Abstract
The architecture of polycations plays an important role in both gene transfection efficiency and cytotoxicity. In this work, a new polymer, sunflower poly(2-dimethyl amino)ethyl methacrylate) (pDMAEMA) were prepared by atom transfer radical polymerization (ATRP) and employed as nucleic acid carriers compared to linear pDMAEMA homopolymer and comb pDMAEMA. The sunflower pDMAEMAs showed higher IC50, greater buffering capacity, and stronger binding capacity towards plasmid DNA than their linear and comb counterparts. In vitro transfection studies demonstrated that sunflower pDMAEMAs exhibited high transfection efficiency as well as relatively low cytotoxicity in complete growth medium. In vivo gene delivery by intraventricular injection to the brain showed that sunflower polymer delivers plasmid DNA more effectively than comb polymer. This study provides a new insight into the relationship between polymeric architecture and gene delivery capability, and as well as a useful means to design potent vectors for successful gene delivery.
Keywords: polycations, sunflower polymers, polymeric architecture, nucleic acid delivery
1. Introduction
Nucleic acid therapies are showing promise for a myriad of both inherited and acquired afflictions.[1] The recent approval of two such therapies, Glybera (a gene therapy) and Kynamro (an antisense therapy) further support the clinical potential of this macromolecular class of drugs. Since the nucleic acid drugs act intracellularly, but are membrane impermeable, efficient drug action generally requires the use of a carrier to assist in delivery.[2] Two classes of carriers, viral-based and nonviral-based, are typically employed to facilitate nucleic acid delivery. While viral carriers, such as adeno-associated virus (AAV) used in Glybera, have seen more early clinical success, nonviral carriers, such as polycations, are being actively developed due to their comparatively favorable safety profiles.[3] Importantly, polycations can be produced relatively affordable at large scale, which is crucial for clinical translation. However, the in vivo transfection efficiency of polycation systems remains relatively low compared to their complex viral counterparts. Therefore, a significant need exists for new materials that achieve efficient delivery in vivo while retaining the safety and manufacturing advantages of synthetic systems.
Many of the polycations used to date, e.g. poly((2-dimethyl amino)ethyl methacrylate) (pDMAEMA),[4] oligoamine modified poly(glycidyl methacrylate),[5] poly(L-lysine),[6] chitosan[7] and poly(β-amino ester)s,[8] have linear structures. However, advanced polymeric architectures have been shown in several cases to offer advantages over linear polymers. For example, previous work by Wang et al,[9] Xu et al.,[10] Patrickios et al.,[11] and our group[12] have revealed the benefits of highly branched, grafted, and cyclic pDMAEMA structures on gene transfer and cell survival compared to linear pDMAEMA structure. Schallon et al. also demonstrated that nucleic acid delivery complexes using branched and star pDMAEMA and polyethylenimine polymers traffic differently inside the cell compared to linear polymers, further emphasizing the role of polymeric structure on gene transfer.[13] Inspired by this cumulative work, we synthesized a new polycation nanostructure with macrocyclic brush architecture,[14] termed “sunflower polycation”. Sunflower polymers were synthesized using a cyclic multimacroinitiator to initiate polymerization of polymer “petals”. We evaluated these materials for gene transfer in comparison to linear and comb materials and report here that sunflower polymers offered the best delivery efficiency to cultured cells in vitro and for gene transfer to mouse brain in vivo through intraventricular injection. Furthermore, specific sunflower structures are able to resist negative serum effects on transfection that is commonly observed with polycation/nucleic acid complexes. Sunflower polymers are therefore promising nanoarchitectures for future developments as gene and drug delivery materials.
2. Results and Discussion
For these studies, we selected pDMAEMA as a model polymer. Since its initial reported use as a gene carrier by the Hennink group in 1996,[15] pDMAEMA has been used as a platform for probing the structure-function relationship of polycations as nucleic acid delivery vehicles, particularly due to the ability to control and define polymer properties using controlled living radical polymerization (CLRP) techniques.[16] The positive correlation between its molecular weight and both transfection efficiency and cytotoxicity has been intensively investigated.[17] Therefore, a cyclized and brominized poly(2-hydroxyethyl methacrylate) (pHEMA) multimacroinitiator was used to synthesize sunflower pDMAEMA by ATRP (atom transfer radical polymerization) of DMAEMA (Scheme 1). For comparison, a linear pDMAEMA290 (L290) was synthesized by ATRP and comb pDMAEMAs were synthesized from the same brominized pHEMA without cyclization.
Scheme 1.
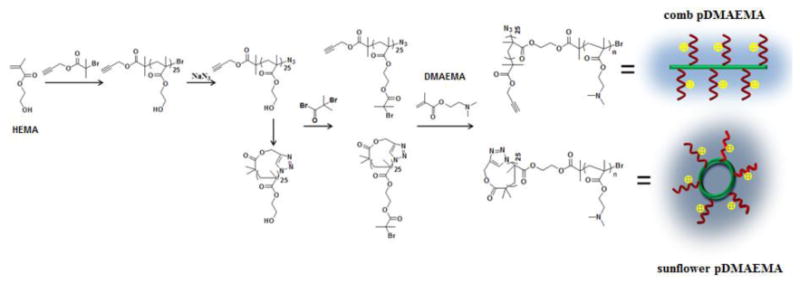
Synthetic routes of comb and sunflower pDAMEMA.
For preparation of multimacroinitiators, cyclic p(2-hydroxylethyl methacrylate) (c-pHEMA) was synthesized by the combination of ATRP and inter-chain click coupling.[18] All polymers were characterized by 1H NMR, FT-IR and GPC. A new signal attributed to the proton in the triazole at 8.5 ppm appeared in NMR spectrum (Figure S1A), and the characteristic band of the azide group in FT-IR spectrum (around 2100 cm-1) disappeared after cyclization (Figure S1B), both of which confirmed the successful inter-chain click reaction between alkynyl and azide group and generation of cyclic polymer. Furthermore, the GPC elution trace of c-pHEMA showed a detectable right-shift towards lower molecular weight compared to its linear counterpart due to the decreased hydrodynamic volume of cyclic polymers (Figure S1C). The <G> value, the apparent peak molar mass (Mp) of c-pHEMA to that of linear p(2-hydroxylethyl methacrylate) (l-pHEMA), was about 0.86, which is within the reported results of other cyclic polymers (0.70-0.97).[19] Thus, all the results confirmed successful synthesis of c-pHEMA. The multimacroinitiators, cyclic-poly(2-(2-bromoisobutyrnyloxy)ethyl methacrylate (c-pHEMA-Br) and linear poly(2-(2-bromoisobutyrnyloxy)ethyl methacrylate (l-pHEMA-Br), were then synthesized by esterification between the HEMA hydroxyl side group and α-bromoisobutyryl bromide, and the complete conversion was confirmed by 1H NMR (Figure S2).
The sunflower and comb polymers were synthesized by ATRP of DMAEMA using the c-pHEMA-Br and l-pHEMA-Br multimacroinitiators, respectively. Polymerization from the locally-concentrated initiating sites of a multimacroinitiator can result in undesired coupling termination during polymerization, yielding cross-linked structures. To minimize this occurrence, a small amount of CuBr2 was added in the catalyst system and a low monomer concentration (0.5 M) was used to reduce the polymerization kinetics. Under such conditions, comb and sunflower pDMAEMAs with different degree of polymerizations (DPs) were prepared with relatively low monomer conversion (< 25%) after polymerization for 20 h, and the polydispersity indices (PDIs) determined by GPC analysis (Table 1) ranged from 1.4 to 1.8 (Table 1). The relatively narrow PDIs indicated negligible crosslinking during polymerization. The average DPs of pDMAEMA were calculated assuming equivalent pDMAEMAs side chain lengths, and the amount of the repeated units per arm for the six polymers is summarized in Table 1. We thus denoted the six comb and sunflower polymers, l-pHEMA25-g-(pDMAEMA12)25, c-pHEMA25-g-(pDMAEMA11)25, l-pHEMA25-g-(pDMAEMA16)25, c-pHEMA25-g-(pDMAEMA16)25, l-pHEMA25-g-(pDMAEMA21)25, c-pHEMA25-g-(pDMAEMA22)25, as C-12, S-11, C-16, S-16, C-21 and S-22, respectively, where “C” and “S” denote the respective comb or sunflower structure, and the number indicates the average DP of DMAEMA units for each petal. The representative 1H NMR spectrum of sunflower pDMAEMA is shown in Figure S3. Comb and sunflower polymers were synthesized with similar DPs for a better comparison, but comb polymers did show narrower PDIs, likely due to the bigger hydrodynamic volume and lower density of initiating sites.
Table 1.
Characterizations of comb and sunflower pDMAEMAs.
| code | DP/arma | Mnb (kDa) | PDIb | IC50 (mg/L) | |
|---|---|---|---|---|---|
| l-pHEMA25-g-(pDMAEMA12)25 | C-12 | 12 | 192 | 1.48 | 10.4 ± 1.0 |
| l-pHEMA25-g-(pDMAEMA16)25 | C-16 | 16 | 263 | 1.67 | 8.5 ± 0.4 |
| l-pHEMA25-g-(pDMAEMA21)25 | C-21 | 21 | 371 | 1.75 | 8.1 ± 0.5 |
| c-pHEMA25-g-(pDMAEMA11)25 | S-11 | 11 | 176 | 1.44 | 13.4 ± 0.6 |
| c-pHEMA25-g-(pDMAEMA16)25 | S-16 | 16 | 287 | 1.81 | 11.2 ± 0.6 |
| c-pHEMA25-g-(pDMAEMA22)25 | S-22 | 22 | 413 | 1.88 | 9.3 ± 0.1 |
Determined by 1H NMR;
Obtained by GPC;
The sizes of the resulting comb and sunflower pDMAEMAs were measured by dynamic light scattering (DLS). As shown in Figure S4, polymer size ranged from 4 to 12 nm, and increased with the increasing of polymer molecular weight. Moreover, sunflower polymers showed reduced size compared to their comb counterparts, consistent with their expected compact morphology.
Polycations, such as pDMAEMA, that contain protonatable amines can improve cytosolic delivery of nucleic acid cargo through the “proton sponge effect”, whereby the buffering of acidifying endosomes by pH-sensitive polymers combined with increased osmotic pressure induces endosomal destabilization and cargo release.[20] The buffering capacities (BCs) of comb and sunflower pDMAEMAs were examined by acid-base titration, and defined as the μmol H+/mg polymer required to decrease the pH value of a polymer solution from 7.4 to 5.0 (Figure 1).[21] The buffering capacity increased with increasing molecular weight of polycation. Intriguingly, sunflower pDMAEMAs showed higher buffering capacity than comb polycations, which may promote endosomal escape of their corresponding polyplexes. However, their BCs were still lower than that of PEI (Figure S5). The star-like structure of sunflower polymers may result in reduced pKa of the tertiary amines closest to the core, as reported previously for higher generation PAMAM dendrimers.[22]
Figure 1.
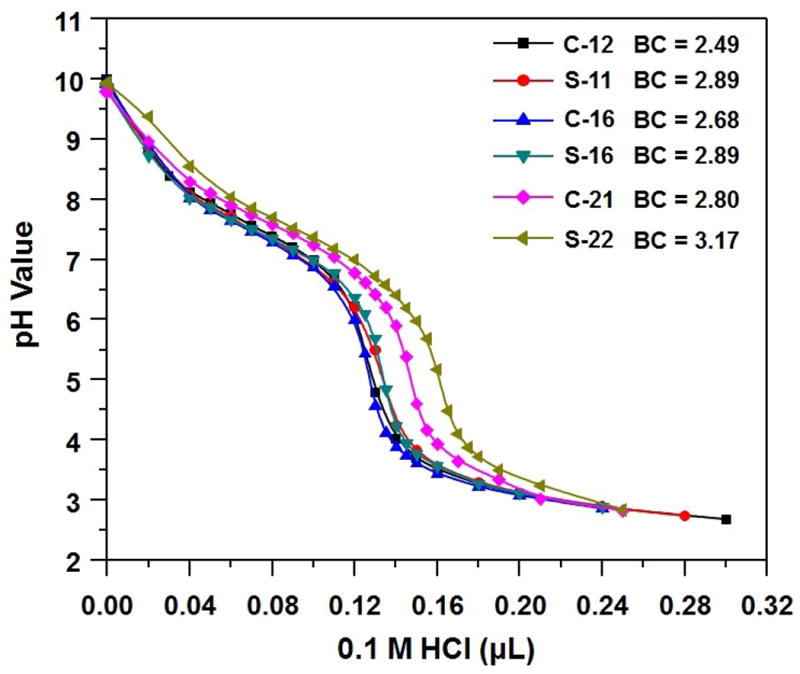
Titration curves of comb and sunflower pDMAEMAs.
Since cytotoxicity of biomaterials is a key factor to be considered in biomedical applications, we examined whether the polymeric architecture affected cell survival. Our previous study revealed that cyclic pDMAEMAs demonstrated improved cell biocompatibility compared to their linear counterparts.12 Therefore, an MTS assay was performed to determine the metabolic activity of mammalian cells treated with the polymers at various concentrations. The IC50 (polymer concentration at 50% cellular inhibition) values of sunflower polymers (9.1-13.4 mg/L) to HeLa cells trended higher than those of the corresponding comb polymers (8.1-10.4 mg/L) and also higher than the industry standard branched polyethylenimine (bPEI 25 kDa, IC50 ∼ 4.6 mg/L).[23] Moreover, L290 with similar DP of DMAEMA as C-12 and S-11 exhibited lower IC50 (9.7 ± 0.6 mg/mL). Thus, sunflower polymers show the least cytotoxicity of all polycations tested. The smaller and constrained size of the sunflower polymers may reduce cell membrane interaction compared to the more elongated comb polymers.
Next, polymer/DNA nanoparticles were prepared by complexing plasmid DNA (pDNA) with polycations via electrostatic interactions, forming condensed structure called “polyplexes”. The plasmid DNA (pDNA) binding ability of comb and sunflower pDMAEMAs was assessed by an electromobility shift assay (EMSA) via agarose gel electrophoresis, which showed that all the polymers could effectively condense pDNA by an amine/phosphate (N/P) charge ratio of 2 (Figure S6). The morphologies of polyplexes were imaged by transmission electron microscope (TEM) at an N/P ratio of 5 (Figure 2A). All the polyplexes were compact structures with relatively uniform spherical shape. Furthermore, two size populations were observed, one with diameter ∼ 60 nm and the second with diameter ∼ 100 nm. The longer “petal” lengths (higher DP) in the polycation correlated with lower fractions of the larger sized particle population in the formulated polyplexes, indicating enhanced condensing capability. In addition, the particle sizes were also characterized by dynamic light scattering (DLS) and the hydrodynamic diameter of all the complexes ranged from 115 to 135 nm (Figure 2B), which is within the desired range for cellular endocytosis,[24] and the size of the S-16 and S-22 polyplexes were smaller than those formed by respective comb pDMAEMAs. Polyplexes formed by S-16 and S-22 also showed lower zeta potential compared to each counterpart, also suggesting more efficient neutralization and compaction of pDNA by sunflower polymers (Figure S7).
Figure 2.
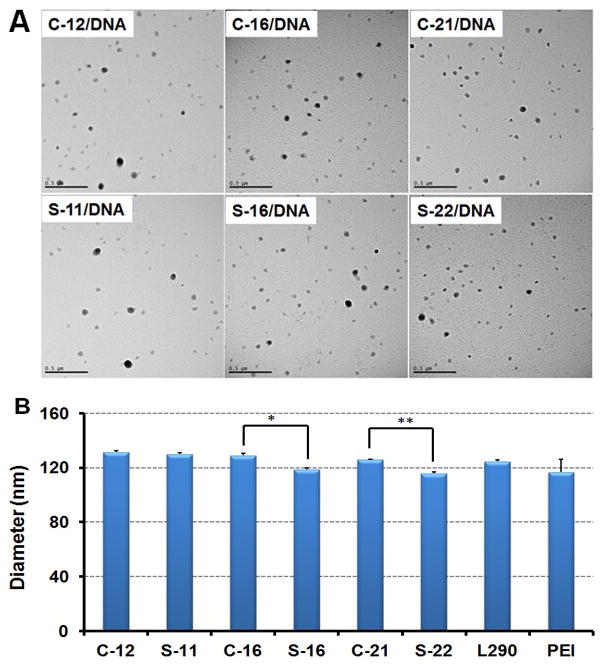
(A). TEM images of comb and sunflower pDMAEMA polyplexes formed at N/P = 5 (scale bar: 500 nm). (B). Hydrodynamic diameters of polyplexes formed at N/P = 5. Data are shown as mean ± SD (n = 3; student's t test, *p <0.01, **p <0.001).
To assess the gene transfer ability, linear, comb, and sunflower polymers were formulated with the luciferase reporter plasmid at different N/P (polymer amine to DNA phosphate) ratios for gene delivery evaluation in the human cervical cancer cell line HeLa. The transfection efficiency increased with the N/P ratio from 3 to 5 (Figure 3A). However, compromised luciferase activity was observed with N/P ratio of 7, likely due to the cytotoxicity caused by the excess, free polycation (cell viability< 60%) (Figure S8). We also employed branched PEI (25 kDa) as comparison, which was previous determined to show optimized transfection efficiency at N/P ratio of 5.[25] As shown in Figure 3A, sunflower polymers mediated improved transfection efficiency compared to their corresponding comb counterpart with N/P ratios of 5 and 7, and this trend was more obvious at the higher N/P ratio. At the optimized N/P ratio of 5, S-22 exhibited the highest luciferase activity of all polymers, and transfected with 3.5-fold, 6.5-fold and 17.4-fold higher efficiencies than polyplexes formed with C-21, L290 and bPEI, respectively. Furthermore, cell viability remained high (>75%) for all sunflower and comb pDMAEMA polyplexes (Figure S8). Thus, the S-22 sunflower polymer may be promising for in vivo gene transfer applications.
Figure 3.
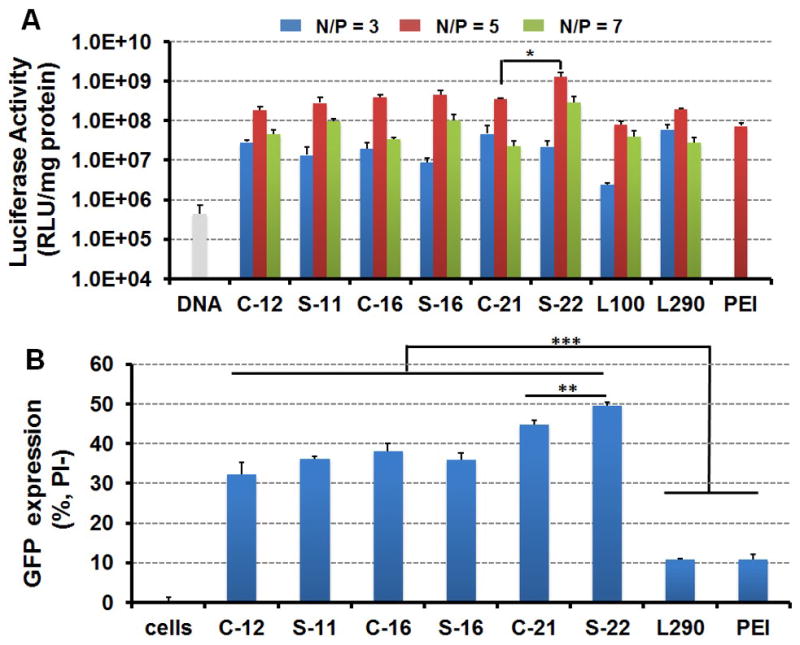
(A) Transfection efficiency of polyplexes to HeLa cells with different N/P ratios (B) GFP expression in live cells (PI negative) treated by different polyplexes analyzed by flow cytometry. Data are shown as mean ± SD (n = 3; student's t-test, *p < 0.05, **p < 0.01, ***p < 0.001).
The green fluorescent protein (GFP) reporter gene was also delivered by pDMAEMA based polycations to HeLa cells in serum-containing media to evaluate transfection (GFP+) and cell viability (propidium iodide, PI-) under optimized N/P ratio of 5 by flow cytometry. Polyplexes formed by comb and sunflower pDMAEMAs had a much higher percentage of cells transfected (30-50% GFP+/PI-) compared with L290 and bPEI polyplexes (∼10% GFP+/PI-), and the S-22 sunflower polymer again showed the highest percentage of transfected of cells (Figure 3C). Moreover, reduced cytotoxicity was also observed in cells treated with sunflower polymers compared to those treated with comb polymers (Figure S9).
To better understand the enhanced transfection efficiency of S-22 polyplexes versus C-21 polyplexes, we assessed the intracellular distribution polyplexes by TEM imaging of HeLa cells transfected for 1 h (Figure 4A-D and Figure S10). Polyplexes were observed being endocytosed (Figure 4B and Figure S10B), within in endosomes (Figure 4C and Figure S10C) and free in the cytoplasm (Figure 4D and Figure S10D). However, no obvious differences were observed in intracellular distribution between comb and sunflower polyplex formulations. Therefore, polyplex unpackaging was assessed by YOYO-1 assay in the presence of anionic biomolecules. The YOYO-1 fluorescence is quenched in compact pDNA such as in polyplexes; however, the fluorescence is recovered as the polyplexes and DNA unpackages. Ideal gene carriers need to strongly condense pDNA and protect pDNA before cellular uptake while also releasing the payload intracellularly, e.g. by competitive displacement, for plasmid expression. Results from the YOYO-1 assay suggest that the sunflower polymer architectures may provide the desired balance between extracellular stability and intracellular DNA release. In water, L290 polyplexes are not tightly complexed, which may leave the nucleic acid cargo susceptible to extracellular nucleases (Figure 4E). In contrast, in the presence of a biological polyanion to trigger competitive displacement (heparin sulfate), S-16 and S-22 polyplexes show preferential unpackaging compared to bPEI polyplexes and polyplexes formed using comb polymer analogues. Thus, S-22 likely provides the highest transfection of the tested polymers due to its efficient polyplex condensation for nuclease protection of DNA, high buffering capacity for promoting endosomal release, and ability to unpackage and release DNA cargo in the presence of competitive polyanions.
Figure 4.
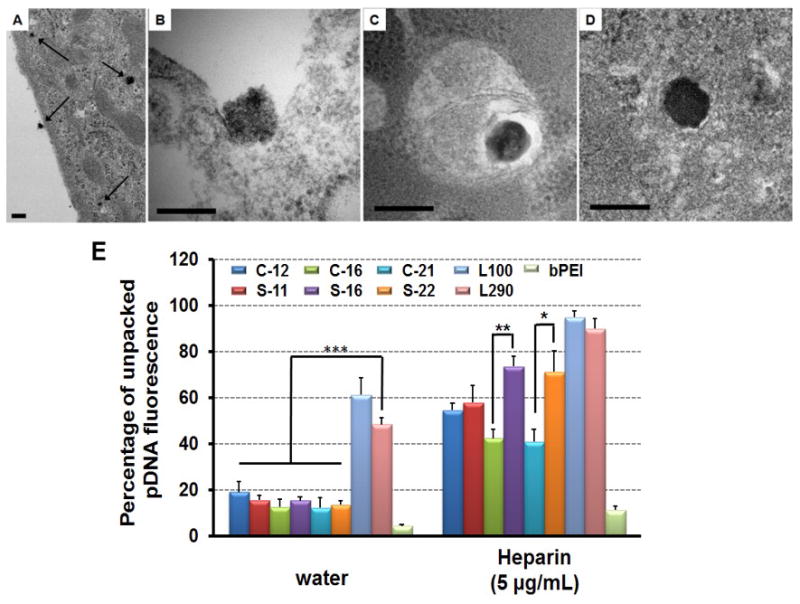
Transmission electron microscopy of polyplexes form by S-22 interacting with HeLa cells, including overview (A), endocytosis (B), entrapped in endosome (C) and locating in cytoplasm (D). HeLa cells were treated with polyplexes for 1 h prior to fixation and preparation for TEM. Scale bar indicates 200 nm. (E) Unpacking study of polyplexes (N/P = 5) in the absence and presence of heparin sulfate (5 μg/mL). Data are shown as mean ± SD (n = 3; student's t-test, *p < 0.05, **p < 0.01, ***p < 0.001).
Based on the promising in vitro transfection results obtained using S-22, we next evaluated S-22 versus C-21 as a vehicle for in vivo gene transfer to the central nervous system of C57/B16 mice. S-22 and C-21 polyplexes (N/P=5 and N/P=10) containing the luciferase reporter gene were therefore injected stereotactically into the right lateral ventricle of mice and brain tissues harvested two days after injection for analysis. S-22 polyplex-treated mice had statistically significant higher luciferase expression (1.8 and 2.6-fold higher luciferase activity at N/P=10 and N/P=5, respectively) in the right brain compared to the mice treated with C-21 at both charge ratios (Figure 5). In addition, all formulations were well tolerated and no significant gross morbidities or mortalities were observed after injection. For comparison, polyplexes formed using our previously reported and optimized block-statistical copolymer, PCL-SS-P[(GMATEPA)-st-OEGMA], for comparison, which showed promising delivery efficiency in vivo.[5b, 26] S-22 mediated similar luciferase activity compared to statistical polymer, indicating outstanding in vivo gene transfer ability.
Figure 5.
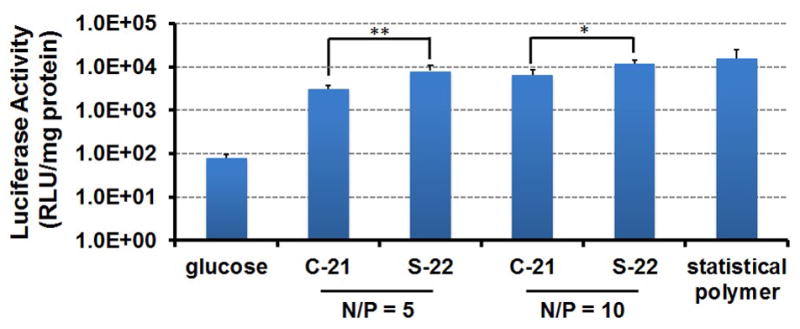
In vivo luciferase plasmid delivery into mouse brains two days after intraventricular injection using glucose, luciferase plasmid complexes formed from PCL-SS-P[(GMATEPA)-st-OEGMA] (block-statistical polymer), comb (C-21) and sunflower pDMAEMA (S-22) at N/P 5 and 10 (n=4-5; student's t-test, *p<0.05, **p<0.01).
3. Conclusion
In summary, we have synthesized and evaluated a new polycation nanoarchitecture, “sunflower” polymers, for its gene transfer ability in comparison to comb and linear polymers. The sunflower polymers offered several advantages over comb and linear polymers: higher buffering capacity, less cytotoxicity, smaller particles, stronger serum-resistance, and, perhaps most importantly, higher gene transfection capability. Sunflower polymers are therefore a promising new architecture for applications in gene and drug delivery.
4. Experimental Section
Materials
2-Hydroxyethyl methacrylate (HEMA) and 2-(dimethylamino)ethyl methacrylate (DMAEMA) were purchased from Sigma-Aldrich. The monomers were purified by passing through a column filled with basic alumina to remove the inhibitor prior to polymerization. α-Bromoisobutyryl bromide (98%), copper (I) bromide (CuBr, 99.999%), N,N,N′,N″,N″-pentamethyldiethylenetriamine (PMDETA, 99%), anisole (99.7%) and propargyl alcohol (99%) were purchased from Sigma-Aldrich and used without further purification. ATRP initiator, propargyl 2-bromoisobutyrate (PBrIB) was synthesized as our previous work.[12] Endotoxin-free plasmid pCMV-Luc (Photinuspyralis luciferase under control of the cytomegalovirus (CMV) enhancer/promoter) was produced with the Qiagen Plasmid Giga kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. YOYO-1 iodide was purchased from Invitrogen (Carlsbad, CA).
Characterization
1H NMR spectra were recorded on a Bruker AV 300 (Bruker Corporation, Billerica, MA) nuclear magnetic resonance (NMR) instrument in deuterated dimethyl sulfoxide (DMSO-d6) or deuterated chloroform (CDCl3). Fourier transform infrared (FT-IR) spectra were recorded on a Bruker Vector 33 FTIR spectrometer. Samples were pressed into potassium bromide (KBr) pellets for measurements. Gel permeation chromatography (GPC) was used to determine molecular weight and polydispersity (Mw/Mn, PDI) of the prepared polymers. SEC Tosoh TSK-GEL R-3000 and R-4000 columns (Tosoh Bioscience, Montgomeryville, PA) were connected in series to a Agilent 1200 series (Agilent Technologies, Santa Clara, CA), refractometer Optilab-rEX and triple-angle static laser light scattering detector miniDAWN TREOS (Wyatt Technology, Santa Barbara, CA). HPCL-grade N,N-dimethylformamide (DMF) containing 0.1 wt% LiBr at 60 °C was used as the mobile phase at a flow rate of 1 mL/min.
Synthesis of cyclic p(2-hydroxyethyl methacrylate) (c-pHEMA)
Cyclic p(2-hydroxyethyl methacrylate) was synthesized by three steps. First, alkylnyl-pHEMA-Br was prepared by typical ATRP. HEMA (2.74 g, 21.1 mmol), PBrIB (165.2 mg, 0.80 mmol) and PMDETA (170 μL) were dissolved in the mixture of DMF and 2-propanol (v/v, 9/1, 3.5 mL), and the solution was purged with nitrogen for 10 min to remove oxygen in the system. Then CuBr (116 mg, 0.80 mmol) was added quickly under the protection of nitrogen flow. The reaction was placed in an oil bath (60 °C). After 30 min, the polymerization was stopped through the exposure of the reaction solution in air for 1 h. The solution was diluted with DMF, and then directly subjected to dialysis against distilled water to remove copper catalyst and unreacted monomer. Alkylnyl-pHEMA-Br was obtained after lyophilization with yield of 85%.
Secondly, alkylnyl-pHEMA-Br (2.0 g, 0.6 mmol) and sodium azide (0.8 g, 12 mmol) were dissolved in the mixed solution of DMF and water (20 mL, v/v, 4/1), and the reactive solution was placed in an oil bath at 45 °C for two days. The azide terminated polymer, alkylnyl-pHEMA-N3 (l-pHEMA) was obtained after dialysis followed by lyophilization. Yield: 90%.
Cyclic pHEMA was synthesized by the intra-molecular click reaction as reported by Grayson. 31 In a typical procedure, 1 L of DMF was degassed with nitrogen in a three-neck flask for an hour, and then CuBr (430 mg, 3.0 mmol) and PMDETA (625 μL, 3.0 mmol) were added under the protection of nitrogen flow. Afterwards, a solution of alkylnyl-pHEMA-N3 (0.5 g, 0.15 mmol) in degassed DMF (10 mL) was added to the catalyst solution via a syringe pump at a rate of 0.4 mL/h. The reaction was carried out in a nitrogen atmosphere at 100 °C for 24 h. After the addition of the polymer solution, the reaction was kept for additional 24 h under same condition. At the end of the cyclization, DMF was removed by vacuum evaporation, and the concentrated residue was transferred to the dialysis tube (MWCO: 3.5 kDa, Fisher Scientific) for dialysis against distilled water for two days. The resulting cyclic polymer, c-pHEMA was obtained after freeze-drying with a yield of 90%.
Bromination of l-pHEMA and c-pHEMA
Brominized l-pHEMA (l-pHEMA-Br) and c-pHEMA (c-pHEMA-Br) were prepared by the same procedure. Typically, c-pHEMA (0.1 g, 0.77 mmol) and TEA (386 μL, 2.77 mmol) were dissolved in 5 mL of DMF, and the mixture was stirred in an ice bath for 0.5 h. Then α-bromoisobutyryl bromide (285 μL, 2.31 mmol) was added dropwise during 15 min. The mixture was kept stirring in an ice bath for additional 30 min and further 24 h in room temperature. The reaction was stopped by pouring into cold water, and the precipitate was collected by centrifuging for 10 min at 5000 rpm. The solid was washed three times with water followed by lyophilization. The brominized polymer, c-pHEMA-Br was obtained as a light yellow powder with yield 95%.
Synthesis of l-pHEMA-g-pDMAEMA (comb pDMAEMA) and c-pHEMA-g-pDMAEMA (sunflower pDMAEMA)
Comb and sunflower pDMAEMA with different degree of polymerizations (DPs) were prepared by ATRP using brominized pHEMA as multimacroinitiator in anisole. Taking synthesis of sunflower pDMAEMA as an example, the procedure was briefly described as followed: c-pHEMA-Br (20 mg, 0.072 mmol –Br), DMAEMA (604 μL, 3.6 mmol), CuBr2 (1.6 mg, 0.0072 mmol), DMF (80 μL, used as internal reference) were dissolved in anisole (7.2 mL), and the mixture was degassed with nitrogen for 10 min, followed by the addition of CuBr (10.3 mg, 0.072 mmol) under the protection of insert atmosphere. Then the reaction was placed in an oil bath at 60 °C for 16 h. The polymerization was quenched by exposing the solution to air, and the mixture was precipitated in to cold hexane. After centrifuging, the product was dissolved in methanol, and dialyzed against distilled water for 2 days. Sunflower pDMAEMA was obtained after freeze-drying with yield of 20%. The other polymers with different DPs were prepared applying the same procedures with similar yields.
Acid-base titration
The buffering capacities of various polymers were tested by acid-base titration over a pH range from 2.0 to 11.0 as our previous work.35 Polymer was firstly dissolved in 150 mM NaCl aqueous solution (0.2 mg/mL), and the pH was brought to 10 as the starting point with addition of 0.1 M NaOH. Then the solution was titrated with 0.1 M HCl using a pH meter. The buffering capacity was determined as μmol of H+ per mg of polymer required to decrease the pH of 0.2 mg/mL polymer solution from 7.4 to 5.0.
Preparation and characterization of DNA polyplexes
All the polymer stock solution were prepared at a nitrogen concentration of 1.52 mM in PBS (0.1X, pH 6.0) and stored at 4 °C. The pCMV-Luc2 plasmid (endotoxin free) with a concentration of 0.1 mg/mL was prepared by diluting the stock solution with double-distilled water, and mixed with an equal volume of polymer solution at different N/P ratios. After gently mixing, the polyplexes were incubated at room temperature for 10 min.
For the gel retardation study, the polyplexes with various N/P ratios were loaded onto a 1% agarose gel containing TAE buffer (40 μmM tris-acetate, 1 mM EDTA) and 5 mg/mL ethidium bromide, and were electrophoresed at 100 V for 40 min. The pDNA was then visualized using a Kodak (Rochester, NK) UV transilluminator (laser-excited fluorescence gel scanner).
The size and surface charge of the polyplexes were tested on a Brookhaven Instruments Corporation (Holtsvile, NY) ZetaPLUS instrument. The samples were prepared by mixing polyplexes (1 μg DNA, 20 μL solution, N/P = 5) with 800 μL ddH2O. The measurements were performed in triplicate.
Transmission Electron Microscopy
The morphology size of polyplex under dried state was imaged by electron microscopy on a hydrophilic surface. Copper/Formvar grids (400-mesh) were treated with glow discharge for 45 s to creat a hydrophilic surface. Twelve microliters of polyplexes (in dH2O) was applied to the Formvar-side of the grid for 30 min. Then the grid was washed three times with dH2O, followed by dipping in 4% (w/v) uranyl acetate (in dH2O) to negatively stain the sample. Excess solution was wicked off the grid with filter paper, and the grid was allowed to dry overnight in a desiccator prior to imaging. Images of the sample grids were taken with a JEOL 1010 transmission electron microscope (Electron Microscopy Facility, Fred Hutchinson Cancer Research Center).
For visualizing polyplex fate by TEM, 5 × 106 HeLa cells were placed into 15-cm plates overnight. The cells were then treated with either C-21 or S-22 polyplexes (containing 100 μg DNA) at N/P 5 in complete growth medium for 1 h at 37 °C, 5% CO2. Cells were washed once with PBS and fixed with 1:1 (v/v) mixture of 1/2 Karnovsky's fixative: complete growth medium for 10 min at room temperature. The fixative was removed, and the cells were then further incubated with full-strength 1/2 Karnovsky's fixative for 1 h. Cells were then scraped off the plate, pelleted, and resuspended in a small volume of fixative for preparation for TEM (Fred Hutchinson Cancer Research Center, Electron Microscopy Services).
Polyplex unpacking
The pCMV-Luc2 was first mixed with the bis-intercalating dye YOYO-1 iodide at a dye to base pair ratio of 1:100 and incubated for 1 hour at room temperature to equilibrate the bis-intercalator complexes with pDNA. Polyplexes were formed at different N/P ratios by mixing YOYO-1 labeled pDNA with different polymers. Afterwards, 10 μL polyplexes solution was added into 96-well plate, followed by 90 uL of 5 μg/mL heparin sulfate or distilled water. The plate was incubated at room temperature for 1 h before test. The fluorescence (ex: 491 nm, em: 509 nm) was measured on a Tecan Safire2 plate reader.
IC50 study
The cytotoxicity of polymers was assessed in vitro using MTS assay. Suspensions of HeLa cells (100 uL, DMEM) were first plated in 96-well plate at a density of 2,500 cells per well, and incubated for 24 h. Then cells were gently washed with PBS, and polymer solutions with different concentrations were added. After incubation for 4 h, the polymer solutions were removed, and cells were rinsed with PBS twice. The medium was replaced with 100 μL fresh complete growth medium. After additional 44 h incubation, 20 μL 3-(4,5-dimethyl thiazol-2-yl)-5-(3-carboxymethoxyphenyl )-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Promega) were added into each well. Cells were then incubated for 2 h before the absorbance measurement (490 nm).
Luciferase plasmid transfection
HeLa cells were seeded at a density of 20,000 cells/well in MEM medium supplemented with 10% FBS and 1% antibiotic/antimicrobial in a 24-well plate. Cells were firstly incubated at 37 °C, 5% CO2 for 24 h. Polyplexes were prepared at different N/P ratios using 1 μg of pCMV-Luc2 pDNA in 20 μL total volume. Each sample was diluted to 200 μL with OptiMEM medium (Gibco) or complete cell culture medium. The cells were rinsed once with PBS, followed by the addition of transfection solution. After incubation for 4 h, cells were washed with PBS twice and the polyplexes solution was replaced with complete cell culture medium. After additional 44 h incubation, luciferase activity was quantified with a luciferase assay kit (Promega Corp, Fitchburg, WI) according to the manufacturer's instruction, except that a freeze-thaw cycle at -80 °C was included after the addition of the lysis buffer to ensure complete cell lysis. Luminescence intensity was measured on the plate reader with integration for 1 s. The total protein content in each well was measured by a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instruction so that the luciferase activity was normalized to the total protein content in each well. Each sample was tested with a sample size (n) = 3.
GFP transfection
HeLa cells were seeded overnight in 24-well plates at a density of 2.5 × 104 cells per well (0.5 mL/well) at 37 °C, 5% CO2. Polyplexes were formulated as described previously and then diluted to 200 μL with complete cell culture media and added to cells after washing twice with PBS. The cells were incubated at 37 °C, 5% CO2 for 4 h, washed and then incubated with complete cell culture media at 37 °C, 5% CO2 for another 44 h. For analysis, cells were washed with PBS, trypsinized and pelleted at 300 × g for 5 min at 4 °C. The pellet was resuspended in 0.3 mL propidium iodide (PI) solution (1 μg/mL in 0.5% BSA in PBS), kept on ice and analyzed using flow cytometry, MACSQuant Analyzer (Miltenyi Biotec Inc., Auburn, CA). Intact cells were identified using the forward and side scatter data. The resulting cell population was gated into GFP+/PI+, GFP+/PI-, GFP-/PI+ and GFP-/PI- based on the green fluorescence and PI intensity from the control samples (cells transfected without the polymers but DNA only) and reported as the mean percentage of cell population that is GFP+/PI- including standard deviation (SD). All experiments were conducted in triplicate.
In vivo plasmid delivery of polymers
All animal procedures were done using protocols approved by the Institutional Animal Care and Use Committee at the University of Washington. Polyplexes were prepared as described above in 5% glucose using 2.5 mg of DNA in 10 μL at desired N/P. 7-9 week old female C57/Bl6 mice from Jackson Laboratories were housed for 1 week prior to experimentation. Mice were anesthetized by an intraperitoneal injection of Avertin (0.23 mL/20 gram). A 1 mm diameter craniotomy was made on the right-side of the skull using a dental drill and 10 μL polyplex or 5% glucose solution was stereotaxically injected at 1mm lateral, 0.5mmcaudal to bregma, and 1.75mm deep from the dura using a 33 gauge 10 μL Hamilton syringe. The injection was made at 1-2 μL/min and the syringe was kept in place for 2 min after injection to prevent backflow.
Brains were harvested from mice two days after injection and collected in lysis buffer supplemented by protease inhibitors (Roche, Nutley, NJ) and three freeze thaw cycles were performed in liquid nitrogen. Tissues were mechanically homogenized and lysate was cleared by spinning at 21,000 g for 15 min at 4 °C. 20 μL of lysate was assayed for luminescence with 100 μL of luciferase substrate. Luminescence was measured and normalized by protein content in the three brain sections, determined using a BCA Protein Assay Kit (Pierce), and reported as relative light units (RLU) per mg brain.
Statistical analysis
All statistical analyses were performed using a two-tailed Student's t-test with unequal variance.
Supplementary Material
Acknowledgments
This work was supported by NIH 2R01NS064464. JKYT is supported by an NSF Graduate Research Fellowship (2011128558).
Contributor Information
Yilong Cheng, Department of Bioengineering and Molecular Engineering & Sciences Institute, University of Washington, Seattle, Washington 98195, United States (USA)
Hua Wei, Department of Chemistry, Lanzhou University, Lanzhou 730000, China
James-Kevin Y. Tan, Department of Bioengineering and Molecular Engineering & Sciences Institute, University of Washington, Seattle, Washington 98195, United States (USA)
David J. Peeler, Department of Bioengineering and Molecular Engineering & Sciences Institute, University of Washington, Seattle, Washington 98195, United States (USA)
Don O. Maris, Department of Neurological Surgery, University of Washington Seattle, WA 98195, (USA)
Drew L. Sellers, Department of Bioengineering and Molecular Engineering & Sciences Institute, University of Washington, Seattle, Washington 98195, United States (USA)
Philip J. Horner, Department of Neurological Surgery, University of Washington Seattle, WA 98195, (USA)
Suzie H. Pun, Email: spun@u.washington.edu, Department of Bioengineering and Molecular Engineering & Sciences Institute, University of Washington, Seattle, Washington 98195, United States (USA).
References
- 1.Kay MA. Nat Rev Genet. 2011;12:316. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 2.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nat Rev Genet. 2014;15:541. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 3.a) Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]; b) Mintzer MA, Simanek EE. Chem Rev. 2009;109:259. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 4.a) Jiang X, Lok MC, Hennink WE. Bioconjugate Chemistry. 2007;18:2077. doi: 10.1021/bc0701186. [DOI] [PubMed] [Google Scholar]; b) Zhu CH, Zheng M, Meng FH, Mickler FM, Ruthardt N, Zhu XL, Zhong ZY. Biomacromolecules. 2012;13:769. doi: 10.1021/bm201693j. [DOI] [PubMed] [Google Scholar]; c) Durmaz YY, Lin YL, ElSayed MEH. Adv Funct Mater. 2013;23:3885. [Google Scholar]
- 5.a) Wei H, Pahang JA, Pun SH. Biomacromolecules. 2013;14:275. doi: 10.1021/bm301747r. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wei H, Volpatti LR, Sellers DL, Maris DO, Andrews IW, Hemphill AS, Chan LW, Chu DSH, Horner PJ, Pun SH. Angew Chem Int Edit. 2013;52:5377. doi: 10.1002/anie.201301896. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ma M, Li F, Chen FJ, Cheng SX, Zhuo RX. Macromol Biosci. 2010;10:183. doi: 10.1002/mabi.200900183. [DOI] [PubMed] [Google Scholar]; d) Yang XC, Niu YL, Zhao NN, Mao C, Xu FJ. Biomaterials. 2014;35:3873. doi: 10.1016/j.biomaterials.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 6.a) Tian HY, Guo ZP, Lin L, Jiao ZX, Chen J, Gao SQ, Zhu XJ, Chen XS. J Control Release. 2014;174:117. doi: 10.1016/j.jconrel.2013.11.008. [DOI] [PubMed] [Google Scholar]; b) Shi J, Choi JL, Chou B, Johnson RN, Schellinger JG, Pun SH. Acs Nano. 2013;7:10612. doi: 10.1021/nn403069n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Ping YA, Liu CD, Tang GP, Li JS, Li J, Yang WT, Xu FJ. Adv Funct Mater. 2010;20:3106. [Google Scholar]; b) Yin L, Song Z, Kim KH, Zheng N, Gabrielson NP, Cheng J. Adv Mater. 2013;25:3063. doi: 10.1002/adma.201205088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eltoukhy AA, Chen DL, Alabi CA, Langer R, Anderson DG. Adv Mater. 2013;25:1487. doi: 10.1002/adma.201204346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao TY, Zhang H, Newland B, Aied A, Zhou DZ, Wang WX. Angew Chem Int Edit. 2014;53:6095. doi: 10.1002/anie.201402341. [DOI] [PubMed] [Google Scholar]
- 10.a) Xu FJ, Zhang ZX, Ping Y, Li J, Kang ET, Neoh KG. Biomacromolecules. 2009;10:285. doi: 10.1021/bm8010165. [DOI] [PubMed] [Google Scholar]; b) Xiu KM, Yang JJ, Zhao NN, Li JS, Xu FJ. Acta Biomater. 2013;9:4726. doi: 10.1016/j.actbio.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Georgiou TK, Phylactou LA, Patrickios CS. Biomacromolecules. 2006;7:3505. doi: 10.1021/bm060657y. [DOI] [PubMed] [Google Scholar]
- 12.Wei H, Chu DSH, Zhao J, Pahang JA, Pun SH. ACS Macro Letters. 2013;2:1047. doi: 10.1021/mz400560y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schallon A, Jérôme V, Walther A, Synatschke CV, Müller AHE, Freitag R. Reactive and Functional Polymers. 2010;70:1. [Google Scholar]
- 14.a) Xia Y, Boydston AJ, Grubbs RH. Angewandte Chemie International Edition. 2011;50:5882. doi: 10.1002/anie.201101860. [DOI] [PubMed] [Google Scholar]; b) Zhang K, Tew GN. ACS Macro Letters. 2012;1:574. doi: 10.1021/mz2001675. [DOI] [PubMed] [Google Scholar]; c) Wei H, Wang CE, Tan N, Boydston AJ, Pun SH. ACS Macro Letters. 2015 doi: 10.1021/asmacrolett.5b00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Cherng JY, van de Wetering P, Talsma H, Crommelin DA, Hennink W. Pharm Res. 1996;13:1038. doi: 10.1023/a:1016054623543. [DOI] [PubMed] [Google Scholar]; b) van de Wetering P, Cherng JY, Talsma H, Hennink WE. J Control Release. 1997;49:59. doi: 10.1016/s0168-3659(97)00248-4. [DOI] [PubMed] [Google Scholar]
- 16.a) Xu FJ, Yang WT. Progress in Polymer Science. 2011;36:1099. [Google Scholar]; b) Chu DSH, Schellinger JG, Shi JL, Convertine AJ, Stayton PS, Pun SH. Accounts Chem Res. 2012;45:1089. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Xu FJ, Ping Y, Ma J, Tang GP, Yang WT, Li J, Kang ET, Neoh KG. Bioconjugate Chemistry. 2009;20:1449. doi: 10.1021/bc900044h. [DOI] [PubMed] [Google Scholar]; b) Guo S, Huang Y, Wei T, Zhang W, Wang W, Lin D, Zhang X, Kumar A, Du Q, Xing J, Deng L, Liang Z, Wang PC, Dong A, Liang XJ. Biomaterials. 2011;32:879. doi: 10.1016/j.biomaterials.2010.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurent BA, Grayson SM. Journal of the American Chemical Society. 2006;128:4238. doi: 10.1021/ja0585836. [DOI] [PubMed] [Google Scholar]
- 19.a) Ge ZS, Zhou YM, Xu J, Liu HW, Chen DY, Liu SY. Journal of the American Chemical Society. 2009;131:1628. doi: 10.1021/ja808772z. [DOI] [PubMed] [Google Scholar]; b) Fan XS, Wang GW, Huang JL. J Polym Sci Pol Chem. 2011;49:1361. [Google Scholar]
- 20.Behr JP. CHIMIA International Journal for Chemistry. 1997;51:34. [Google Scholar]
- 21.Shi J, Schellinger JG, Johnson RN, Choi JL, Chou B, Anghel EL, Pun SH. Biomacromolecules. 2013;14:1961. doi: 10.1021/bm400342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cakara D, Kleimann J, Borkovec M. Macromolecules. 2003;36:4201. [Google Scholar]
- 23.Johnson RN, Chu DSH, Shi J, Schellinger JG, Carlson PM, Pun SH. J Control Release. 2011;155:303. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Wetering P, Moret EE, Schuurmans-Nieuwenbroek NME, van Steenbergen MJ, Hennink WE. Bioconjugate Chemistry. 1999;10:589. doi: 10.1021/bc980148w. [DOI] [PubMed] [Google Scholar]
- 25.Chu DSH, Johnson RN, Pun SH. J Control Release. 2012;157:445. doi: 10.1016/j.jconrel.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi JL, Tan J-KY, Sellers DL, Wei H, Horner b PJ, Pun SH. Biomaterials. 2015;54:87. doi: 10.1016/j.biomaterials.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


