Abstract
Methicillin resistant Staphylococcus aureus (MRSA) is a public health threat worldwide. Although the mobile genomic island responsible for this phenotype, called SCC, was labeled non-replicative, we predicted DNA replication-related functions for some of their conserved proteins. We show that one of these, Cch, is homologous to the self-loading initiator helicases of an unrelated family of genomic islands, that it is an active 3’ to 5' helicase, and that the adjacent ORF encodes an ssDNA-binding protein. Our 2.9Å crystal structure of intact Cch shows that it forms a hexameric ring. Cch belongs to the pre-sensor II insert clade of AAA+ ATPases, as do the archaeal and eukaryotic MCM-family replicative helicases. Additionally, we find that SCC elements are part of a broader family of mobile elements that all encode a replication initiator upstream of their recombinases. Replication after excision would enhance the efficiency of horizontal gene transfer.
Introduction
Staphylococcal Cassette Chromosome, or SCC elements, are a family of genomic islands found in S. aureus and closely related species. SCC elements that carry the mecA gene are called SCCmec and render S. aureus methicillin-resistant, creating the MRSA strains that are a major public health problem1. SCC elements are integrated into and excised from the host genome by site-specific DNA recombinases2,3, but very little is known about the events between excision from one genome and appearance in another, and no other core “housekeeping” genes have been characterized. To better understand the lifestyle of SCC elements, we undertook to define their most conserved genes, and to propose functions for the proteins they encode.
SCC elements are highly mosaic but share two defining features: their specific insertion site in the S. aureus chromosome, and their recombinase gene complex. The latter contains 1 or 2 site-specific DNA recombinases, termed Ccr (cassette chromosome recombinase) and additional conserved genes whose functions have not been described. Plasmid-based overexpression of the recombinases results in excision of SCCmec elements as circular molecules4,5. Presumably because replication of such artificially triggered circles was not detected, SCCmec elements were cataloged as non-replicative mobile elements4,6. However, our bioinformatic analysis of the conserved genes surrounding those of the recombinases found that several probably function in DNA replication. This work focuses on the large ATPases encoded immediately upstream of the recombinases. As described below, our bioinformatic analysis showed that they are related to the self-loading initiator helicases (“Rep” proteins) of S. aureus pathogenicity islands (SaPIs). Furthermore, our biochemical and structural analysis of one, Cch from SCCmec type IV, showed that it is enzymatically active and that it is unexpectedly similar to the MCM helicases of archaeal and eukaryotic DNA replication (for simplicity, we refer to both as MCM although the eukaryotic version is a heterohexamer of related subunits).
The SaPIs are a different family of genomic islands whose molecular biology is better understood than that of the SCC elements. They carry toxins and other superantigens, causing food poisoning, toxic shock syndrome and necrotizing fasciitis7. Like SCCs, SaPIs integrate into specific sites in their hosts’ chromosomes, but the SaPI integrases belong to the tyrosine family of site-specific recombinases whereas the SCC integrases belong to the “large serine” recombinase family8. The SaPIs hijack bacteriophages for horizontal transfer. After activation by a helper phage, the SaPIs excise, replicate, and package their own newly generated DNA into phage capsids7,9,10. SaPI replication is supported by a module composed of adjacent (sometimes fused) genes encoding a primase and a Rep protein, and the origin of replication that is immediately downstream of the rep gene11. Although conserved, the primase is not strictly required for SaPI replication9,11. The most extensively characterized SaPI Rep, that of SaPIBov1, has been shown to have helicase activity in vitro and to use its C-terminal domain to recognize 11 short sequence motifs within its cognate origin, where it opens a bubble between the two strands without the aid of any additional proteins11,12.
Self-loading helicases are encoded by a variety of bacteriophages and eukaryotic viruses11,13,14. These enzymes can recognize sequences in dsDNA that mark the relevant origin of replication, separate the two strands to open a bubble, and then somehow become active ring-shaped helicases that encircle a single strand of DNA and drive progression of the replication fork. In contrast, cellular organisms use a series of different proteins to perform these functions. Although the simplicity of self-loading helicases makes them good systems for understanding the mechanics of replication initiation, modeling the process has been hampered by limited structural information for intact self-loading helicases.
Results
Analysis of SCC Core Genes Suggests Replication
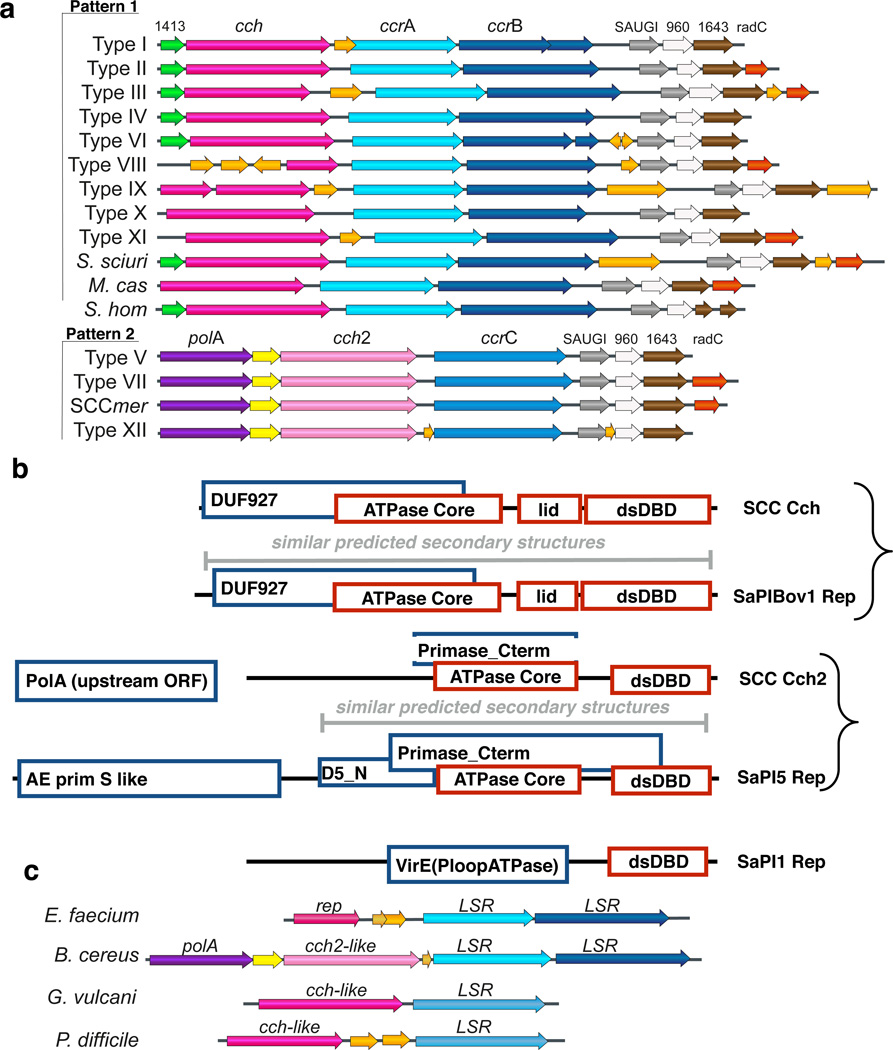
The recombinase gene complex consists of the recombinase gene(s) flanked by a few additional conserved open reading frames (ORFs). This complex comprises the only set of genes that are conserved across all SCC elements, and is thus likely to encode their core machinery. However, the literature does not describe the boundaries of this complex or the functions of the additional ORFs. By comparing the sequences and predicted conserved domains15 of the recombinase-flanking ORFs within the prototypes of all staphylococcal SCCmec types defined to date as well as several additional SCC elements, we defined two related patterns of conserved genes (Fig. 1a).
Figure 1. Cartoon of SCC and related mobile elements’ conserved core.
a. Two different patterns are observed in SCC elements. Arrows represent open reading frames in the recombinase locus of the twelve SCCmec elements identified to date plus elements from S. sciuri, S. hominis and M. caseolyticus. SCCmer encodes mercury rather than methicillin resistance. Conserved genes are labeled above each set according to known or putative function, or database annotation (domain of unknown function (DUF) number for 1413, 960, and 1643, and radC for the semi-conserved red gene). Large serine recombinase genes are in blue, putative replication initiators in different shades of pink according family, and non-conserved open reading frames orange. Note that SAUGI (uracil DNA glycosylase inhibitor) genes are usually annotated as DUF95016.
b. Cch and Cch2 are homologous to different SaPI Reps. Annotations in blue boxes are from NCBI. Annotations in red boxes are based on our structure for Cch, and our predictions combined available biochemical data for other proteins11. Of the 3 different types of SaPI Rep (exemplified by SaPIBov1, SaPI5 and SaPI1), Cch is homologous to the 1st and Cch2 to the 2nd. Cch2 lacks the N-terminal primase domain of SaPI5 Rep, but there is a putative primase (“PolA”)19 upstream in the same predicted operon.
c. Selected SCC-like elements found inserted in the same target site as SCC in other Gram-positive bacteria. Color-coding is as in part a. LSR: Large Serine Recombinase (similar to CcrA, B, and C). The rep gene of the E. faecium element shown here belongs to the Rep_Trans superfamily of nickase/relaxase proteins. See Supplementary Table 1 for accession numbers.
Both patterns include the same three small ORFs downstream of the recombinases. These contain domains of unknown function (DUFs) 950, 960 and 1643. The first, containing DUF950, was previously characterized as SAUGI, a uracil-DNA glycosylase (UDGase) inhibitor, although its context on a mobile genetic element was not noted16. That SAUGI is always present in SCC elements suggests that they can replicate because the only other characterized UDGase inhibitors are encoded by phages to protect their newly replicated DNA from the base excision activity of their host’s UDGase17. The roles of the other two ORFs remain unknown. A fourth ORF, annotated as RadC, is partially conserved, often truncated, and is further discussed in the Supplementary Information. We noted that the ACME element that is sometimes inserted adjacent to SCCmec also contains the DUF 950-960-1643 trio. ACME depends on SCCmec-encoded recombinases for mobilization, but the presence of these 3 ORFs suggests that ACME may be a degenerate SCC element that has lost the other conserved core genes.
Both patterns of SCC elements encode a predicted helicase upstream of the recombinases, but the type varies (Fig. 1a). Cch, found in pattern 1,18 contains DUF927, whereas in pattern 2 the predicted helicase, which we call Cch2, contains a “primase_Cterm” domain instead (named after the C-terminus of phage P4’s fused primase-helicase protein13). Cch is usually preceded by a small (~100 aa) positively charged protein that we provisionally named LP1413 (Little Protein with DUF1413) and that we found has high affinity for ssDNA (see below). In all pattern 2 elements, cch2 is preceded by two ORFs. The first is annotated as PolA-like and contains only the polymerase domain of an A-family polymerase (similar to that of DNA Pol I and T7 RNA polymerase). This protein was included in a new family, “TV Pol,” of probable primases identified through bioinformatics that are found on transposons and viruses19. Between PolA and Cch2 is a ~122aa ORF that is not homologous to LP1413, and carries no recognizable conserved domains. Differences in the helicase-encoding operons correspond to two other features: (1) the recombinases and (2) insertion orientation relative to the host's chromosomal origin of replication. Pattern 1 SCC elements encode two recombinases (ccrA and ccrB, in one operon), whereas pattern 2 elements encode a single recombinase (ccrC). The host’s chromosomal origin of replication is downstream (to the right as drawn in Figure 1a) for pattern 1 elements and upstream (or to the left) for pattern 2 elements.
Both predicted helicases, Cch and Cch2, have homologs among SaPI Rep proteins (7; Fig. 1b). Although the sequences of Cch and SaPIBov1 Rep are only 17.6% identical, the similarity extends across the entirety of both proteins, both contain DUF927, and their predicted secondary structures align nearly perfectly (Supplementary Fig. 1). As structure and function are often more highly conserved than primary sequence, this strongly suggests that Cch and SaPIBov1 Rep have similar functions (e.g. the catalytic domains of Flp and Cre recombinases are only 13% identical, yet they have nearly-identical structures and functions20). There are 3 families of SaPI Rep proteins, and Cch2 has homology to a different one, exemplified by SaPI5 Rep (Supplementary Fig. 2). SaPI5 Rep contains an N-terminal primase domain that Cch2 lacks (Fig. 1b), which might be functionally replaced by the separate “PolA” ORF upstream of Cch2.
Additional database searching revealed that SCC elements are part of a larger family of mobile genetic elements found in Gram-positive bacteria, a few examples of which are shown in Fig. 1c. These elements are always inserted into the same target site as SCC (the 3’ end of the coding region for an rRNA methyltransferase, rlmH) and encode one or two Ccr-like large serine recombinases downstream of a probable Rep. While the Rep-recombinase functional pairing seems conserved, the type of Rep varies: some are similar to those of SCC elements, but the Enterococcus faecium21 mobile element carries a Rep from the nickase family of rolling circle initiators.
These lines of in silico evidence all suggest that Cch and Cch2 have functions related to DNA replication. To further examine our predictions through structure and biochemistry, we focused on Cch from SCCmec type IV (a pattern 1 SCC element) because this type has been found in multiple unrelated genetic backgrounds22 suggesting its promiscuity in horizontal transfer, and because it is harbored by an epidemic community-associated MRSA strain (USA300) in the US6,23.
Overall Architecture of Cch
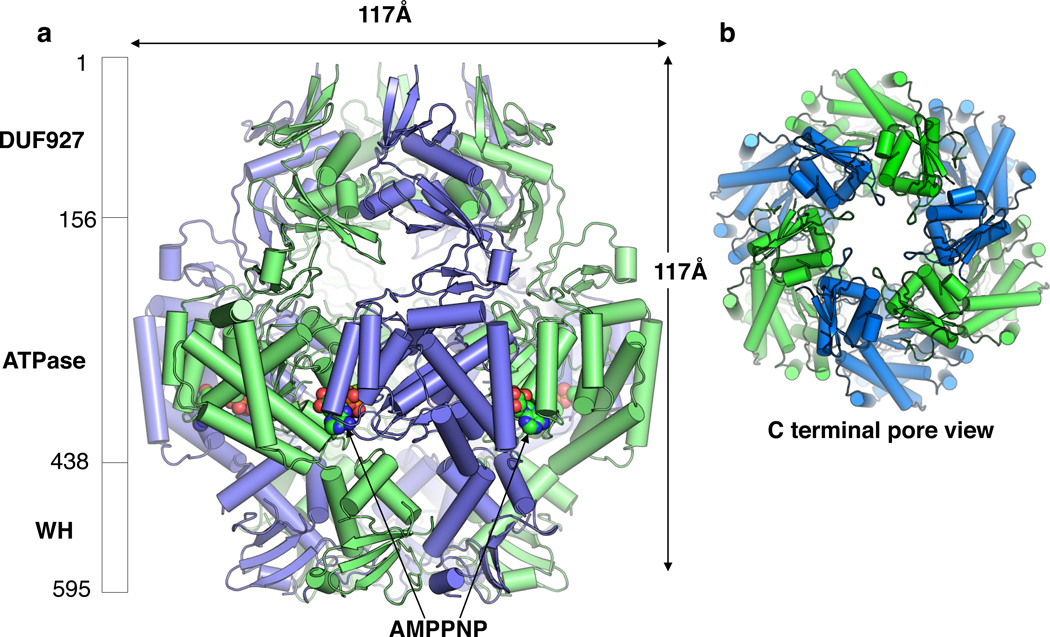
We crystallized intact Cch in the presence of a non-hydrolyzable ATP analog (AMPPNP) and magnesium. We determined phases by single-wavelength anomalous dispersion (SAD) using selenomethionine-containing protein, and refined the final model to an Rwork-Rfree of 22.10%–26.11% (Supplementary Fig. 3, Table 1). The Cch-AMPPNP complex model is shown in Fig. 2. There are two nearly-identical subunits in the asymmetric unit, packed around a crystallographic 3-fold axis to form a hexameric ring with approximate 6-fold symmetry. The AMPPNP is sandwiched between monomers (Fig. 2a). The barrel-shaped hexamer is ≈117Å long and ≈117Å at its widest point. The ring’s lumen diameter is narrowest at the openings (≈18 – 20Å, measured between C-alphas) (Fig. 2b). The barrel has three layers corresponding to the three domains present in each subunit. The N-terminal domain (residues 1–156) corresponds to the first half of DUF927 and according to the Dali server24 has a unique fold (Supplementary Fig. 4). The central domain (residues 157–438) contains an AAA+ ATPase fold, and the C-terminal domain (residues 439–595) includes a WH motif typical of many dsDNA-binding proteins. All three domains contribute to the inter-subunit interactions that probably stabilize the hexamer. Some of these contacts may also coordinate conformational changes among subunits in response to ATP binding and hydrolysis and DNA binding. Each domain contacts the same domain of the neighboring subunits, and each AAA+ domain also interacts with the N- and C-terminal domains of its neighbors to the left (as shown in Fig. 2a).
Table 1.
Data collection, phasing and refinement statistics
| SeMet Cch (5DGK) |
|
|---|---|
| Data collection | |
| Space group | R3:H |
| Cell dimensions | |
| a, b, c (Å) | 110.14 110.14 302.76 |
| α, β, γ (°) | 90 90 120 |
| Peak | |
| Wavelength | 0.9792 |
| Resolution (Å)a | 50 − 2.9 (2.95 − 2.9)a |
| Rmeas | 0.117 (0.771) |
| I/σ(I) | 19 (1.12) |
| CC1/2 | (0.623) |
| Completeness (%) | 92.19 (64.16) |
| Redundancy | 19 (1.12) |
| Refinement | |
| Resolution (Å) | 50 − 2.9 |
| No. reflections | 28098 |
| Rwork / Rfree | 0.2210/0.2611 |
| No. atoms | |
| Protein | 8198 |
| Ligand | 64 |
| Water | 17 |
| B factors | |
| Protein | 36.30 |
| Ligand | 20.80 |
| Water | 23 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.96 |
One crystal was used to determine the structure of Cch.
SeMet, selenomethionine.
Values in parentheses are for highest-resolution shell.
Figure 2. Cch forms a hexameric ring.
a. Crystal structure of Cch. Bound AMPPNP (spheres) is sandwiched between subunits. Alternating subunits are colored green and blue. The three domains and their boundaries are denoted on the left (WH = winged helix turn helix).
b. View down the central axis from the C-terminal end. The lumen diameter is similar when viewed from the N-terminal end. Measured from alpha carbons=18Å
Cch also forms oligomers that are most likely hexamers in solution, as determined size exclusion chromatography (SEC) and blue native polyacrylamide gel electrophoresis (Supplementary Fig. 5). Although the molecular weights determined by these techniques are subject to confounding factors such the protein’s deviation from a sphere and its interactions with dye, the results were consistent with a mixture of monomers and hexamers, with the presumed hexamer predominating in the blue native gel (~3µM Cch in low salt) and the monomer more prominent in the SEC experiments (32µM Cch in 0.5M NaCl). When ssDNA was added in the blue native gel experiment, the smaller band diminished and the larger bind split slightly, suggesting that ssDNA stabilizes the hexameric form.
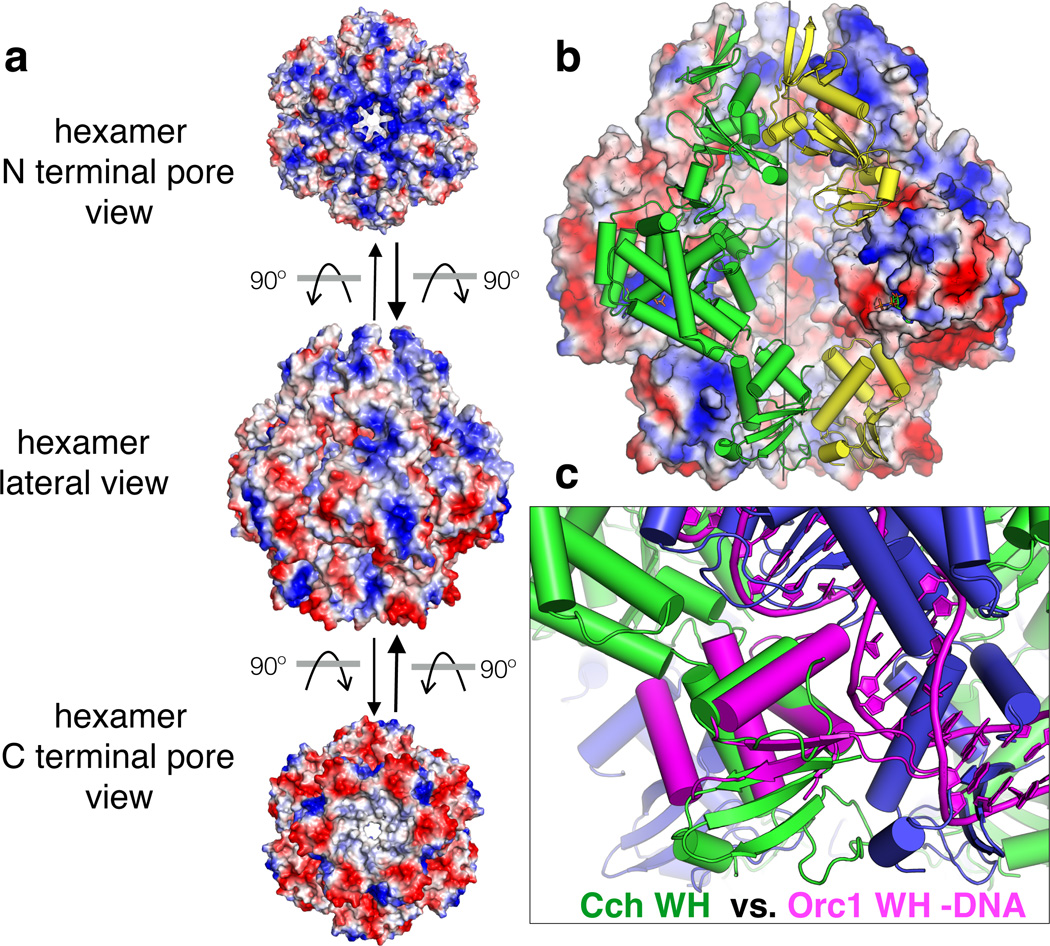
The ring-shaped architecture strongly implies that Cch has processive helicase activity, threading a single DNA strand through the pore as other replicative helicases have been shown to do25. The distribution of electrostatic potential is consistent with DNA binding on the inside as well as possibly the outside of the barrel (Fig. 3a–b). Two loops that are expected to bind DNA within the pore, residues 269–271 and 288–296 (Supplementary Fig. 1), could not be modeled but would contribute an additional net charge of +3 per subunit. Another patch of positive potential corresponds to the recognition helix of the WH motif. Although this face of WH motifs usually binds dsDNA, it is occluded by a neighboring subunit in our structure, suggesting that a Cch hexamer is unable to bind dsDNA unless the WH domain swings outward (Fig. 3b, c). This may be a recurring theme in replication initiation, as the C-terminal WH domains of the eukaryotic origin recognition complex (ORC) also make protein-protein contacts between subunits (Supplementary Fig. 6 and 26).
Figure 3. Potential DNA binding surfaces of Cch.
a. Three views of the Cch hexamer with the surface colored according to vacuum electrostatic potential (blue positive and red negative).
b. View inside the barrel. Two subunits are shown in cartoon rather than surface mode, with the ATPase domain of the yellow subunit removed. Positively charged but disordered loops extending from the ATPase domain toward the center of the barrel are not shown.
c. The WH motif of archaeal Orc1and bound DNA (magenta) (from a heterodimeric Cdc6/Orc1 – DNA complex, PDBid 2QBY)36 is superimposed onto the WH domain of one Cch subunit (green). The dsDNA-binding surface of Cch is blocked by the adjacent subunit (blue) within the hexamer.
Cch’s ATPase domain is closest to MCM’s
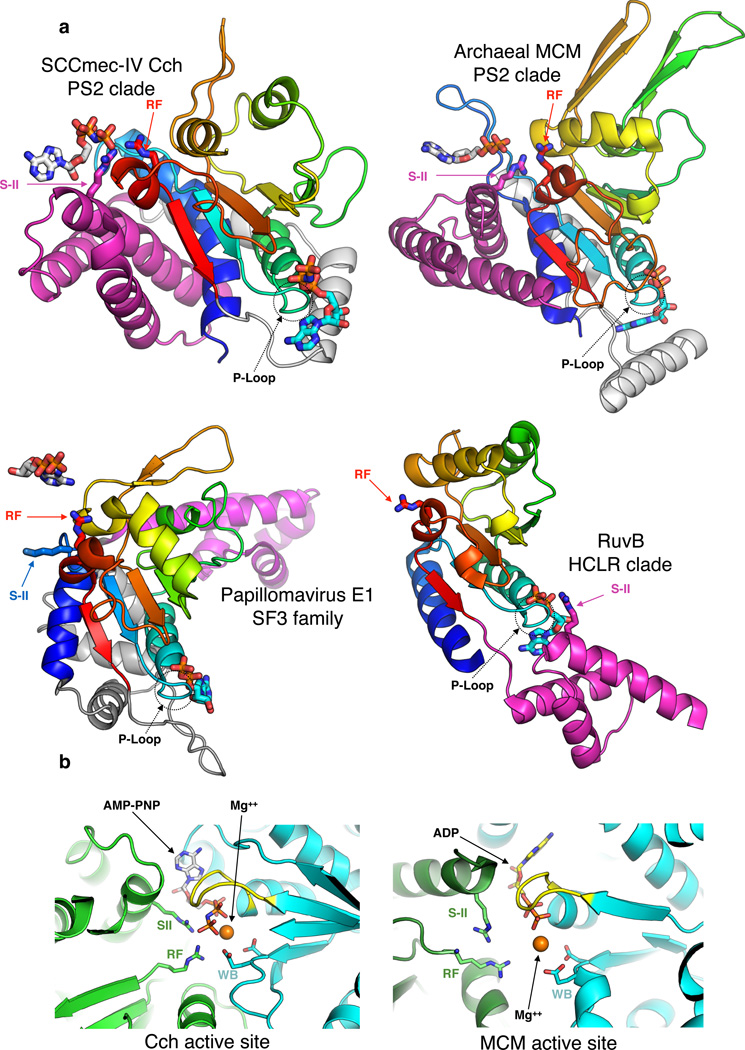
AAA+ family proteins are defined by a structurally conserved ATP-binding and hydrolysis module and are active as oligomers27. ATP binds at the interface between adjacent subunits, and conformational changes triggered by ATP binding and hydrolysis lead to substrate translocation or remodeling. Following the classification schemes of references27,28, Cch belongs to the Pre-Sensor I insert superclade, where a beta hairpin is inserted between helix 3 and strand 427 (Fig. 4 and Supplementary Fig. 1). In helicases belonging to this superclade, this hairpin binds ssDNA and plays a critical role in ATP-dependent translocation14,29. In Cch, this insertion protrudes into the central pore but is disordered, possibly due to the absence of DNA. Although many viral self-loading helicases belong to the superfamily III helicase clade within this superclade, Cch does not have the helical bundle characteristic of that group. Instead, it has an insertion preceding the sensor-II motif that reorients helices 6 and 7 to the other side of the ATPase domain and positions the sensor II Arg395 to act in trans. (Fig. 4, Supplementary Fig. 1, reference27). This architecture places it into the “pre-sensor II insert” clade that includes the MCM family of replicative helicases. Furthermore, according to the DALI server24 the closest structural homolog to the Cch ATPase domain is an archaeal MCM helicase (Fig. 4, Z-score=11.1).
Figure 4. Comparison of DNA helicase / translocase AAA+ domains.
a. The ATPase domains of Cch, archaeal MCM helicase (PDBid 4R7Y)42, papillomavirus E1 helicase (PDBid 2GXA)29 and DNA translocase RuvB (PDBid 1IN4)45 are shown in similar orientations. The core ATP-binding folds are shaded from blue (helix 0) to red (strand 5). C-terminal lid subdomains for Cch, MCM and RuvB and the N-terminal helical bundle of E1 are magenta, and insertions between the ATP binding core and lid or helical bundle are gray. Nucleotide cofactors bound to this subunit are shown with cyan carbons and those bound to the neighboring subunit with white carbons. The conserved arginine finger (RF) always found before strand 5 is in red, and the sensor II (SII) arginine, found in all but E1, in magenta. E1 uses a lysine (blue) instead. RF always interacts with the nucleotide cofactor in trans, while SII is in cis for RuvB but trans for Cch and MCM.
b. Close-up view of Cch and MCM active sites. Two conserved catalytic residues, the arginine finger (RF) and Sensor-II (S-II) are donated in trans to the ATPase site of the cyan subunit. The P loop is colored yellow; The two most important residues of Walker B (WB), nucleotides, RF and S-II are displayed as sticks; magnesium as orange sphere.
Cch has helicase activity with 3’ to 5’ polarity
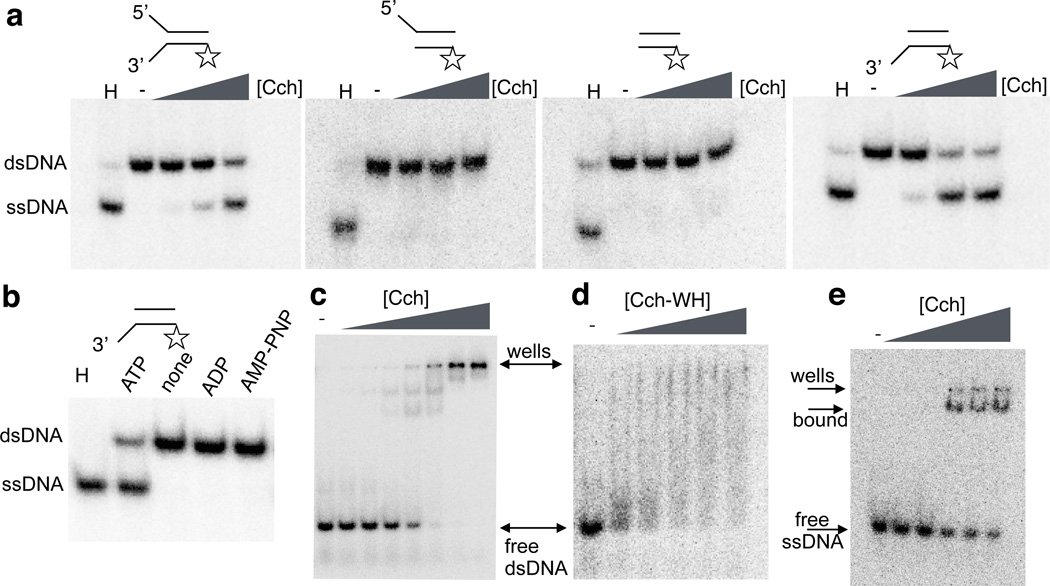
Our model strongly suggests that Cch has ATP-dependent helicase activity, which we confirmed in vitro. Our helicase assay was based on that described in30, but used a radiolabeled 54bp duplex with 6nt mismatched extensions (Fig. 5a and b, methods section). Cch separated the DNA strands in the presence of ATP but not AMPPNP or ADP, showing that ATP hydrolysis is required (Fig. 5b).
Figure 5. Helicase and DNA binding activities of Cch.
a. Helicase assays. 54bp duplexes with 6nt ssDNA extensions as cartooned were incubated with Cch, ATP, and excess unlabeled bottom strand to trap strand separation products. Open star indicates 32P label. H, heat-treated control with no Cch. Cch concentrations were 0, 5, 100 and 200 nM.
b. Helicase assays with different nucleotide cofactors. 360 nM Cch was used with ATP, no nucleotide, ADP and the non-hydrolyzable ATP analog AMPPNP.
c. EMSA of Cch binding to a 60bp duplex. Cch concentrations: 0, 75, 100, 150, 200, 300, 400, 500 nM.
d. EMSA of the isolated C-terminal WH domain of Cch binding to the same duplex as in c. protein concetrations were: 0, 100, 300, 600, 800, 1000 nM. Rapid on-off rates for the isolated domain probably cause the smearing of the shifted DNA.
e. EMSA of intact Cch (0, 10, 20, 50, 80, 100 nM) binding to ssDNA.
Experiments were repeated at least three times.
Most helicases translocate along a single strand, displacing its partner. However, they vary in the direction of translocation: for example, E. coli DnaB and bacteriophage T7 gene 4 translocate in the 5’ to 3’ direction but, bacteriophage P4 helicase and the archaeal and eukaryotic MCM helicases show the opposite polarity31–34. To test the polarity of Cch, we compared its activity on substrates with 3’ or 5’ single stranded tails as well as blunt ends. Cch’s helicase activity required a 3’single stranded end (Fig. 5a), indicating that Cch has 3’ to 5’ polarity, sharing not only structural but also functional similarities with archaeal and eukaryotic MCM replicative helicases.
Cch binds dsDNA and ssDNA
ATP-binding replication initiators such as DnaA, Orc proteins, and SaPI Reps all have C-terminal sequence-specific dsDNA binding domains11,35–37. Fig. 5c–d shows that intact Cch and its isolated C-terminal WH domain bind dsDNA, although latter dissociates during the EMSA experiment giving a smear. By analogy to DnaA and the SaPI Reps, one might expect that the origin for SCC replication lies near the 3’ end of the SCC gene, but our attempts to identify a strongly preferred binding sequence were inconclusive. Specificity might have been masked by non-specific binding of dsDNA in the central Cch pore, or weak specificity may be compensated for in vivo by the high local concentration of Cch near the 3’ end of its own gene due to translation being coupled with transcription. As expected for a helicase, intact Cch has strong affinity for ssDNA (Fig. 5e).
LP1413 binds ssDNA
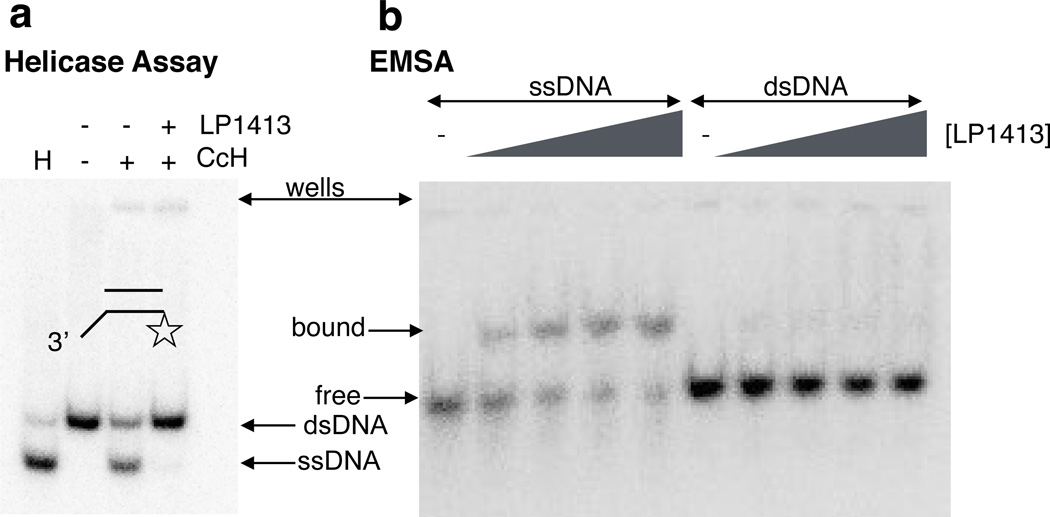
We also cloned and purified LP1413, the small protein encoded just upstream of Cch, which appears to be in the same operon, as the open reading frames overlap by a single nucleotide. LP1413 bound ssDNA but not dsDNA with nanomolar affinity (Fig. 6), and it inhibited Cch’s helicase activity in assays with short model substrates, presumably by blocking access to the 3’ ssDNA tail that Cch requires for loading. Single-stranded DNA binding proteins (SSBs) are important components of replication forks in vivo, where they prevent reannealing of the unwound strands and protect the single-stranded DNA from damage, and we hypothesize that LP1413 also does so in vivo, particularly as it is apparently co-expressed with a helicase.
Figure 6. LP1413 binds ssDNA.
a. Helicase assays in the presence and absence of LP1413. The substrate was the same as that used in Fig. 5a, right panel. Protein concentrations, when added, were 200 nM for both Cch and LP1413.
b. EMSA of LP1413 binding to ssDNA (left) and dsDNA (right). 0, 20, 50, 80, and 100 nM LP1413 was tested with 2 nM 40 nt or bp DNA.
Experiments were repeated at least three times.
Discussion
Our work suggests a previously unsuspected DNA replication activity in the lifestyle of SCC elements. Our in silico analysis of the conserved recombinase gene complex found that the recombinase genes are flanked by ones encoding replication-related proteins: a putative initiator helicase related to those of the SaPIs (Cch or Cch2) on the upstream side, and a uracil DNA glycosylase inhibitor (SAUGI), on the downstream side. Furthermore, whenever cch2 is present, there is also a putative primase belonging to the TV-Pol subfamily of PolA-type polymerases19. Our structural and biochemical analyses further support the idea that SCC elements can replicate: Cch is structurally related to MCM, it is an active helicase, and the other protein in its operon (LP1413) is a single-stranded DNA binding protein.
What role would DNA replication play in the biology of SCC elements? It would presumably be repressed until after the recombinases (CcrAB or CcrC) have catalyzed excision of the element as a covalently closed circle. Replication of that circle would render horizontal transfer of SCC elements to new hosts more efficient by supplying multiple copies to start with. While horizontal transfer of SCC elements has been difficult to demonstrate in the lab, sequencing data clearly show that in the wild, at least some variants such as SCCmec types II, IV and V do spread to new strains and even to different species of staphylococci6,38–40. Replication could also help prevent loss of the element by providing more substrates for reintegration after excision. Our analysis and new biochemical and structural data provide a framework for designing experiments to address these questions in vivo.
We also found that SCC-related mobile genetic elements are widespread among Gram-positive bacteria (see Fig. 1c for representative examples). These elements have three features in common with SCCs: (1) they are inserted into the 3’ end of a putative rlmH-encoding gene (2) they encode one or two large serine recombinases and (3) they carry a putative replication initiator upstream of the recombinase(s). Although there is a correlation within the SCCmec elements between the type of Rep protein and the presence of one vs. two recombinases, that correlation breaks down in the more distantly-related SCC-like elements listed in Fig. 1c. Overall, the genetic pattern of rep – recombinase genes in SCC-like elements appears to be widely conserved even though the type of Rep protein is not. These observations suggest that SCC and SCC-like elements have been subjected to extensive modular evolution and that the rep-recombinase gene pairing has been conserved for functional reasons.
Although both Cch and Cch2 found in SCCmec elements are homologous to replication initiators from the SaPIs, we found no evidence for further similarity between these two types of genomic islands. After replication, SaPIs are packaged into the modified capsids of specific helper phages for transduction to new strains. However, no specific helper phages have been identified for SCCs, and we found no homologs for the machinery used by SaPIs to exploit their helper phages. Additionally, their DNA recombinases are unrelated.
The crystal structure of Cch shows that it belongs to an unusual clade of AAA+ proteins that place the catalytic residue Sensor-II (Arg395) in trans rather than in cis (the PS2 or pre-sensor II insert clade27). The PS2 family is quite different from the RecA-like group that E. coli DnaB and phage T7 helicases belong to, and from the superfamily 3 that many viral helicases, such as SV40 large T antigen and papilloma virus E1 belong to. Surprisingly, the only other helicase of known structure belonging to the PS2 clade is the archaeal and eukaryotic MCM replicative helicase. Cch and MCM also both translocate in the 3’ to 5’ direction and both have a C-terminal WH domain. We could find only one other reported prokaryotic MCM-like helicase, BcMCM from a Bacillus cereus bacteriophage, which was identified by its sequence homology to MCMs41. Although Cch, BcMCM and archaeal MCM all have different N-terminal domains, their helicase and WH domains probably share a common ancestor. Presumably Cch and BcMCM entered Gram-positive eubacteria through the cross-kingdom transfer of mobile genetic elements. In support of this, we found that BLAST does retrieve Cch homologs from archaea, and at least one of these is surrounded by putative DNA recombinases, suggesting that it is carried on a mobile element (NCBI Reference Sequence: WP_011024025.1).
Cch and the related SaPIbovI-type Rep proteins may provide simple model systems for understanding how a hexameric AAA+ ATPase of this clade can progress from binding dsDNA to encircling and translocating along a single strand. For MCMs, the loading and activation processes require multiple additional proteins that facilitate exquisite control of DNA replication initiation. In contrast, the SaPI Reps are self-loading.
MCM is initially loaded as a double hexamer with each ring surrounding duplex DNA. Details of how an initial bubble is opened are unclear but it may involve concerted motor action of the two interacting helicases. SaPIBov1 Rep, the best-studied Cch homolog, is also probably loaded as two hexamers. The SaPIBov1 origin contains 5–6 iterons on each side of a central AT-rich region where melting occurs. Binding by SaPIbovI Rep produced a single shifted band for each half of the origin, suggesting that the full origin is bound by two preformed hexamers12. It is unknown whether or not this Rep’s self-loading process involves an intermediate in which dsDNA is threaded through the hexamer’s central pore. The fact that bubble opening by SaPIBov1 Rep could be detected in the absence of ATP hydrolysis suggests that the systems may differ in this respect, but the diameters of the MCM and Cch ATPase rings are similar: the distances between the beta phosphates of the nucleotide cofactors bound to opposite sides of each ring are 69 Å in Cch, 65 Å in archaeal MCM42 and 60–64 Å in a recent cryo-EM structure of a eukaryotic MCM2–7 heterohexamer43. The N- and C-terminal portions of the Cch ring are narrower but dsDNA might be accommodated, especially if some loops were moved or subdomains shifted. For the eukaryotic version in its active helicase form, most of the WH domains are disordered, but two of them appear to narrow the central channel such that only ssDNA can fit44.
MCM is recruited to the DNA by the ORC complex, whereas SaPI Reps recruit themselves via sequence-specific binding of their WH domains to iterons in their cognate origins. In our hexameric Cch structure the WH domain’s positively charged recognition helix is blocked by the neighboring subunit, suggesting that it must swing outward to interact with dsDNA. Alternatively, iterons may be initially bound by monomers that later assemble into hexamers due to high local concentration. In either case, interactions between hexamers must somehow underwind the central region to favor strand separation. Transient opening of the protein ring to trap DNA in the pore may be triggered by two factors: strain in the dsDNA due to wrapping it around the hexamer when the WH domains bind the iterons, and weakening of the intersubunit contacts within the hexamer due to the WH domains swinging outward to bind the dsDNA. More work is needed to fully understand these processes.
In summary, we have uncovered strong evidence SCC elements encode proteins that support their own DNA replication, and that they are part of a larger family of replicative mobile genetic elements found in Gram-positive bacteria outside the Staphylococceae. Our structure of Cch, the self-loading helicase encoded by SCCmec type IV, is to our knowledge the first of an intact self-loading helicase. Cch most closely resembles the MCM replicative helicases of archaea and eukaryotes, and provides a simple, self-contained model system for studying helicase loading and translocation.
Online Methods
Bioinformatics
We initially discovered the relationship between SCC helicases and SaPI Reps by using PSI-BLAST to find sequence homologs of Cch and then scanning the results for manually-annotated entries46. We also found the Phyre server47 very helpful in assessing the possible function of Cch. Threading of the Cch sequence onto known structures by the Phyre server identified an AAA+ (ATPases Associated with diverse cellular Activities) ATPase module followed by a winged helix-turn-helix (WH) domain, but no good templates were found for the N-terminal region containing DUF927. The AAA+ - WH domain organization is shared by several proteins involved in DNA replication, including those that recognize the origin in both prokaryotes (DnaA) and eukaryotes (ORC proteins and cdc6), and by the MCM helicase of eukaryotes27. Geneious was used extensively in examining SCC element sequences48.
Cch cloning, expression and purification
The SAUSA300_0039 gene (GeneID: 3913650) was amplified from Staphylococcus aureus strain USA300. The resulting sticky-end PCR product, using oligonucleotides ccHF1 (5’- tgagttacacattgtttgaaattggcc); ccHR1 (5’-tcgaggaaatcttcgtgaactcctg), ccHF2 (5’-tatgagttacacattgtttgaaattggcc); ccHR2 (5’-ggaaatcttcgtgaactcc tg), was cloned into the NdeI/XhoI sites in pet21a (Novagen). The cloning process added the following tag to the C-terminus: LEHHHHHH. Protein was expressed in Rosetta (DE3)pLysS cells (Novagen). The cells were grown in Luria–Bertani medium with 100 µg ml−1 ampicillin at 37°C until they reached OD600 ≈ 0.6 and then were induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After induction, cells were grown for 4 h at 37°C and harvested by centrifugation at 8000 rpm in an F10S rotor for 10 min. Cell pellets were kept at −80°C until needed. Cell pellets were resuspended in buffer A without salt (42.5 mM Na2HPO4, 7.5 mM NaH2PO4, 0.5 mM EDTA, 5% Glycerol, 1 mM dithiothreitol (DTT) pH 7.5–8.0) containing protease inhibitor cocktails (Complete Mini, Roche Diagnostics GmbH). Lysozyme was added to 200 µg ml−1, and the mixture was incubated at 37° for 30 min, sonicated and centrifuged at 18000 rpm in an SS-34 rotor at 4°C for 1 h. Optionally, cell pellets were resuspended again in buffer A without salt, sonicated and centrifuged at 18000 rpm in an SS-34 rotor at 4°C for 1 h. Finally, cell pellets were resuspended in buffer A (50 mM sodium phosphate, pH 7.5 –1M NaCl-5% Glycerol-1 mM DTT) sonicated and centrifuged at 18000 rpm in an SS-34 rotor at 4°C for 1 h. Supernatant was loaded onto His Trap™ HP (GE Healthcare) and eluted with a gradient of buffer B (buffer A + 500 mM imidazole). Pooled Cch-containing fractions were dialyzed into buffer A2 (20 mM MES, 0.5 mM EDTA, 5% Glycerol, 100 mM NaCl, ph=5.5), loaded onto Heparin FF (GE Healthcare) and eluted with a gradient of buffer B2 (20 mM MES, 0.5 mM EDTA, 5% Glycerol, 2M NaCl, ph=5.5). Pooled fractions were then dialyzed into buffer A2, loaded onto MonoS™ 5/50 GL (GE Healthcare), and eluted with a gradient of B2. Purified Cch-6xHis was then concentrated up to ≈4.3 mg ml−1, dialysed overnight into stock buffer (20 mM Tris, 1M NaCl, 0.5 mM EDTA, 5% glycerol, pH 8) at 4°C, aliquoted and stored at −80°C until needed.
Selenomethionine-derivative Cch protein purification
Cells were grown in 20ml of M9 media plus additives (0.4% Glucose, 10 mM NaCl, 0.1 mM CaCl2, 2 mM MgSO4, 1 µg ml−1 thiamine) and 100µg ml−1 ampicillin at 37°C, 300 rpm for 1 hour. They were spun down at 8000rpm in an F10S rotor at 4°C and resuspended with new 20ml M9 media plus additives and 100µg ml−1 ampicillin and used as starter culture. Starter culture was added to 1L of M9 media plus additives and 100µg ml−1 ampicillin (initial concentration OD600<0.1) and grown at 37°C, 300rpm up to OD600 0.4–0.5 when an amino acid cocktail containing equal amounts each of L-isoleucine, L-leucine, L-lysine, L-phenylalanine, L-threonine, and L-valine was added to a final concentration of 100 mg/liter. Selenomethionine was added to a final concentration of 60 mg/liter and the culture was grown for another 15 min, at which point the cells were induced with 0.5 mM IPTG, grown for 4 hr and harvested via centrifugation. The protein was purified, concentrated, and stored exactly as the native protein except all buffers contained 10 mM DTT.
CchWH cloning expression and purification
The untagged Carboxy terminal domain of Cch (CchWH)(from S436 to F595) was cloned into the NdeI/XhoI sites in vector pet21a (Novagen) using sticky end PCR products with primers: Cch DBD-F1 (5’ tatgtctgaacgctcattagc), Cch DBD-F2 (5’tgtctgaacgctcattagc), Cch DBD-R1 (5’ gttagaaatcttcgtgaactcc), Cch DBD-R2 (5’ tcgagttagaaatcttcgtgaactcc). CchWH protein was expressed in Rosetta (DE3)pLysS cells (Novagen). The cells were grown in Luria–Bertani medium with 100 µg ml−1 ampicillin at 37°C until they reached OD600 ≈ 0.6 and then were induced with 0.5 mM IPTG. After induction, cells were grown for 4 h at 37°C and harvested by centrifugation at 8000 rpm in an F10S rotor for 10 min. Cell pellets were kept at −80°C until needed. Cell pellets were resuspended in buffer containing 100 mM Tris pH8, 10% sucrose, 10% glycerol, 1M NaCl, 1 mM EDTA, and sonicated. Proteins were precipitated consecutively with 30%, 50% and 80% ammonium sulfate (w/v). Precipitated proteins from the last fraction were resuspended in buffer A2 (20 mM MES, 0.5 mM EDTA, 5% Glycerol, pH 5.5) loaded onto columns Heparin FF (GE Healthcare) and eluted with a gradient of buffer B2 (20 mM MES, 0.5 mM EDTA, 5% Glycerol, 2M NaCl, ph=5.5). Pooled fractions were then dialyzed into buffer A2, loaded onto MonoS™ 5/50 GL (GE Healthcare), and eluted with a gradient of B2. Purified untagged Cch-WH was then concentrated up to ≈7 mg ml−1, dialyzed overnight into stock buffer (20 mM Tris, 100 mM NaCl, 0.5 mM EDTA, 5% glycerol, pH 8) at 4°C, aliquoted and stored at −80°C until needed.
LP1413 cloning expression and purification
LP1413 (GeneID: 3912801) from USA300 strain was cloned (without tags) into NdeI/XhoI sites in pet21a vector (Novagen) using sticky end PCR products obtained with oligos LP1413-F1 (5’ tatgaataagaaatctaagcagcaag), LP1413-F2 (5’tgaataagaaatctaagcagcaag), LP1413-R1 (5’gttagcaatcacctcctttcgaagg), LP1413-R2 (5’tcgagttagcaatcacctcctttcgaagg). The cells were grown in Luria–Bertani medium with 100 µg ml−1 ampicillin at 37°C until they reached OD600 ≈ 0.7 and then were induced with 0.5 mM IPTG. After induction, cells were grown for 3 h at 37°C and harvested by centrifugation at 8000 rpm in an F10S rotor for 10 min. Cell pellets were resuspended in buffer containing 100 mM Tris pH8, 60% sucrose, 10% glycerol, 1M NaCl, 1 mM EDTA, 1 mM DTT, lysed with a microfluidizer (model NO. LV1-30K, Microfluidics Newton, Massachusetts USA) and spun down with an SS-34 rotor for 1 hour at 18000rpm, 4°. Supernatant was mixed with 50% ammonium sulfate (w/v) and proteins were spun down using an SS-34 rotor at 18000rpm for 1 hour at 4°. Pellet was dissolved in 50mL of buffer A1 containing 25 mM Hepes pH7, 0.5 mM EDTA, 5% glycerol, 1 mM DTT, and loaded onto HiPrep™ 16/10 SP FF column (GE Healthcare). Protein was eluted with a gradient of buffer B1 containing 25 mM Hepes pH7, 2M NaCl, 0.5 mM EDTA, 5% glycerol and 1 mM DTT. Positive fractions were mixed, diluted ½ with buffer A1 and loaded again to the same column. After elution with buffer B1, positive fractions were mixed and dialyzed over night with 10%B1. Dialyzed proteins were loaded onto MonoS™ 5/50 GL (GE Healthcare), eluted with buffer B1. Positive fractions were mixed, concentrated and dialyzed at 4° overnight into storage buffer containing 20 mM Tris pH8, 0.5 mM EDTA, 200 mM NaCl, 20% Glycerol and 1 mM DTT.
Cch Crystallization and Structure Determination
Native and Selenomethionine-derivatized Cch-6xHis protein were crystallized by the hanging drop vapor diffusion method. AMP-PNP and MgCl2 were added to the protein vial to a final concentration of 2 mM each. Then the complex was mixed in a 1:2 ratio with well solution containing 32% PEG 200, 0.1M HEPES, pH 7.4. Hexagonal plates grew to full size in ≈2 days but were thin, fragile, and tended to be macroscopically twinned. Microseeding was used to obtain thicker and more single-looking hexagonal plates. Multiple data sets were collected at the SBC beamline 19-ID at the Advanced Photon Source (Argonne, IL). Processing with HKL-200049 revealed that many crystals were microscopically twinned: a few data sets could only be scaled in space group R3, but many with similar unit cell dimensions scaled well in higher-symmetry space groups (some P6 and some R32). The best data set was from a microseeded SeMet crystal and was not twinned according to xtriage. Phases and an experimental map were generated using Phenix Autosol50 (Supplementary Fig. 3). The asymmetric unit contains two copies of Cch that are nearly identical, and the central axis of the hexamer is coincident with a crystallographic 3fold axis. We modeled our protein with Coot51 and refined with Phenix Refine50. After refinement, Ramachandran favored (%), outliers (%) and clashcore values were 93, 0 and 3.65 respectively. The C-terminal Thr557-Phe595 residues and the following loops were not visible in the electron density: Met1-Tyr3; Pro24-Glu31; Thr269-Arg271; Arg288-Lys296; Lys526-Leu539.
Blue Native Polyacrylamide Gel Electrophoresis
BN-PAGE was performed as in reference52, with some modifications. 1µl of Cch (64.5µM) was added to 20µl of buffer containing (20 mM Tris, 5 mM AMP-PNP, 5 mM MgSO4, pH 7.8) and left at room temperature for 30 minutes. 5µl of 80% glycerol was added to the sample, gently mixed, loaded in 4–12% Tris-Glycine gel and run for 3–4 hours at 100V in running buffer (25 mM Tris-glycine pH 7.5). Cathode buffer contains 0.01% Coomasie Brilliant blue G-250 (Bio-Rad). Gels were cleared and stained again in order to remove background signal. When needed, ssDNA (25-mer Ts) was added to the sample at final concentration of 10µM. For all assays described, DNA molarities are given with respect to entire chains or duplexes, rather than nucleotides or base pairs.
Electrophoretic Mobility Shift Assays (EMSAs)
Cch: 2 nM of substrate DNA (3’end or control) was incubated with Cch in reaction buffer (20 mM Tris pH 8, 100 mM NaCl, 5% glycerol, 50ng µl−1 BSA, 5 mM DTT, 2.5 mM AMP·PNP, 2.5 mM MgSO4, 1ng µl−1 of salmon sperm DNA as competitor DNA). The final volume of the reaction was 20µl. Cch was diluted in protein dilution buffer (20 mM Tris pH8, 0.5 mM EDTA, 100 mM NaCl, 5% glycerol, 25ng µl−1 BSA, 5 mM DTT) up to 10× the desired final concentration (see Fig. 5), and then 2 µl were added to a final volume of 20 µl containing the reaction buffer and DNA. Oligonucleotides scc-emsa-3F (5’ttttaaatccgctaatgaagggataggcccttcaggagttcacgaagatttctaattgtc) and scc-emsa-4R (5’gacaattagaaatcttcgtgaactcctgaagggcctatcccttcattagcggatttaaaa) correspond to 3’end of the Cch coding region. Oligonucleotides scc-emsa-3F and HELA8-R at 1µM concentration were 5’ end 32P-phosphorylated with T4 kinase, then 10× excess of F and R strands were added and annealed. See Fig. 5 for final concentrations of Cch and CchWH. For ssDNA binding assays with Cch, 2 nM of radiolabeled oligonucleotide HELA-8R was used as substrate. Reactions were performed in 20µl of buffer containing 20 mM Tris pH 8, 100 mM NaCl, 5% glycerol, 50ng µl−1 BSA, 5 mM DTT. Cch was diluted in the same protein dilution buffer than above up to desired concentrations (see Fig. 5). In all cases, after 30 min at room temperature, 5µl of 80%glycerol was added to the reaction and the whole volume was loaded on 5% non-denaturing polyacrylamide gel. Gels were run in 0.5× TBE and bands were visualized by phosphor-imaging (BioRad Personal Molecular Imager).
LP1413: 1uM oligonucleotide iter-int-1F (5’gagtaatgatatgtggctatgtcttaatatatgtgaagaa) was 5’ end 32P-phosphorylated with T4 kinase, then 10× excess of iter-int-1F and inter-int-2R (5’ ttcttcacatatattaagacatagccacatatcattactc) were mixed and annealed to make dsDNA substrate. To make ssDNA substrate, the process was repeated without inter-int-2R oligonucleotide. LP1413 protein was diluted in dilution buffer (20 mM Tris pH 8, 200 mM NaCl, 5% glycerol, 50ng µl−1 BSA, 5 mM DTT, up to the desired concentration. Reactions were performed in reaction buffer containing 20 mM Tris pH 8, 200 mM NaCl, 5% glycerol, 50ng µl−1 BSA, 5 mM DTT and 2 nM DNA substrate (ssDNA or dsDNA) at room temperature. After 30 minutes, 5uL of 80% glycerol was added and the entire volume was loaded on 5% non-denaturing polyacrylamide gel. Gels were run in 0.5× TBE and bands were visualized by phosphor-imaging (BioRad Personal Molecular Imager).
Helicase Assay
Helicase assays were performed as in paper reference 30 with some modifications. Oligonucleotide sequences used in this experiment: HELA-5F (5’ ggatatgtatggcatacaactggaagtggtaagacgttgacttcttttaaa gcgagtcag), HELA-6R (5’ gactgacgctttaaaagaagtcaacgtcttaccacttccagttgta tgccatacatatcc), HELA-7R (5’ ctgactcgctttaaaagaagtcaacgtcttaccacttccag ttgtatgccatacatatcc), HELA-8R (5’ cgctttaaaagaagtcaacgtcttaccacttccagt tgtatgccatacatatcc), HELA-9F (5’ ggatatgtatggcatacaactggaagtggtaagacg ttgacttcttttaaagcg). Oligonucleotides HELA-5F and HELA-9F (top strands) at 1µM were 5’ end 32P-phosporylated using T4 kinase. Then 10× of unlabeled top and bottom strands were added and annealed as follows: HELA-5F/HELA-6R (6bp mismatched); HELA-5F/HELA-8R (6bp 3’ tail); HELA-9F/HELA-7R (6bp 5’ tail); HELA-9F/HELA-8 (blunt ended duplex). Protein was diluted in dilution buffer (20 mM Tris pH8, 0.5 mM EDTA, 100 mM NaCl, 5% glycerol, 25ng µl−1 BSA, 5 mM DTT) up to desired concentration (see Fig. 5). Reaction was performed in 20µl (containing 20 mM Tris pH 8, 100 mM NaCl, 5% glycerol, 50ng µl−1 BSA, 5 mM DTT, 5 mM ATP, 5 mM MgSO4, 2 nM radiolabelled duplex DNA) at 37°C. Once reaction was initiated, 3 minutes after, an excess of cold HELA-5F or HELA-9F was added. The reaction was stopped 27 minutes later by adding stop buffer (0.25% SDS, 15 mM EDTA, 5% glycerol) and products were separated on 12% non-denaturing PAGE. Bands were visualized by a phosphor-imaging (BioRad Personal Molecular Imager). Helicase Assay in presence of LP1413: Cch and LP1413 were incubated together for 30’ on ice or room temperature. Reaction was performed at 37° in 20ul (containing 20 mM Tris pH7.8, 10 mM KCl, 4% sucrose, 50ng µl−1 BSA, 5 mM DTT, 5 mM ATP, 5 mM MgSO4, 2 nM radiolabelled duplex DNA HELA-5F and HELA-8R). Once the reaction was initiated, 3 minutes after, an excess of cold HELA-5F was added. The reaction was stopped 27 minutes later by adding 3ul of 10% SDS and products were separated on 12% non-denaturing PAGE. Bands were visualized by a phosphor-imaging (BioRad Personal Molecular Imager).
SEC-MALS
Data were calculated following reference53.
Figures displaying structural data were made with Pymol (http://www.pymol.org/).
Original images of gels, autoradiographs and blots used in this study can be found in Supplementary Data Set 1.
Supplementary Material
Acknowledgments
We thank Robert Daum from the Department of Pediatrics and MRSA Center at The University of Chicago for providing USA300 MRSA strain 923 from which we cloned Cch and LP1413, JR. Penadés and MR. Boocock for insightful discussions, J. Herrou for assistance with crystallizations, Y-L. Chan for biochemical advice, the CCP4 summer school for crystallographic advice and the staff of the Structural Biology Center at Argonne National Laboratory for assistance with data collection. This work was funded by R21AI117593 to PAR.
Footnotes
Accession code
Coordinates and structure factors have been deposited at the RCSB under accession code 5DGK, and raw data have been deposited with SBgrid, doi:10.15785/SBGRID/166.
Author Contributions
PAR conceived and directed the project and carried out most of the bioinformatics. IMS carried out all of the crystallography and biochemistry shown in the figures as well as some of the bioinformatics. SBV advised on Staphylococcal molecular biology, suggested examining the cch gene, and carried out the initial cloning of Cch. AM contributed to the cloning and initial stages of the Cch biochemistry, CAR worked out the purification and initial characterization of LP1413, and YZP helped with protein purification and crystallization. IMS and PAR wrote the paper.
Competing Financial
The authors declare no competing financial interests.
References
- 1.Moellering RC. MRSA: the first half century. J. Antimicrob. Chemother. 2012;67:4–11. doi: 10.1093/jac/dkr437. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misiura A, et al. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol. Microbiol. 2013;88:1218–1229. doi: 10.1111/mmi.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katayama Y, Ito T, Hiramatsu K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito T, et al. Novel Type V Staphylococcal Cassette Chromosome mec Driven by a Novel Cassette Chromosome Recombinase, ccrC. Antimicrob. Agents Chemother. 2004;48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, Tsubakishita S, Kuwahara-Arai K, Han X, Hiramatsu K. Staphylococcal Cassette Chromosome(SCC) : A Unique Gene Transfer System in Staphylococci. Chapter 18 in Bacterial Integrative Mobile Genetic Elements. In: Roberts AP, Mullany P, editors. Landes Bioscience. CRC Press; 2013. 2013. [Google Scholar]
- 7.Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 9.Úbeda C, et al. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol. Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 10.Mir-Sanchis I, et al. Control of Staphylococcus aureus pathogenicity island excision. Mol. Microbiol. 2012;85:833–845. doi: 10.1111/j.1365-2958.2012.08145.x. [DOI] [PubMed] [Google Scholar]
- 11.Úbeda C, Barry P, Penadés JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc. Natl. Acad. Sci. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubeda C, Tormo-Más MÁ, Penadés JR, Novick RP. Structure–function analysis of the SaPIbov1 replication origin in Staphylococcus aureus. Plasmid. 2012;67:183–190. doi: 10.1016/j.plasmid.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegelin G, Linderoth NA, Calendar R, Lanka E. Domain structure of phage P4 alpha protein deduced by mutational analysis. J. Bacteriol. 1995;177:4333–4341. doi: 10.1128/jb.177.15.4333-4341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman AB, Dyda F. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 2005;15:77–85. doi: 10.1016/j.sbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H-C, et al. Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor. Nucleic Acids Res. 2014;42:1354–1364. doi: 10.1093/nar/gkt964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano-Heras G, Bravo A, Salas M. Phage phi29 protein p56 prevents viral DNA replication impairment caused by uracil excision activity of uracil-DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19044–19049. doi: 10.1073/pnas.0808797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lina G, et al. Staphylococcal chromosome cassette evolution in Staphylococcus aureus inferred from ccr gene complex sequence typing analysis. Clin. Microbiol. Infect. 2006;12:1175–1184. doi: 10.1111/j.1469-0691.2006.01548.x. [DOI] [PubMed] [Google Scholar]
- 19.Iyer LM, Abhiman S, Aravind L. A new family of polymerases related to superfamily A DNA polymerases and T7-like DNA-dependent RNA polymerases. Biol. Direct. 2008;3:39. doi: 10.1186/1745-6150-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 21.Bjorkeng EK, et al. ccrABEnt serine recombinase genes are widely distributed in the Enterococcus faecium and Enterococcus casseliflavus species groups and are expressed in E. faecium. Microbiology. 2010;156:3624–3634. doi: 10.1099/mic.0.041491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 23.David MZ, Daum RS. Community-Associated Methicillin-Resistant Staphylococcus aureus: Epidemiology and Clinical Consequences of an Emerging Epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 25.Trakselis MA. Structural Mechanisms of Hexameric Helicase Loading, Assembly, and Unwinding. F1000Research. 2016;5 doi: 10.12688/f1000research.7509.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 28.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 30.Soni RK. Functional characterization of Helicobacter pylori DnaB helicase. Nucleic Acids Res. 2003;31:6828–6840. doi: 10.1093/nar/gkg895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Araki H. Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells. 2013;18:266–277. doi: 10.1111/gtc.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 33.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 34.Ziegelin G, Lanka E. Bacteriophage P4 DNA replication. FEMS Microbiol. Rev. 1995;17:99–107. doi: 10.1111/j.1574-6976.1995.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 35.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 37.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural Basis of DNA Replication Origin Recognition by an ORC Protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 38.Mkrtchyan HV, Xu Z, Cutler RR. Diversity of SCCmec elements in Staphylococci isolated from public washrooms. BMC Microbiol. 2015;15 doi: 10.1186/s12866-015-0451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth DS, Wong A, Robinson DA. Cross-species spread of SCCmec IV subtypes in staphylococci. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011;11:446–453. doi: 10.1016/j.meegid.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crossley KB, Jefferson KK, Archer GL, Fowler VG. Staphylococci in Human Disease. John Wiley & Sons; 2009. [Google Scholar]
- 41.Samuels M, et al. A biochemically active MCM-like helicase in Bacillus cereus. Nucleic Acids Res. 2009;37:4441–4452. doi: 10.1093/nar/gkp376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JM, Arachea BT, Epling LB, Enemark EJ. Analysis of the crystal structure of an active MCM hexamer. eLife. 2014;3:e03433. doi: 10.7554/eLife.03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 44.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Putnam CD, et al. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
Methods References
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 48.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinforma. Oxf. Engl. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode", Methods in Enzymology. Vol. 276. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wittig I, Braun H-P, Schägger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 53.Nedelkov Dobrin, Nelson Randall W, Folta-Stogniew Ewa. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. New and Emerging Proteomic Techniques. 2006:97–112. doi: 10.1385/1-59745-026-X:97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.