Abstract
Recent work suggests that DNA replication origins are regulated by the number of multiple Mini-Chromosome Maintenance (MCM) complexes loaded. Origins are defined by the loading of MCM - the replicative helicase which initiates DNA replication and replication kinetics determined by origin’s location and firing times. However, activation of MCM is heterogeneous; different origins firing at different times in different cells. Also, more MCMs are loaded in G1 than are used in S phase. These aspects of MCM biology are explained by the observation that multiple MCMs are loaded at origins. Having more MCMs at early origins makes them more likely to fire, effecting differences in origin efficiency that define replication timing. Nonetheless, multiple MCM loading raises new questions, such as how they are loaded, where these MCMs reside at origins and how their presence affects replication timing. In this review, we address these questions and discuss future avenues of research.
Keywords: MCM, ORC, Replication Origin, Replication Timing, Nucleosome
Introduction
Although the biochemistry of DNA replication origin establishment and activation in eukaryotes is increasingly well understood, how origin firing is regulated to produce the observed patterns of replication timing remains a major question (1). Replication timing is the cumulative result of individual origin firing kinetics, and contributes to the efficient and robust replication of the genome (2–4). Origins in budding yeast are defined by a 17 bp AT-rich consensus sequence, but in multicellular eukaryotes, origins are not well defined (5). Nonetheless, in both cases origins are established in G1 by loading of a head-to-head dimer of the ring-shaped MCM replicative helicase complex (6–10). MCM activation in S phase leads to origin unwinding, polymerase loading and the initiation of bidirectional DNA replication. The multi-subunit Origin Recognition Complex (ORC) binds a nucleosome-free region (NFR) of origin DNA and in concert with Cdc6 and Cdt1 loads the MCM complex (11) (Figure 1A). MCMs are loaded at many more origins across the genome, and in much greater number, than are activated in any one S phase (12–14). There are estimated to be 5,000–10,000 MCM complexes per cell in budding yeast (15), which have 500–1000 origins (16). The presence of such excess MCMs has been referred to as the MCM paradox (13, 17). Excess MCMs are loaded at inefficient origins, sometimes referred to as dormant origins, that normally fire in only a small subset of cells, but which are crucial for providing backup origin capacity during replicative stress (18–20). However, accumulating evidence suggests that excess MCMs can also be loaded in the form of multiple MCMs at early efficient origins (12, 21,22) (Figure 1B). Having multiple MCMs at origins raises interesting questions about how MCM loading is regulated, where MCMs are positioned at origins and how having multiple MCMs loaded affects the robustness of DNA replication.
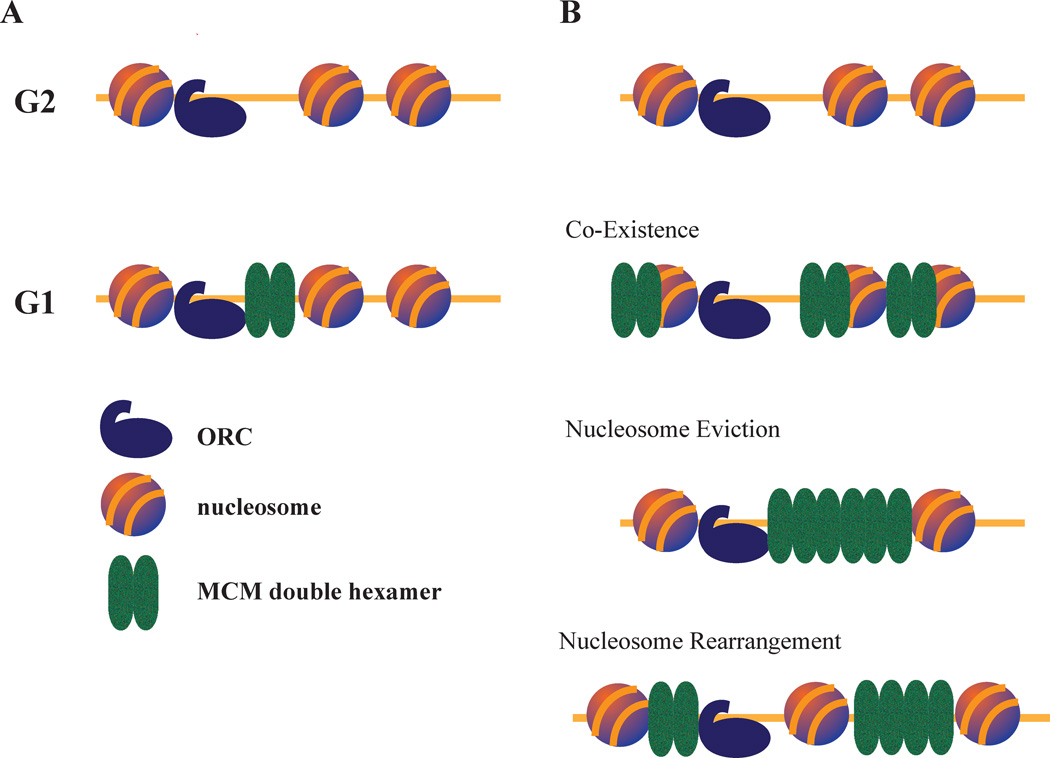
Figure 1. MCM loading at origins of DNA replication.
A. Single MCM Loading Model. ORC binds origins throughout the cell cycle. During G1, in collaboration with other proteins (not shown), it loads a single double-hexamer constituting the MCM complex in the adjacent NFR.
B. Multiple MCM Loading Model. During G1, ORC loads multiple MCMs at each origin. Having multiple MCMs loaded requires MCMs to move away from the NFR and interact with surrounding nucleosomes. Such interaction could include nucleosome eviction, nucleosome rearrangement or the co-existence of MCM complexes and nucleosomes. Recent data supports the latter possibility (28).
Evidence for Multiple MCMs at Individual Origins
Several lines of evidence support the idea that multiple MCM complexes are loaded at individual origins. Initial evidence came from in vitro experiments using Xenopus egg extracts showing that many copies of MCM could be loaded on short pieces of DNA that seemed to bind only one copy of ORC (12). The idea that a single ORC complex could processively load multiple MCMs was supported by in vitro experiments in budding yeast. In these experiments, purified yeast ORC was shown to load multiple copies of MCM on individual origins in yeast extract in the presence of excess origin DNA (21). A fully purified in vitro system also demonstrated multiple MCM loading, but at low frequency, suggesting that loading in that particular system is not processive (7). Independent support for the idea that multiple MCMs can be loaded at individual origins came from a computational analysis of budding yeast replication kinetics that lead to a quantitative model of replication in which loading of multiple MCMs at origins explains the variation in genome-wide replication timing (23). We directly tested this model with MCM ChIP-seq and quantitative ORC and MCM westerns on purified plasmids. We found strong evidence for multiple MCMs at origins, and observed that more MCMs are loaded on early origins than on late ones (22). On average, about three MCM double hexameric complexes were found to be loaded on single early firing origins isolated from yeast nuclei (Figure 1B).
Despite these lines of evidence that multiple MCMs are loaded at origins, there are several results that can be interpreted to support the contention that a single MCM complex is loaded at each origin. In vivo footprinting data showing a strong footprint next to ORC during G1 is consistent with a single, well-positioned MCM complex in the origin NFR (24,25). However, extension of the ORC footprint is not due to MCM loading, but instead is dependent on the binding of Cdc6 (26). In vivo replication initiation point mapping identified a single replication initiation site in the origin NFR next to ORC (27), suggesting loading and activation of a single, well-positioned MCM. However, that study examined only about 150 bp of sequence in the origin NFR, precluding the identification of other distal initiation sites. Moreover, recent data indicates that loaded MCMs are in fact not found in the NFR, but present on either side of the origin overlapping with the +1 and/or −1 nucleosomes (28). That study concluded that each origin primarily has one MCM loaded, in association with either the +1 or −1 nucleosome, but the averaged nature of the data does not exclude the presence of other less-well-positioned MCMs.
Multiple MCM Loading indicates that Origin Organization is More Complicated than Previously imagined
The current biochemical understanding of origin licensing supports a simple model for the organization of an origin in vivo, in which ORC binds in a NFR and loads a single MCM next to it (Figure 1A). However, having multiple MCMs loaded at an origin requires this model to be re-imagined (Figure 1B).
The most basic question addresses where the multiple MCMs would go (Figure 1B). Would they stay in the (possibly expanded) NFR, or would they diffuse into the surrounding nucleosomes? If the latter is true, would they displace nucleosomes or co-exist with them? A surprising finding of a recent MCM-mapping study is that MCMs do not appear to inhabit the origin NFR, but rather associate with the flanking nucleosomes (28). It is known that MCMs can slide on DNA in vitro (7, 8), and recent work shows that they can also be pushed at least a kilobase in vivo by RNA Pol II and retain function (29). In this case, nucleosomes must be evicted for MCMs to slide past. So, it is possible to imagine that at origins, as nucleosomes unbind and rebind during normal nucleosome exchange, MCMs can slide past as they diffuse away from their ORC-proximal loading site. Nucleosomes could then rebind, or be excluded by MCMs. Recent work suggests that MCMs tightly associate with origin-flanking nucleosomes (28), favoring the possibility that nucleosomes rebind (the co-existence model in Figure 1B).
In the context of thinking about nucleosomes unbinding to let MCMs slide by, it is interesting to note that in both budding yeast and flies the rate of nucleosome exchange correlates with origin timing (30,31). Nucleosomes at early origins exchange more quickly than those at late origins. It is possible that high rates of nucleosome exchange allow MCMs to quickly diffuse away from the origin and, in turn, allow more MCMs to be loaded or nucleosome dynamics could play a more direct role in origin-firing kinetics. Alternatively, it is possible that increased MCM loading destabilizes nucleosomes, increasing their exchange rates. Finally, the two effects could be caused by a third, independent, factor, such as the presence of nucleosome-modifiers. Nonetheless, the correlation suggests an intimate relationship between MCM loading, nucleosome stability and replication timing.
How is Multiple MCM Loading Regulated?
Another fundamental question is raised by these observations: Why do some origins load more MCMs than others? A simple possibility is that ORC has a higher affinity for some origins, and the resulting higher ORC occupancy leads to more MCM loading (Figure 2A). This possibility is supported by the observation that ORC occupancy, as measured by ORC ChIP-seq (32), correlates with MCM ChIP-seq, suggesting that ORC occupancy does affect MCM loading (22). Increased ORC affinity could be due to a higher affinity for origin DNA. However, ORC affinity for origin DNA in vitro only correlates well with ORC occupancy in vivo at some origins, suggesting that chromatin context or other factors can also affect ORC affinity for origins (33). Another possibility for why some origins load more MCMs is that chromatin context constrains MCM loading and determines how many are loaded (Figure 2C). Early origins tend to have both larger NFRs and higher rates of nucleosome exchange (30, 34), both of which could lead to increased MCM loading. Finally, a number of trans-acting factors, such as Rif1, Rpd3 and Fkh1, have been shown to regulate origin timing (35–38)(Figure 2D). These factors could be involved in the regulatory mechanisms described above by affecting ORC binding or activity, either directly or via chromatin modifications. They also could affect replication timing by regulating the activation of MCMs during S phase, after they have been loaded.
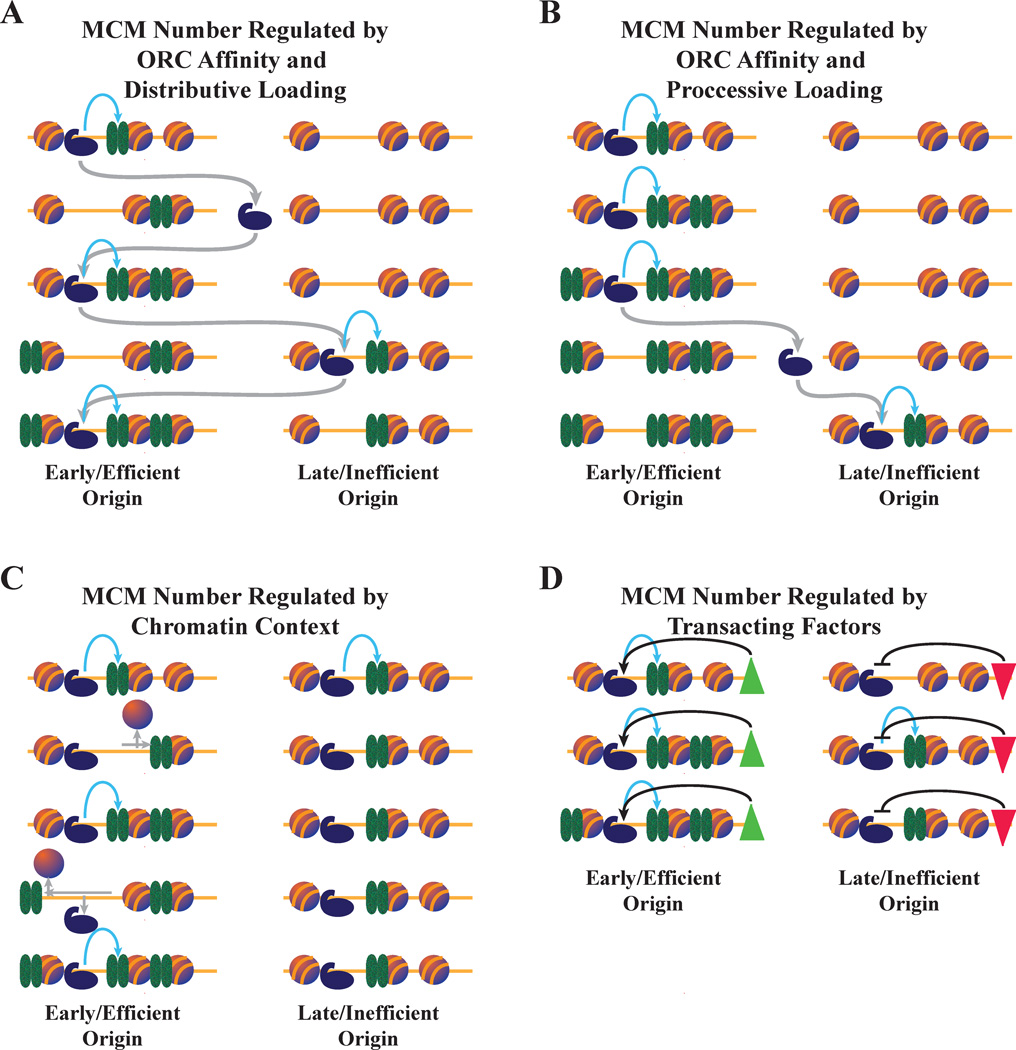
Figure 2. Possible mechanisms for regulating the number of MCMs loaded.
A. MCM number regulated by ORC affinity and distributive loading. ORC spends more time at origins for which it has higher affinity, and thus loads more MCMs there. In this case, ORC has a fast off rate and is thus unlikely to load more than one MCM complex during each binding event, leading to distributive MCM loading. Gray arrows represent ORC unbinding and diffusion. Blue arrows represent MCM loading. ORC binding events that do not result in MCM loading are not shown.
B. MCM number regulated by ORC affinity and processive loading. As in A, except that ORC at the efficient origin has a slow off rate and loads multiple MCMs during each binding event, leading to processive MCM loading.
C. MCM number regulated by chromatin context. Left: At origins with high rates of nucleosome exchange, unbinding of nucleosomes allows MCMs to diffuse into adjacent chromatin, making room for more MCMs to be loaded. Right: At origins with low rates of nucleosome exchange, MCMs are confined to the origin NFR, preventing further MCM loading.
D. MCM loading regulated by trans-acting factors. MCM loading is regulated by trans-acting factors that either promote (green triangle) or repress (red triangle) MCM loading. The factors could act directly through the regulation of ORC or indirectly through chromatin effects.
The loading of multiple MCMs at origins also raises the question of whether loading is processive or distributive. In processive loading, one ORC would bind an origin and load successive MCMs (Figure 2B). In distributive loading, one ORC would load one MCM at an origin, and then unbind and move to a different origin before loading another MCM (Figure 2A). The distinction is most significant in terms of the distribution of MCM within individual cells. Processive loading would lead to many MCMs loaded at the origins where ORC binds first, favoring a heterogeneous distribution of MCM numbers at a given origin across a population of cells. Distributive loading would favor a more homogeneous distribution of MCM loading across a population of cells. Early biochemical results notwithstanding (21), more recent biochemistry (7) and the short half-life of ORC on chromatin in vivo (39) suggest that MCM loading is likely to be distributive. Distributive loading also explains how MCM can end up on either side of the ORC binding site (28). During each ORC exchange cycle, MCM would have the opportunity to slide across the ORC-binding site (Figure 2C).
Multiple MCM Loading Regulates Replication Timing
The possibility that different numbers of MCMs are loaded at different origins provides a new way of thinking about the regulation of origin timing (40, 41). If there is one MCM at each origin, an MCM at an early origin must behave differently than an MCM at a late origin; in particular, it must have a higher probability of firing earlier (Figure 3A). However, if early origins have more MCMs loaded than late origins, all MCMs can behave the same, with no need for regulation of their individual firing times or firing probabilities (Figure 3B). Instead, the simple fact that earlier origins have more MCMs loaded means that early origins are likely to have MCMs fire before late origins, thus ensuring that, on average, early origins will fire earlier.
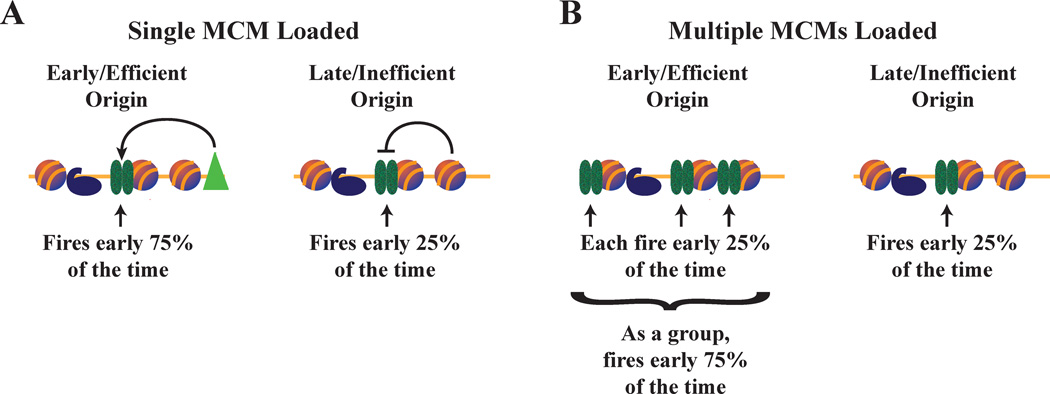
Figure 3. Implications of multiple MCMs in the regulation of replication timing.
A. If each origin has a single MCM complex bound, MCMs loaded at early origins must fire earlier than those loaded at late origins. Left: A trans-acting factor could advance origin firing time by increasing the probability of MCM firing. Right: In the absence of such factors, chromatin context could inhibit MCM firing.
B. If multiple MCMs are loaded at origins, each individual MCM can behave in the same way. Simply having more MCMs loaded at an origin increases the probability of one of those MCMs firing, increasing the efficiency of the origin and advancing its average firing time. In this simplistic, two origin model, in which only one of the four MCMs fire, the left origin is three times more likely to fire than the right. In a real cell, more MCMs loaded would lead to a greater probability of firing, but the relationship between the number of MCMs loaded and the probability of firing would be complicated and non-linear.
This distribution of MCMs guarantees that, in addition to the early, efficient, origins with more MCMs loaded, there are plenty of late, inefficient origins with few MCMs loaded, which usually will not fire before they are passively replicated. Such origins, also known as dormant origins, contribute little to replication kinetics in unperturbed S phases. However, they become essential when replication forks stall at high frequencies, because they are required to initiate replication between two stalled forks (17–20). For such a backup system to work, MCMs must be fairly evenly distributed across the genome, and that evenness must be in every cell, not just on average. Since there appear to be about only one third as many ORC complexes as there are potential origins (42, 43), it is not sufficient for an ORC to bind and load MCMs at only one origin during G1. Instead, it is likely that each ORC would need to cycle through a number of origins during G1, loading MCMs at each location. This model is consistent with the short half-life of ORC on chromatin in vivo (39) and the idea (discussed above) that MCM loading is likely to be distributive.
Future Directions
The idea that multiple MCMs can be loaded at individual replication origins provides solutions to old puzzles and contributes to a framework for understanding the regulation of replication timing, but it raises a host of new questions. There are two major immediate challenges for the field. The first challenge is to better map MCMs in space and time. Current data does not sufficiently constrain models for MCM localization or how MCM localization evolves over time. Furthermore, if the concept of multiple MCM loading is to become generally applicable, these studies need to be extended to metazoans. Techniques that can map individual MCMs on DNA, such as DNA adenine methyltransferase identification (DamID) (44), would prove extremely useful for understanding the organization of MCMs at origins, especially in conjunction with long-read sequencing technology, which would allow multiple MCMs to be mapped over kilobases of sequence. Such assays could elucidate the heterogeneity of MCM loading, providing a more realistic understanding of the dynamics of MCM in origin licensing. Another solution to many of these questions would be in vivo imaging, but the resolution of light microscopy and the complexity of the nuclear milieu makes this a daunting task. Nonetheless, small steps have been taken towards that goal, including super resolution imaging of replication factories (45) and in vivo fluorescent localization of replication complexes in yeast and bacteria (46, 47), so there is hope that new creative approaches are possible.
The second major challenge is to experimentally test the significance of MCM distribution. Uniformly reducing the number of MCMs loaded has little effect on replication timing, consistent with a competition model in which the relative number of MCMs per origin is more important for timing than the absolute number (40, 48). We showed that reducing the number of MCMs loaded at a particular origin delays the average firing time of that origin (22). The next step is to manipulate MCM loading across the genome to investigate its direct effect on replication timing and indirect effect on replication efficiency and genome stability. Such experiments are plausible in budding yeast, where origins are well defined and whole genome manipulation is being pioneered (49), and fission yeast, where the site-specific DNA binding domain of ORC is known (50), such experiments are plausible. Alternatively, MCM loading could be redistributed by manipulating nucleosome positioning or turnover, albeit with more pleiotropies. The prospects of such genome-wide engineering in metazoans are also improving with modern genome-editing technologies. How wholesale remodeling of an organism’s replication timing profile would affect its nuclear metabolism is a wide-open and fascinating question.
Acknowledgments
We thank members of the Rhind lab for helpful suggestions about this work and comments on this manuscript. This work was supported by NIH grant GM098815 to NR.
References
- 1.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson D, Wang X, Rudner DZ. Spatio-temporal organization of replication in bacteria and eukaryotes (nucleoids and nuclei) Cold Spring Harb Perspect Biol. 2012;4:a010389. doi: 10.1101/cshperspect.a010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K, Poveda A, Pasero P. Time to be versatile: regulation of the replication timing program in budding yeast. J Mol Biol. 2013;425:4696–4705. doi: 10.1016/j.jmb.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Donley N, Thayer MJ. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, et al. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 6.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remus D, Beuron F, Tolun G, Griffith JD, et al. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evrin C, Clarke P, Zech J, Lurz R, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem. 2011;286:11855–11864. doi: 10.1074/jbc.M110.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N, Zhai Y, Zhang Y, Li W, et al. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 11.Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- 12.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, et al. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 13.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 14.Alver RC, Chadha GS, Blow JJ. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst) 2014;19:182–189. doi: 10.1016/j.dnarep.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S, Huh WK, Bower K, Howson RW, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 16.Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:682–686. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das M, Singh S, Pradhan S, Narayan G. MCM Paradox: Abundance of Eukaryotic Replicative Helicases and Genomic Integrity. Mol Biol Int. 2014;2014:574850. doi: 10.1155/2014/574850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodward AM, Gohler T, Luciani MG, Oehlmann M, et al. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman KM, Jones RM, Petermann E, Jeggo PA. Diminished origin-licensing capacity specifically sensitizes tumor cells to replication stress. Mol Cancer Res. 2013;11:370–380. doi: 10.1158/1541-7786.MCR-12-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Das SP, Borrman T, Liu VW, Yang SC, et al. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015;25:1886–1892. doi: 10.1101/gr.195305.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SC, Rhind N, Bechhoefer J. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol. 2010;6:404. doi: 10.1038/msb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 25.Cocker JH, Piatti S, Santocanale C, Nasmyth K, et al. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 26.Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 27.Bielinsky AK, Gerbi SA. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- 28.Belsky JA, MacAlpine HK, Lubelsky Y, Hartemink AJ, et al. Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev. 2015;29:212–224. doi: 10.1101/gad.247924.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gros J, Kumar C, Lynch G, Yadav T, et al. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2–7 Sliding along DNA. Mol Cell. 2015;60:797–807. doi: 10.1016/j.molcel.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dion MF, Kaplan T, Kim M, Buratowski S, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 31.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eaton ML, Galani K, Kang S, Bell SP, et al. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoggard T, Shor E, Muller CA, Nieduszynski CA, et al. A Link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet. 2013;9:e1003798. doi: 10.1371/journal.pgen.1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soriano I, Morafraile EC, Vazquez E, Antequera F, et al. Different nucleosomal architectures at early and late replicating origins in Saccharomyces cerevisiae. BMC Genomics. 2014;15:791. doi: 10.1186/1471-2164-15-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, et al. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012;26:137–150. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lian HY, Robertson ED, Hiraga S, Alvino GM, et al. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell. 2011;22:1753–1765. doi: 10.1091/mbc.E10-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knott SR, Peace JM, Ostrow AZ, Gan Y, et al. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonneville R, Querenet M, Craig A, Gartner A, et al. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012;28:374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhind N, Yang SC, Bechhoefer J. Reconciling stochastic origin firing with defined replication timing. Chromosome Res. 2010;18:35–43. doi: 10.1007/s10577-009-9093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marguerat S, Schmidt A, Codlin S, Chen W, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Yanagisawa Y, Tsankov AM, Hart C, et al. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol. 2012;13:R27. doi: 10.1186/gb-2012-13-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 45.Baddeley D, Chagin VO, Schermelleh L, Martin S, et al. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 2010;38:e8. doi: 10.1093/nar/gkp901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011;30:4805–4814. doi: 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Annaluru N, Muller H, Mitchell LA, Ramalingam S, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci U S A. 2001;98:13589–13594. doi: 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]