SUMMARY
The immune checkpoint receptor PD-1 and its ligand, PD-L1, have emerged as key regulators of anti-tumor immunity in humans. Recently, we reported an ultra high-affinity PD-1 mutant, termed HAC PD-1, which shows superior therapeutic efficacy in mice compared to antibodies. However, the molecular details underlying the action of this agent remain incompletely understood, and a molecular view of PD-1/PD-L1 interactions in general is only beginning to emerge. Here, we report the structure of HAC PD-1 in complex with PD-L1, showing it binds PD-L1 using a unique set of polar interactions. Biophysical studies and long-timescale molecular dynamics experiments reveal the mechanisms by which ten point mutations confer a 35,000-fold enhancement in binding affinity, and offer atomic-scale views of the role of conformational dynamics in PD-1/PD-L1 interactions. Finally, we show that the HAC PD-1 exhibits pH-dependent affinity, with pseudo-irreversible binding in a low pH setting akin to the tumor microenvironment.
eTOC
Pascolutti et al. describe the biophysical basis for enhanced binding of a mutant PD-1 to its ligand PD-L1, regulators of anti-tumor immunity in humans. The authors also characterize wild-type and mutant PD-1 using long-timescale molecular dynamics. The mutant PD-1/PD-L1 interaction is enhanced in acidic pH, suggesting utility as a microenvironment-targeted therapeutic.
INTRODUCTION
Programmed cell death protein 1 (PD-1), or CD279, is an important regulator of immune tolerance and T-cell exhaustion, and it has recently emerged as a key target in the treatment of cancer. PD-1 is a type I transmembrane protein containing an Ig Variable-type (V-type) amino-terminal extracellular domain, a transmembrane region, and a cytoplasmic tail with an immunoreceptor tyrosine-based inhibitory motif (ITIM) and an immunoreceptor tyrosine-based switch motif (ITSM) (Ishida et al., 1992). PD-1 belongs to the CD28/CTLA-4 (cytotoxic T lymphocyte antigen) family of co-receptors, which are generally poorly conserved in sequence, sharing only 21 – 33% amino-acid sequence identity within the family. PD-1 is expressed on T and B cells upon receptor engagement (Agata et al., 1996). In these cells, the PD-1 pathway maintains tolerance to self-antigens to limit inappropriate autoimmune responses. However, cancer and chronic infections exploit this pathway to evade host immunity (reviewed in (Wherry, 2011) and (Keir et al., 2008)).
Two ligands have been described for PD-1, called PD-L1 (CD274) and PD-L2 (CD273). The binding of PD-L2 to PD-1 exhibits 2 – 6 fold higher affinity and shows different association/dissociation kinetics compared to the interaction PD-1/PD-L1 (Youngnak et al., 2003). While PD-L1 is widely expressed on mouse B and T cells, macrophages, dendritic cells (DCs), bone marrow-derived mast cells and mesenchymal stem cells (Yamazaki et al., 2002), PD-L2 expression is restricted to activated macrophages and DCs (Latchman et al., 2001). Of the two PD-1 ligands, PD-L1 is thought to be the primary mediator of cancer immune evasion, although PD-L2 may play a role in certain cases (Nguyen and Ohashi, 2015).
PD-1 and its ligands have limited conservation between human and mouse orthologs. Comparison between human and murine amino-acid sequences shows 60% and 77% identity for PD-1 and PD-L1 respectively (Vibhakar et al., 1997) (Freeman et al., 2000). Moreover, murine PD-1 includes an additional beta strand (residues 53 – 57) not present in the human receptor, and structures of human and mouse PD-1 show substantial divergence in loops flanking the ligand-binding surface, including backbone deviations of as much as 8 Å. It has been shown that human and murine PD-1 can bind PD-L1 of either species with similar affinities in vitro (Lin et al., 2008). Until recently, only the structures of the complex of mPD-1/hPD-L1 (Lin et al., 2008), and mPD-1/mPD-L2 (Lazar-Molnar et al., 2008), had been solved. Very recently, the structure of hPD-1 in complex with the V-set Ig domain fragment of hPD-L1 was reported (Zak et al., 2015).
Recently, we developed an ultra high affinity mutant of PD-1, termed High Affinity Consensus (HAC) PD-1 (Maute et al., 2015). This molecule exhibits ~35,000-fold enhanced affinity for PD-L1 relative to the wild-type protein, and shows superior efficacy compared to antibodies in mouse cancer treatment models. To better understand the mechanistic basis for the actions of this molecule we undertook X-ray crystallographic studies of the HAC PD-1/hPD-L1 complex. Using the structure as a guide, we designed a series of biophysical experiments and molecular dynamics simulations to probe the basis for high affinity as well as the molecular details of PD-1 interaction with its ligand. These results offer insights into general principles of affinity maturation and provide insight into the distribution of conformations underlying the PD-1/PD-L1 interaction.
RESULTS
Overall structure of the HAC PD-1/PD-L1 complex
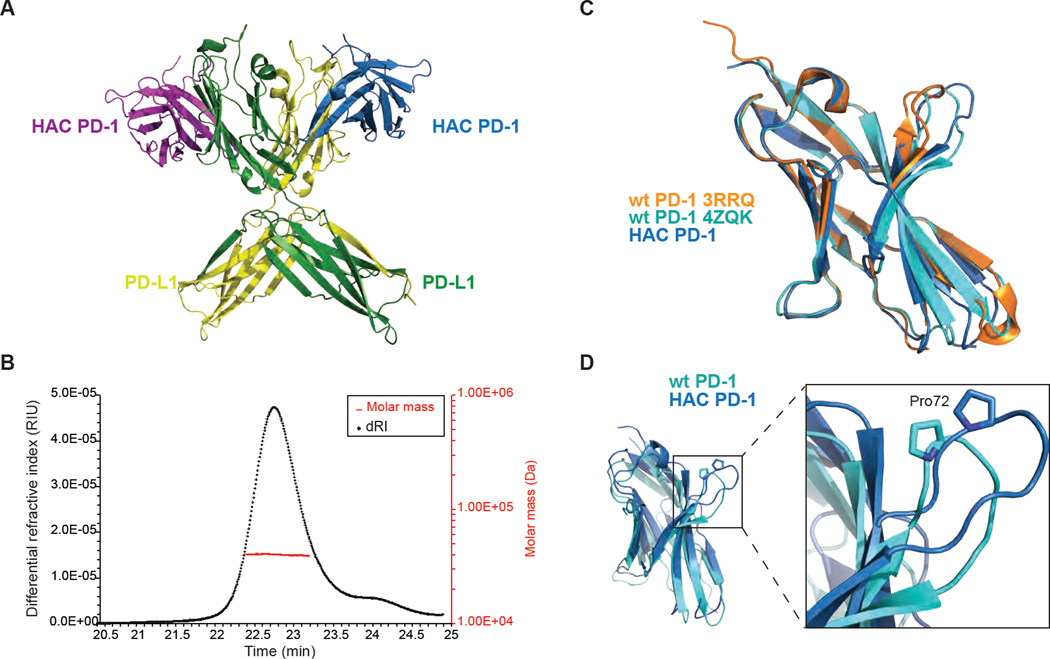
To explore the basis for hPD-1/hPD-L1 interaction we employed the PD-1 HAC variant described previously (Maute et al., 2015). This mutant was obtained by directed evolution and contains a total of 10 point substitutions compared to wild-type, including buried and surface residues. Most surface residue mutations either are expected to increase hydrophilicity or have little effect on hydrophobicity overall. Together, these mutations confer a 35,000-fold enhancement in binding affinity, allowing formation and purification of a highly stable complex for structural study (Figure S1). Crystallization was straightforward, and a 2.9 Å resolution structure was obtained for the complex (Table S1). The asymmetric unit contains two copies of each molecule, with swapping of the carboxy-terminal β-strand between the C2-type Ig domains of the PD-L1 molecules (Figure 1A). This feature is reminiscent of a similar domain swap seen previously in the structure of PD-L1 homolog B7-H3 (Vigdorovich et al., 2013). This swapping could imply a 2:2 binding stoichiometry, in contrast to previous reports of 1:1 binding for the wild-type proteins (Lin et al., 2008) (Cheng et al., 2013). To assess the oligomerization state of the HAC PD-1/PD-L1 complex in solution, we used size exclusion with multi-angle light scattering (SEC-MALS) to measure the size of the complex. The results of this experiment indicated a molecular weight of approximately 40 kDa, confirming that the complex is 1:1 in stoichiometry in solution (Figure 1B) and that the domain swapping is an artifact of crystallization.
Figure 1.
HAC PD-1 and PD-L1 structure. (A) The overall structure of HAC PD-1 in complex with PD-L1 shows two copies of each molecule in each asymmetric unit, with domain swapping in PD-L1. HAC PD-1 is depicted in purple and blue, while PD-L1 is represented in green and yellow. The complex is shown in ribbon presentation. (B) SEC-MALS analysis shows that the two molecules interact in a 1:1 complex. (C) Superimposition of wild-type PD-1 apo (3RRQ), wild-type PD-1 (4ZQK) and HAC PD-1 (blue) shows slight differences in conformations. (D) Wild-type PD-1 (PDB ID: 4ZQK) and HAC PD-1 are depicted in cyan and blue respectively. Superimposition of HAC PD-1 with wild-type PD-1 shows a major conformational change in the loop containing Pro72.
The interaction between HAC PD-1 and PD-L1 is similar overall to that of the recently reported complex between wild-type human PD-1 and a fragment of human PD-L1 (PDB ID: 4ZQK). The PD-L1 V-set Ig domain is virtually identical in the two structures, with an all atom RMSD of 0.6 Å. In contrast, HAC PD-1 adopts a unique conformation slightly different from that seen in wild-type hPD-1 alone (PDB: 3RRQ) or in complex with hPD-L1 (Zak et al., 2015) (Figure 1C). Most notably, the loop containing Pro72 and the two adjacent β-strands are rotated substantially between the two structures, with HAC PD-1 presenting a distinct interaction surface to PD-L1 (Figure 1D). In addition, the loops centered on Ser60 and Pro130 are also significantly rearranged in HAC PD-1 compared to wild-type PD-1 (Figure S2). With these exceptions, the rest of the molecule is virtually identical between the two structures.
Binding of HAC PD-1 to PD-L1 is enthalpically driven
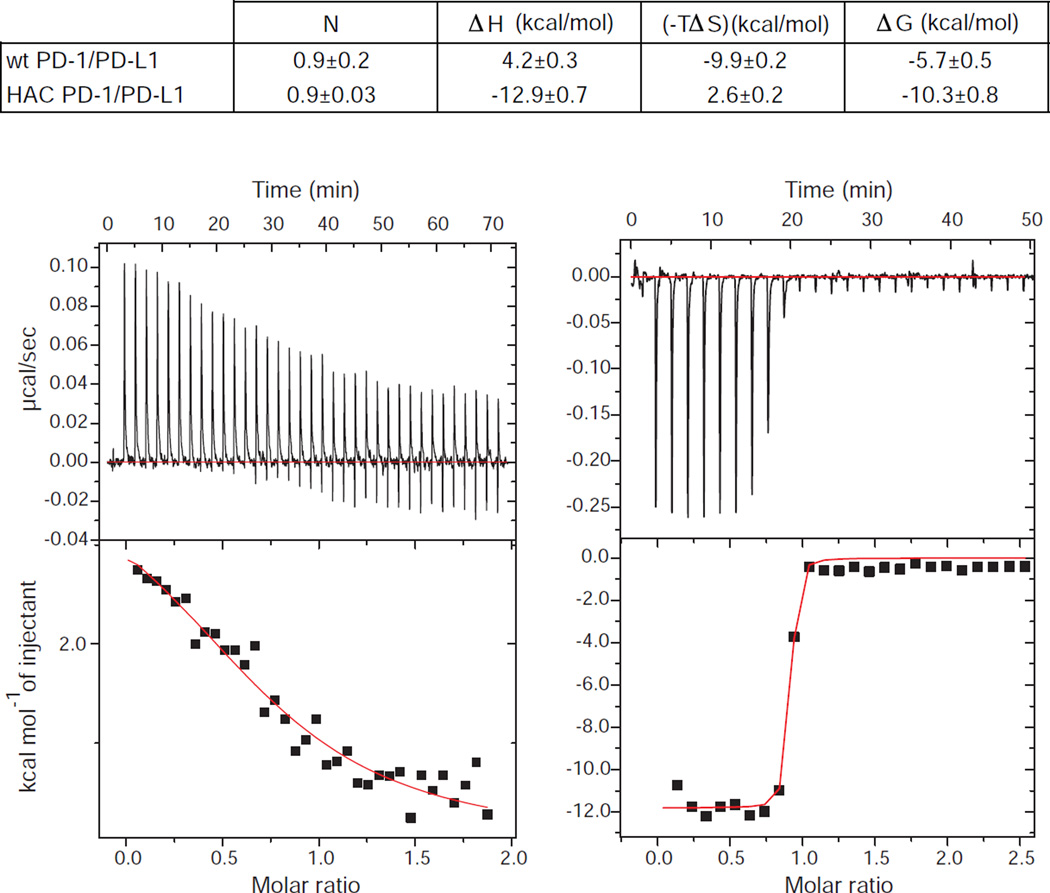
To better understand the molecular basis for the exceptionally high affinity of the HAC PD-1 variant, we performed isothermal titration calorimetry (ITC) experiments to investigate the thermodynamics of the wild-type and mutant PD-1 binding to its ligand PD-L1. For the wild-type proteins, we observed somewhat different results than those reported previously (Cheng et al., 2013), where the driving force of the interaction between wild-type PD-1 and PD-L1 showed a major entropic component. While our measured affinity of PD-1 and PD-L1 closely resembles previously reported values, we consistently observe different relative enthalpic and entropic contributions to binding energy as compared to the previous report (Figure 2). However, our experiments were performed with a wild-type monomeric PD-1 construct, while the previous report used an Fc fusion in which an unpaired cysteine (Cys93) was mutated to serine. Both the use of fusion proteins and the mutation could contribute to the observed differences in binding energies. In contrast to our results for the wild-type proteins, the HAC PD-1 mutant shows strongly enthalpic binding (−12.96 ± 0.72 kcal/mol) counteracting an unfavorable entropic contribution to binding energy (2.625 ± 0.16 kcal/mol) (Figure 2). Inspection of the structure reveals an increase in the number of polar interactions (Figure S3), consistent with enhancements in binding enthalpy driving higher affinity.
Figure 2.
ITC measurements of wild-type PD-1 or HAC PD-1 binding to PD-L1. The values for ΔH and ΔS were obtained by fitting a single binding site model to the ITC data and are shown with standard error values. The top panel shows examples of raw data indicating the titration of wild-type PD-1 (left) or HAC PD-1 (right) into an isothermal calorimetry cell containing PD-L1. The bottom panels represent plots of heat released during the isothermal reaction vs. molar ratio for the interaction of wild-type PD-1 (left) or HAC PD-1 (right) with PD-L1. The values for ΔG and ΔS in HAC/PD-L1 interaction should be interpreted with some caution due to the sharp slope of the curve caused by high binding affinity.
M70E, Y68H and K78T stabilize the interface between HAC PD-1 and PD-L1
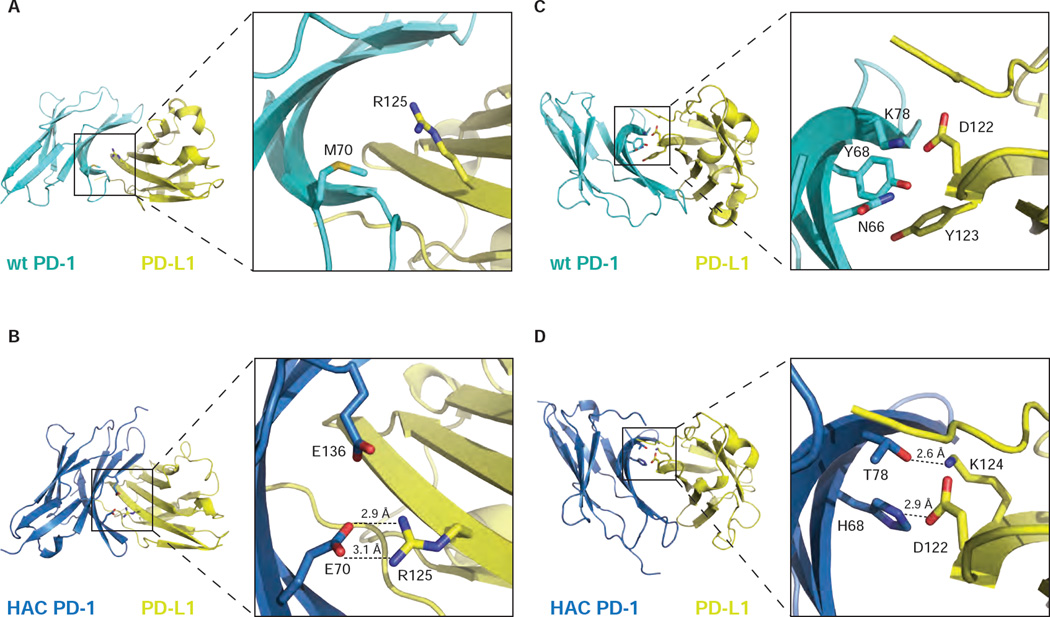
The structures offer possible explanations for several of the mutations. In particular, the mutation Met70Glu in HAC PD-1 allows the formation of two hydrogen bonds and a salt-bridge with Arg125 in PD-L1 (Figure 3B). In addition, Glu136 establishes a charge-charge interaction with Arg125. In the wild-type complex, however, the side chain of Met70 in PD-1 points towards the inner side of the loop, preventing the formation of favorable contacts with PD-L1 (Figure 3A). Other mutations include Tyr68His and Lys78Thr (Figure 3C and D): the side chains of these amino acids form hydrogen bonds with Asp122 and Lys124 respectively in PD-L1, while the wild-type residues form a polar pocket with Asn66 in PD-1, decreasing the possibility to stabilize contacts with the surface of PD-L1.
Figure 3.
Close-up views of HAC PD-1 mutations at the interface with PD-L1. Within the complex structure, wild-type PD-1 and PD-L1 (PDB: 4ZQK) are represented in cyan and yellow respectively (A and C). HAC PD-1 and PD-L1 are depicted in blue and yellow respectively (B and D). The complexes are shown in ribbon presentation. Mutation M70E leads to the formation of new hydrogen bonds (B) compared to the wild-type structure (A), stabilizing the interface of interaction with PD-L1. A similar behavior is shown by the mutations Y68H and K78T in HAC-PD-1 (D) compared to the wild-type protein (C).
We were interested to explore the relative contributions of these individual mutations to binding affinity, and prepared point mutants on a wild-type PD-1 background: Tyr68His, Met70Glu, Lys78Thr, and a triple mutant incorporating all three substitutions. Unfortunately, these mutants were not biochemically tractable with the exception of Met70Glu, which showed affinity equal to that of wild-type PD-1. Taken together, these results indicate that the mutations in HAC are likely to act in a synergistic manner with respect to protein stability and PD-L1 binding affinity. Since the selection procedure used to identify the HAC variant originally sampled mutations in all possible combinations, this result is perhaps not entirely surprising. As discussed below, specific receptor-ligand interactions do appear to contribute to binding stability, but the affinity enhancement appears to derive from the concerted activity of all or most of the single point substitutions.
HAC PD-1 is less flexible than wild-type PD-1
While the structure of HAC PD-1/PD-L1 and the previously reported structure of wild-type hPD-1 in complex with a hPD-L1 fragment offer important insights into the molecular basis for ligand/receptor interactions, the role of conformational disorder in PD-1 function has not yet been explored. To address this, we performed molecular dynamics simulations to understand ligand binding in both the wild-type and HAC PD-1 mutant. Simulations, summarized in Table S2, included HAC PD-1 and wild-type PD-1 in isolation, as well as simulations of the high affinity HAC PD-1/PD-L1 complex and the low affinity wild-type complex.
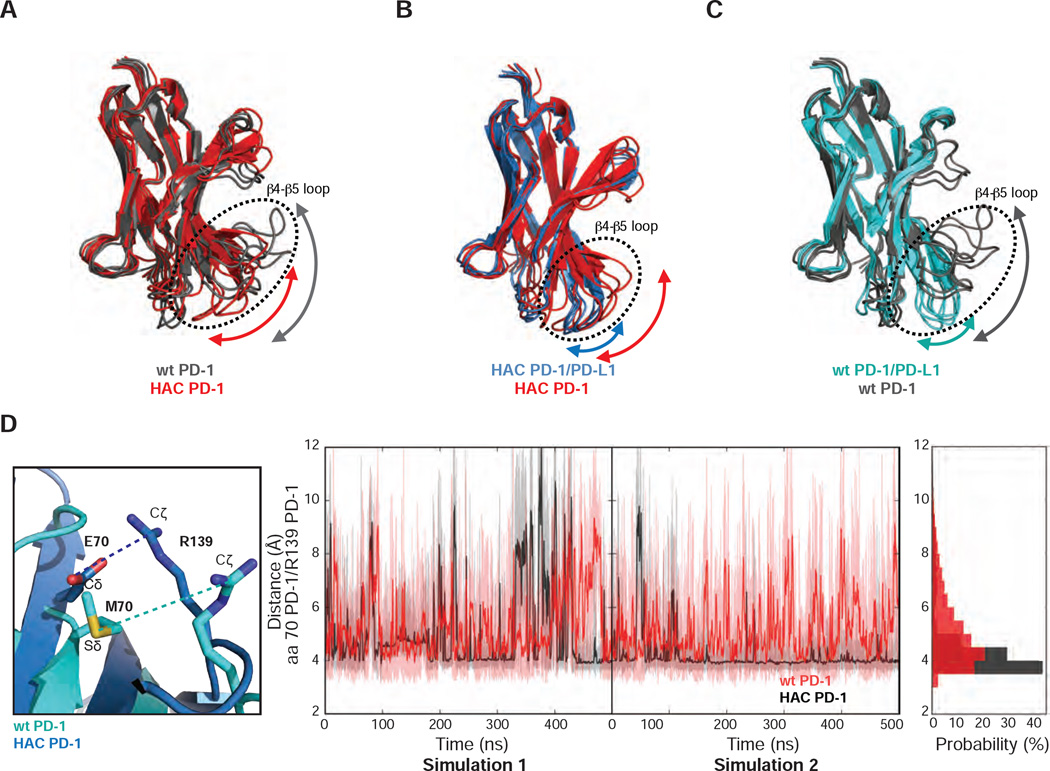
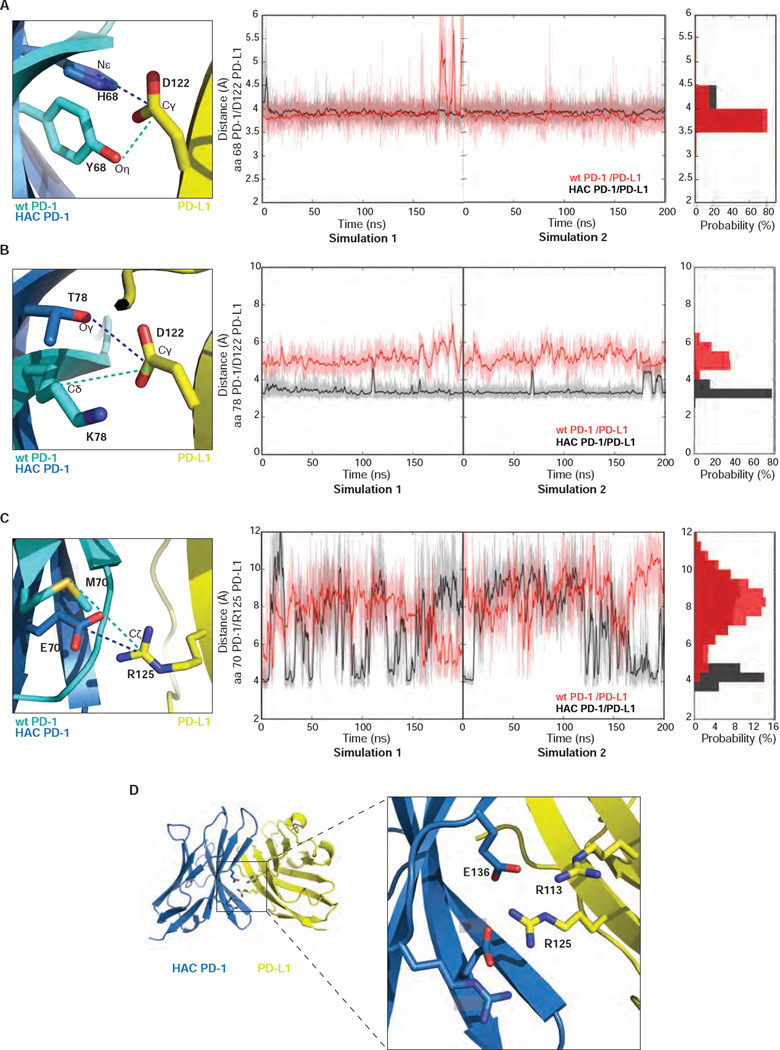
Simulations with isolated proteins revealed a much greater degree of conformational disorder in wild-type PD-1 compared to HAC PD-1, particularly in loop regions. In the β4–β5 loop (residues 70 – 78) the HAC PD-1 mutant shows moderate conformational heterogeneity, while the wild-type receptor exhibits a more disordered loop as indicated by the arrow in Figure 4A. This loop in both HAC PD-1 and wild-type PD-1 becomes conformationally restricted upon binding to PD-L1 (Figure 4B and C). Within this loop, the M70E mutation in HAC PD-1 appears to be a contributor to improved stability, altering the distribution of distances between Glu70-Arg139 in HAC PD-1 and Met70-Arg139 in wild-type PD-1 (Figure 4D). The distance between the Cδ of Glu70 and Cζ of Arg139 in HAC PD-1 peaks around 4 Å, suggesting the presence of a salt-bridge. For wild-type PD-1, the distance between Met70 and Arg139 peaks at about 4.5 Å as shown in the probability distribution of the distances. However, this distance is distributed evenly from 3.5 Å to 8 Å, suggesting a weak interaction.
Figure 4.
M70E seems to be the main factor to improve stability. (A) Five most populated conformations of HAC PD-1 and wild-type PD-1 shown respectively in red and grey. (B) Five most populated conformations of HAC PD-1 when bound to PD-L1 (blue) and HAC PD-1 free (red). (C) Five most populated conformations of wild-type PD-1/PD-L1 and wild-type PD-1 free depicted respectively in cyan and grey. (D) Temporal evaluations of the distances of M70 Sδ - R139 Cζ in wild-type PD-1 and E70 Cδ - R139 Cζ in HAC PD-1.
Molecular dynamics analysis of PD-L1 interaction with PD-1
To assess the role of individual mutations in conformational stabilization of the HAC PD-1/PD-L1 interface, we tracked dynamics of the mutant residues, including Tyr68His, Lys78Thr and Met70Glu. Given that His68 was likely protonated in the crystal structure based on its apparent hydrogen bond to Asp122 in PD-L1, we performed simulations with this residue fixed in the protonated state for this analysis. The distance between the Nε of His68 on HAC PD-1 and the Cγ of Asp122 on PD-L1 is stabilized at 3 – 3.5 Å in the simulations of HAC PD-1/PD-L1 (Figure 5A). Lys78Thr allows HAC PD-1 to engage in a stable hydrogen bond with Asp122, restraining an otherwise slightly flexible region in the molecule (Figure 5B). The corresponding residue to Thr78 in wild-type PD-1/PD-L1 is Lys78. Although electrostatic interactions between Lys78 (wild-type PD-1) and Asp122 (PD-L1) can form in the simulations, this electrostatic interaction is less stable than the hydrogen bond between Thr78 and Asp122 in HAC PD-1/PD-L1. The side chain of Glu70 in HAC PD-1 is engaged in a charge-charge interaction with Arg139 throughout the simulations, restricting mobility of the β4–β5 loop. Glu70 is also at the center of a charge-charge “zipper” running along the length of the HAC PD-1/PD-L1 interface in the complex structure. However, because of the enrichment of the charged residues in the HAC PD-1/PD-L1 interface, the distance between Glu70 and Arg139 fluctuates widely (Figure 5C and D), suggesting this interaction is not as stable as the interactions introduced by the Tyr68His and Lys78Thr mutations.
Figure 5.
Y68H and K78T mutations contribute to stability, but at lower extent. (A) Temporal evolution of distances between the side chains of residue 68 on PD-1 and the Cγ of D122 on PD-L1. For HAC PD-1/PD-L1, Nε on H68 (PD-1) and the Cγ on D122 (PD-L1) have been used to calculate the distances. For the wild-type PD-1/PD-L1 system, side chain Oη on Y68 (PD-1) and Cγ on D122 (PD-L1) were used. (B) Temporal evolution of distances between the side chains of residue 78 on PD-1 and the Cγ of D122 on PD-L1. For the HAC PD-1/PD-L1 system, Oγ on T78 (PD-1) and the Cγ on D122 (PD-L1) were considered to calculate the distances. For the wild-type PD-1/PD-L1 system, Cδ on K78 (PD-1) and Cγ on D122 (PD-L1) were studied. (C) Temporal evolution of distances between the side chains of residue 70 on PD-1 and the Cζ of R125 on PD-L1. For HAC PD-1/PD-L1, Cδ on E70 (PD-1) and Cζ of R125 (PD-L1) were used to calculate the distances. For the wild-type PD-1/PD-L1 system, side chain Sδ on M70 (PD-1) and Cζ of R125 (PD-L1) were used. (A–C) In all the panels, HAC PD-1/PD-L1 is represented in black and wild-type PD-1/PD-L1 in red. (D) Graphical representation of the interactions undertaken by E70 in HAC PD-1/PD-L1. HAC PD-1 is depicted in blue, while PD-L1 is represented in yellow. The proteins are shown in ribbon presentation.
In the course of these simulations, we observed that the Tyr68His mutation is of particular interest, as the salt bridge with PD-L1 Asp122 is contingent on the His68 protonation state. To further assess the effect of His68 protonation in the HAC PD-1 mutant, we performed simulations with both the protonated and unprotonated forms. As shown in Figure S4, the protonation of His68 in HAC PD-1 dramatically improves the stability of the complex, suggesting that the interaction should be stabilized at low pH.
HAC PD-1/PD-L1 interaction is essentially irreversible at low pH
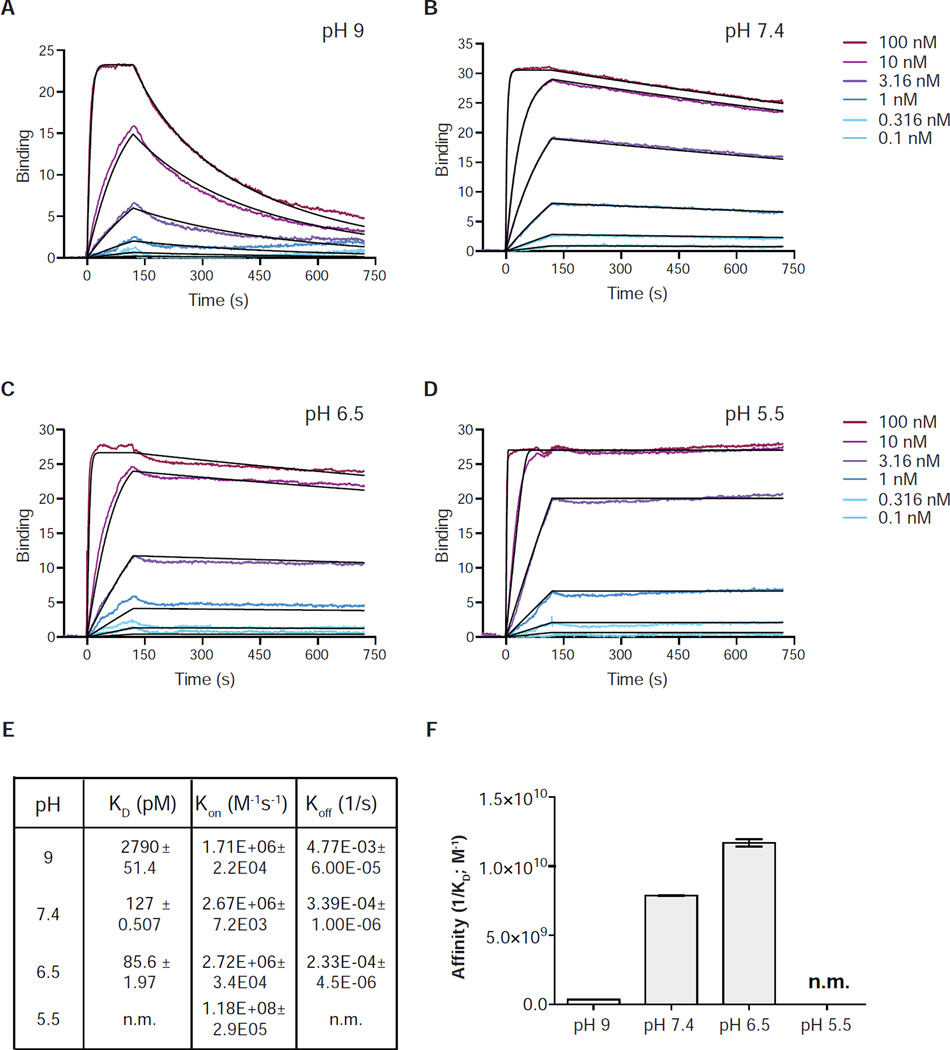
The possibility of pH-dependent stabilization for HAC PD-1 is particularly relevant in view of the fact that the extracellular environment of tumors often exhibits significantly lower pH (5.5 – 7.0) than normal tissues (Gatenby and Gillies, 2004) (Vaupel et al., 1989). In order to empirically assess the prediction that low pH would stabilize the HAC PD-1/PD-L1 interaction, we performed surface plasmon resonance (SPR) under conditions that would promote or inhibit the protonation of HAC PD-1. The experiments were carried out by conducting full association/dissociation experiments at various pH values ranging from 9.0 to 5.5 (Figure 6 A–D). Strikingly, the affinity of HAC PD-1 to PD-L1 increases by 10-fold at pH 6.5 compared to neutral conditions and by 100-fold compared to the basic environment (Figure 6E and F), confirming that the interaction shows strong pH-dependence as predicted by molecular dynamics simulations. At pH 5.5 the interaction became so strong as to be essentially irreversible, with no detectable dissociation observed in the 600 seconds experimental timeframe.
Figure 6.
pH-dependent interaction between HAC PD-1 and PD-L1. (A–D) Representative surface plasmon resonance (SPR) sensorgrams of HAC PD-1 binding to immobilized PD-L1. (E–F) KD values extrapolated from SPR analysis at different pH are represented with their s.e. For the experiment carried out at pH 5.5, the KD was not measurable (n.m.)
DISCUSSION
The T-cell exhaustion-associated receptor PD-1 and its ligand PD-L1 have emerged as critical regulators of T cell biology and important targets in cancer immunotherapy. Here, we report the structure of human PD-L1 in complex with an ultra high-affinity engineered PD-1 mutant, HAC. The structure closely resembles that of the recently reported complex between hPD-1 and a fragment of hPD-L1, but nonetheless differs in key areas including the β4–β5 loop. The HAC PD-1 mutant engages in a much more extensive polar contact network with PD-L1 than does the wild-type protein, likely accounting for the strong enthalpic contribution to binding energy. Interestingly, similar enthalpic enhancements have been seen in other affinity matured protein-protein interactions, indicating that enthalpic stabilization may be a generally useful approach to engineering high affinity protein variants. For instance, structural and molecular dynamics analysis show that the evolved mutations in the H9 IL-2 “superkine” led to conformational stabilization of the protein in receptor-binding competent conformation and showed a substantial enhancement in binding enthalpy offsetting an unfavorable change in binding entropy (Levin et al., 2012). For the most part, the affinity enhancing effects of the individual mutations can be readily explained by inspection of the structures, which show the formation of new hydrogen bonds and salt bridges between PD-1 and its ligand.
In molecular dynamics simulations, wild-type PD-1 was found to be less ordered, with loops adopting a wide range of conformations in addition to those seen previously in static crystal structures. In contrast, the HAC PD-1 mutant shows more modest flexibility, especially in the β4–β5 loop. This result appears to arise from a single non-conservative substitution, Met70Glu, which holds open the β4–β5 loop in a binding-competent conformation via a stable salt bridge with Arg139. In addition, this residue participates in an extended charge-charge interaction network along the PD-1/PD-L1 interface, likely contributing to enthalpic enhancements in binding affinity. In simulations of receptor/ligand complexes, the conformational disorder of wild-type and mutant PD-1 is significantly restricted, allowing the formation of stable interactions throughout the receptor/ligand interface. Nonetheless, differences between wild-type and mutant receptors were still apparent, with polar interactions showing greater conformational stability in the HAC PD-1 mutant.
Moreover, molecular dynamics simulations show how the electrostatic interactions between His68 on HAC PD-1 and Asp122 on PD-L1 can facilitate pH-dependent high-affinity binding, contingent on the protonation state of the His68 imidazole ring. SPR analysis confirmed this pH-dependent interaction, revealing a substantial stabilization of the HAC PD-1/PD-L1 interaction at low pH, to the extent that the interaction shows no measurable dissociation rate at pH 5.5. The tumor microenvironment exhibits extracellular pH values in the range of 5.5 to 7 (Justus et al., 2013), suggesting that pH-dependent binding of HAC PD-1 may serve to preferentially promote tumor binding in vivo. Although in the case of HAC PD-1, the observed pH dependent-binding is fortuitous, future efforts could use similar principles to rationally design immunotherapeutic agents to selectively target the tumor microenvironment. Such approaches could be particularly important for the safe targeting of biological pathways with a high degree of expected on-target toxicity in non-tumor tissues.
In summary, these studies provide the first molecular description of PD-1/PD-L1 interaction dynamics and offer a detailed structural view of an ultra high-affinity PD-1 mutant in complex with its receptor. As with other affinity-matured proteins, high affinity binding is driven by enthalpic gains that more than compensate for poorer binding entropy compared to the wild-type protein, suggesting this is likely to be a generalizable principle in protein affinity engineering. Moreover, we show that wild-type PD-1 is less ordered relative to the HAC mutant, with the latter adopting a conformation resembling the bound state even in the absence of PD-L1.
EXPERIMENTAL PROCEDURES
Protein production
Wild-type and mutant human PD-1 and wild-type human PD-L1 proteins were produced in Trichoplusiani (Tni) insect cells (Expression Systems; Davis, CA). In brief, the gene encoding the ectodomain for each of the proteins was cloned into expression vector pAcGP67a. Baculovirus was produced by co-transfection of transfer vector and linearized “BestBac” viral DNA in Spodoptera frugiperda Sf9 cells (Expression Systems, Davis, CA). Viruses were amplified twice in Sf9 cells and the resulting P2 virus stock was used to infect Tni cells for protein production. Cells were infected at a density of 3 × 106 million/mL and cultured for 72 hours following infection prior to protein harvest.
Protein was harvested by centrifugation to remove cells, and the supernatant was treated with 50 mM Tris pH 8, 1 mM nickel chloride, and 5 mM calcium chloride. The medium was centrifuged again and then filtered with a 0.45 micron filter to remove precipitated material. Clarified supernatant was loaded by gravity flow over nickel-bound chelating sepharose resin (GE Healthcare). The column was washed with HEPES-buffered saline (20 mM HEPES pH 7.5, 150 mM sodium chloride, 20 mM imidazole), and protein was then eluted in the same buffer supplemented with 250 mM imidazole. Eluted protein was incubated with 1:100 (w:w) carboxypeptidase A (Sigma Aldrich) and carboxypeptidase B (Roche) in order to remove the c-terminal 6 × His tag. Protein was dialyzed against 20 mM HEPES pH 7.5, 150 mM sodium chloride prior to calorimetry assays.
Calorimetry
Isothermal Titration Calorimetry (ITC) experiments were performed using the MicroCal iTC-200 system (GE Healthcare) and data analysis was performed with Origin™ software using fitting models to calculate the stoichiometry (N), the binding constant (Ka), enthalpy (ΔH), and entropy (ΔS) of the interaction. The experiments for measuring HAC PD-1/PD-L1 binding were performed using 0.2 mM HAC PD-1 added in 30 1ul-injections to a 0.01 mM solution of PD-L1 at 25°C (confirmed also in the opposite direction). The measurements of wild-type PD-1/PD-L1 binding were performed using 0.5 mM PD-L1 added in 30 1ul-injections to a 0.05 mM solution of wild-type PD-1 at 4°C and confirmed using 0.5 mM wild-type PD-1 added in 20 1.5 ul-injections to a 0.05 mM solution of PD-L1 at 4°C. The resulting data were fitted after subtracting the heats of dilution originated from the addition of PD-L1 or wild-type PD-1 or HAC PD-1 to the buffer, obtained in separate control experiments. The data were fitting using a non-linear least square curve-fitting algorithm with three variables: N, Ka, and ΔH. The data were analyzed by fitting the binding to a single independent binding site model through the Origin™ software. A complete thermodynamic characterization of an interaction can be achieved on the basis of the correlation between temperature (T), standard free energy (ΔG), observed enthalpy and entropy (ΔS) changes from unbound to bound states. The experiments were performed using insect purified proteins and were performed in triplicates.
Crystallography
Protein for crystallographic studies was produced in Escherichia coli BL21 (DE3), ensuring absence of glycosylation, which could otherwise compromise crystallographic studies. HAC PD-1 was expressed as a periplasmic fusion to amino-terminal maltose binding protein (MBP), purified by nickel affinity chromatography, and then digested with 3C protease to remove the MBP tag. Cleaved protein was repurified by nickel affinity to remove cleaved MBP. Complete digest was confirmed by SDS-PAGE. Protein was digested with carboxypeptidase A and B as described above, and dialyzed into 20 mM HEPES pH 7.5, 150 mM sodium chloride. PD-L1 was expressed in inclusion bodies and refolded as described previously (Li et al., 2004). Properly refolded material was isolated by ammonium sulfate precipitation followed by size exclusion purification. Finally, purified HAC PD-1 and PD-L1 proteins were mixed in a 2:1 ratio and the complex was isolated by size exclusion chromatography. Eluted complex was concentrated to 21 mg/mL prior to crystallization.
HAC PD-1 in complex with PD-L1 was crystallized by vapor diffusion by mixing 100 nL of protein with 100 nL of a precipitant solution consisting of 0.1 M bis-TRIS pH 6.4, 17% PEG MME 5000, 2 mM LiCl. Crystals grew in 2/3 days. Crystals were highly branched and mostly unsuitable for data collection. To address this, crystals were cut using MicroTools (Hampton Research; Aliso Viejo, CA) to isolate single crystal fragments for data collection. Crystals were transferred to a solution of mother liquor supplemented with 20% (v/v) glycerol for cryoprotection prior to freezing in liquid nitrogen.
Data collection was performed at Advanced Photon Source beamline 24ID-C (NE-CAT) using a single crystal. Data reduction and scaling were carried out using XDS (Kabsch, 2010), and the structure was solved by molecular replacement in Phaser (McCoy et al., 2007) using hPD-1 (PDB ID: 3RRQ) and hPD-L1 (PDB ID: 3SBW) as search models. Refinement was conducted using Phenix 1.9 (Afonine et al., 2012) and with manual building in Coot (Emsley and Cowtan, 2004). Structure quality was assessed using MolProbity (Chen et al., 2010). All data processing software was installed and supported by the SBGrid consortium (Morin et al., 2013). The refined structure has been deposited in the Protein Data Bank under accession code 5IUS.
SEC-MALS
For size exclusion chromatography coupled with multi angle light scattering (SEC-MALS) analysis, the sample (70 ul at 2 mg/mL) was previously filtered in a 0.45 µm pore size filter and loaded onto a TSKgel BioAssist G4SWxl column (inner diameter 7.8 mm; length 30 cm; beads size 8 µm) (Tosoh Bioscience LLC) and passed through a Wyatt DAWN Heleos II EOS 18-angle laser photometer coupled to a Wyatt Optilab rEX refractive index detector at 25°C. Elution was monitored inline by absorption at 280 nm. Typical conditions were as follows: buffer 100 mM NaCl, 20 mM Hepes pH7.5 degassed; flow rate of 0.5 ml/min; 100 µL injection volume; analysis time per sample, 35 min. Data were analyzed using Astra 6.1 software (Wyatt Technology Corp., CA, USA).
Structure preparation for molecular dynamics simulations
We used our crystal structure of HAC PD-1/PD-L1 complex and the recently published structure of wild-type PD-1/PD-L1 complex (PDB ID: 4ZQK) (Zak et al., 2015) as the initial conformations to set up our simulations. The structure of HAC PD-1/PD-L1 contains the residues ranging from 18 to 229 for PD-L1. In the crystal structure of wild-type PD-1/PD-L1, PD-L1 is crystallized for residues from 18 to 132, which constitute the entire binding interface between PD-1 and PD-L1. To save computational resources and to make our simulations consistent with each other, we removed the C2 domain (residues 133 – 229) of PD-L1 in the simulation of HAC PD-1/PD-L1. Additionally, Asp122 on PD-L1 is positioned adjacent to His68 on HAC PD-1. The close distance between Asp122 and His68 can lead to the protonation of the side chain on His68. We thus considered both His68 on HAC PD-1 to be protonated (two NH groups exist on the imidazole ring) and His68 at natural condition (one NH group exists on ε position of the imidazole ring). For the wild-type PD-1/PD-L1, we added the missing side chains on this complex structure with Pymol and used it as the initial conformation for the simulations of the wild type complex. For the simulation of PD-1 in the ligand-free state, the initial structures for HAC PD-1 and wild-type PD-1 are obtained by directly removing PD-L1 from the complex. Based on all the structures aforementioned, we built the systems as listed in Table S2.
Molecular dynamics simulations
Molecular dynamics (MD) simulations used a similar protocol to that described in previous work (Sun et al., 2015) (Sun et al., 2014) (Bowman, 2016). In brief, simulations were carried out using Gromacs 5.1 (Abraham et al., 2015) with the Amber03 force field (Duan et al., 2003). The TIP3P water model (Jorgensen et al., 1983) was used to solvate the protein in a dodecahedron box that extended 10 Å beyond the protein in every dimension. Thereafter, sodium or chloride ions were added to produce neutral systems. Each system was subjected to energy minimization with 1000 kJ/mol/nm as the force threshold. Afterwards, each system was relaxed by an MD simulation of 100 ps with 1 fs as the time step using the NVT ensemble. Each system further underwent an NPT MD simulation for 1 ns with the time step of 2 fs for equilibration.
After the equilibration runs, each system was simulated using the NPT ensemble with the temperature and pressure set to 300 K and 1 bar, respectively. The V-rescale thermostat and Parrinello−Rahman pressure coupling were applied during the simulation. The bonds containing hydrogen atoms were constrained with the LINCS algorithm, and virtual sites are applied to allow us to use a time step of 4 fs. Periodic boundary conditions were applied, and the cut-offs for the electrostatic and van der Waals interactions were set to 9 Å, with the long-range electrostatic interaction recovered by the particle mesh Ewald summation.
The representative structures were picked using the hybrid k-centers/k-medoids method embedded in MSMBuilder 2.8 (Bowman et al., 2009) (Beauchamp et al., 2011). With this method, all the conformations were clustered with k-centers algorithm based on the root mean square deviation of Cα atoms in the protein. The cutoff value for the k-centers clustering was set to 2.5 Å, which lead to the maximum distance between data point in the cluster and the cluster centers less than 2.5 Å. Then, 10 update iterations were used to center the clusters on the densest regions of conformational space. The center structures for the five most populated clusters were used to make the figures to describe the dynamic behaviors of the protein in the simulations. The figures were generated using Pymol.
Surface Plasmon Resonance
Experiments were performed on a Biacore X100 (GE Healthcare) and carried out at 25°C. Biotinylated PD-L1 protein was immobilized onto a Biacore SA sensor chip (GE Healthcare) at an Rmax of ~91 RU. A control protein (biotinylated mouse SIRPA) was immobilized onto the chip reference cell at an approximately matching RU value to control for nonspecific protein-protein interactions. Measurements were made using serial dilutions of the recombinant human PD-1 variant, HAC, in individual buffers as per the pH requirements of the run. pH 9.0 buffer was composed of 20 mM Tris-HCl, 150 mM NaCl, and 0.0005% Tween-20 (vol/vol), and titrated with NaOH. pH 7.4 buffer was composed of 10 mM Hepes pH7.4, 150 mM NaCl and 0.005% surfactant P20 (vol/vol), and titrated with NaOH as appropriate. pH 6.5 and pH 5.5 buffers were composed of 20 mM MES pH 6.0, 150 mM NaCl and 0.0005% Tween-20, titrated with NaOH or HCl as appropriate. The PD-L1 surface was regenerated by one 60 s injection of 50% (vol/vol) ethylene glycol and 100 mM glycine pH 9.5. All data were analyzed with the Biacore X100 evaluation software with a 1:1 Langmuir binding model. Raw data and fit curves were exported and plotted in GraphPad Prism.
Supplementary Material
Highlights.
Structure of engineered PD-1 receptor with ligand PD-L1 shows basis for high affinity
Polar interaction increases conformational stability of the complex
Mutant PD-1 binding affinity is strongly pH dependent
Binding is essentially irreversible in acidic conditions
Acknowledgments
G.R.B. holds a Career Award at the Scientific Interface from the Burroughs Welcome Fund. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the GTX Titan X GPU used for our simulations. X-ray crystallographic data were collected at NE-CAT beamline 24ID-C. We would like also to recognize Dr. K. Arnett for assistance in SEC-MALS data collection and analysis and Dr. B. Zimmerman for helpful experimental discussion. This work was supported in part by NIH grant 1DP5OD021345 to A.C.K. The authors J.K. and R.M. are paid employees of Ab Initio Biotherapeutics, Inc., which holds the license to a patent on the HAC PD-1 mutant. R.M., A.M.R., and A.C.K. are founders of Ab Initio Biotherapeutics, and A.M.R and A.C.K. are members of the Scientific Advisory Board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
R.P. performed protein expression, purification, and structure determination, as well as designing and carrying out ITC experiments. X.S. performed and analyzed molecular dynamics simulations with supervision and input from G.R.B. SPR experiments were performed by J.K. and R.M., and data analysis was conducted by R.P. Experimental design and data analysis throughout the project was conducted with input from R.M., G.R.B., A.M.R. and A.C.K. The manuscript was written by A.C.K. and R.P. with input from the other authors.
REFERENCES
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- Beauchamp KA, Bowman GR, Lane TJ, Maibaum L, Haque IS, Pande VS. MSMBuilder2: Modeling Conformational Dynamics at the Picosecond to Millisecond Scale. J Chem Theory Comput. 2011;7:3412–3419. doi: 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR. Accurately modeling nanosecond protein dynamics requires at least microseconds of simulation. J Comput Chem. 2016;37:558–566. doi: 10.1002/jcc.23973. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Huang X, Pande VS. Using generalized ensemble simulations and Markov state models to identify conformational states. Methods. 2009;49:197–201. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Veverka V, Radhakrishnan A, Waters LC, Muskett FW, Morgan SH, Huo J, Yu C, Evans EJ, Leslie AJ, Griffiths M, Stubberfield C, Griffin R, Henry AJ, Jansson A, Ladbury JE, Ikemizu S, Carr MD, Davis SJ. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288:11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- Justus CR, Dong L, Yang LV. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013;4:354. doi: 10.3389/fphys.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, Greenfield EA, Bourque K, Boussiotis VA, Carter LL, Carreno BM, Malenkovich N, Nishimura H, Okazaki T, Honjo T, Sharpe AH, Freeman GJ. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- Lazar-Molnar E, Yan Q, Cao E, Ramagopal U, Nathenson SG, Almo SC. Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc Natl Acad Sci U S A. 2008;105:10483–10488. doi: 10.1073/pnas.0804453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, Moraga I, Raeber ME, Bowman GR, Novick P, Pande VS, Fathman CG, Boyman O, Garcia KC. Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'. Nature. 2012;484:529–533. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Yu JF, Chen YJ, Ma HB, Wang ZF, Zhu YB, Zhang XG. Refolding and characterization of recombinant human GST-PD-1 fusion protein expressed in Escherichia coli. Acta Biochim Biophys Sin (Shanghai) 2004;36:141–146. doi: 10.1093/abbs/36.2.141. [DOI] [PubMed] [Google Scholar]
- Lin DY, Tanaka Y, Iwasaki M, Gittis AG, Su HP, Mikami B, Okazaki T, Honjo T, Minato N, Garboczi DN. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci U S A. 2008;105:3011–3016. doi: 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute RL, Gordon SR, Mayer AT, Mccracken MN, Natarajan A, Ring NG, Kimura R, Tsai JM, Manglik A, Kruse AC, Gambhir SS, Weissman IL, Ring AM. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A. 2015;112:E6506–E6514. doi: 10.1073/pnas.1519623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Ohashi PS. Clinical blockade of PD1 and LAG3--potential mechanisms of action. Nat Rev Immunol. 2015;15:45–56. doi: 10.1038/nri3790. [DOI] [PubMed] [Google Scholar]
- Sun X, Agren H, Tu Y. Microsecond Molecular Dynamics Simulations Provide Insight into the Allosteric Mechanism of the Gs Protein Uncoupling from the beta Adrenergic Receptor. J Phys Chem B. 2014 doi: 10.1021/jp506579a. [DOI] [PubMed] [Google Scholar]
- Sun X, Cheng J, Wang X, Tang Y, Agren H, Tu Y. Residues remote from the binding pocket control the antagonist selectivity towards the corticotropin-releasing factor receptor-1. Sci Rep. 2015;5:8066. doi: 10.1038/srep08066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- Vibhakar R, Juan G, Traganos F, Darzynkiewicz Z, Finger LR. Activation-induced expression of human programmed death-1 gene in T-lymphocytes. Exp Cell Res. 1997;232:25–28. doi: 10.1006/excr.1997.3493. [DOI] [PubMed] [Google Scholar]
- Vigdorovich V, Ramagopal UA, Lazar-Molnar E, Sylvestre E, Lee JS, Hofmeyer KA, Zang X, Nathenson SG, Almo SC. Structure and T cell inhibition properties of B7 family member, B7-H3. Structure. 2013;21:707–717. doi: 10.1016/j.str.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll DM, Okumura K, Azuma M, Yagita H. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol. 2002;169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- Youngnak P, Kozono Y, Kozono H, Iwai H, Otsuki N, Jin H, Omura K, Yagita H, Pardoll DM, Chen L, Azuma M. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Commun. 2003;307:672–677. doi: 10.1016/s0006-291x(03)01257-9. [DOI] [PubMed] [Google Scholar]
- Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Domling A, Dubin G, Holak TA. Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure. 2015;23:2341–2348. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.