Abstract
Allogeneic hematopoietic cell transplantation (HCT) offers the potential to cure hematologic malignancies. Absent an HLA matched donor, HLA mismatched unrelated donors may be used although risks of graft-versus-host disease (GvHD) and treatment related mortality (TRM) are higher. Identification and avoidance of amino acid substitutions and position types (AASPT) conferring higher risks of TRM and GvHD would potentially improve the success of transplantation from single HLA mismatched unrelated donors. Using random forest and logistic regression analyses, we identified 19 AASPT associated with greater risks for at least one adverse transplant outcome: grade III-IV acute GvHD, TRM, lower disease free survival or worse overall survival relative to HLA matched unrelated donors and to other AASPT. When tested in an independent validation cohort of 3,530 patients, none of the AASPT from the training set were validated as high-risk, however. Review of the literature shows that failure to validate original observations is the rule and not the exception in immunobiology and emphasizes the importance of independent validation prior to clinical application. Our current data do not support avoiding any specific class I AASPT for unrelated donors. Additional studies should be performed to fully understand the role of AASPT in HCT outcomes.
Introduction
In the absence of an HLA identical sibling, an HLA-A, -B, -C, and -DRB1 matched unrelated donor (8/8 URD) is the best-established donor option for patients with high-risk hematological malignancies who need hematopoietic cell transplantation (HCT). However, approximately 30% of Caucasian and 70%-94% of African-American patients are unable to find an 8/8 URD (1). A single antigen or allele HLA mismatch results in approximately 10% worse absolute 5-year survival and greater risk of graft-versus-host disease (GvHD) for patients with early-stage hematological malignancies compared to an 8/8 URD-HCT (2-5). Alternatives to mismatched unrelated donors include haploidentical donors and umbilical cord blood (6-8) but these may have higher rates of relapse and delayed engraftment, respectively.
Considerable interest has emerged in identifying more tolerable or permissive HLA mismatches or, conversely, HLA mismatches to avoid between patients and their unrelated donors to minimize adverse consequences of HLA mismatching. Numerous factors may determine the effect of specific HLA mismatches, considering that alleles differ in the number, type, and location of amino acid substitutions (AAS) in the structure of the HLA molecule, and their impact on peptide binding and T-cell allorecognition may be critical in determining transplant outcomes. Non-permissive AAS have been identified using both in vitro structure-function studies (9, 10) and outcome studies (11-14).
We hypothesized that we could establish a subset of AAS associated with worse HCT outcomes. In 2012, our group reported a set of AAS that were associated with worse 100 day survival in single HLA class I mismatched recipient-donor pairs compared to 8/8 URD pairs (15). We now extend the analysis to include all AAS positions and as well as amino acid substitution types (AASPT) and 1 year transplant and grades III-IV GvHD outcomes in separate training and validation datasets.
Materials and Methods
Patients
Patients had early or intermediate risk hematologic malignancies [acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), and myelodysplastic syndromes (MDS)]. Patients and adult unrelated donors were either 8/8 matched at HLA-A, -B, -C and -DRB1 or 1 allele mismatched at class I by high resolution typing. The training set underwent transplant from 1988-2003 (n=2,107) whereas the validation set underwent transplantation from 2004-2011 (n=3,349). Table 1 describes baseline characteristics of patients in each dataset.
Table 1. Characteristics of the training and testing cohorts.
| Training Dataset n=2,107 | Validation Dataset n=3,349 | Training versus Validation Datasets | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-class I mismatched URD | 8/8 URD | p-value | HLA-class I mismatched URD | 8/8 URD | p-value | HLA-class I mismatched URD | |||||
| n | % | n | % | n | % | n | % | p-value | |||
| Number of Donor/Recipient pairs | 600 | 100 | 1,507 | 100 | 745 | 100 | 2,604 | 100 | <0.001 | ||
| HLA-A mismatch | 179 | 30 | 299 | 40 | |||||||
| HLA-B mismatch | 88 | 15 | 160 | 21 | |||||||
| HLA-C mismatch | 333 | 55 | 286 | 38 | |||||||
| Age at Transplant | 0.0018 | ||||||||||
| Median (range) | 31 | (1-66) | 34 | (1-63) | <0.001 | 32 | (1-70) | 38 | (1-73) | <0.001 | |
| Gender Donor/Recipient | 0.36 | <0.001 | 0.79 | ||||||||
| Male/Male | 207 | 35 | 572 | 38 | 253 | 34 | 1,046 | 40 | |||
| Female/Male | 119 | 20 | 276 | 18 | 160 | 21 | 376 | 14 | |||
| Female/Female | 129 | 21 | 288 | 19 | 147 | 20 | 439 | 17 | |||
| Male/Female | 145 | 24 | 371 | 25 | 185 | 25 | 743 | 29 | |||
| Disease | 0.03 | 0.003 | <.0001 | ||||||||
| ALL | 155 | 26 | 352 | 23 | 307 | 41 | 895 | 34 | |||
| AML | 172 | 29 | 370 | 25 | 381 | 51 | 1,497 | 57 | |||
| CML | 256 | 43 | 717 | 48 | 0 | 0 | 0 | 0 | |||
| MDS | 17 | 2 | 68 | 4 | 57 | 8 | 212 | 8 | |||
| Disease Status | 0.001 | 0.001 | <.0001 | ||||||||
| Early | 282 | 47 | 834 | 55 | 445 | 60 | 1,721 | 66 | |||
| Intermediate | 318 | 53 | 673 | 45 | 300 | 40 | 883 | 34 | |||
| Conditioning Regimen | 0.03 | ||||||||||
| Myeloablative | 591 | 99 | 1499 | 99 | 745 | 100 | 2604 | 100 | |||
| Non-myeloablative | 9 | 1 | 8 | 1 | |||||||
| GvHD Prophylaxis | 0.01 | <0.001 | <.0001 | ||||||||
| Tacrolimus ± Other | 121 | 20 | 298 | 20 | 492 | 64 | 1912 | 73 | |||
| Cyclosporine A+ MTX ± Other | 324 | 54 | 890 | 59 | 200 | 27 | 494 | 19 | |||
| Cyclosporine A ± Other | 13 | 2 | 57 | 4 | 30 | 4 | 79 | 3 | |||
| MTX ± Other | 5 | 1 | 7 | 0 | 4 | <1 | 12 | <1 | |||
| T-Cell Depletion | 137 | 23 | 254 | 17 | 0 | 0 | |||||
| Other | 0 | 0 | 1 | 0 | 19 | 2 | 107 | 4 | |||
| Stem Cell Source | 0.91 | 0.09 | <.0001 | ||||||||
| Bone Marrow | 559 | 93 | 1402 | 93 | 284 | 38 | 906 | 35 | |||
| PBSC | 41 | 7 | 105 | 7 | 461 | 62 | 1698 | 65 | |||
| Year of Transplant | 0.25 | 0.06 | |||||||||
| 1988 – 1992 | 65 | 11 | 212 | 14 | |||||||
| 1993 – 1996 | 174 | 29 | 410 | 27 | |||||||
| 1997 – 2000 | 241 | 40 | 597 | 40 | |||||||
| 2001 – 2004 | 120 | 20 | 288 | 19 | |||||||
| 2004 – 2007 | 343 | 46 | 1100 | 42 | |||||||
| 2008 – 2011 | 402 | 54 | 1504 | 58 | |||||||
Data Source
Clinical and outcome data from study participants were obtained from the Center for International Blood and Marrow Transplant Research (CIBMTR®) database. The CIBMTR is a research collaboration between the NMDP/Be The Match Registry and the Medical College of Wisconsin. All patients and donors provided informed consent for inclusion in the CIBMTR research program, and the NMDP Institutional Review Board approved the study protocol.
Study end Points
End points of this study were overall survival (OS), disease-free survival (DFS) and treatment-related mortality (TRM) at one year and severe (grades III-IV) acute GvHD by day 100. These outcomes were dichotomized for the random forest and logistic regression analyses and treated as time-to-event outcomes in the survival analyses as described below.
Statistical Analyses
Random forest analysis
The random forest method, which grows a collection of classification trees, is a combination classifier and has been previously described (15, 16). Briefly, the random forest (RF) method is an extension of CART (Classification and Regression Trees), which is used to discover relationships among a large number of predictor variables and a categorical or continuous outcome variable. Models were created that included four patient-donor clinical characteristics generally known to be associated with transplant outcomes (recipient age, gender match, type of disease, and disease stage) and all HLA-A, -B, and -C AASPT (n=389 candidate AASPT) as predictor variables. The procedure ranks all predictor variables in terms of an importance score (IS). In our previous publication (15) we chose an IS cutoff of ≥3 to indicate potentially important amino acid substitutions; here a more conservative IS of 5 or greater was selected as the threshold. Note that the IS does not, by itself, indicate the statistical significance nor even the direction of the effect. The analysis was performed using commercially available software (Salford Systems, Version 6, San Diego, CA).
Multivariate logistic regression analysis
To determine the direction and statistical significance of the effect of each AASPT on each clinical outcome, we performed multivariate logistic regression adjusted for the same four clinical variables mentioned above, i.e. each model included five predictor variables. To avoid sparse data, we limited the logistic regression to the AASPT present in at least 10 individuals, thereby reducing the number of candidate AASPT from 389 to 155. The odds ratio (OR) for the given AASPT relative to 8/8 URD was determined and the average marginal effect for each outcome was estimated by computing a marginal probability from the fitted logistic regression model using Stata (version 12.1, Stata, College Station, TX). AASPT were categorized into “high-risk” or “non-high-risk” on the basis of the logistic regression and random forest results.
Survival Analysis
The Kaplan-Meier (17) method was used to estimate OS and DFS rates according to risk groups, [high-risk (presence of one or more high-risk AASPT), non-high-risk (mismatch but no high-risk AASPT), and 8/8 URD], defined on the basis of the random forest and multivariate logistic regression analyses as described above. Comparison among the three groups was performed using the Cox proportional-hazards (PH) model (18). All patients without an event were censored at one year. Cox regression was also performed to estimate cause-specific hazard ratios for TRM and GvHD. For TRM, patients who died from disease relapse were censored at the time of death and for acute GvHD all deaths in the absence of prior GvHD were treated as censored observations. Finally, cumulative incidence curves (19) were generated to estimate the incidence rates of acute GvHD, with death in the absence of acute GvHD as a competing event. All models were adjusted for the four patient-donor clinical characteristics.
Validation analyses
The validation analysis tested the AASPT discovered in the analysis of the training set directly in the validation cohort. Of note, identified high-risk AASPT varied for each outcome in the training dataset. The AASPT were therefore tested in the validation cohort in two ways. First, outcome-specific AASPT were created with a trichotomous indicator variable for whether the donor/recipient pair had a high-risk AASPT, non-high-risk AASPT or were 8/8 matched for the specific outcome. A second test of the AASPT was based on an aggregated AASPT indicator variable which defines “high-risk” as presence of any of the AASPT identified as important for any of the four outcomes. This aggregated AASPT variable reflected the clinical reality that, if possible, transplant centers are likely to avoid donors with a high-risk AASPT for any adverse outcome.
We used the Cox PH model to evaluate the association between risk groups and each outcome controlling for outcome-specific significant clinical variables that were identified based on the validation cohort instead of limiting adjustment to the previously identified four clinical variables. The outcome-specific models were constructed using a stepwise selection procedure with a threshold of 0.05 for both entry and removal, and the final set of adjustment clinical variables differed from the training cohort. As above, both the outcome-specific and aggregate definitions of high-risk AASPT were tested separately based on these models. Because of multiple testing, P<0.01 was considered statistically significant. All reported p-values are two-sided. Analyses were performed using SAS version 9.3.
Results
Identifying high-risk AASPT and risk groups in the training set
Based on the criteria of AASPT present in at least 10 individuals (n>=10), IS>=5 from random forest, OR>1 (i.e. a detrimental effect) and P<=0.01 from logistic regression, the analysis discovered 19 high-risk AASPT, shown in Table 2. Among them, only C97_WR was classified as high-risk for all four outcomes. Given each list, we then stratified patients into three categories: 1) high-risk group included patients with at least one of the identified high-risk AASPT for that outcome; 2) non-high-risk group if they had any of the remaining AASPT including AASPT where n<10; and 3) 8/8 URD. The complete list of AASPT with IS>=5 or P<=0.01, including predicted event rates, is provided in the Supplementary Appendix, ST1.
Table 2. High-risk amino acid substitution position and type identified in the training set, ORs adjusted for patient age (continuous), sex matching, disease type and disease risk.
| Variable | 1Y Overall Survival | 1Y Disease-Free Survival | 1Y Treatment-Related Mortality | Acute Graft-versus-Host Disease | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IS | Odds Ratio | p-value | IS | Odds Ratio | p-value | IS | Odds Ratio | p-value | IS | Odds Ratio | p-value | ||
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | ||||||||||
| Clinical Factors | In Peptide Binding Region | ||||||||||||
| AASPT | |||||||||||||
| C97_WR | 23.2 | 2.70 (1.59-4.59) | <0.001 | 13. 9 | 2.43 (1.40-4.23) | 0.002 | 6.4 | 2.07 (1.27-3.39) | 0.004 | 13.7 | 2.25 (1.37-3.69) | 0.001 | Yes |
| C80_NK | 18.0 | 2.09 (1.32-3.32) | 0.002 | 14.0 | 2.07 (1.28-3.33) | 0.003 | Yes | ||||||
| C77_SN | 12.9 | 2.09 (1.32-3.32) | 0.002 | 8.0 | 2.07 (1.28-3.33) | 0.003 | Yes | ||||||
| C156_ RW | 13.6 | 4.07 (1.47-11.31) | 0.007 | 16.7 | 4.50 (1.73-11.66) | 0.002 | Yes | ||||||
| C156_QR | 17.4 | 15.70 (1.95-126.26) | 0.010 | Yes | |||||||||
| A167_WG | 9.6 | 5.04 (1.59-15.98) | 0.006 | Yes | |||||||||
| A166_ED | 13.9 | 5.04 (1.59-15.98) | 0.006 | Yes | |||||||||
| A43_RQ | 7.8 | 5.88 (1.53-22.57) | 0.010 | Outside | |||||||||
| C116_YL | 11.0 | 5.73 (1.55-21.21) | 0.009 | Yes | |||||||||
| C116_YS | 7.5 | 3.11 (1.34-7.23) | 0.008 | Yes | |||||||||
| A9_FY | 6.5 | 3.08 (1.31-7.21) | 0.010 | Yes | |||||||||
| C11_SA | 6.9 | 2.28 (1.39-3.73) | 0.001 | Yes | |||||||||
| C24_AS | 5.2 | 1.96 (1.20-3.22) | 0.007 | Yes | |||||||||
| A76_EV | 18.3 | 4.57 (1.83-11.41) | 0.001 | Yes | |||||||||
| C116_SY | 30.7 | 3.41 (1.59-7.32) | 0.002 | Yes | |||||||||
| C21_RH | 10.6 | 1.97 (1.19-3.25) | 0.008 | Outside | |||||||||
| C97_RW | 10.9 | 1.86 (1.17-2.96) | 0.009 | Yes | |||||||||
| C80_KN | 9.9 | 1.73 (1.15-2.62) | 0.009 | Yes | |||||||||
| C77_NS | 6.4 | 1.73 (1.15-2.62) | 0.009 | Yes | |||||||||
Abbreviations: AAS, high-risk AASPT defined by the training dataset; IS, importance score from random forest analysis
High-risk AASPT for OS
We observed that 7 AASPT were associated with significantly increased mortality as compared with the 8/8 URD group (HR 1.92; 95% CI, 1.55 to 2.37) and the non-high-risk group (HR 1.58; 95% CI, 1.25 to 2.01) as shown in Figure 1a (left-hand side).
Figure 1.
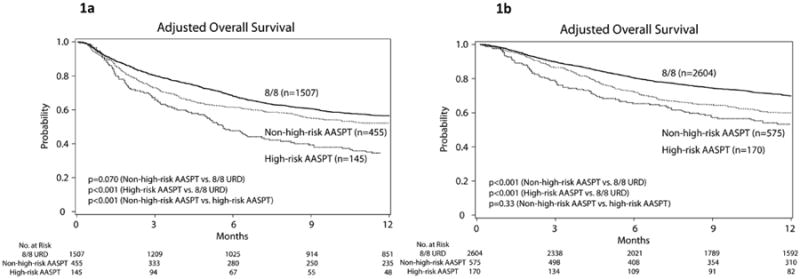
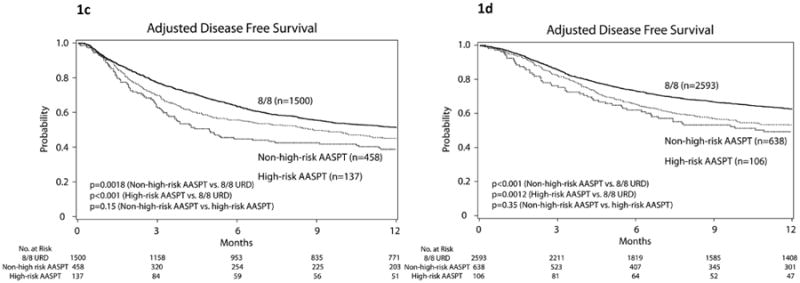
Adjusted estimates of overall survival (1a and 1b) and disease free survival (1c and 1d) by 12 months after transplantation according to the three (matched, high-risk and non-high-risk) groups. Training cohort (panels 1a and 1c), validation cohort (panels 1b and 1d).
High-risk AASPT for DFS
The estimated 1-year event rate for 8/8 URD pairs was 50.7%. Only 3 AASPT met high-risk criteria. Figure 1c shows Kaplan Meier curves according to AASPT risk groups.
High-risk AASPT for TRM
Eight AASPT were significantly associated with an increased risk of TRM. High-risk group had significantly higher TRM than the non-high-risk group (HR 1.42; 95% CI, 1.13 to 1.78) and 8/8 URD group (HR 1.70; 95% CI, 1.40 to 2.06).
High-risk AASPT for acute GvHD grades III-IV
A total of 7 AASPT were identified as risk factors for severe acute GvHD by day 100. Amino acid substitution C97_WR was again identified as a high-risk factor similar to the results for the other three HCT outcomes whereas the other 6 AASPT were uniquely identified as risk factors for acute GvHD. The HR for the high-risk group relative to the 8/8 URD was 1.72 (95% CI, 1.38 to 2.15; P<0.0001) and the HR for the non-high-risk group relative to 8/8 URD was 1.40 (95% CI, 1.15 to 1.70; P=0.0009). Figure 2a shows the cumulative incidence curves according to patient-stratifications.
Figure 2.
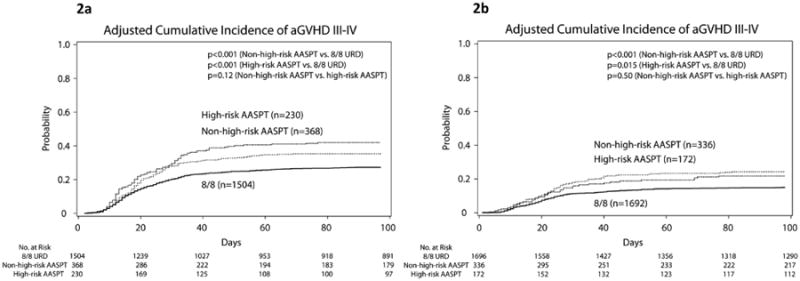
Adjusted cumulative incidence estimates of acute GvHD by day 100 after transplantation according to the three (matched, high-risk and non-high-risk) groups. Panel 2a: training cohort; panel 2b: validation cohort.
HLA class I mismatched alleles comprising high-risk and non-high-risk AASPT in the training set
The mismatched HLA-class I allele (n>=10) comprising high-risk AASPT associated with all outcomes was HLA-C*01:02/02:02 (n=11). In addition, HLA-C*02:02/01:02 (n=14), and C*04:01/16:01 (n=11) were identified in the TRM and acute GvHD analyses and the HLA-C*15:02/14:02 (n=11) allele combination was identified in the acute GvHD analysis. The HLA-C*03:03/03:04 (n=20) and HLA-C*03:04/03:03 (n=30) were the most common mismatched allele combinations comprising all non-high-risk AASPT. These alleles differ by one amino acid at position 91 (HLA-C*03:03→91A, HLA-C*03:04→91G).
Validation of high-risk AASPT
Of 155 AASPT present in at least 10 individuals in the training set, a total of 19 were identified as high-risk across all four outcomes. Only these 19 AASPT were considered when analyzing the validation set. Of these 19, all but two (C156_QR and C116_YL) were present in at least 10 individuals in the validation set. Note, however, that in the validation analysis, patients were grouped as “high-risk” if they had any one of the high-risk AASPT found in the training set.
Outcome specific analysis in the validation cohort
Table 3 shows the results for the outcome-specific analyses in the validation cohort. Presence of at least one high-risk AASPT relative to HLA mismatch without a high-risk AASPT was not associated with OS, DFS, TRM or grade III-IV acute GvHD (all P>0.33). As expected, HLA mismatch had inferior outcomes relative to 8/8 MUD, irrespective of presence or absence of high-risk AASPT defined by the training dataset. Figure 1b shows survival and Figure 1d shows DFS Kaplan Meier curves according to AASPT risk groups. Figure 2b shows cumulative incidence curves for the validation cohort. All curves are adjusted for recipient age, gender match, type of disease, and disease stage.
Table 3. Outcome-specific analyses of high-risk amino acid substitution position and type in the validation cohort.
| Hazard Ratio | 95% Confidence Interval | p-value | ||
|---|---|---|---|---|
| Overall Survival1 | No AASPT (n=575) vs. 8/8 | 1.38 | 1.22-1.57 | <0.0001 |
| At least 1 AASPT (n=170) vs. 8/8 | 1.55 | 1.26-1.91 | <0.0001 | |
| At least 1 AASPT vs. no AASPT | 1.12 | 0.89-1.41 | 0.33 | |
| Disease-Free Survival2 | No AASPT (n=638) vs. 8/8 | 1.33 | 1.18-1.49 | <0.0001 |
| At least 1 AASPT (n=106) vs. 8/8 | 1.50 | 1.17-1.93 | 0.0012 | |
| At least 1 AASPT vs. no AASPT | 1.13 | 0.87-1.47 | 0.35 | |
| Treatment-Related Mortality3 | No AASPT (n=539) vs. 8/8 | 1.61 | 1.37-1.91 | <0.0001 |
| At least 1 AASPT (n=205) vs. 8/8 | 1.75 | 1.38-2.22 | <0.0001 | |
| At least 1 AASPT vs. no AASPT | 1.09 | 0.83-1.42 | 0.54 | |
| Grades III-IV Acute GvHD4 | No AASPT (n=336) vs. 8/8 | 1.74 | 1.36-2.22 | <0.0001 |
| At least 1 AASPT (n=172) vs. 8/8 | 1.52 | 1.08-2.14 | 0.015 | |
| At least 1 AASPT vs. no AASPT | 0.88 | 0.60-1.29 | 0.50 |
Adjusted for patient age, CMV match, conditioning, donor age, disease status, KPS; stratified by graft type.
Adjusted for patient age, CMV match, donor age, disease status, KPS, year of transplantation; stratified by graft type and disease type.
Adjusted for patient age, donor age, disease, disease status, KPS; stratified by graft type
Adjusted for patient sex; stratified by graft type.
Abbreviations: AAS, high-risk AASPT defined by the training dataset; GvHD, graft-versus-host disease; CMV, cytomegalovirus
Aggregate analysis in the validation cohort
Table 4 shows the aggregate analysis results in the validation cohort comparing high-risk AASPT for any outcome to only non-high-risk AASPT. Presence of at least one high-risk AASPT for any outcome was not associated with OS, DFS, TRM, or grade III-IV acute GvHD relate to non-high-risk AASPT.
Table 4. Aggregate analyses of all high-risk amino acid substitution position and type in the validation cohort.
| Hazard Ratio | 95% Confidence Interval | p-value | ||
|---|---|---|---|---|
| Overall Survival1 | No AAS (n=390) vs. 8/8 | 1.41 | 1.21-1.64 | <0.0001 |
| At least 1 AAS (n=354) vs. 8/8 | 1.43 | 1.23-1.66 | <0.0001 | |
| At least 1 AAS vs. no AAS | 1.01 | 0.83-1.23 | 0.91 | |
| Disease-Free Survival2 | No AAS vs. 8/8 | 1.33 | 1.15-1.54 | <0.0001 |
| At least 1 AAS vs. 8/8 | 1.37 | 1.18-1.59 | <0.0001 | |
| At least 1 AAS vs. no AAS | 1.03 | 0.85-1.24 | 0.76 | |
| Treatment-Related Mortality3 | No AAS vs. 8/8 | 1.65 | 1.37-1.99 | <0.0001 |
| At least 1 AAS vs. 8/8 | 1.66 | 1.36-2.01 | <0.0001 | |
| At least 1 AAS vs. no AAS | 1.00 | 0.79-1.28 | 0.97 | |
| Grades III-IV Acute GvHD4 | No AAS vs. 8/8 | 1.75 | 1.34-2.30 | <0.0001 |
| At least 1 AAS vs. 8/8 | 1.58 | 1.18-2.10 | <0.002 | |
| At least 1 AAS vs. no AAS | 0.90 | 0.63-1.28 | 0.55 |
Adjusted for patient age, CMV match, conditioning, donor age, disease status, KPS; stratified by graft type.
Adjusted for patient age, CMV match, donor age, disease status, KPS, year of transplantation; stratified by graft type and disease type.
Adjusted for patient age, donor age, disease, disease status, KPS; stratified by graft type.
Adjusted for patient sex; stratified by graft type.
Abbreviations: AAS, high-risk AASPT defined by the training dataset; GvHD, graft-versus-host disease; CMV, cytomegalovirus
Discussion
We sought to identify and validate high-risk class I HLA AASPT for one-year outcomes and grade III-IV acute GvHD after allogeneic transplants. To accomplish this, we used two independent recipient-donor populations, training dataset 1988-2004 predominantly bone marrow and a more recent validation dataset (2004-2011) predominantly peripheral blood stem cell as donor sources and employed a novel statistical approach using RF and logistic regression methods in a large and homogeneous group of patients with hematological malignancies to define AASPT associated with worse outcomes. AASPT C97_WR (patient-donor respectively) showed greater risks across all outcomes and AASPT C80_NK, C77_SN, and C156_RW conferred greater risks in more than one endpoint. These results are consistent with day 100 survival results previously reported from our group (15), Table 5, as well as with previous reports from the literature (11, 12) (14). The training set was the same as our prior analysis but extended to 1 year outcomes. We tested our results in an independent cohort from a more recent era for validation. Although a similar trend in the validation results was observed, Figures 1 and 2, we did not confirm that the specific AASPT found in the training set were significantly associated with any of the tested outcomes in the validation cohort.
Table 5.
Comparison of the high-risk amino acid substitutions (AAS) identified in this report with the high-risk AAS identified in our previous report (Marino et al, BMT 2012). High-risk AAS identified in both studies are highlighted and bolded.
| This study | Previous study (Marino et al, BMT 2012) | |
|---|---|---|
| HLA-C | - | HLA-C6 |
| - | HLA-C9 | |
| HLA-C11 | HLA-C11 | |
| - | HLA-C14 | |
| HLA-C21 | HLA-C21 | |
| HLA-C24 | - | |
| - | HLA-C66 | |
| HLA-C77 | HLA-C77 | |
| HLA-C80 | HLA-C80 | |
| - | HLA-C95 | |
| HLA-C97 | HLA-C97 | |
| - | HLA-C99 | |
| HLA-C116 | HLA-C116 | |
| HLA-C156 | HLA-C156 | |
| - | HLA-C163 | |
| - | HLA-C173 | |
| HLA-A | HLA-A9 | HLA-A9 |
| HLA-A43 | HLA-A43 | |
| - | HLA-A62 | |
| - | HLA-A63 | |
| HLA-A76 | HLA-A76 | |
| - | HLA-A77 | |
| - | HLA-A95 | |
| - | HLA-A97 | |
| - | HLA-A114 | |
| - | HLA-A116 | |
| - | HLA-A152 | |
| - | HLA-A156 | |
| HLA-A166 | HLA-A166 | |
| HLA-A167 | HLA-A167 | |
| HLA-B | - | HLA-B97 |
| - | HLA-B positon 109 | |
| - | HLA-B position 116 | |
| - | HLA-B position 156 |
Evaluation of permissiveness of HLA mismatches has been an area of interest in the field of HCT for over a decade and by using approaches based on a biological hypothesis and experimental data, some permissive HLA alleles have been identified and validated in independent datasets, i.e. HLA-DPB1 T-cell-epitope groups (20-22) as well as HLA-C*03:03 versus HLA-C*03:04 (23, 24).
Several groups have studied HLA non-permissiveness using other approaches. In regards to high-risk AASPT at HLA-class I loci, review of the literature indicates that no classification scheme has been validated in an independent dataset (11, 12, 14) (25-28). It is noteworthy that our results from the training dataset were consistent with results from Ferrara (11) and Kawase (12) even though those studies were performed in heterogeneous populations that included both patients with advanced hematological malignancies and patients with non-malignant diseases (12). In particular AAS HLA-C-116, located in the F pocket of the peptide binding region (29) has been consistently been identified as high-risk in outcome studies (11, 12, 14, 15) as well as in cellular assays (30). A number of other CIBMTR studies testing proposed algorithms for categorizing mismatches were unsuccessful when applied to HCT data (25-28). Approaches to categorizing mismatches varied using serologically defined cross-reactive epitopes (25), HLA Matchmaker, (26), traditional statistical methods (27), and the Histocheck algorithm (28, 31) (32).
Therefore, at the present time despite substantial effort, suitability of HLA-mismatched unrelated donors cannot be predicted with a few exceptions. We believe that the main reasons why results from training datasets fail to be validated in independent datasets can be grouped into two categories: 1) differences in analyzed datasets, 2) complexity of the problem, and/or inadequacy of statistical approaches used. First, in the case of our study, the training and testing datasets differed because transplant technologies evolved, i.e. predominantly bone marrow transplants in the initial dataset versus peripheral blood stem cells in the discovery dataset, and different conditioning regimens and GvHD prophylaxis were used. Because of how the study evolved, the training and testing sets were derived from two different eras of transplantation. Lack of validation of previous results using more modern datasets has been reported (33, 34). Although a randomized analysis would remove population heterogeneity as a potential cause of failure to validate, it also fails to acknowledge that the goal of the testing set is to prove that results are robust enough to be applicable to future patients. Therefore, established algorithms of non-permissible mismatches need to be re-tested as clinical and transplant conditions evolve.
The second reason that initial observations are not validated in independent cohorts may be that robust identification of permissive or non-permissive HLA mismatches may not be a tractable problem given the size of the available analytic population, the analytic methods currently available, the blunt outcomes available to study, and the complexity of the problem. It is likely that a combination of AASPT rather than single residues is associated with specific outcomes. In addition, our statistical approach is not based on a specific biological hypothesis and assumes that any AAS could potentially have comparable impact on outcomes. With multiple AASPT being responsible for outcomes, our ability to use traditional statistical models may not be sufficient due to the high number of variables and low number of cases for each of them. It is possible that genome wide diversity affecting immune responses and other determinants of alloreactivity may also play a role on HCT outcomes. Therefore, investigation of novel methodological approaches is a highly desirable goal.
The complexity of donor-recipient incompatibility is the result of the exponential number of combinations that must be considered to evaluate risk stratification. Our group has explored a molecular modeling based approach to simulate binding of peptides among mismatched recipient-donor pairs to predict high versus non-high-risk groups (35). Simulations such as these may be used to propose hypothesis driven molecular modeling approaches, and results could then be validated experimentally, and statistically and then translated into clinical practice.
This study has some limitations. Only patients with HLA-A, B, C, and DRB1 typing have been studied. Therefore, the impact of the cumulative effects of HLA disparities in low expression class II HLA loci (36) on HCT outcomes is unknown. In addition, the effect of non-permissive DPB1 T-cell epitope mismatching (21) has not been studied.
Based on our results and review of the literature, we strongly recommend that initial observations are tested in independent cohorts and validation results follow the original observations. Although identification of an algorithm that can help stratify risk of specific recipient-donor mismatches is a desirable goal, it has proved difficult and elusive to date. Our results do not suggest that any particular AASPT in class I should be avoided based on OS, DFS, relapse or grade III-IV acute GvHD. Additional studies should be performed to fully understand the role of AASPT in HCT outcomes.
Supplementary Material
Acknowledgments
This study was partially supported by the University of Chicago Cancer Research Center, Chicago, Illinois (Fund-6-33573 [SRM]). The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U10HL069294 from NHLBI and NCI; a contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-13-1-0039 and N00014-14-1-0028 from the Office of Naval Research; and grants from Actinium Pharmaceuticals; Allos Therapeutics, Inc.; Amgen, Inc.; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Celgene Corporation; Chimerix, Inc.; Fred Hutchinson Cancer Research Center; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Gentium SpA; Genzyme Corporation; GlaxoSmithKline; Health Research, Inc. Roswell Park Cancer Institute; HistoGenetics, Inc.; Incyte Corporation; Jeff Gordon Children's Foundation; Kiadis Pharma; The Leukemia & Lymphoma Society; Medac GmbH; The Medical College of Wisconsin; Merck & Co, Inc.; Millennium: The Takeda Oncology Co.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Onyx Pharmaceuticals; Optum Healthcare Solutions, Inc.; Osiris Therapeutics, Inc.; Otsuka America Pharmaceutical, Inc.; Perkin Elmer, Inc.; Remedy Informatics; Sanofi US; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; St. Baldrick's Foundation; StemCyte, A Global Cord Blood Therapeutics Co.; Stemsoft Software, Inc.; Swedish Orphan Biovitrum; Tarix Pharmaceuticals; TerumoBCT; Teva Neuroscience, Inc.; THERAKOS, Inc.; University of Minnesota; University of Utah; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
Footnotes
Conflicts of Interest: The authors declare no competing financial interests.
References
- 1.Gragert L, Eapen M, Williams E, Freeman J, Spellman S, Baitty R, et al. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014;371(4):339–48. doi: 10.1056/NEJMsa1311707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–30. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–83. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 4.Woolfrey A, Klein JP, Haagenson M, Spellman S, Petersdorf E, Oudshoorn M, et al. HLA-C Antigen Mismatch Is Associated with Worse Outcome in Unrelated Donor Peripheral Blood Stem Cell Transplantation. Biol Blood Marrow Transplant. 2011;17(6):885–92. doi: 10.1016/j.bbmt.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horan J, Wang T, Haagenson M, Spellman SR, Dehn J, Eapen M, et al. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood. 2012;120(14):2918–24. doi: 10.1182/blood-2012-03-417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Rich ES, Godley L, Odenike O, Joseph L, Marino S, et al. Reduced-intensity conditioning with combined haploidentical and cord blood transplantation results in rapid engraftment, low GVHD, and durable remissions. Blood. 2011;118(24):6438–45. doi: 10.1182/blood-2011-08-372508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Besien K, Liu H, Jain N, Stock W, Artz A. Umbilical cord blood transplantation supported by third-party donor cells: rationale, results, and applications. Biol Blood Marrow Transplant. 2013;19(5):682–91. doi: 10.1016/j.bbmt.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunstein CG, Fuchs EJ, Carter SL, Karanes C, Costa LJ, Wu J, et al. Alternative donor transplantation after reduced intensity conditioning: results of parallel phase 2 trials using partially HLA-mismatched related bone marrow or unrelated double umbilical cord blood grafts. Blood. 2011;118(2):282–8. doi: 10.1182/blood-2011-03-344853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischhauer K, Kernan NA, O'Reilly RJ, Dupont B, Yang SY. Bone marrow-allograft rejection by T lymphocytes recognizing a single amino acid difference in HLA-B44. N Engl J Med. 1990;323(26):1818–22. doi: 10.1056/NEJM199012273232607. [DOI] [PubMed] [Google Scholar]
- 10.Keever CA, Leong N, Cunningham I, Copelan EA, Avalos BR, Klein J, et al. HLA-B44-directed cytotoxic T cells associated with acute graft-versus-host disease following unrelated bone marrow transplantation. Bone Marrow Transplant. 1994;14(1):137–45. [PubMed] [Google Scholar]
- 11.Ferrara GB, Bacigalupo A, Lamparelli T, Lanino E, Delfino L, Morabito A, et al. Bone marrow transplantation from unrelated donors: the impact of mismatches with substitutions at position 116 of the human leukocyte antigen class I heavy chain. Blood. 2001;98(10):3150–5. doi: 10.1182/blood.v98.10.3150. [DOI] [PubMed] [Google Scholar]
- 12.Kawase T, Morishima Y, Matsuo K, Kashiwase K, Inoko H, Saji H, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110(7):2235–41. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 13.Kawase T, Matsuo K, Kashiwase K, Inoko H, Saji H, Ogawa S, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009;113(12):2851–8. doi: 10.1182/blood-2008-08-171934. [DOI] [PubMed] [Google Scholar]
- 14.Pidala J, Wang T, Haagenson M, Spellman SR, Askar M, Battiwalla M, et al. Amino acid substitution at peptide-binding pockets of HLA class I molecules increases risk of severe acute GVHD and mortality. Blood. 2013;122(22):3651–8. doi: 10.1182/blood-2013-05-501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marino SR, Lin S, Maiers M, Haagenson M, Spellman S, Klein JP, et al. Identification by random forest method of HLA class I amino acid substitutions associated with lower survival at day 100 in unrelated donor hematopoietic cell transplantation. Bone Marrow Transplant. 2012;47(2):217–26. doi: 10.1038/bmt.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breiman L. Random Forests Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 17.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–81. [Google Scholar]
- 18.Cox DR. Regression Models and Life-Tables. Journal of the Royal Statistical Society Series B. 1972;34(2):187–220. [Google Scholar]
- 19.Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Statistics in Medicine. 1999;18:695–706. doi: 10.1002/(sici)1097-0258(19990330)18:6<695::aid-sim60>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 20.Zino E, Vago L, Di Terlizzi S, Mazzi B, Zito L, Sironi E, et al. Frequency and targeted detection of HLA-DPB1 T cell epitope disparities relevant in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2007;13(9):1031–40. doi: 10.1016/j.bbmt.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366–74. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pidala J, Lee SJ, Ahn KW, Spellman S, Wang HL, Aljurf M, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124(16):2596–606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oudshoorn M, Doxiadis II, van den Berg-Loonen PM, Voorter CE, Verduyn W, Claas FH. Functional versus structural matching: can the CTLp test be replaced by HLA allele typing? Hum Immunol. 2002;63(3):176–84. doi: 10.1016/s0198-8859(01)00384-6. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Vina MA, Wang T, Lee SJ, Haagenson M, Aljurf M, Askar M, et al. Identification of a permissible HLA mismatch in hematopoietic stem cell transplantation. Blood. 2014;123(8):1270–8. doi: 10.1182/blood-2013-10-532671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade JA, Hurley CK, Takemoto SK, Thompson J, Davies SM, Fuller TC, et al. HLA mismatching within or outside of cross-reactive groups (CREGs) is associated with similar outcomes after unrelated hematopoietic stem cell transplantation. Blood. 2007;109(9):4064–70. doi: 10.1182/blood-2006-06-032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duquesnoy R, Spellman S, Haagenson M, Wang T, Horowitz MM, Oudshoorn M. HLAMatchmaker-defined triplet matching is not associated with better survival rates of patients with class I HLA allele mismatched hematopoietic cell transplants from unrelated donors. Biol Blood Marrow Transplant. 2008;14(9):1064–71. doi: 10.1016/j.bbmt.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baxter-Lowe LA, Maiers M, Spellman SR, Haagenson MD, Wang T, Fernandez-Vina M, et al. HLA-A disparities illustrate challenges for ranking the impact of HLA mismatches on bone marrow transplant outcomes in the United States. Biol Blood Marrow Transplant. 2009;15(8):971–81. doi: 10.1016/j.bbmt.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spellman S, Klein J, Haagenson M, Askar M, Baxter-Lowe LA, He J, et al. Scoring HLA class I mismatches by HistoCheck does not predict clinical outcome in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2012;18(5):739–46. doi: 10.1016/j.bbmt.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329(6139):512–8. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 30.Sugimoto K, Murata M, Terakura S, Naoe T. CTL clones isolated from an HLA-Cw-mismatched bone marrow transplant recipient with acute graft-versus-host disease. J Immunol. 2009;183(9):5991–8. doi: 10.4049/jimmunol.0804310. [DOI] [PubMed] [Google Scholar]
- 31.Elsner HA, Blasczyk R. Sequence similarity matching: proposal of a structure-based rating system for bone marrow transplantation. Eur JImmunogenet. 2002;29(3):229–36. doi: 10.1046/j.1365-2370.2002.00301.x. [DOI] [PubMed] [Google Scholar]
- 32.Risler JL, Delorme MO, Delacroix H, Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988;204(4):1019–29. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 33.Kanda Y, Kanda J, Atsuta Y, Fuji S, Maeda Y, Ichinohe T, et al. Changes in the clinical impact of high-risk human leukocyte antigen allele mismatch combinations on the outcome of unrelated bone marrow transplantation. Biol Blood Marrow Transplant. 2014;20(4):526–35. doi: 10.1016/j.bbmt.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Kanda Y, Kanda J, Atsuta Y, Maeda Y, Ichinohe T, Ohashi K, et al. Impact of a single human leucocyte antigen (HLA) allele mismatch on the outcome of unrelated bone marrow transplantation over two time periods. A retrospective analysis of 3003 patients from the HLA Working Group of the Japan Society for Blood and Marrow Transplantation. Br J Haematol. 2013;161(4):566–77. doi: 10.1111/bjh.12279. [DOI] [PubMed] [Google Scholar]
- 35.Binkowski TA, Marino SR, Joachimiak A. Predicting HLA class I non-permissive amino acid residues substitutions. PLoS One. 2012;7(8):e41710. doi: 10.1371/journal.pone.0041710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Vina MA, Klein JP, Haagenson M, Spellman SR, Anasetti C, Noreen H, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121(22):4603–10. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


