Abstract
Although clonal studies of lineage potential have been extensively applied to organ specific stem and progenitor cells, much less is known about the clonal origins of lineages formed from the germ layers in early embryogenesis. We applied lentiviral tagging followed by vector integration site analysis (VISA) with high-throughput sequencing to investigate the ontogeny of the hematopoietic, endothelial and mesenchymal lineages as they emerge from human embryonic mesoderm. In contrast to studies that have used VISA to track differentiation of self-renewing stem cell clones that amplify significantly over time, we focused on a population of progenitor clones with limited self-renewal capability. Our analyses uncovered the critical influence of sampling on the interpretation of lentiviral tag sharing, particularly among complex populations with minimal clonal duplication. By applying a quantitative framework to estimate the degree of undersampling we revealed the existence of tripotent mesodermal progenitors derived from pluripotent stem cells, and the subsequent bifurcation of their differentiation into bipotent endothelial/hematopoietic or endothelial/mesenchymal progenitors.
Keywords: Pluripotent stem cells, Vector integration site analysis, Mesoderm, Lineage tracing, Clonal tracking, Lentiviral vectors, Hematopoiesis
Introduction
During the earliest stages of embryogenesis, a morphologic region called the primitive streak (PS) contains cells committed to form either mesoderm or definitive endoderm. Fate-mapping experiments in vertebrates show that mesoderm patterning in the PS strictly correlates with the place and time of mesoderm induction, specifying posterior PS (extraembryonic mesoderm, lateral plate mesoderm), anterior PS (cardiac mesoderm, definitive endoderm, axial mesoderm) and late PS (presomitic mesoderm) [1]. This dramatic period of morphogenesis, and the dynamic transcriptional and signaling events that shape mesoderm patterning, suggest that the PS is a rapidly differentiating population in which self-renewal may be limited or non-existent.
Given the inaccessibility of early human embryonic tissues, modeling with human pluripotent stem cells (hPSC) has become an essential tool for studying the complex cellular events of human germ layer commitment [2]. An early PS-like population has been identified during hPSC differentiation using transcriptional reporters [3] and by expression of cell surface markers of epithelial to mesenchymal transition [4]. Recently, cardiac and paraxial mesoderm, subtypes derived from the anterior PS and late PS respectively, were shown to be specified through distinct BRACHYURY+ mesoderm progenitors during exit from pluripotency [1].
Transcriptome profiling of single cells often reveal a surprising level of heterogeneity within purified populations that contain apparently identical cells. The differential expression of lineage specific genes between individual cells is often inferred as evidence that those cells intrinsically possess different types of lineage potential. Such analyses provide a snapshot of the transcriptional status of individual mesoderm progenitors, but do not definitively prove the clonal relationship of the lineages that will be ultimately produced from each progenitor. The functional interrogation of lineage output at a clonal level is crucial to understand the process of lineage commitment, and can provide valuable insights in how to guide stepwise generation of therapeutically relevant tissues and organs from hPSC. Clonal analyses of lineage potential have been extensively applied to adult tissues and to fetal cells isolated from tissues after germ layer differentiation. Although PS-like populations can be differentiated into various lineages [3, 4], little is known about the clonal relationship of cells that initiate early embryonic development. We have previously published a differentiation and isolation strategy that captures the earliest stage of mesodermal commitment from hPSC, comprising a population that displays broad lateral plate and cardiac mesoderm potential [4]. In this study, we used these human embryonic mesoderm progenitors (hEMP) as the starting population for clonal tracking of mesoderm derivatives, specifically examining the hematopoietic, endothelial and mesenchymal lineage potential of single cells by high throughput sequencing of lentiviral tags. A fundamental challenge in applying this approach to heterogeneous progenitors undergoing rapid differentiation is that clonal output must be detected within a highly complex population with limited clonal duplication. To meet this challenge, we applied a “mark-recapture” statistical approach to estimate clonal abundance [5–7] and developed a mathematical model to accurately interpret datasets from high complexity samples that cannot be exhaustively characterized.
Through high throughput sequencing and analysis of lentiviral integration sites combined with attention to the impact of sampling and the examination of essential control populations, we demonstrated the presence of a tripotent mesodermal progenitor and uncovered the bifurcation of the hematopoietic and mesenchyme lineages early in lateral plate mesoderm commitment. We propose that the critical concept of undersampling and the mathematical approaches used here are highly relevant to other studies of transitional populations, including those that attempt to uncover the role of genetically diverse malignant subclones during disease evolution.
Materials and Methods
Vector Constructs and Production
FUGW (carrying the EGFP reporter) [8] was used in all transductions except for the negative control experiment (Fig. 4D) in which mCitrine, mCerulean and mStrawberry were expressed in pCCLc-UBC-reporter-PRE-FB-2xUSE [9]. Vector production and titer were as described [10].
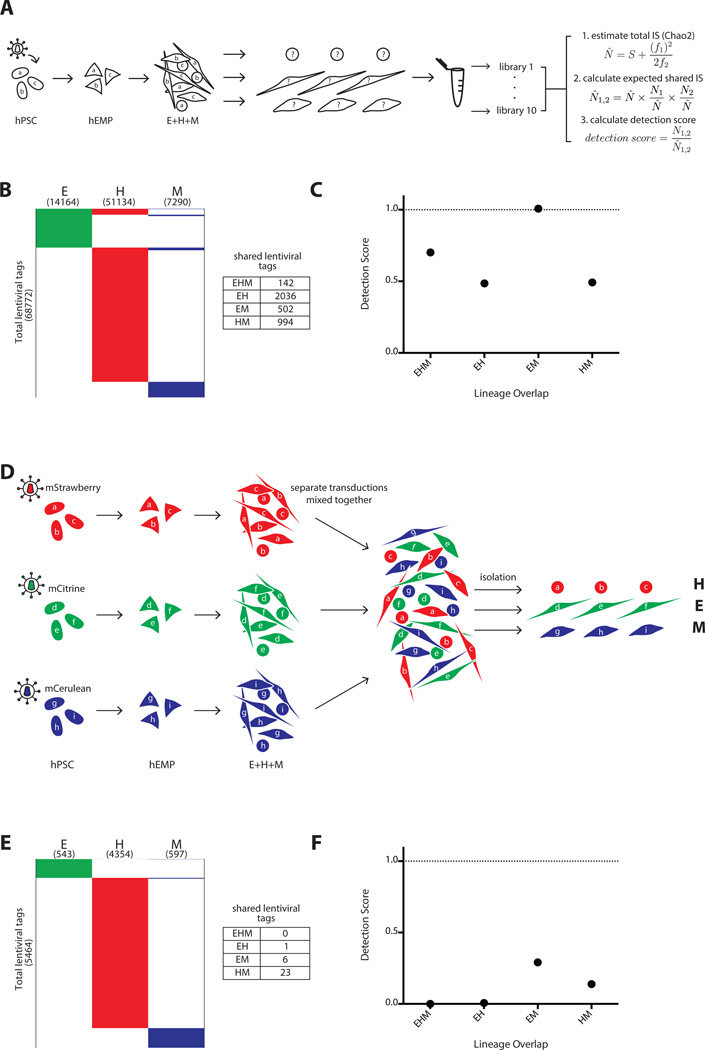
Figure 4.
Positive and negative control experiments to determine the maximal and minimal expectations for lentiviral tag sharing. (A–C) Positive control, and (D–F) negative control for clonal detection limits. (A): Schema of polyclonal hPSC tagging experiment performed to define upper boundary of detection. A pool of hPSC were transduced, expanded briefly without selection, and induced to become hEMP, which were then differentiated and analyzed via non-restrictive linear amplification-mediated PCR (nrLAM-PCR) and HTS. For each lineage, 10 separate DNA samplings were taken for nrLAM-PCR and sequencing, with each sampling labeled by a unique barcode. These data were used for Chao2 estimation of total tag count in each lineage, and this information was used in our model to calculate a shared tag “detection score” (N̂ = estimated total number of lentiviral tags, S = observed number of total tags, f1 = number of tags observed only once, f2 = number of tags observed only twice). N̂ 1,2 = expected shared tags between population 1 and 2, N1,2 = observed shared tags between population 1 and 2 (see also Supporting Information Figs. 3, 4 and Methods for Chao2 explanation and model). (B): Lentiviral tags identified in each lineage derived from a polyclonal pool of transduced hPSC. A total of 68,772 tags were detected in one or more lineage. Each horizontal line in the schematic heatmap represents a specific lentiviral tag and tags are clustered based on their presence in lineages; tags identified in two or more lineages are shown in the same horizontal position. The total number of tags sequenced from each lineage is listed in parentheses below the lineage label. 142 tags were shared among all three lineages. 2036, 502 and 994 shared tags were found between EH, EM, and HM, respectively. The majority of tags (65,098) were detected in only one lineage, illustrating the compounded effect of sampling during cell harvest and sequencing preparation. (C): The numbers of expected shared tags were compared to the observed shared tags in the form of a detection score (observed/expected). The detection score for EHM, EH, EM, and HM were 0.7, 0.5, 1.0, and 0.5, respectively. Since hPSC are assumed to be pluripotent, the detection score between any set of lineages represents the maximum tag sharing one can detect in this experimental system. (D): Three separate pools of hPSCs were transduced with lentiviral vectors expressing distinct fluorescent markers and differentiated in parallel on OP9. The three pools of differentiated cells were combined, and each lineage was then isolated from this pool based on the sets of lineage markers (Fig. 1A), each of which was paired with a distinct fluorescent marker (mStrawberry hematopoietic; mCitrine endothelium; mCerulean mesenchyme). This negative control served to determine the frequency of false clonal overlap by chance and sorting errors. (E): Only 1, 6 and 23 shared tags were found between EH, EM, and HM, respectively and no tags were shared with all lineages (EHM). (F): The detection score for EHM, EH, EM, and HM were 0, 0.007, 0.3, and 0.1, respectively. These scores represent the false positive rate of tag sharing in this experimental system. Abbreviations: hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell.
Mesoderm Differentiation from hPSC and hEMP
The hESC line H1 (WiCell, Madison, WI, http://www.wicell.org/) was maintained and expanded on irradiated primary mouse embryonic fibroblasts (EMD Millipore, Billerica, MA, http://www.emdmillipore.com/). Mesoderm commitment was induced as previously described [4]. CD326−CD56+ embryonic mesoderm progenitors were isolated by flow cytometry at day 3.5 (Fig. 1A) and cocultured on OP9 stroma for trilineage (hematopoietic, endothelial, and mesenchymal) differentiation over the next 13 days (see Supporting Information Methods).
Figure 1.
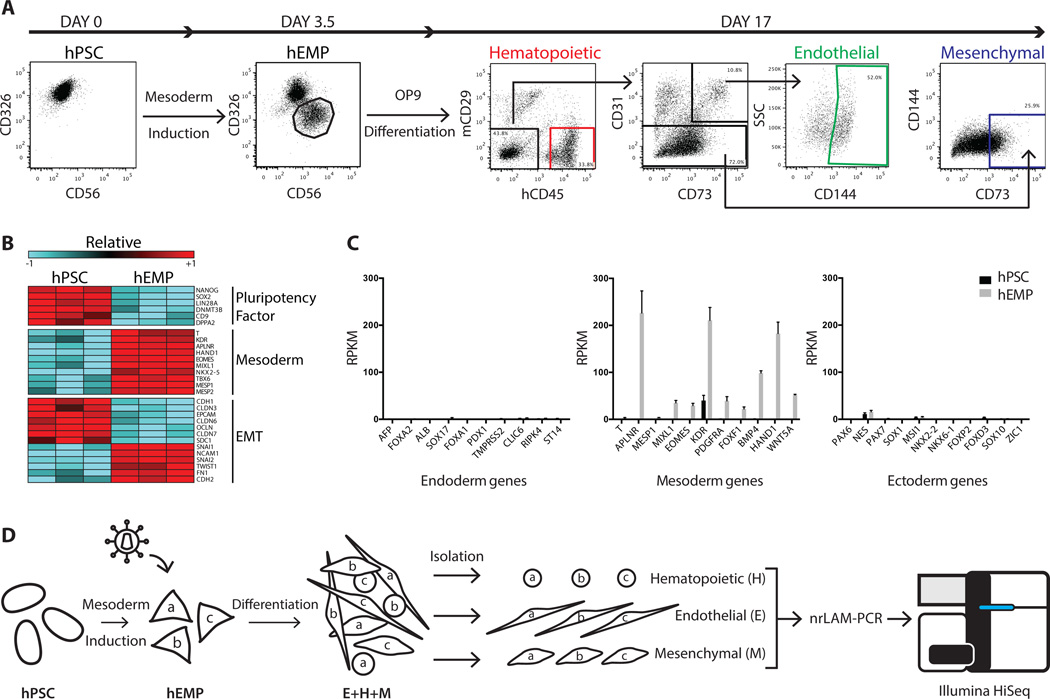
Generation of a human embryonic mesodermal progenitor population from hPSC with hematopoietic, endothelial, and mesenchymal potential. (A): Flow cytometry analysis of EPCAM/CD326 and NCAM/CD56 expression in undifferentiated hPSC and hEMP generated after 3.5 days of sequential morphogen induction with Activin A, BMP4, VEGF and bFGF. hEMP were FACS isolated as the CD326−CD56+ population. Further differentiation on the murine stromal line OP9 generated hematopoietic, endothelial, and mesenchymal cells after 2 weeks. After exclusion of mCD29+ murine cells, the cell surface marker CD45 was used to define human hematopoietic cells. The endothelial lineage was isolated by the markers CD144+CD31+CD73+CD45−, a phenotype that defines non-hemogenic endothelium [14]. The mesenchymal lineage was isolated by the markers CD73+CD31−CD45−CD144−. (B): Transcriptome comparison via RNA-Seq of hPSC and hEMP showed downregulation (blue) of pluripotency factors, upregulation (red) of mesoderm genes and changes in gene expression indicative of EMT. Both FDR < 0.01 and fold change > 2-fold were applied as filters. Color scale in the heatmap shows the relative expression for each gene using its min/max moderate expression estimates as reference. (C): Gene expression in hPSC (black) and hEMP (gray) showed marked upregulation of mesodermal genes enrichment in hEMP without ectoderm or endoderm gene expression. (D): Schematic of lentiviral tagging approach to interrogate lineage potential of hEMP. After mesodermal induction of hPSC, hEMP were isolated at day 3.5, and transduced with a lentiviral vector. For the purposes of illustration, three distinct lentiviral tags created by transduction are shown as “a,” “b,” and “c.” These transduced hEMP were cocultured on OP9 stroma with conditions supporting differentiation of hematopoietic, endothelial, and mesenchymal cells; these lineages as well as lin neg cells were FACS isolated after two weeks (as in Fig. 1A). Integration sites in each lineage (E, H, and M) were identified by nrLAM-PCR followed by high throughput sequencing on an Illumina HiSeq. Abbreviations: EMT, epithelial-mesenchymal transition; hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell; nrLAM-PCR, non-restrictive linear amplification-mediated PCR; RPKM, Reads Per Kilobase of transcript per Million mapped reads.
Transduction of hPSC, hEMP, and K562 Cells
A full description of transduction methods for each cell type is found in Supporting Information Methods. In brief, hPSC were transduced in mTESR on matrigel for 24 hours, yielding ~50% GFP + cells. Isolated hEMP were transduced for 12 hours, yielding 80% GFP + cells. To limit further viral integration during hEMP differentiation, integrase inhibitor (raltegravir, Merck & Co, White House Station, NJ, http://www.merck.com/index.html) was added at 1 µM at 12 hours when OP9 coculture was initiated and was supplemented throughout the 13 days of differentiation. K562 cells (American Type Culture Collection, http://www.atcc.org/) were transduced at MOI 4 and raltegravir added at various time points between 6 hours and 3 days. After 3 weeks of further expansion, DNA was extracted to measure vector copy number/cell by droplet digital PCR (see Supporting Information Methods).
Flow Cytometry and Cell Sorting
Identification and isolation of lineages was performed using the following gating sequence: murine CD29-APC-Cy7 was used to exclude murine cells. CD45-PE-Cy7 identified the hematopoietic population (CD45+). From the non-hematopoietic compartment (CD45−), the CD31-APC and CD73-PE-Cy7 coexpressing cells were first gated, and CD144-PerCP-Cy5.5 positivity was then used to define the endothelial population (CD45−CD31+ CD73+CD144+). From the non-hematopoietic, non-endothelial compartment (CD45−CD31−CD144−), the mesenchymal population was identified based on CD73-PE-Cy7 positivity (CD45−CD31−CD144−CD73+). Lineage negative cells were mCD29− cells that could not be assigned to a lineage based on immunophenotype (mCD29−CD45−CD31−CD73−). See Supporting Information Methods for details.
Lentiviral Tag Sequencing
Genomic DNA was isolated from cells using the PureLink Genomic DNA Mini kit (Invitrogen, https://www.thermofisher.com/us/en/home/brands/invitrogen.html) or NucleoSpin Tissue XS kit (Clontech, Mountain View, CA, https://www.clontech.com/), depending on starting cell number. Five microliters of DNA (or a maximum of 75,000 lentiviral tags/reaction) was used as starting input for non-restrictive linear amplification-mediated PCR (nrLAM-PCR) [11]. Four to ten independent nrLAM-PCR reactions were performed on the genomic DNA from each population. PCR products were mixed and quantified by probe-based droplet digital qPCR and appropriate amounts were used to load Illumina v3 flow cells. See Supporting Information Methods for PCR primer sequences and conditions.
Paired-end 50- or 100-bp sequencing was performed on an Illumina HiSeq 2000 using a custom read 1 primer (GAGATCTACACTGATCCCTCAGACCCTTTTAGTC). Sequence reads were required to begin with the end of the vector LTR sequence and have no more than two mismatches with the LTR sequence. To map lentiviral tags within the human genome, LTR sequences and Illumina adapter sequences were trimmed from the reads, which were then aligned to the hg19 build of the human genome with Bowtie2 [12]. Alignments were condensed and annotated by a custom Python wrapper script. Conservative cutoffs were set for calling integration sites and demultiplexing (i.e., calling lentiviral tags as detected within samples marked by different indexes) to control for spurious tag sharing due to sequencing artifacts or FACS impurity. Four reads were required to call a lentiviral tag detected, and the power law function (threshold) = 0.0874 * (total readcount among samples)0.7025 was used to establish a readcount threshold for calling a tag detected in a sample. The result of this processing was a set of lentiviral tags (integration sites denoted by chromosome number, strand and nucleotide position) that were detected in each sampling of DNA from the various differentiated lineages. The numbers of sequence reads obtained for the samples analyzed are detailed in Supporting Information Fig. 6.
Estimation of Total Tag Count and Calculation of Detection Scores
From the set of lentiviral tags detected among all samplings from all lineages in an experiment, the total number of tags (the union) was taken, along with the number of tags detected in only one sample and the number of tags detected in exactly two samples. These three values were used to calculate a Chao2 lower bound on the total lentiviral tags among all lineages. This estimate was taken to reflect the number of tags generated during hEMP transduction at the beginning of the experiment that survived through the end of the experiment (see Supporting Information Fig. 3). These calculated lower bounds were used in the model described in Supporting Information Fig. S4 to calculate the expected number of shared lentiviral tags between all possible pairs and trios of lineages, assuming that all tagged cells were multipotent and that the probabilities of a tag being detected in the lineages are independent. We model lineage commitment of a clone and detection of a tag within that clone together as a random draw of a lentiviral tag from the set of tags inferred in the transduced hEMP population.
Graphical and Statistical Analysis
Graphs were generated and statistics analyzed using GraphPad Prism software. Student’s two-tailed t tests were used to calculate p-values, except in Fig. 5C, where an extreme value test (using the cumulative distribution function) was performed using a normal distribution with mean and standard deviation calculated from the three hEMP experiment replicates. p < .05 was considered statistically significant.
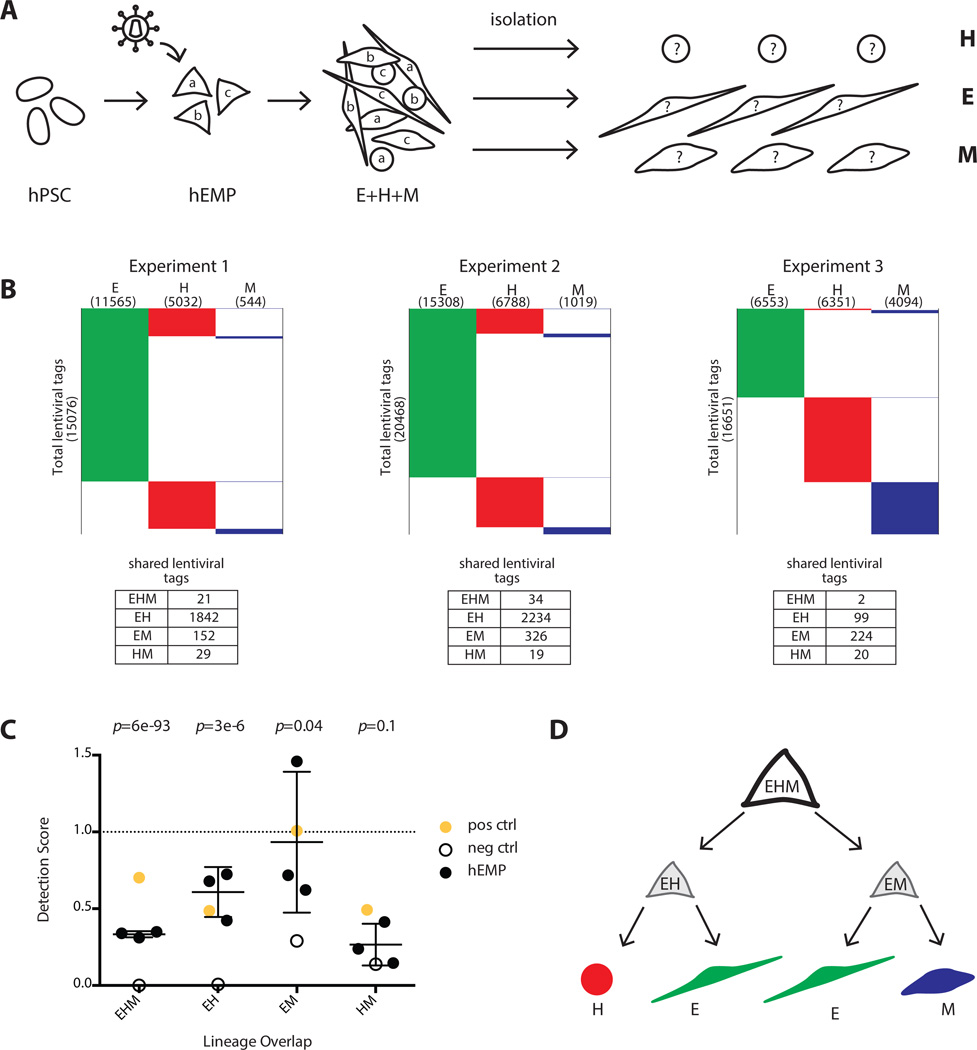
Figure 5.
Lentiviral tagging of human embryonic mesodermal progenitors reveal subpopulations with bipotent and tripotent potential. (A): Experimental design to track clonal output of a pool of tagged mesoderm progenitors (hEMP) differentiated into H, E, and M lineages. (B): Data from three independent experiments showing number of lentiviral tags identified in each lineage. Experiment 1 was performed with the integrase inhibitor raltegravir added after 12 hour of transduction while experiments 2 and 3 were performed without raltegravir. The addition of raltegravir did not alter the detection score of shared lentiviral tags. (C): The detection scores from marking mesoderm progenitors are shown (black dots) as well as the maximum detection expectation (set by positive control, yellow dot) and the false positive rate (set by negative control, empty circle). The negative control detection scores for EHM, EH, and EM tag sharing were significantly lower than the detection scores in the mesoderm progenitor tagging experiments (p-values 6 × 10−93, 3 × 10−6 and .04, respectively for EHM, EH, and EM). The HM detection scores were similar between the progenitor tagging experiments and the negative control (p-value = .1). p-value calculation used an extreme value test performed using the cumulative distribution function of a normal distribution with mean and standard deviation calculated from progenitor experimental data. (D): Proposed model of the developmental potential of human mesodermal progenitor defined by unbiased lentiviral marking. During mesodermal differentiation from hPSC, a tripotent progenitor (EHM) with limited self-renewal ability rapidly generates either endothelial/hematopoietic progenitors (EH) or endothelial/mesenchyme progenitor (EM). The lentiviral tagging data do not support the existence of a bipotent HM progenitor. Abbreviations: E, endothelial; H, hematopoietic; hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell; M, mesenchymal.
RESULTS
The CD326−CD56 + Population Marks Early Mesoderm Commitment from Human PSC
Our previous studies described an early stage of mesoderm commitment during hPSC differentiation that corresponds to the onset of epithelial-mesenchymal transition (EMT) in the primitive streak, and is marked by loss of CD326 (EpCAM) and acquisition of CD56 (NCAM) expression (Fig. 1A) [4]. The CD326−CD56 + hEMP population partially overlaps with the APLNR + population [13] and first emerges as early as day 2 of differentiation [4] before cell surface expression of more lineage-specific markers (CD43, CD34, VE-Cadherin, CD235) [14–17]. Transcriptome profiling of hPSC and day 3.5 hEMP by RNA-Seq demonstrated the onset of mesoderm commitment with significant upregulation (FDR<0.01, >2-fold changes) of genes known to be involved in primitive streak formation (MIXL1, EOMES, T, and MESP1) and EMT (CDH2, FN1, TWIST1 and SNAI2), with concomitant downregulation of cell-cell adhesion molecules such as CDH1 (which encodes E-Cadherin) and claudins (Fig. 1B, Supporting Information Methods). The marked downregulation of pluripotency factors in CD326−CD56 + cells (Fig. 1B) matched the functional loss of teratoma-forming ability previously seen in vivo [4]. Using established differentiation conditions, we have previously shown the ability of CD326−CD56 + cells to give rise to all mesodermal lineages tested including hematopoietic, endothelial, mesenchymal (bone, cartilage, fat, fibroblast), smooth muscle, and cardiomyocyte [4]. The lack of endoderm and ectoderm gene expression (Fig. 1C) and the inability to generate these germ layers in vitro [4] further confirmed that the hEMP population is specifically committed to mesoderm fate.
While multiple mesoderm lineages can be generated from the hEMP population, it is not known whether hEMP represent a homogenous group of multipotent progenitors or a heterogeneous mixture of more lineage-restricted bipotent and/or unipotent progenitors. In more committed cell types (e.g., hemangioblasts and hematopoietic progenitors) lineage potential has been examined by cloning single cells and examining the lineage composition of the progeny [13, 18–20]. However, we found that a rigorous and quantitative assignment of clonal lineage potential from single hEMP in culture was not feasible due to technical limitations (e.g., temporal variability in proliferation and differentiation from single cells, the need for stromal cocultivation obscuring readout and unreliable lineage discrimination from low-frequency events).
We therefore turned to a lentiviral genetic labeling strategy to trace cellular genealogy in the mixture of clones present in differentiating bulk cultures. Lentiviral vectors integrate in a semi-random fashion throughout the cellular genome and the resulting integration sites are replicated along with the cellular genome. These integration sites can therefore be treated as unique sequence tags (hereafter referred to as “lentiviral tags”) marking all progeny of a tagged clone (Fig. 1D). We developed differentiation conditions that could generate hematopoietic, endothelial and mesenchymal cells from bulk populations of hEMP in one culture vessel, so that clones would not be disturbed after lentiviral tagging and would therefore be free to proliferate and populate multiple lineages. Under these conditions, tri-lineage output was reliably detected based on cell surface marker expression after two weeks of differentiation (Fig. 1A).
Anticipating that multipotent progenitors may represent a rare subset of the total hEMP population, we chose to maximize the complexity of integrations by labeling a high starting number of cells and by using high titer vectors, achieving 60%–80% transduction efficiency. To retrieve lentiviral tags from the differentiated cells with high efficiency, we amplified the vector-genome junction sequences via nrLAM-PCR [11, 21, 22] and sequenced them on an Illumina HiSeq 2000 (Supporting Information Fig. 1). nrLAM-PCR circumvents the restriction digest required in standard LAM-PCR, allowing for less biased and more comprehensive integration site amplification [23].
Efficient Detection of Lentiviral Tagging of Tri-Lineage Output from Monoclonal hPSC
As an initial proof of concept, we assessed lentiviral tag sharing in cells differentiated from a single transduced and expanded hPSC (Fig. 2A). The undifferentiated monoclonal transduced hPSC line contained 25 distinct lentiviral tags, as determined by high-throughput lentiviral tag sequencing (Fig. 2B). The tagged hPSC clone was subjected to mesoderm induction to generate a population of hEMP, which was then differentiated into hematopoietic, endothelial and mesenchymal lineages (Fig. 2A). After 2 weeks, each of the three lineages was isolated by flow cytometry based on cell surface markers (Fig. 1A).
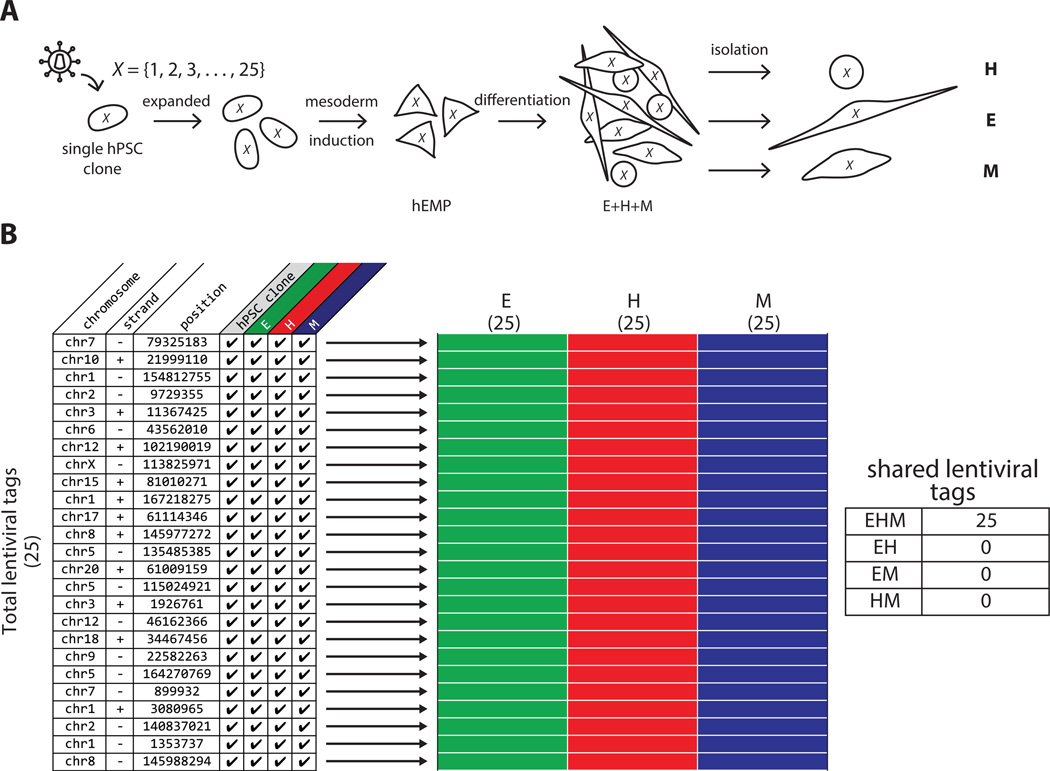
Figure 2.
Lentiviral tagging and HTS demonstrates trilineage mesoderm differentiation of monoclonal hPSC. (A): Schematic of monoclonal hPSC lineage tagging. A single transduced hPSC with 25 lentiviral tags was clonally expanded, induced to form embryonic mesoderm progenitors (hEMP) and differentiated into H, E, and M lineages, which were then isolated as separate populations by FACS and subjected to vector integration site analysis by HTS. X is the set of all 25 lentiviral tags found in the initial hPSC clone. (B): Table listing all 25 lentiviral tags detected by HTS in the hPSC clone and the three lineages differentiated from hEMP generated from the hPSC clone, with chromosome, strand and position information. Checkmarks indicate that a particular tag was detected in the lineage indicated at the top of the column. Each row in schematic heatmap to the right represents a distinct lentiviral tag, equivalent to the rows of the table on left. A filled in box indicates the presence of that tag in the population, and the three populations (E, H, and M) are annotated as green, red and blue colors respectively for clarity. Numbers in parentheses under each lineage label refer to the total number of tags found in each population, and numbers on the y-axis indicate the total number of tags found among all populations. All 25 lentiviral tags were detected as shared among the three lineages, as indicated in the table to the right of the heatmap. Abbreviations: E, endothelial; H, hematopoietic; hEMP, human embryonic mesoderm progenitor; hPSC, human pluripotent stem cell; M, mesenchymal.
HTS of nrLAM-PCR products from each lineage was performed on an Illumina HiSeq. The same twenty-five tags were detected in DNA from the original expanded, undifferentiated PSC clone and from all three lineages that were generated from isolated hEMPs derived from the hPSC clone; no additional lentiviral tags were detected (Fig. 2B). This experiment demonstrated the ability of the methodology to reliably detect lentiviral tags shared among multiple lineages differentiated from a tagged population of minimum complexity.
Lentiviral Tagging of Polyclonal Pluripotent Populations and Estimation of Undersampling of Shared Tags
We anticipated that even if the lentiviral system were applied to study a population of progenitor cells that were all multipotent, it would fail to fully detect all lentiviral tags in the multiple differentiated lineages of interest due to undersampling arising from practical limitations inherent in our experimental system, such as the infeasibility of collecting the genomic DNA of all cells in the culture vessel, of preparing sequencing libraries containing all of the lentiviral tags present in the isolated genomic DNA, and of sequencing all of the lentiviral tags present in the sequencing libraries. In experiments attempting to detect the same clones in multiple cell populations, the impact of undersampling is amplified multiplicatively, leading to a dramatic underestimate of the frequency of sites shared between lineages.
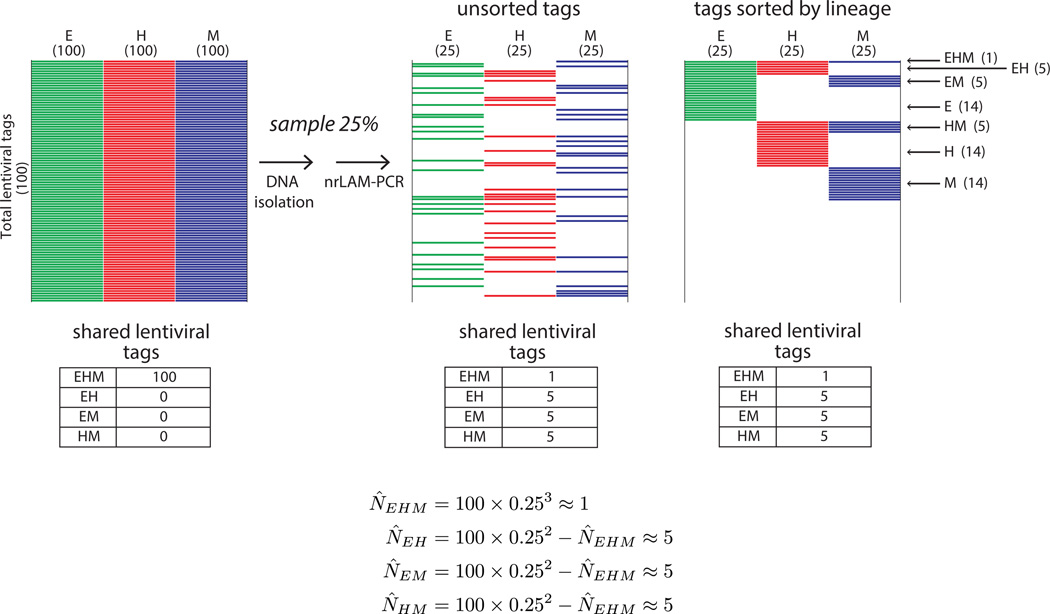
To illustrate this issue, we present a theoretical situation in which cellular populations of three target lineages all contain the same 100 lentiviral tags (Fig. 3). Using an arbitrary scenario in which only 25% of the lentiviral tags were recovered and sequenced at random from each lineage (from compounded undersampling during processing), we can predict the expected number of shared tags between E, H, and M assuming independence in the detection events in the three lineages (Fig. 3). Each tag has a probability of .25 of being detected in a single lineage, and only a probability of .252, or .0625, of being detected in two lineages. Therefore, the most likely result is that 6 tags would be detected in both the H and E lineages. This issue compounds with each population that is added to the analysis; in the case of three populations, the probability falls to .253, or only ~.016; thus the most likely result is to detect only one to two shared tags of the 100 that are actually present. Interpreting such a result without considering the impact of sampling would lead one to conclude that most of the tagged cells were not multipotent, when in reality, all of them were. In this theoretical setting, the expectation can be calculated because of the initial assertion that 100 clones were present, but in an experimental setting, the total number of clones is not known. Repeated sampling will discover a larger proportion of lentiviral tags in the populations and thus increase the chance of detecting shared tags, but hundreds of sequencing runs would be needed to uncover the entire pool of lentiviral tags present in a highly complex population (Supporting Information Fig. 2). This requirement is both technically and financially challenging to satisfy.
Figure 3.
Theoretical effect of sampling on shared lentiviral tag detection. Schematic heatmaps are shown for the theoretical scenario of a population in which all cells are multipotent and marked by a total of 100 lentiviral tags. Each multipotent cell goes on to form E, H, and M progeny, and all three lineages therefore contain all 100 tags in the culture vessel (left panel). We show how the results of this experiment appear using a model in which only 25% of tags are found from each lineage because of cumulative undersampling during cell and DNA processing. The tags are unsorted in the middle panel to reflect the randomness of the sampling; in the right panel the same tags are sorted according to lineage to aid visualization. With this degree of undersampling (25%), an average experiment will only detect 58 of the 100 lentiviral tags, only 15 will be shared by two lineages, and most importantly, only one tag will be detected as shared between the three lineages. Abbreviations: E, endothelial; H, hematopoietic; M, mesenchymal; nrLAM-PCR, non-restrictive linear amplification-mediated PCR.
The issue of sampling has been well studied in the field of ecology, and has stimulated the development of various mark-recapture based statistical methods to address the “unseen species” problem. We chose to use one of these methods, the Chao2 estimator, to estimate the size of the sampling problem described above in our experiments. Whereas this estimator has been used previously in the context of viral tagging to estimate the number of tagged cells [5–7], we go further by using these estimates to determine the degree of undersampling and calculate dataset-specific expectations. The Chao2 estimator was chosen as it requires only presence/absence information for species in multiple samples taken from a population [24] and is thus appropriate for use with sequencing of nrLAM-PCR based products. The Chao2 formula uses the frequency of observing rare tags detected only once (f1) or twice (f2) to estimate the number of unsequenced tags (Supporting Information Fig. 3). We applied the Chao2 estimator to estimate the number of lentiviral tags in the parental population of the E, H and M lineages by repeatedly sampling DNA from the sorted lineages as well as from cells that could not be assigned to a lineage based on immunophenotype (“lineage negative”), which were also captured during FACS sorting. Estimates of the total tags in each hEMP transduction were subsequently used in our model to calculate expectations for tag sharing between the lineages (Supporting Information Fig. 4).
We first examined how important the consideration of sampling would be in a real experimental situation by genetically tagging a large population of hPSC, a cell population with known lineage potential (Fig. 4A). In this experiment, the pluripotent hPSC clones were tagged before mesodermal commitment, and we therefore reasoned that they should individually be capable of producing cells of all three mesodermal lineages irrespective of whether the hEMP stage through which they differentiate is multipotent. Any inability to detect lentiviral tags shared between lineages in this experiment could therefore be attributed to technical limitations of the experimental system rather than an inherent restriction of lineage potential of the target cells.
A pool of approximately 7 million hPSC was transduced and expanded briefly without selection (Fig. 4A). The polyclonal pool of hPSC was then placed into mesoderm induction conditions from which hEMP were isolated at day 3.5, and then replated onto OP9 stroma for trilineage differentiation. After 14 days, hematopoietic (H), endothelial (E), mesenchymal (M) and lineage negative (CD45−CD31−CD73−) cells were isolated as separate populations by FACS, from which genomic DNA was isolated and subjected to nrLAM-PCR and HTS. Importantly, for each lineage, 10 independent samples of the genomic DNA were taken for nrLAM-PCR, and each of these sequencing libraries incorporated a distinct sequencing index that allowed for post hoc determination of which lentiviral tags were detected in which samples. The estimated sample recovery after cell collection, lineage isolation by FACS, DNA isolation and nrLAM-PCR library preparation was 2%–9% of the initial input (Table 1), demonstrating the effect of compounded undersampling (as modeled in Fig. 3).
Table 1.
Approximation of the degree of undersampling in transduced polyclonal hPSC
| Total cell # harvested |
% of each lineage from FACS |
Cell # yield expecteda |
Cell # yield obtained from FACS |
gDNA quant. (uc378/ul)b |
vol of gDNA eluted (ul) |
total cell equivalent after gDNA extractionc |
vol/ nrLAM PCR (ul) |
# nrLAM PCR reactions |
total cell equivalent for nrLAM PCR libraries inputd |
% recovery after FACSe |
% recovery after DNA isolationf |
% recovery after library prepg |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3.10E+07 | H | 22.41% | 6.95e6 | 6.44e6 | 27600 | 100 | 1.38e6 | 1 | 10 | 1.38e5 | 92.64% | 21.44% | 2.14% |
| E | 2.78% | 8.60e5 | 8.57e5 | 11280 | 50 | 2.82e5 | 1 | 10 | 5.64e4 | 99.67% | 32.89% | 6.58% | |
| M | 0.98% | 3.04e5 | 2.26e4 | 2580 | 50 | 6.45e4 | 1.5 | 10 | 1.94e4 | 74.10% | 28.60% | 8.58% |
Cell yield expected is calculated as [Total cell# harvested from OP9 week2] æ [% of each lineage from FACS]
Absolute gDNA quantification is performed by droplet digital PCR using uc378 as a diploid reference target locus
Total cell equivalent after gDNA extraction is calculated as ([gDNA quantification (uc378/ul)] æ [volume of gDNA eluted (ul)])¼2, assuming that each diploid cell contains two copies of uc378
Total cell equivalent for nrLAM PCR libraries input is calculated as ([volume/nrLAM PCR] æ [#nrLAM PCR] æ [total cell equivalent after gDNA extraction])¼[volume of gDNA eluted (ul)]
%recovery after FACS is calculated as ([Cell yield obtain from FACS sorter]¼[Cell yield expected]) æ 100
%recovery after DNA isolation is calculated as ([total cell equivalent after gDNA extraction]¼[Cell yield obtain from FACS sorter]) æ 100
%recovery after library prep is calculated as ([total cell equivalent for nrLAM PCR libraries input]¼[Cell yield obtain from FACS sorter]) æ 100
A total of 68,772 lentiviral tags were recovered from HTS of all samples from all three lineages. 142 of these were found in all three lineages, and 3,532 were found in only two lineages. The vast majority of tags (65,098) were found in only one lineage (Fig. 4B). The Chao2 estimator was applied to these data and the resulting “lower bound” for total lentiviral tags in the transduced hEMP population was used to calculate expected tag sharing between lineages (Fig. 4A, Supporting Information Figs. 3 and 4). We compared the observed number of shared lentiviral tags to these expected values and henceforth refer to these observed-to-expected ratios as “detection scores” (Fig. 4A). The average detection score among all of the two- and three-lineage combinations was 0.7. The detection scores for tags shared between endothelial-hematopoietic lineages (EH), endothelialmesenchymal lineages (EM) and hematopoietic-mesenchymal lineages (HM) were 0.5, 1.0, and 0.5, respectively, and the detection score for tags shared among endothelial-hematopoietic-mesenchymal lineages (EHM) was 0.7 (Fig. 4C). These results from the transduction of polyclonal hPSC define the maximum expectation of the system and set a standard for comparison with datasets from our experiments with transduced mesoderm progenitors in which the existence of multipotent cells is unknown.
Spurious Tag Sharing is Rare with Lentiviral Tagging and FACS Isolation Strategy
We next designed an assay to estimate the potential for erroneously identifying common lentiviral tags between two lineages that do not share a common origin. Such false positives could occur if the vector inserted into the same site in the genome in two or more independent events [6, 7], or if cells from one lineage contaminated cells of another lineage due to errors during FACS isolation. For example, if one hematopoietic cell contaminated the endothelial population, it would yield a shared lentiviral tag incorrectly indicating a bipotent hematopoietic-endothelial clone.
To assess the magnitude of these two effects in our experimental system, three separate pools of hPSC were transduced, each with one of three lentiviral vectors expressing distinct fluorescent proteins (Fig. 4D). Mesoderm progenitors were generated and isolated from each transduced pool and differentiated in parallel cultures into hematopoietic, endothelial and mesenchymal lineages. After 2 weeks, cells from the three differentiated cultures were mixed, and specific lineages were isolated by FACS with the added requirement that each lineage express a distinct fluorescent vector marker: mCitrine+CD31+CD73+CD144+CD45− endothelial cells; mCerulean+CD73+CD31−CD45− mesenchymal cells; mStrawberry+CD45+CD31−CD73− hematopoietic cells. Because the hPSC that generated each lineage were transduced separately prior to differentiation and isolation, the differentiated lineages would not be expected to share any common tags.
When the lentiviral tags were identified from each lineage, the numbers of shared tags between EH, EM and HM lineages were 1, 6 and 23 out of a total of 5,464 tags sequenced, and no sites were shared between all three lineages (Fig. 4E). These events produced an average detection score of 0.1 (Fig. 4F). These negative control data demonstrate a clear distinction between false positive and true positive lentiviral tag sharing. False positives were predominantly seen in tags shared by the hematopoietic and mesenchymal lineages, with no false positive trilineage events (Fig. 4E, 4F). Hereafter, we use the detection scores from this experiment as a background cutoff/threshold for considering lentiviral tag sharing to be the result of biological events rather than technical artifacts.
Lentiviral Tagging Reveals Tripotent and Bipotent Progenitors Within hEMP
With the upper and lower limits of the experimental system defined, we applied the lentiviral tagging approach to interrogate the lineage potential of hEMP. hEMP were isolated at day 3.5 of mesoderm induction and then transduced with a single addition of lentiviral vector. After 12 hours, cells were washed and replated into trilineage differentiation conditions (Fig. 5A). Because lentiviral vectors continue to integrate into host DNA for at least 48 hours in culture [25–27] and because multipotent progenitors within the hEMP population might make fate decisions soon after transduction, we chose to demarcate the window of lentiviral integration in our system. To this end, we performed experiments with or without the HIV-1 integrase inhibitor raltegravir added after 12 hours of transduction, the timing of which was established using the K562 erythroleukemia cell line (Supporting Information Fig. 5).
When lentiviral tags were sequenced from hematopoietic, endothelial and mesenchymal cells isolated from tagged and differentiated hEMPs (n = 3 experiments), the detection scores were analyzed in comparison to the hPSC tagging experiments in Fig. 4. Using an extreme value test, the negative control result (Fig. 5C hollow dot) was found to deviate significantly from the detection scores from hEMP tagging (Fig. 5C black dots). This indicates that tag sharing in the experimental samples was not detected by chance.
Specifically, the detection scores of tag sharing in EHM, EH, and EM in the negative control experiment were significantly lower than in the hEMP transduction experiments (p-values 6 × 10−93, 3 × 10−6 and .04). Shared tags among all three lineages had an average detection score of 0.3 (n = 3), compared with a detection score of 0.7 for hPSC. The detection scores between EH lineages and between EM lineages were 0.6 and 0.9, similar to the upper limits defined from tagged hPSC (0.5 and 1.0, Fig. 5C yellow dots). This indicates that the hEMP differentiates through an EH and EM bipotent stage. On the other hand, the HM detection score in the negative control experiment was indistinguishable from the hEMP experiment scores (p = .1), indicating that differentiation does not proceed through an HM intermediate stage.
Addition of raltegravir to hEMP transduction decreased the total number of lentiviral tags but did not influence detection scores. By combining these methods of lentiviral tagging, HTS and quantitative analysis, we conclude that the day 3.5 hEMP population contains tripotent (EHM) progenitors that rapidly differentiate into bipotent EH and EM progenitors, and that strictly bipotent HM progenitors are not generated in this system (Fig. 5D).
Discussion
Genetic labeling via heritable genetic barcodes, transposon insertions, or retroviral integrations has long been informative in systems with low or moderate clonal diversity and extensive clonal amplification. Typically, these studies have been used to track long-term hematopoietic stem cell behavior after transplantation in either experimental models or clinical gene therapy trials [21, 28–33]. The capacity of hematopoietic stem cells to self-renew in vivo after transplantation allows hematopoiesis to be sustained by relatively few clones, particularly after the initial burst of progenitor output has disappeared. Recent intriguing studies using transposon insertions and inducible genetic labeling to track endogenous murine hematopoiesis have revealed that a far greater clonal contribution during steady state is attributable to lineage-restricted progenitors than was inferred from transplantation models [21, 34–36]. The framework we have presented extends the capabilities of these approaches to cell populations that have much greater clonal complexity and/or little clonal amplification, due to either temporal or biological constraints. Unlike barcoding methods, the high complexity labeling strategy used here does not provide quantitative information on clone size [21, 28, 30, 37], but can nonetheless be used to interrogate a population of unknown multipotency.
As a proof of concept, we examined a transient population of mesodermal progenitors derived from hPSC and demonstrated by lentiviral tagging that at least some of this population possess trilineage (hematopoietic, endothelial, and mesenchymal) potential. The finding that trilineage detection scores from labeling of mesoderm progenitors were lower than those from labeling of pluripotent cells, presumably reflects the shorter timeframe for clonal amplification from the progenitors and/or the possibility that tripotent progenitors are only a rare subset of the total progenitor population.
Unlike genetic tagging of the pluripotent hESC, in which shared lentiviral tags were readily detected between any two given downstream lineages, genetic tagging of mesoderm progenitors revealed significant shared tags only between all three lineages (EHM) or between either endothelial and hematopoietic (EH), or endothelial and mesenchymal (EM) lineages. The level of lentiviral tag sharing between the hematopoietic and mesenchymal (HM) lineages was indistinguishable from the negative control (i.e., false positive events). This data lead us to conclude that hematopoietic and mesenchymal cells, while sharing a common tripotent progenitor with endothelial cells, do not branch off from a common bipotent (HM) progenitor but rather arise from mutually exclusive pathways downstream from the tripotent EHM.
Studies based on dual-lineage colony formation assays from others independently confirm the lineage bifurcation pattern observed in our data. Modification of standard hematopoietic progenitor colony forming assays has led to identification of a common precursor for hematopoietic and endothelial cell (hemangioblast) [18, 19, 38] as well as for mesenchymal stem and endothelial cells (mesenchymangioblast) [13]. However, these reports did not identify the earlier tripotent stage of mesoderm because of the technical limitations of the clonal culture systems.
An additional insight from our findings is that endothelial specification occurs through two mutually exclusive pathways. It was recently shown that the hemogenic endothelium and the arterial vascular endothelium derived from hESC represent non-overlapping populations [39]. However, the developmental origin of these two types of endothelium was not explored. Further studies will be needed to elucidate if endothelium from different clonal origins revealed in our system inherit distinct functional fates.
We observed fewer shared lentiviral tags than initially expected from genetically tagged hPSC and found that this discrepancy can be explained by undersampling in this experimental system. Various mark-recapture approaches have been adopted in the gene therapy setting to estimate the size of the gene-corrected cell pool [5–7]. We applied one of these methods, the Chao2 estimator, in a novel way not only to estimate the number of genetically tagged cells, but also to estimate and correct expectations for the magnitude of undersampling inherent in our HTS-based clonal tracking studies. Of note, even after use of the Chao2 method to estimate the number of undetected lentiviral tags within these large populations, the calculation of expected shared tags will most likely still be higher than observations. The Chao2 estimator for unseen species only guarantees a lower bound for the total number of species, meaning that the true number of species is always greater than or equal to the estimate. The lower bound of expected events also assumes a situation where species have a perfectly uniform abundance distribution, and in most experimental systems, clones amplify in a non-uniform fashion. Furthermore, even though the nrLAM-PCR strategy used for HTS library preparation is less biased than previous methods, it still cannot amplify all lentiviral tags uniformly, and current technologies cannot sequence and map all lentiviral tags with uniform efficiency. The more different the species abundance distribution is from uniform, the lower the Chao2 estimate will be relative to the true species count. In our model, underestimating the total number of species (N̂2) leads to an over-estimation of our sampling (N2/N̂), which in turn yields a greater expected number of shared lentiviral tags (N̂ 1,2) in our calculations and a lower detection score.
Additionally, our model assumes that clones proliferate sufficiently to populate all three lineages with their progeny. The number of cellular divisions that occur between genetic labeling and clonal fate decision is not known, and it is therefore possible that some clones do not populate all target lineages even though they have the biological potential to do so, or that a clone that initially populates a lineage is extinguished before the end of the experiment. Notably, this assumption is also problematic when analyzing the lineage output of single cell cultures. A more thorough understanding of these technical and biological effects would extend the improvements in the interpretation of genetic labeling datasets made in this study.
Conclusion
In summary, our clonal tracking approach has revealed the existence of tripotent mesodermal progenitors derived from pluripotent stem cells and the importance of consideration of sampling during analysis. As genetic labeling strategies and transplantation studies continue to become more efficient and sensitive, we anticipate that the consideration of sampling will become increasingly essential for the accurate interpretation of results. We believe that the quantitative framework presented here represents significant progress toward defining and addressing this need. Moreover, our strategy allows current clonal tracking methods to be extended to experimental systems in which only limited clonal amplification is possible, whether because of limited time for clonal expansion or because clones are actively transitioning and committing into different cellular fates at the time of genetic labeling. We propose that this approach also has potential applications beyond vector integration studies, for example, in the tracking of subclones during the evolution of leukemia and other malignancies.
Supplementary Material
Significance Statement.
We used lentiviral tagging and high throughput sequencing to examine, at a clonal level, the lineage potential of embryonic mesoderm progenitors (EMP) as they differentiate from human pluripotent stem cells. Our data demonstrate that EMP, which represent the in vitro counterparts of the primitive streak, contain not only the previously reported bipotent (hemato-endothelial and mesenchymal-endothelial) progenitors but also multipotent progenitors with differentiation potential of all three lineages. These studies highlight an underappreciated challenge in the clonal tracking of complex populations with limited self-renewal, and offer a mathematical framework to more accurately estimate the important effect of undersampling in such studies.
Acknowledgments
This work was supported by the UCLA BSCRC and by the following external grants: CIRM Basic Biology Award (RB3-05217) (GMC, DBK and DE); CIRM Training Grant (TG2-01169) (CJC); International Fulbright Science and Technology Award (CJC); Philip J. Whitcome Predoctoral Fellowship from the UCLA Molecular Biology Institute (ARC); Ruth L. Kirschstein National Research Service Award (GM007185) (ARC); NIH K01AR061415, DOD grant OR120161 and CIRM Basic Biology Award (RB5-07230) (DE). We thank Felicia Codrea and Jessica Scholes (UCLA BSCRC Flow Cytometry core), the BSCRC High Throughput Sequencing Core and the UCLA Clinical Microarray Core for their technical support, and Dr. Jerome A. Zack for providing raltegravir.
Footnotes
Author Contributions
C.J.C., A.R.C.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; G.R.L.: collection and/or assembly of data, data analysis and interpretation, manuscript writing; D.E.: conception and design; Y.Z., C.B.H.: collection and/or assembly of data; D.C.: data analysis and interpretation; M.P., D.B.K.: conception and design, data analysis and interpretation; G.M.C.: conception and design, manuscript writing, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
See www.StemCells.com for supporting information available online.
References
- 1.Mendjan S, Mascetti VL, Ortmann D, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Davis RP, Ng ES, Costa M, et al. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 4.Evseenko D, Zhu Y, Schenke-Layland K, et al. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci USA. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang GP, Berry CC, Malani N, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 8.Lois C, Hong EJ, Pease S, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin K, Urbinati F, Romero Z, et al. Enrichment of human hematopoietic stem/progenitor cells facilitates transduction for stem cell gene therapy. Stem Cells. 2015;33:1532–1542. doi: 10.1002/stem.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper AR, Patel S, Senadheera S, et al. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J Virol Methods. 2011;177:1–9. doi: 10.1016/j.jviromet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paruzynski A, Arens A, Gabriel R, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B, Schatz MC, Lin J, et al. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vodyanik MA, Yu J, Zhang X, et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi KD, Vodyanik MA, Togarrati PP, et al. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slukvin II. Hematopoietic specification from human pluripotent stem cells: Current advances and challenges toward de novo generation of hematopoietic stem cells. Blood. 2013;122:4035–4046. doi: 10.1182/blood-2013-07-474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy M, Awong G, Sturgeon CM, et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Choi K, Kennedy M, Kazarov A, et al. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy M, D’Souza SL, Lynch-Kattman M, et al. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohn LA, Hao QL, Sasidharan R, et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat Immunol. 2012;13:963–971. doi: 10.1038/ni.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Candotti F, Shaw KL, Muul L, et al. Gene therapy for adenosine deaminase-deficient severe combined immune deficiency: Clinical comparison of retroviral vectors and treatment plans. Blood. 2012;120:3635–3646. doi: 10.1182/blood-2012-02-400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero Z, Urbinati F, Geiger S, et al. β-globin gene transfer to human bone marrow for sickle cell disease. J Clin Invest. 2013;123:3317–3330. doi: 10.1172/JCI67930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel R, Eckenberg R, Paruzynski A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 24.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- 25.Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 27.Munir S, Thierry S, Subra F, et al. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology. 2013;10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naik SH, Perié L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 29.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 30.Gerrits A, Dykstra B, Kalmykowa OJ, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 31.Brenner S, Ryser MF, Choi U, et al. Polyclonal long-term MFGS-gp91phox marking in rhesus macaques after nonmyeloablative transplantation with transduced autologous peripheral blood progenitor cells. Mol Ther. 2006;14:202–211. doi: 10.1016/j.ymthe.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Kim N, Presson AP, et al. Dynamics of HSPC repopulation in nonhuman primates revealed by a decade-long clonal-tracking study. Cell Stem Cell. 2014;14:473–485. doi: 10.1016/j.stem.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken MN, Gschweng EH, Nair-Gill E, et al. Long-term in vivo monitoring of mouse and human hematopoietic stem cell engraftment with a human positron emission tomography reporter gene. Proc Natl Acad Sci USA. 2013;110:1857–1862. doi: 10.1073/pnas.1221840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busch K, Klapproth K, Barile M, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 36.Naik SH, Schumacher TN, Perié L. Cellular barcoding: A technical appraisal. Exp Hematol. 2014;42:598–608. doi: 10.1016/j.exphem.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Lu R, Neff NF, Quake SR, et al. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber TL, Kouskoff V, Fehling HJ, et al. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 39.Ditadi A, Sturgeon CM, Tober J, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.