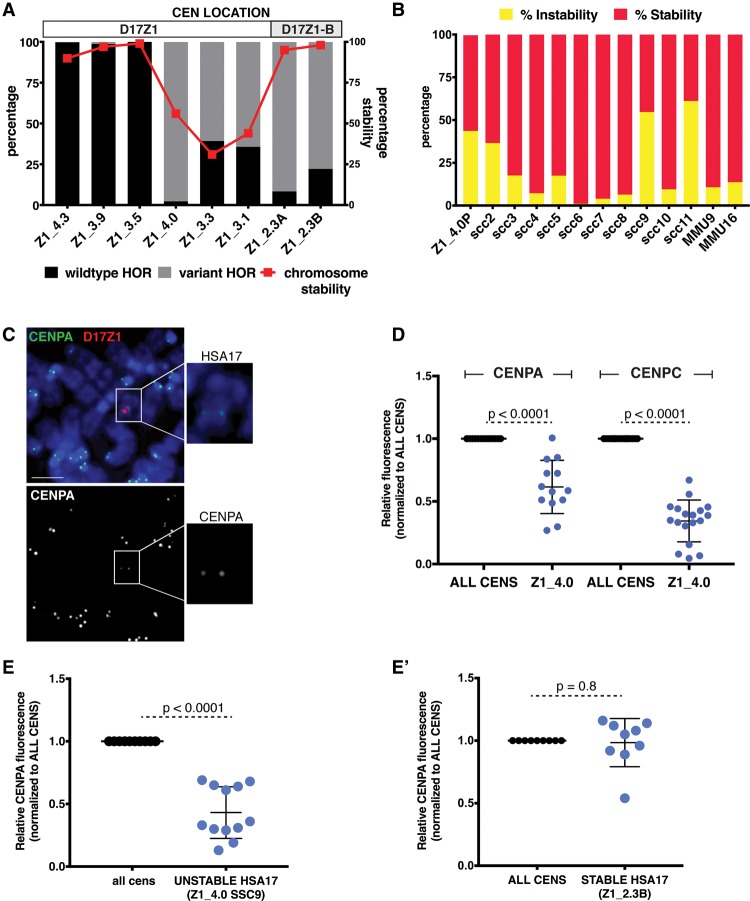
Larger D17Z1 arrays tend to be more homogeneous and are the site of centromere assembly. (A) Although the sizes of many D17Z1 arrays in our data set were already known (Maloney et al. 2012), D17Z1 arrays in new somatic cell hybrid lines were molecularly sized. D17Z1 array sizes were estimated using restriction digestion with enzymes that cut infrequently within the alpha satellite array, followed by resolution of large DNA fragments by pulsed field gel electrophoresis and Southern blotting. Representative Southern blot shows hybridization with a D17Z1-specific DNA probe p17H8. The parental diploid line shows many large DNA fragments from both HSA17 homologs. Individual HSA17 array sizes could only be resolved by moving each HSA17 homolog from the diploid line into the somatic cell hybrid background. Multiple bands were added to estimate the final array sizes. In this example, D17Z1 array size on Homolog 1 is 2.3 Mb and 4.0 Mb on H2. Each sample is shown in duplicate, along with a D17Z1 sizing control (0.7 Mb) for Southern blotting. (B) Because D17Z1-B is a relatively recently identified array, less is known about array size. We measured D17Z1-B array sizes on 12 different HSA17s using stretched DNA fibers and FISH with probes specific to D17Z1 and D17Z1-B (Supplemental Fig. S4). The size of D17Z1 was used as a normalizer to calculate D17Z1-B array size from fluorescent signals on DNA fibers; sizes of both arrays for individual HSA17s were plotted as shown. D17Z1 array sizes ranged from 2.3 to 4.3 Mb, while D17Z1-B sizes ranged from 0.7 to 1.6 Mb. The smaller D17Z1 arrays were associated with HSA17s in which D17Z1-B, not D17Z1, was the functional centromere. HSA17s are named and organized along the x-axis by D17Z1 array size (largest to smallest). Location of the centromere is denoted above the graph. (C) To investigate the correlation between array size and variation, D17Z1 array size and the proportion of wild-type and variant HORs (size + SNP) were plotted, revealing that inactive D17Z1 arrays have higher proportions (>80%) of variant HORs. Centromere location for each HSA17 is denoted above the plot. Z1_4.0 exhibited extensive D17Z1 variation but assembled the centromere at D17Z1. Z1_3.3, Z1_3.1, and Z1_2.6 exhibited moderate variation (∼60%) but still assembled the centromere at D17Z1.
Alpha satellite array size and sequence variation are not necessarily independent variables, due to the nature of repetitive arrays. The range of array sizes among different individuals has been attributed to expansion and contraction over iterative rounds of unequal exchange, due to recombination or, more likely, gene conversion (Waye and Willard 1986a; Warburton and Willard 1992; Warburton et al. 1993). This process should be more efficient on homogenous arrays, as it relies on internal homology to expand arrays. More variant arrays would be expected to be smaller due to inefficient expansion processes. In our data set, there was a weak correlation between D17Z1 array size and the proportion of the array composed of wild-type HORs (R2 = 0.21, P = 0.13, Pearson correlation = 0.4622, Spearman correlation = 0.3638) (Supplemental Fig. S3). However, the data appear to be skewed by the single, extremely large and variant D17Z1 array in line Z1_4.0 that also displays a high degree of HSA17 instability (see below). If this line is excluded from analysis due to its unusual behavior, the correlation between D17Z1 size and homogeneity greatly increases (R2 = 0.57; Pearson correlation = 0.7557, Spearman correlation 0.69337), supporting our hypothesis that larger D17Z1 arrays are more likely to be homogeneous and the site of centromere assembly. Overall, our data suggest that a significant factor that distinguishes the two functional centromeric states (active versus inactive) of D17Z1 is the amount of variation with the D17Z1 array.
Variant, active D17Z1 arrays are associated with chromosome instability
The array size and variation analyses identified a few notable HSA17s that did not obviously fit the pattern of a large, invariant D17Z1 array correlating with centromere location (Supplemental Fig. S3). Specifically, four HSA17s (Z1_4.0, Z1_3.3, Z1_3.1, and Z1_2.6) assembled their centromeres at D17Z1 arrays that contained a large proportion of HOR variation (60%–98%) (Fig. 3C). We asked if the centromeres on these variant D17Z1 arrays were functionally normal. First, we monitored chromosome stability as a marker for proper centromere function. We defined chromosome stability as the ability of the chromosome to maintain its ploidy over time. FISH was used to analyze somatic cell hybrid lines containing individual HSA17s for the number of HSA17s (< or > 1 HSA17/cell). HSA17s in which the centromeres formed at large, invariant D17Z1 arrays (Z1_4.3, Z1_3.9, Z1_3.5) were extremely stable (Fig. 4A; Supplemental Fig. S4C). Likewise, HSA17s that assembled the centromere at D17Z1-B and contained extremely variant D17Z1 arrays were also very stable. However, HSA17s that assembled the centromere at D17Z1 arrays that had moderate to extreme HOR variation exhibited chromosome instability (Fig. 4A; Supplemental Fig. S4D).
Figure 4.
Centromeres assembled at variant D17Z1 arrays are less stable than homogeneous wild-type arrays. (A) The proportion of variation (wild-type versus variant HORs) in a subset of HSA17s is plotted with stability of the HSA17 (red line). For each line, chromosome stability was determined using FISH with D17Z1 probes and counting the number of HSA17s in 200 cells (stability was defined by maintenance of HSA17 ploidy in each line). When the centromere formed on a large, homogeneous array of D17Z1, such as in Z1_4.3, Z1_3.9, and Z1_3.5, the HSA17 was extremely stable in mitosis. Similarly, when the centromere assembled at D17Z1-B in lines Z1_2.3A and Z1_2.3B (highly variant D17Z1 arrays), HSA17 was very stable. However, when the centromere was assembled on D17Z1 arrays that had moderate or extreme variation, HSA17 was mitotically unstable. Centromere location (D17Z1—white, D17Z1-B—gray) on each HSA17 is denoted above the plot. (B) Line Z1_4.0 had the most variant (98%), yet active, D17Z1 array in our data set, and this HSA17 exhibited chromosome instability. The parental Z1_4.0 (Z1_4.0P) was subcloned to produce multiple, independent versions of the HSA17 (single-cell clones, SCC); subcloning could also account for aberrant behavior in a single cell line that did not reflect inherent behavior of the HSA17. The single-cell clones showed varying levels of chromosome instability, indicating that the unstable phenotype was inherent to this HSA17. The stability of two mouse chromosomes (MMU9, MMU16) was measured to account for genetic background effects that might alter the stability of all chromosomes. (C) CENPA and CENPC (Supplemental Fig. S4E) immunostaining (green) was combined with FISH using D17Z1 probe p17H8 (red) to quantitate the amount of centromere proteins on unstable Z1_4.0. Insets show the HSA17 alone and a single channel image of CENPA staining on the HSA17. Scale bar, 15 µm. (D) The amount of CENPA and CENPC on unstable Z1_4.0 was plotted compared to all other centromeres in the cell. Fluorescence from all centromeres (ALL CENS) was normalized to one, and the fluorescence at Z1_4.0 was calculated according to this normalized value. The amount of CENPA at the Z1_4.0 centromere was half of the amount at all other centromeres in the cell; CENPC was reduced by more than 50%. (E) The unstable Z1_4.0 single cell clone SSC9 also showed reduced amounts of CENPA. By comparison, the amount of CENPA on Z1_2.3B, a stable HSA17 that has a variant D17Z1 array but assembles the centromere at D17Z1-B, was comparable to all the other centromeres in the cell (E′).
We extended our stability studies of each HSA17 by analyzing multiple versions of the same HSA17 in clonal cell lines derived by dilution and single cell cloning from the parental somatic cell hybrid (Fig. 4B; Supplemental Fig. S4). This approach accounted for random mutations in a single cell line that might affect HSA17 chromosome stability. The stability of two mouse chromosomes was also measured to rule out a general chromosome instability phenotype within a given single cell clone (SCC), since all HSA17s were transferred to the same mouse background. In these analyses of multiple versions of the same HSA17, we observed that HSA17s with large, invariant (wild-type) D17Z1 arrays were largely stable (Supplemental Fig. S4C). However, single cell clones of different HSA17s that had substantial amounts of variation (>50%) showed considerable chromosome instability, suggesting that the unstable phenotype was inherent to the specific HSA17 (Fig. 4B; Supplemental Fig. S4D). These results imply that centromeres formed on variant D17Z1 arrays are functionally deficient and that this has long-term consequences for overall HSA17 mitotic stability.
Heterogeneous D17Z1 has reduced amounts of centromere protein A (CENPA)
We wanted to understand the functional basis for why the highly variant D17Z1 centromeres were associated with HSA17 instability. We compared amounts of two centromere proteins (CENPs) at specific HSA17s to normal centromeres within the same cells. CENPA is a histone H3 variant that is an important epigenetic marker of functional centromeres (Warburton et al. 1997; Black et al. 2007), creating a specialized type of chromatin (centromeric chromatin) that serves as the foundation of the kinetochore (Shelby et al. 1997; Ando et al. 2002; Foltz et al. 2006). CENPC is a member of the constitutive centromere-associated network (CCAN), a multiprotein network that mechanically links CENPA chromatin to the outer kinetochore where spindle microtubules attach (Hori et al. 2013; Suzuki et al. 2014; Klare et al. 2015). We measured the amounts of CENPA and CENPC at D17Z1 on unstable HSA17s using IF-FISH (Fig. 4C). The centromere of Z1_4.0, the most unstable HSA17 in our cohort of highly heterogeneous D17Z1 arrays, had approximately half the amount of CENPA and <50% the amount of CENPC compared to all other centromeres (Fig. 4C,D). Conversely, stable HSA17s, including those in which the centromere was assembled at D17Z1-B, had amounts of the CENPA that were comparable to other chromosomes in the cells (Fig. 4E,E′). These results indicate that centromeres formed on variant D17Z1 arrays are functionally defective and compositionally different than centromeres formed on invariant D17Z1 and D17Z1-B arrays and on alpha satellite arrays on other chromosomes.
Discussion
Genomic variation is a source of functional diversity and is broadly studied in the genic and noncoding regulatory regions of the genome. However, few studies have ventured into the ∼10% of the human genome that is highly repetitive and remains unassembled and uncharacterized to explore the extent of variation and its functional consequences. Alpha satellite DNA comprises at least 5% of the genome and has a well-established role in centromere function. Centromere proteins are concentrated at alpha satellite arrays on all endogenous human centromeres (Vafa and Sullivan 1997; Ando et al. 2002); moreover, alpha satellite DNA is the only sequence capable of forming de novo centromeres in human artificial chromosomes assays (Harrington et al. 1997; Grimes et al. 2002; Maloney et al. 2012). Structural variation within alpha satellite (i.e., variations in overall array size among individuals) has been well known for decades (Wevrick and Willard 1989; Miga et al. 2014). However, the extent of sequence variation is less understood, since contiguous alpha satellite regions have not been assembled and compared among large numbers of individuals. The situation is further complicated by the fact that half of human chromosomes have more than one alpha satellite array within their centromere regions, thereby potentially compounding the amount of variation per individual (Pironon et al. 2010; Hayden et al. 2013; Miga et al. 2014; see UCSC Genome Browser centromere track). Other human chromosomes in addition to HSA17 exhibit centromeric epialleles (Maloney et al. 2012; SM McNulty and BA Sullivan, unpubl.), making it imperative to understand how the site of centromere formation is chosen when two or more adjacent genomic regions are available for centromere assembly. Although centromere identity is thought to have a stronger epigenetic basis, our results support a connection between genomic variation within alpha satellite DNA and centromere location. Our data also highlight the functional consequences of genomic variation within the repetitive portion of the human genome that remains unassembled and uncharacterized.
Our study raises several questions and areas for future study. The data presented here point to an association between large alpha satellite array size and centromere function at D17Z1. This correlation is less apparent for D17Z1-B arrays, perhaps because they vary less in size. Although array length cannot absolutely predict where the centromere will be assembled, on HSA17 we observed a trend toward an active D17Z1-B array being closer in size to the neighboring D17Z1 array. In the context of HSA17 centromeric epialleles, our data suggest a stochastic model of competition for centromere proteins, in which larger alpha satellite arrays may have an advantage at recruiting a critical mass of CENPA nucleosomes and centromere proteins to establish centromere identity (Sullivan et al. 2011). This model might also explain why certain centromeres of dicentric chromosomes are more often inactivated (Sullivan et al. 1994).
Our finding that variant D17Z1 arrays are associated with reduced numbers of centromere proteins was surprising and suggests that alpha satellite arrays with HOR size and sequence variants cannot effectively recruit CENPs. Alternatively, the wide range in stability of HSA17s with variant D17Z1 arrays in our data set may indicate that variant HORs can recruit CENPs but are unable to maintain a critical number of molecules or cannot organize them into a properly structured kinetochore. Differentiating between these hypotheses will be important for understanding the impact of alpha satellite variation in de novo centromere assembly versus centromere inheritance. In addition to CENPA and CENPC, many other proteins are constitutively associated with the centromere (Perpelescu and Fukagawa 2011). One of these proteins is CENPB, a DNA-binding protein that binds to the alpha satellite at the CENPB box, a 17-bp sequence motif that is found in a subset of monomers on all human chromosomes except the Y (Masumoto et al. 1989; Muro et al. 1992; Haaf and Ward 1994; Ikeno et al. 1994; Ando et al. 2002). Historically, CENPB was not thought to play a functional role in centromeric chromatin, since it is present at both active and inactive alpha satellite arrays (Earnshaw et al. 1989; Sullivan and Schwartz 1995). However, de novo centromere assembly of HACs depends on CENPB-box-containing alpha satellite DNA (Ohzeki et al. 2002; Okada et al. 2007), and CENPB is thought to position CENPA nucleosomes and stabilize CENPA and CENPC within centromeric chromatin (Yoda et al. 1998; Okada et al. 2007; Hasson et al. 2013; Fachinetti et al. 2015). CENPB binding sites (i.e., the number of CENPB boxes) within an array might determine how well an array can recruit centromere proteins and achieve the three-dimensional structure required for kinetochore assembly and centromere function. Shorter D17Z1 arrays with more HOR variants may have fewer CENPB binding sites, making them functionally inferior to a D17Z1-B array that is similar in size but contains invariant HORs. Because CENPB binding sites are present in every human alpha satellite array regardless of centromere function, it is technically difficult to measure and compare numbers of CENPB boxes within closely spaced arrays like D17Z1 and D17Z1-B that are on the same chromosome. Improvements in the resolution of chromatin fibers and long single-molecule sequencing of alpha satellite arrays will be necessary to achieve the chromosome-specific identification needed to test this hypothesis.
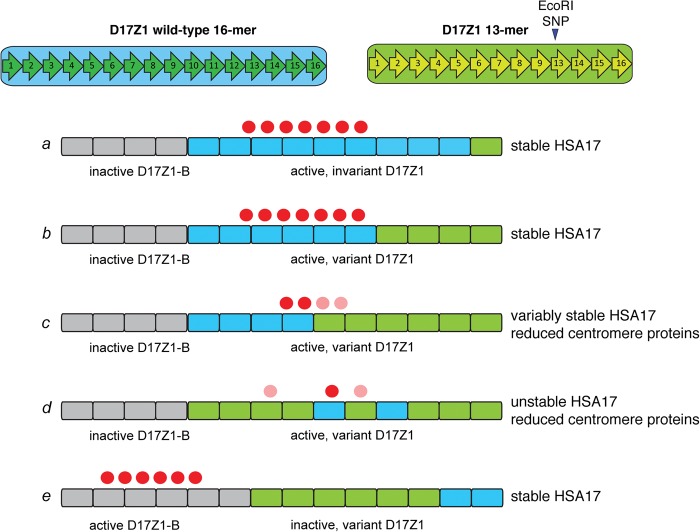
Long-range organization of HORs may also determine the functional potential of an alpha satellite array. Prior investigation of three HSA17s suggested that HOR variants within D17Z1 are clustered into subdomains (Warburton and Willard 1990). How the HOR size variants are organized on the HSA17s with stable active, unstable active, and inactive D17Z1 arrays in our data set is unknown. If centromere establishment relies on a homogeneous subdomain of sufficient size, then a highly variant array may appear as many small subarrays consisting of groups of 13-mers and/or groups of 13-mers with the EcoRI SNP that may each be of insufficient size to attract or retain a critical number of centromere proteins to achieve proper kinetochore structure (Fig. 5, scenario d or e). Such irregularity of HOR organization in D17Z1 might make homogenous D17Z1-B a more attractive location for centromere function (Fig. 5, scenario e).
Figure 5.
Models for epiallele choice on HSA17 based on CENPB box number or D17Z1 long-range organization. Centromere assembly on HSA17 appears to occur predominantly at large D17Z1 arrays that contain wild-type (invariant, blue blocks) HORs (scenario a). When D17Z1 contains variant HORs (green blocks), centromere assembly either occurs at D17Z1-B (gray blocks, scenario e) and the HSA17 is stable, or at variant D17Z1 and the HSA17 is unstable due to reduction in CENPs (red circles). It is not clear if variant arrays cannot recruit or cannot maintain the appropriate number of CENPs, and the molecular basis for the reduction in CENP molecules is unknown. Long-range organization of D17Z1 might affect CENP recruitment and binding. Large arrays with moderate variation may provide a sufficiently sized domain of homogenous wild-type HORs for centromere assembly (scenario b). However, in the cases of HSA17s that build their centromeres on variant HSA17 arrays and are unstable, CENPA may be distributed across wild-type and variant HOR domains, the latter of which may be less efficient at CENP recruitment/maintenance (scenario c, light red circles). Moreover, irregularity in wild-type and variant HOR organization (i.e., interspersed subarrays of variant and wild-type HORs) may negatively affect centromere function or CENP recruitment (scenario d). It will be important to experimentally discriminate between these organizational scenarios, particularly on HSA17, in order to better understand the spatial relationship between centromere proteins and long-range alpha satellite organization.
On partially stable HSA17s that contained moderately variant (40%–60%) D17Z1 arrays, it is possible that the wild-type and variant domains may be distinctly separated (Fig. 5, scenarios b and c). Since CENPA chromatin is only formed on a portion of an alpha satellite (Sullivan et al. 2011), on these HSA17s, centromere proteins might be preferentially associated with the domain of wild-type rather than variant HORs (Fig. 5, scenario b). Alternatively, CENPA chromatin might equivalently or unequally straddle the two domains, wild-type and variant, perhaps contributing to the reduction in HSA17 stability if centromere proteins were inefficiently maintained on the variant HORs (Fig. 5, scenario c). Of course, the centromere formed at a given variant D17Z1 array might reflect several of these organizational scenarios. We recently showed that CENPA chromatin is assembled and maintained at the same general region of alpha satellite DNA (Ross et al. 2016), so a key question is if D17Z1 array variation disrupts the largely static placement of centromeric chromatin on HSA17.
From these studies, we conclude that centromere identity on at least one human chromosome is linked to the genomic composition of alpha satellite DNA and that centromere function is particularly sensitive to structural and sequence variation within an array. Given that HSA17 is often unstable in many cancers and also contains several oncogenes and tumor suppressor genes (Mitelman 2000; Garcia et al. 2003), our results suggest that, like variation within genic and regulatory regions, DNA variation within repetitive sequences has functional consequences and could predispose human chromosomes to instability and aneuploidy. In the future, it will be important to functionally connect distinct long-range alpha satellite configurations on other chromosomes with centromere assembly and chromosome stability. Our work also highlights the need for genomic resources to be directed toward achieving longer, accurate sequence data and assemblies for the repetitive regions of the human genome. Such information will provide insight into how variation in these essential noncoding regions affects basic chromosome function and, ultimately, human disease.
Methods
Cell lines and culture conditions
Diploid cell lines included HT1080, HDF, HCT116, RPE1, CEPH 1345 lymphoblastoid cell lines (LCL), and somatic cell hybrids lines that have been described previously (Willard et al. 1987; Dausset et al. 1990; Warburton and Willard 1995; Maloney et al. 2012). The haploid line HAP1, derived from KDM-7 cells, was used because it contained only one HSA17 in a human background. Cells were grown at 37°C in a humidified chamber with 5% CO2. Fibroblasts (HT1080, HDF) and somatic cell hybrids were grown in minimum essential medium (MEM) alpha medium (Gibco) supplemented with 10% fetal bovine serum (FBS). Somatic cell hybrids were also grown in the presence of 1× hypoxanthine, aminopterin, thymidine (HAT), and 1× ouabain (2 × 10−6 M). LCLs were grown in RPMI (Gibco) supplemented with 15% FBS. HCT116 and U-2 OS cells were grown in McCoy's media supplemented with 10% FBS. HAP1 cells were grown in IMDM supplemented with 10% FBS. All complete media contained 1× antibiotic-antimycotic (Gibco). Each human-mouse somatic cell hybrid was verified to contain a single HSA17 using chromosome arm polymorphisms (Maloney et al. 2012).
Pulsed field gel electrophoresis
Alpha satellite array sizes were estimated by PFGE and Southern blotting using established methods (Wevrick and Willard 1989; Mahtani and Willard 1990; Sullivan et al. 2011). High molecular weight DNA was embedded in low melting point agarose plugs and digested with restriction enzymes that cut infrequently within alpha satellite DNA and released the arrays as one or a few large fragments. For D17Z1 sizing, digested plugs were run on 1% agarose gel in 1× TAE buffer. Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Hansenula wingei chromosomes embedded in agarose were used as size standards (Bio-Rad CHEF DNA Size Markers). Gels were run at 3 V/cm for 50 h at 14°C in 1× TAE buffer, using switch times of 250 sec (initial)–900 sec (final). Cell lines containing previously sized D17Z1 were used as controls, including a derivative HSA17 containing a partially deleted D17Z1 array and completely lacking the D17Z1-B array (Wevrick et al. 1990).
After electrophoresis, gels were stained with ethidium bromide and imaged using a UV light source. Gels were rinsed briefly with distilled water, depurinated with 0.25 M HCl or irradiated at 600 µJ to nick large DNA, and incubated in denaturing buffer (1.5 M NaCl, 0.5 M NaOH). DNA was transferred to HyBond-N+ membrane (GE Healthcare/Amersham) for 16–40 h in denaturing buffer. Dried membranes were UV-crosslinked (auto-crosslink setting on Stratagene Stratalinker) and either used immediately or stored in sealed plastic until hybridization.
Southern blotting
Probes were labeled by nick translation with digoxygenin-11-dUTP or biotin-12-dUTP. Membranes were prehybridized for 30–60 min in ExpressHyb buffer (Clontech) at the hybridization temperature. For D17Z1 sizing, membranes were hybridized with 125–150 ng of labeled plasmid p17H8 (a generous gift from H.F. Willard) at 69.5°C for 16–18 h. Membranes were washed at the hybridization temperature twice for 12–15 min in 2× SSC/0.1% sodium dodecyl sulfate (SDS), followed by a single high stringency wash in 0.2× SSC/0.1% SDS. Membranes were blocked in 1× Western blocking reagent (Roche) in maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5) for 2 h at room temperature, then incubated for 30 min in blocking buffer with anti-digoxygenin-alkaline phosphatase (Roche, 1:1000–1:2000) or streptavidin-alkaline phosphatase conjugate (Roche, 1:1000). Chemiluminescent detection was performed using the CDP-Star Reagent (New England Biolabs; 1:500 in 1× CDP-Star Dilution Buffer). In a few early experiments, membranes were exposed (using multiple exposure times) to BIOMAX XAR film (Carestream) that were scanned for quantitative analyses. Most of the Southern blots in this study were digitally captured on the G:Box CHEMI XT4 using GeneSys software (Syngene) for direct image analysis. Images were adjusted (leveled to curves) and labeled in Adobe Photoshop. For presentation purposes only, blot images were reversed (black bands on white background).
D17Z1 polymorphisms identified using PCR-restriction digestion
Genomic DNA from diploid human cell lines or somatic cell hybrid lines was amplified with primers (17-1A, 17-2A) that amplify multiple bands representing different higher order repeat units within polymorphic D17Z1 arrays (Warburton et al. 1991). Each sample of gDNA (50–100 ng) was amplified in a 25-µL reaction under the following conditions: 1 cycle of 95°C for 4 min; 20 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 2 min; 1 cycle of 72°C for 7 min). PCR products from 2–3 separate reactions per sample were ethanol precipitated and resuspended in 10–15 µL of molecular grade water. To visualize HOR size variants only, 3–5 µL of purified PCR product were run on a 1.2% agarose gel. To visualize both HOR size and SNP variants, 1.5 µg of the purified PCR products were digested with EcoRI and separated on a 1.2% agarose gel. Digital images of gels stained with EtBr or SYBR Safe were captured at different exposure times using a G:BOX XT4 imaging system (Syngene). Quantitative analyses of individual gel bands (white bands or black background), equating to the proportion of each variant in a given array, was done using the GeneSys software (Syngene). After calibrating to the molecular weight marker, the software automatically defined gel bands and calculated the band intensity. Band intensities within a single lane, representing the D17Z1 array from a single individual or single HSA17, were converted to proportions that reflect amounts of each HOR variant within the specific D17Z1 array. For presentation purposes only, blot images were reversed (black bands on white background).
Quantitation of CENPA and CENPC at centromeres using IF-FISH
To measure the amount of CENPA and CENPC on individual HSA17s, antibodies to CENPA (custom rabbit polyclonal D601AP, Quality Controlled Biochemical QCB) and CENPC (mouse monoclonal 2159C5a, Abcam, ab50974) were applied to metaphase chromosomes that were cyto-spun onto slides. Primary antibodies were detected using Alexa Fluor 488 or Alexa Fluor 649 secondary antibodies (Invitrogen), followed by FISH with the directly labeled (AF594-dUTP, Invitrogen/Thermo-Scientific) probe p17H8 that recognizes D17Z1 (Maloney et al. 2012). Images from individual metaphase spreads were collected at the same exposure time on the DeltaVision Elite microscope using a 60× (PlanApo N.A. 1.42) or 100× (UPlanSApo N.A. 1.40) objective. Deconvolved images (conservative ratio, 10 iterations) were projected and saved as TIFF and/or Photoshop files. TIFF images were opened in ImageJ and using a custom macro, each CENPA and CENPC signal in the entire metaphase was segmented after background subtraction. Integrated densities for each CENPA/C spot were exported to Excel, and CENPA/C pairs (double dots) representing each sister kinetochore of a single centromere were matched and added to arrive at the CENPA/C integrated density per centromere. Average integrated densities for all centromere pairs except the HSA17 were averaged, and this value was normalized to one. Fluorescent intensity of the CENPA/C pair for HSA17 was compared to the average of all pairs to obtain a normalized ratio of HSA17 CENPA/C to the amount of CENPA at all the other chromosomes. Values were imported into GraphPad Prism 7 and visualized as dot plots. A Student's t-test was used to determine significant differences between all CENPA/C and HSA17 CENPA/C groups.
Supplementary Material
Acknowledgments
We thank Hunt Willard for the generous gift of his entire cell line repository containing many of the single HSA17s and diploid lines included in this study. We thank So Young Kim, Director of the Duke Functional Genomics Core for sharing the HAP1 (Z1_3.5) cell line, and Alexandra Young (Duke University, Class of 2013) for technical support. Kristin Scott and Shannon McNulty provided critical feedback on the data and the manuscript. This research was supported in part by research grant #1-FY13-517 from the March of Dimes Foundation and National Institutes of Health grant 1R01-GM98500-01A1.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.206706.116.
References
- Alexandrov IA, Mitkevich SP, Yurov YB. 1988. The phylogeny of human chromosome specific alpha satellites. Chromosoma 96: 443–453. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB. 1993. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res 21: 2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. 2001. Alpha-satellite DNA of primates: old and new families. Chromosoma 110: 253–266. [DOI] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. 2002. CENP-A, -B, and -C chromatin complex that contains the I-type α-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B, Brock M, Bedard S, Woods V Jr, Cleveland D. 2007. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci 104: 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. 1990. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6: 575–577. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H III, Stetten G. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98: 1–12. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. 2015. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev Cell 33: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR III, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469. [DOI] [PubMed] [Google Scholar]
- Garcia J, Duran A, Tabernero MD, Garcia Plaza A, Flores Corral T, Najera ML, Gomez-Alonso A, Orfao A. 2003. Numerical abnormalities of chromosomes 17 and 18 in sporadic colorectal cancer: incidence and correlation with clinical and biological findings and the prognosis of the disease. Cytometry B Clin Cytom 51: 14–20. [DOI] [PubMed] [Google Scholar]
- Grimes B, Rhoades A, Willard H. 2002. α-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 5: 798–805. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward D. 1994. Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum Mol Genet 3: 697. [DOI] [PubMed] [Google Scholar]
- Hamilton BA. 2002. Variations in abundance: genome-wide responses to genetic variation and vice versa. Genome Biol 3: reviews1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraksingh RR, Snyder MP. 2013. Impacts of variation in the human genome on gene regulation. J Mol Biol 425: 3970–3977. [DOI] [PubMed] [Google Scholar]
- Harrington J, Van Bokkelen G, Mays R, Gustashaw K, Willard H. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355. [DOI] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol 20: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. 2013. Sequences associated with centromere competency in the human genome. Mol Cell Biol 33: 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. 2013. The CCAN recruits CENPA to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200: 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T. 1994. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human chromosome 21. Hum Mol Genet 3: 1245–1257. [DOI] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. 2015. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol 210: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani MM, Willard HF. 1990. Pulsed-field gel analysis of α-satellite DNA at the human X chromosome centromere: high-frequency polymorphisms and array size estimate. Genomics 7: 607–613. [DOI] [PubMed] [Google Scholar]
- Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA. 2012. Functional epialleles at an endogenous human centromere. Proc Natl Acad Sci 109: 13704–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH. 2015. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res 23: 421–426. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24: 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Eisenhart C, Kent WJ. 2015. Utilizing mapping targets of sequences underrepresented in the reference assembly to reduce false positive alignments. Nucleic Acids Res 43: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F. 2000. Recurrent chromosome aberrations in cancer. Mutat Res 462: 247–253. [DOI] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. 1992. Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 116: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. 2011. The ABCs of CENPs. Chromosoma 120: 425–446. [DOI] [PubMed] [Google Scholar]
- Pironon N, Puechberty J, Roizes G. 2010. Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21. BMC Genomics 11: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JE, Woodlief KS, Sullivan BA. 2016. Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenetics Chromatin 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M, Willard H. 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet 20: 529–533. [DOI] [PubMed] [Google Scholar]
- Rudd M, Wray G, Willard H. 2006. The evolutionary dynamics of α-satellite. Genome Res 16: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol 136: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA. 2009. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet 5: e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev VA, Uralsky LI, Alexandrov AA, Yurov YB, Rogaev EI, Alexandrov IA. 2015. Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom Data 5: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4: 2189–2197. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Wolff DJ, Schwartz S. 1994. Analysis of centromeric activity in Robertsonian translocations: implications for a functional acrocentric hierarchy. Chromosoma 103: 459–467. [DOI] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. 2011. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res 19: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED. 2014. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell 30: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. 1997. Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr Biol 7: 897–900. [DOI] [PubMed] [Google Scholar]
- Warburton P, Willard HF. 1990. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol 216: 3–16. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. 1992. PCR amplification of tandemly repeated DNA: analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human alpha satellite DNA. Nucleic Acids Res 20: 6033–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. 1995. Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: evidence for concerted evolution along haplotypic lineages. J Mol Evol 41: 1006–1015. [DOI] [PubMed] [Google Scholar]
- Warburton P, Greig G, Haaf T, Willard H. 1991. PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis. Genomics 11: 324–333. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Waye JS, Willard HF. 1993. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 13: 6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. 1997. Immunolocalization of CENPA suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7: 901–904. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. 1986a. Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res 14: 6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. 1986b. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol 6: 3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. 1989. Long-range organization of tandem arrays of α satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci 86: 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Earnshaw WC, Howard-Peebles PN, Willard HF. 1990. Partial deletion of alpha satellite DNA associated with reduced amounts of the centromere protein CENP-B in a mitotically stable human chromosome rearrangement. Mol Cell Biol 10: 6374–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF. 1985. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet 37: 524–532. [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Waye JS. 1987. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 25: 207–214. [DOI] [PubMed] [Google Scholar]
- Willard HF, Greig GM, Powers VE, Waye JS. 1987. Molecular organization and haplotype analysis of centromeric DNA from human chromosome 17: implications for linkage in neurofibromatosis. Genomics 1: 368–373. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. 1998. In vitro assembly of the CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells 3: 533–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.