Abstract
Background
Mountain-cultivated ginseng (MCG) and cultivated ginseng (CG) both belong to Panax ginseng and have similar ingredients. However, their pharmacological activities are different due to their significantly different growth environments.
Methods
An ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS/MS)-based approach was developed to distinguish MCG and CG. Multivariate statistical methods, such as principal component analysis and supervised orthogonal partial-least-squares discrimination analysis were used to select the influential components.
Results
Under optimized UPLC-QTOF-MS/MS conditions, 40 ginsenosides in both MCG and CG were unambiguously identified and tentatively assigned. The results showed that the characteristic components of CG and MCG included ginsenoside Ra3/isomer, gypenoside XVII, quinquenoside R1, ginsenoside Ra7, notoginsenoside Fe, ginsenoside Ra2, ginsenoside Rs6/Rs7, malonyl ginsenoside Rc, malonyl ginsenoside Rb1, malonyl ginsenoside Rb2, palmitoleic acid, and ethyl linoleate. The malony ginsenosides are abundant in CG, but higher levels of the minor ginsenosides were detected in MCG.
Conclusion
This is the first time that the differences between CG and MCG have been observed systematically at the chemical level. Our results suggested that using the identified characteristic components as chemical markers to identify different ginseng products is effective and viable.
Keywords: cultivated ginseng, identification, mountain-cultivated ginseng, OPLS-DA, UPLC-QTOF-MS/MS
1. Introduction
Ginseng is a slow-growing perennial plant belonging to the genus Panax of the family Araliaceae that has been used as a tonic and functional food to prevent various diseases in China, Korea, and Japan for thousands of years [1], [2], [3]. It has positive effects on the endocrine, cardiovascular, immune, and central nervous systems, and aids in the prevention of fatigue, oxidative damage, mutagenicity and cancer [4], [5], [6], [7]. The effective components of ginseng contain polysaccharides, ginsenosides, volatile oils, etc., with the main bioactive constituents considered to be ginsenosides, which exhibit antioxidant, anti-inflammatory, anticancer, anti-apoptotic, and immune-stimulatory pharmacological activities [2], [8], [9], [10], [11]. Even the rare ginsenosides have significant pharmacological activities [12].
The quality and properties of ginseng products vary greatly due to their different growth environments. According to the growth environment and the cultivation method, ginseng can be divided into three types: cultivated ginseng (CG), mountain-wild ginseng (MWG), and mountain-cultivated ginseng (MCG). CG is the artificial planting ginseng that has a short growth period and rapid weight increment. MWG grows in the mountains with no artificial management and only under natural conditions throughout the entire growing period [13]. MCG, whose seeds are sowed artificially, is cultivated in the natural environment and returns to the wild state before being used clinically [14]. Generally speaking, most CG is harvested after 4–5 year. Meanwhile, MCG is collected at least after 10–20 year or longer, with the age of MWG being much older than that of MCG [15]. Due to excessive excavation, MWG has been nearing extinction. Presently, there are only two primary types of ginseng on the market: one is CG and the other is MCG. Pharmacopoeia of the People's Republic of China also classified ginseng into CG and MCG groups [16]; however, a practicable criterion for distinguishing between CG and MCG in Chinese Pharmacopoeia (2010 Edition) does not exist.
MCG can be considered as a mimic of wild ginseng, which is of better quality than CG. In the past decades, Fourier transform infrared spectroscopy (FT-IR), secondary derivative IR spectra, two-dimensional correlation infrared spectroscopy (2D-IR), and FT-IR microspectroscopy have been applied to discriminate between MCG and CG rapidly, effectively, and nondestructively [17], [18]. HPLC analytical technology was applied to analyze MCG [19], focusing on the differences in the major ginsenosides and malonyl ginsenosides. For many years, MCG has been much more expensive and effective than CG. It is no surprise that only the decoction of mountain wild ginseng is being used in clinical applications. Pharmacological researchers have also revealed that MCG has greater anti-cancer activities than CG [20], [21]; however the difference in chemical components between CG and MCG has not been studied systematically. Thus, it is important to find the different chemical markers and identify the structures between CG and MCG. This will also be helpful in explaining the different pharmacological activities and controlling the quality of CG and MCG.
Recently, ultra-high-performance liquid chromatography (UPLC) coupled with multivariate statistical analysis (MVA) was used to identify ginseng products, such as the white and red ginseng, ginseng of different ages, and white ginseng of different origins [22], [23], [24]. This strategy has the advantages of rapid analysis time, high resolution, selectivity, and sensitive analysis of components in complex medicinal herb mixtures. Moreover it can identify the different marker components and their chemical structures in order to explain the subtle differences between samples. In our study, we developed a sample-profiling strategy combining UPLC orthogonal acceleration time-of-flight mass spectrometry (UPLC/oa-QTOF MS) with MVA to compare the chemical contents of CG and MCG. Our results constitute the first time that the differences between CG and MCG have been observed systematically from the chemical components.
2. Experimental
2.1. Ginseng samples and samples processing
Forty-five ginseng samples, of which 36 CG samples 4- to 7-year of age were collected from the Ji'an, Fusong and Tonghua of Jilin provinces of China, and 9 MCG samples were collected from Kuandian county of Liaoning province and Ji'an city of Jilin province of China, were collected for analysis. The details of the samples are shown in Table 1. All of these samples were identified by Professor Xiangri Li (School of Chinese Materia Medica, Beijing University of Chinese Medicine) and deposited in the specimen cabinet of traditional Chinese medicine at Beijing University of Chinese Medicine.
Table 1.
Details of CG and MCG samples
| No. | Group | Growth year | Classification | Origin |
|---|---|---|---|---|
| J01-J05 | CG | 4 | 4-yr L | Ji'an |
| F01-F05 | CG | 4 | 4-yr L | Fusong |
| J06-J10 | CG | 5 | 5-yr L | Ji'an |
| T01-T04 | CG | 5 | 5-yr | Tonghua |
| J11-J19 | CG | 6 | 6-yr | Ji'an |
| J20-J27 | CG | 7 | 7-yr | Ji'an |
| J28-J32 | MCG | 15 | LX L | Ji'an |
| K01-K04 | MCG | 15 | LX L | Kuandian |
CG, cultivated ginseng; MCG LX, mountain-cultivated ginseng; L, vacuum freeze drying
2.2. Sample preparation
All samples were pulverized into powder of over 65 mesh, then the fine ginseng powder was accurately weighed (0.4 g) and extracted with 50 mL methanol in an ultrasonic bath for 30 min. After cooling to room temperature, the loss of weight was replenished with methanol and then filtrated. We accurately drew subsequent filtrate (25 mL) and concentrated it into residue, which was then dissolved in methanol in a 10-mL volumetric flask. The extraction solution was filtered through a 0.22-μm filter membrane and injected directly into the UPLC system.
2.3. Reagents
Fisher Optima-grade acetonitrile, methanol, and isopropanol were obtained from Thermo Fisher Scientific. (Waltham, MA, USA). Formic acid and leucine enkephaline were purchased from Sigma-Aldrich (St Louis, MO, USA). Deionized water was obtained in our laboratory via a Milli-Q water purification system (Millipore Corporation, Bedford, MA, USA). Ginsenoside Rg1, Re, Rb1, Rf, Rb2, and Rb3 standards were purchased from the National Institute for Pharmaceutical and Biological Products (Beijing, PR China). Ginsenoside Rc and Rg2 standards were obtained from Beijing Xiantong era Pharmaceutical Co. Ltd. (Beijing, PR China). The standards were dissolved in methanol and stored at 4°C.
2.4. UPLC-QTOF conditions
2.4.1. LC conditions
LC separation was performed on an ACQUITY UPLC system (Waters Corporation, Milford, MA, USA) with an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm). The column temperature was set to 40°C, and the flow rate was 0.4 mL/min. Mobile phases A and B consisted of water with 0.1% formic acid and acetonitrile, respectively. The UPLC elution conditions were optimized as follows: linear gradient from 81% to 50% A (0–7 min), 50% to 4% A (7–12 min), 4% to 2% A (12–13 min), 2% to 2% A (13–25 min), 2% to 81% A (25–26 min), and 81% to 81% A (26–29 min). The total run time was 29 min, and the injection volume was 2 μL.
2.4.2. MS conditions
MS detection was performed on a Synapt MS System (Waters Corporation), with the data acquisition mode set to MSE and the ionization mode set for positive electrospray (ESI+). The source temperature was 120°C, the desolvation temperature was 350°C, and the desolvation gas flow was 480.0 L/h. The lock mass compound used was leucine enkephaline, the capillary and cone voltages were 3,000 V and 20 V, respectively, and the cone gas was 50 L/h. The collision energies were 5 eV for low-energy scans and 20–30 eV for high-energy scans. The LC–MS data acquisition was controlled by Mass Lynx 4.1 Mass Spectrometry Software (Waters Corporation).
2.5. Data processing procedure
Post-acquisition data processing, including the MVA, was performed by Marker Lynx XS, which is an application manager for the Mass Lynx software. The structural elucidation was performed by the Mass Fragment tool provided by Mass Lynx Waters Corporation).
2.5.1. Principal component analysis (PCA)
From the chromatographic trace, we acquired a three-dimensional data point representing the retention time, m/z, and intensity. We converted each data point into an exact mass retention time (EMRT) pair using the Marker Lynx XS software. After the EMRT 2D matrix was obtained, the MVA interface was launched with all EMRT information automatically imported, enabling the extended statistics module PCA to be completed.
2.5.2. The scatter plot from orthogonal projections from latent structures discriminant analysis (OPLS-DA)
We acquired the loading plot (S-plot) of every group of pairs by OPLS-DA. In the S-plot, the leading contributing EMRT pairs were captured selectively, resulting in a list of top contributing markers from each sample group being generated and saved as a text file.
2.5.3. The elemental composition calculation for the target markers
We calculated the matched elemental composition with the exact mass of the markers, and searched an existing database to acquire the chemical structure. Once the identity of a marker was tentatively identified, its fragment ion was obtained by going back to the raw data file to investigate the high capillary electrophoresis (CE) scan of the samples. The fragment ions obtained using the Mass Fragment tool in the Mass Lynx software was used for elucidating the structure.
3. Results and discussion
3.1. MS analysis
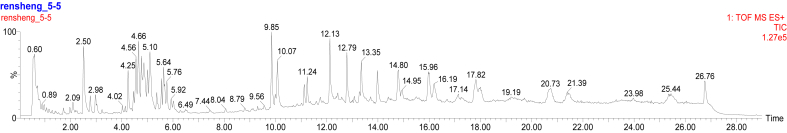
As shown previously [25], the ACQUITY BEH C18 column has frequently been used to separate ginsenosides from various Panax herbs. Figure 1 shows the based-peak intensity (BPI) chromatogram obtained from the analysis of CG in positive-ion mode. There are many peaks in the BPI, indicating that the components of the ginseng samples were complex. The ginsenoside retention times were mainly within 7.5 min according to the BPI chromatogram (Fig. 2). Forty ginsenosides, including panaxatriol and panaxadiol, were identified in MCG and CG. As presented in Figure 2, eight compounds were assigned by comparing them with standard ginsenosides, and 32 ginsenosides were identified by comparing their retention times and mass spectra data with the reference compounds. The compounds were further confirmed through ion-fragmentation patterns. As illustrated in Table 2, ginsenosides were detected as protonated ions [M+H]+, sodium adduct ions [M+Na]+, and/or ammonium adduct ions [M+NH4]+ in the positive-ion mode.
Fig. 1.
Representative base peak intensity chromatograms of cultivated ginseng sample.
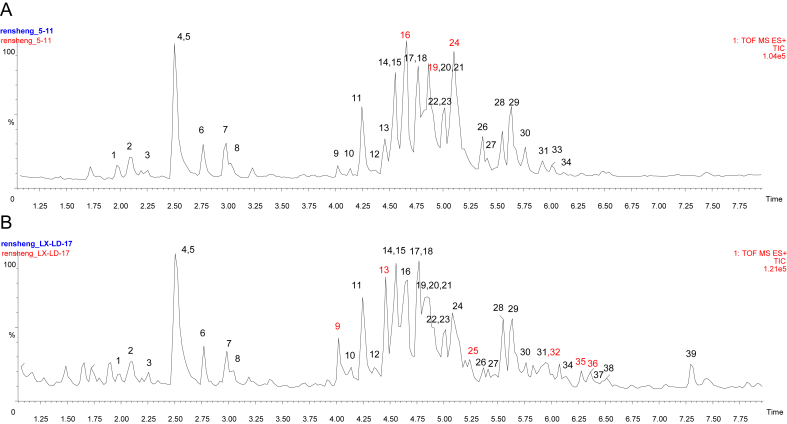
Fig. 2.
Total ion-current chromatograms of ginseng samples within 7.5 min using UPLC-QTOF-MS. (A) cultivated ginseng and (B) mountain cultivated ginseng. Peak definitions: 1, ginsenoside Re4; 2, notoginsenoside R3 isomer; 3, notoginsenoside R1; 4, ginsenoside Re; 5, ginsenoside Rg1; 6, malonyl-ginsenoside Rg1; 7, malonyl-ginsenoside Re; 8, malonyl-ginsenoside Re isomer; 9, ginsenoside Ra3; 10, malonyl-ginsenoside Ra3; 11, ginsenoside Rf; 12, malonyl-notoginsenoside R4; 13, ginsenoside Ra2; 14, ginsenoside Rb1; 15, malonyl-notoginsenoside Fa; 16, malonyl-ginsenoside Rb1; 17, ginsenoside Rc; 18, ginsenoside Ra1; 19, malonyl-ginsenoside Rc; 20, malonyl-ginsenoside Ra1/Ra2; 21, malonyl-ginsenoside Rb1 isomer; 22, ginsenoside Rb2; 23, ginsenoside Rb3; 24, malonyl-ginsenoside Rb2; 25, quinquenoside R1; 26, malonyl-ginsenoside Rb3; 27, malonyl-ginsenoside Rb3 isomer; 28, ginsenoside Rd; 29, malonyl-ginsenoside Rd; 30, ginsenoside Rs1; 31, malonyl-ginsenoside Rd isomer; 32, gypenoside XVII; 33, ginsenoside Rs2; 34, ginsenoside Ra7; 35, notoginsenoside Fe; 36, ginsenoside Ra8; 37, ginsenoside F4; 38, vinaginsenoside R16; 39, ginsenoside Rg3. UPLC-QTOF-MS, ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry.
Table 2.
Characterization of ginsenosides in MCG and CG using UPLC-QTOF-MS
| No. | tR(min) | Precursor ion and/or adduct ions | Exact mass [M+H]+ | Error (ppm) | Formula | Identification |
|---|---|---|---|---|---|---|
| 1 | 1.99 | 933.5476 [M+H]+ | 933.5423 | 5.6 | C47H80O18 | Ginsenoside Re4 |
| 2 | 2.08 | 963.5582 [M+H]+, 980.5865 [M+NH4]+ | 963.5529 | 5.5 | C48H82O19 | Notoginsenoside R3 isomer |
| 3 | 2.11 | 933.5474 [M+H]+ | 933.5423 | 5.4 | C47H80O18 | Notoginsenoside R1 |
| 4 | 2.50 | 947.5628 [M+H]+ | 947.5579 | 5.1 | C48H82O18 | Ginsenoside Re |
| 5 | 2.50 | 801.5038 [M+H]+ | 801.5000 | 4.7 | C42H72O14 | Ginsenoside Rg1 |
| 6 | 2.77 | 887.5040 [M+H]+, 904.5305 [M+NH4]+ | 887.5004 | 4.0 | C45H74O17 | Malonyl-ginsenoside Rg1 |
| 7 | 2.98 | 1,033.5633[M+H]+ | 1,033.5583 | 4.8 | C51H84O21 | Malonyl-ginsenoside Re |
| 8 | 3.03 | 1,033.5630 [M+H]+ | 1,033.5583 | 4.5 | C51H84O21 | Malonyl-ginsenoside Re isomer |
| 9 | 4.02 | 1,241.6609 [M+H]+ ,1,258.6971 [M+NH4]+ | 1,241.6530 | 6.3 | C59H100O27 | Ginsenoside Ra3/ notoginsenoside R4 / notoginsenoside Fa |
| 10 | 4.13 | 1,327.6656[M+H]+, 1,327.6980 [M+NH4]+ | 1,327.6534 | 9.1 | C62H102O30 | Malonyl-ginsenoside Ra3 |
| 11 | 4.25 | 801.5033 [M+H]+ | 801.5000 | 4.1 | C42H72O14 | Ginsenoside Rf |
| 12 | 4.37 | 1,327.6666[M+H]+ | 1,327.6534 | 9.9 | C62H102O30 | Malonyl-notoginsenoside R4 |
| 13 | 4.45 | 1,211.6492[M+H]+, 1,228.6871 [M+NH4]+ | 1,211.6425 | 5.5 | C58H98O26 | Ginsenoside Ra2 |
| 14 | 4.56 | 1,109.6176 [M+H]+, 1,126.6500 [M+NH4]+ | 1,109.6108 | 6.1 | C54H92O23 | Ginsenoside Rb1 |
| 15 | 4.60 | 1,327.6655[M+H]+ | 1,327.6534 | 9.1 | C62H102O30 | Malonyl-notoginsenoside Fa |
| 16 | 4.65 | 1,195.6194 [M+H]+ | 1,195.6112 | 6.8 | C57H94O26 | Malonyl-ginsenoside Rb1 |
| 17 | 4.77 | 1,079.6058 [M+H]+ | 1,079.6002 | 5.1 | C53H90O22 | Ginsenoside Rc |
| 18 | 4.77 | 1,211.6507[M+H]+, 1,228.6721 [M+NH4]+ | 1,211.6425 | 6.7 | C58H98O26 | Ginsenoside Ra1 |
| 19 | 4.85 | 1,165.6094 [M+H]+ | 1,165.6006 | 7.2 | C56H92O25 | Malonyl-ginsenoside Rc |
| 20 | 4.85 | 1297.6490 [M+H]+ | 1,297.6429 | 4.7 | C61H100O29 | Malonyl-ginsenoside Ra2/Ra1 |
| 21 | 4.91 | 1,195.6187 [M+H]+, 1,212.9451 [M+NH4]+ | 1,195.6112 | 6.2 | C57H94O26 | Malonyl-ginsenoside Rb1 isomer |
| 22 | 4.99 | 1,079.6069 [M+H]+, 1,096.6310 [M+NH4]+ | 1,079.6002 | 6.2 | C53H90O22 | Ginsenoside Rb2 |
| 23 | 4.99 | 1,079.6069[M+H]+, 1,096.6312 [M+NH4]+ | 1,079.6002 | 6.2 | C53H90O22 | Ginsenoside Rb3 |
| 24 | 5.10 | 1165.6088 [M+H]+ ,1182.641 [M+NH4]+ | 1165.6006 | 7.0 | C56H92O25 | Malonyl-ginsenoside Rb2 |
| 25 | 5.23 | 1151.6284[M+H]+ ,1168.6471 [M+NH4]+ | 1151.6213 | 6.1 | C56H94O24 | Quinquenoside R1 |
| 26 | 5.37 | 1165.6067 [M+H]+ ,1182.641 [M+NH4]+ | 1165.6006 | 5.2 | C56H92O25 | Malonyl-ginsenoside Rb3 |
| 27 | 5.41 | 1165.6085 [M+H]+ ,1182.641 [M+NH4]+ | 1165.6006 | 6.7 | C56H92O25 | Malonyl-ginsenoside Rb3 isomer |
| 28 | 5.55 | 947.5621 [M+H]+ ,964.5913 [M+NH4]+ | 947.5579 | 4.4 | C48H82O18 | Ginsenoside Rd |
| 29 | 5.64 | 1033.5644 [M+H]+ ,1050.590 [M+NH4]+ | 1033.5583 | 5.9 | C51H84O21 | Malonyl-ginsenoside Rd |
| 30 | 5.76 | 1121.6008 [M+H]+ | 1121.6108 | −8.2 | C55H92O23 | Ginsenoside Rs1 |
| 31 | 5.92 | 1033.5653[M+H]+ ,1050.590 [M+NH4]+ | 1033.5583 | 6.7 | C51H84O21 | Malonyl-ginsenoside Rd isomer |
| 32 | 5.95 | 947.5623[M+H]+ | 947.5579 | 4.6 | C48H82O18 | Gypenoside XVII |
| 33 | 6.01 | 1121.6180 [M+H]+ | 1121.6108 | 5.9 | C55H92O23 | Ginsenoside Rs2 |
| 34 | 6.12 | 1147.6347[M+H]+ | 1147.6264 | 7.2 | C57H94O23 | Ginsenoside Ra7 |
| 35 | 6.28 | 917.5440 [M+H]+ | 917.5474 | −3.7 | C47H80O17 | Notoginsenoside Fe |
| 36 | 6.36 | 1147.6348[M+H]+ | 1147.6264 | 7.3 | C57H94O23 | Ginsenoside Ra8 |
| 37 | 6.40 | 767.4987[M+H]+ | 767.4946 | 5.3 | C42H70O12 | Ginsenoside F4 |
| 38 | 6.51 | 917.5518[M+H]+ | 917.5474 | 4.7 | C47H80O17 | Vinaginsenoside R16 |
| 39 | 7.29 | 785.5082[M+H]+ | 785.5051 | 3.9 | C42H72O13 | Ginsenoside Rg3 |
| 40 | 16.68 | 663.4530[M+H]+ ,685.4382[M+Na]+ | 663.4472 | 8.7 | C38H62O9 | Ginsenosde Rs6/Rs7 |
CG, cultivated ginseng; MCG, mountain-cultivated ginseng; UPLC-QTOF-MS, ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry
3.2. PCA
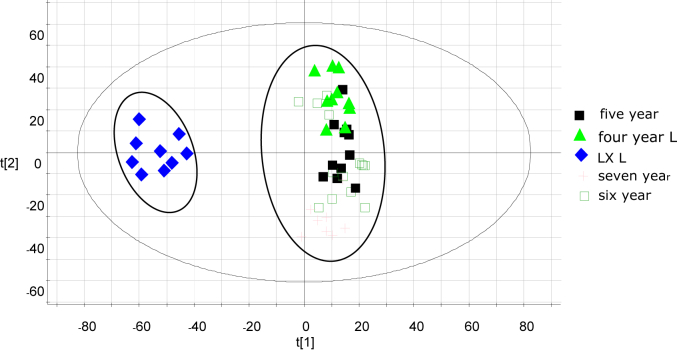
It was difficult to identify the MCG and CG from the BPI chromatograms. In this case, an effective approach for discerning their differences was MVA, which has been widely used in the metabolomics field in recent years to elucidate extremely complex samples [26]. We were able to distinguish between CG and MCG from the PCA and S-plot analysis.
A two-component PCA score plot of UPLC-QTOF-MS data was utilized to depict the general variation of components among the Panax ginseng samples (Fig. 3). The PCA score plot in Fig. 3 was divided into two clusters, with the CG samples of different years clustered into one group, while the MCG samples were clustered into another group. The MCG and CG samples were clearly separated by principal component 1 (PC1), indicating that their components differed between CG and MCG.
Fig. 3.
Principal component analysis of mountain-cultivated ginseng and cultivated ginseng.
3.3. Marker-ion analysis
It was evident that samples were clustered into two groups, with one MCG and the other CG, confirming that MCG and CG components differed in level and occurrence.
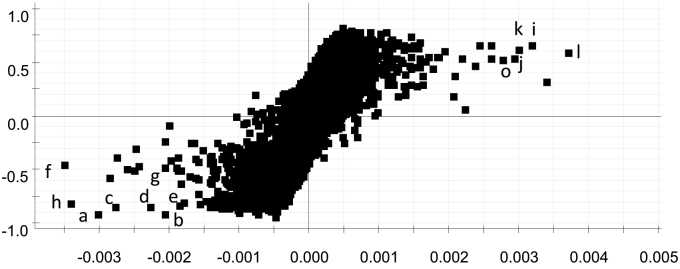
To explore the potential chemical markers that contributed to the differences between MCG and CG, UPLC-QTOF-MS/MS data from MCG and CG samples were processed by supervised OPLS-DA. In the S-plot (Fig. 4), each point represented an EMRT pair (a marker). The x-axis shows the variable contributions. The further away a data point is from the 0 value, the more it contributes to sample variance. The y-axis shows the correlations within the same sample group. The further away an EMRT pair is from the 0 value, the greater its correlation from injection to injection. As shown in the S-plot in Fig. 4, the first eight ions, a (tR 16.68 min, m/z 685.4382), b (tR 16.67 min, m/z 686.4420), c (tR 5.23 min, m/z 1,151.6284), d (tR 6.12 min, m/z 1,147.6347), e (tR 6.28 min, m/z 917.5518), f (tR 4.45 min, m/z 1,211.6492), g (tR 4.02 min, m/z 1,241.6509), and h (tR 5.95 min, m/z 947.5623) at the lower left of the “S” were the ions from MCG that contributed mostly to the differences between MCG and CG. Analogously, the first five ions, j (tR 4.85 min, m/z 1,165.6094), i (tR 8.05 min, m/z 295.2263), k (tR 11.18 min, m/z 355.2849), l (tR 4.65 min, m/z 1,195.6194), and o (tR 5.10 min, m/z 1,165.6088) in the top right corner of the “S”, were ions from CG that contributed mostly to the differences between MCG and CG. These ions could be used as potential chemical markers to distinguish MCG from CG.
Fig. 4.
The S-plot of mountain-cultivated ginseng and cultivated ginseng; a ion (tR 16.68 min, m/z 685.4382), b ion (tR 16.67 min, m/z 686.4420), c ion (tR 5.23 min, m/z 1,151.6284), d ion (tR 6.12 min, m/z 1,147.6347), e ion (tR 6.28 min, m/z 917.5518), f ion (tR 4.45 min, m/z 1,211.6492), g ion (tR 4.02 min, m/z 1,241.6509), and h ion (tR 5.95 min, m/z 947.5623); j ion (tR 4.85 min, m/z 1,165.6094), i ion (tR 8.05 min, m/z 295.2263), k ion (tR 11.18 min, m/z 355.2849), i ion (tR 4.65 min, m/z 1,195.6194), and o ion (tR 5.10 min, m/z 1,165.6088).
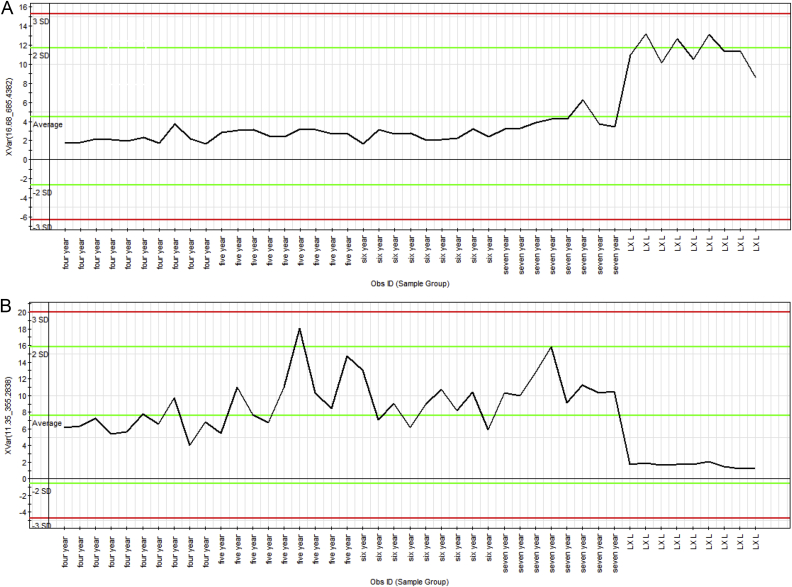
Additionally, we confirmed these spectral variables using the ion intensity plot. The ion intensity plot (Fig. 5) generated by the Marker Lynx software was a convenient instrument for profiling marker ions. The marker ion tR 16.68 min, m/z 685.4382 (Fig.5A) was ginsenoside Rs6/Rs7 from the MCG, and the marker ion tR 11.18 min, m/z 355.2849 (Fig. 5B) was ethyl linoleate from the CG. The representative ion intensity plot illustrated the abundance of marker ions tR 16.68 min, m/z 685.4382 and tR 11.18 min, m/z 355.2849 among the 45 samples. The ions fulfilled the criteria of marker ions, because they were found at higher levels in one herb, but not others.
Fig. 5.
Representative ion-intensity plot for marker ions over 45 samples. (A) Ginsenoside Rs6/Rs7 at m/z 685.4382 (tR 16.68 min) and (B) ethyl linoleate at m/z 355.2849 (tR 11.18 min).
3.4. Component assignments
After obtaining the potential markers, element-composition calculation was performed for the target markers. The molecular formula of markers was obtained by comparing the accurate masses. The next step was to search in a database and correlate references in order to identify marker structures, which were illuminated by the fragments which appeared in the high CE scan. The results are summarized in Table 3.
Table 3.
Characterization of different variable ions from MCG and CG
| Mark ion | Identification | tR (min) | Molecular formula | Ion | Mean measured mass |
Theoretical exact mass |
Mass accuracy (ppm) |
Fragment ions | Classification |
|---|---|---|---|---|---|---|---|---|---|
| l | Malonyl-ginsenoside Rb1 | 4.65 | C57H94O26 | M+H+ | 1,195.6194 | 1,195.6112 | 6.8 | 1,109, 1,015, 853, 785, 425 | CG |
| j | Malonyl-ginsenoside Rc | 4.85 | C56H92O25 | M+H+ | 1,165.6094 | 1,165.6006 | 7.5 | 1,079, 871, 853, 411 | CG |
| o | Malonyl-ginsenoside Rb2 | 5.10 | C56H92O25 | M+H+ | 1,165.6088 | 1,165.6006 | 7.0 | 1,079, 871, 853, 411 | CG |
| i | Palmitoleic acid | 8.05 | C16H30O2 | M+H2O+Na+ | 295.2263 | 295.2249 | 4.7 | 277, 170 | CG |
| k | Ethyl linoleate | 11.18 | C20H36O2 | M+HCOOH+H+ | 355.2849 | 355.2848 | 0.3 | 309, 263, 268 | CG |
| g | Ginsenoside Ra3 /isomer | 4.02 | C60H100O27 | M+H+ | 1,241.6509 | 1,241.6630 | 6.3 | 1,109, 1,079, 947, 767 | MCG |
| f | Ginsenoside Ra2 | 4.45 | C58H98O26 | M+H+ | 1,211.6492 | 1,211.6425 | 5.5 | 1,193, 1,031, 887, 869 | MCG |
| c | Quinquenoside R1 | 5.23 | C56H94O24 | M+H+ | 1,151.6284 | 1,151.6213 | 6.1 | 1,109, 785 | MCG |
| h | Gypenoside XVII | 5.95 | C48H82O18 | M+H+ | 947.5623 | 947.5579 | 4.7 | 785, 767, 605, 443 | MCG |
| d | Ginsenoside Ra7 | 6.12 | C57H94O23 | M+H+ | 1,147.6347 | 1,147.6264 | 7.2 | 917, 835, 755 | MCG |
| e | Notoginsenoside Fe | 6.28 | C47H80O17 | M+H+ | 917.5518 | 917.5474 | 4.7 | 899, 785, 737, 605 | MCG |
| a, b | Ginsenoside Rs6/Rs7 | 16.68 | C38H62O9 | M+Na+ | 685.4382 | 685.4292 | 13.1 | 663 | MCG |
CG, cultivated ginseng; MCG, mountain-cultivated ginseng
By matching the retention times and accurate masses with the published compounds, ions a (tR 16.68 min, m/z 685.4382) and b (tR 16.67 min, m/z 686.4420) in the MCG samples were both tentatively assigned as ginsenoside Rs6/Rs7. Ions c (tR 5.23 min, m/z 1,151.6284), d (tR 6.12 min, m/z 1,147.6347), e (tR 6.28 min, m/z 917.5518), f (tR 4.45 min, m/z 1,211.6492), g (tR 4.02 min, m/z 1,241.6509), and h (tR 5.95 min, m/z 947.5623) in the MCG samples were identified as quinquenoside R1, ginsenoside Ra7, notoginsenoside Fe, ginsenoside Ra2, ginsenoside Ra3 /isomer, and gypenoside XVII, respectively. Ions j (tR 4.85 min, m/z 1,165.6490), i (tR 8.05 min, m/z 295.2263), k (tR 11.18 min, m/z 355.2849), l (tR 4.65 min, m/z 1,195.6194) and o (tR 5.10 min, m/z 1,165.6088) in the CG samples were confirmed to be malonyl-ginsenoside Rc, palmitoleic acid, ethyl linoleate, malonyl-ginsenoside Rb1, and malonyl-ginsenoside Rb2, respectively.
From the different components between MCG and CG, we learned that CG have more malony ginsenosides, and the MCG have more minor ginsenosides. The malonyl ginsenosides are the original type of ginsenoside and occur naturally in ginseng. It is reasonable to deduce that growth circumstances make the malonyl ginsenosids transform into minor ginsenosides through hydrolysis, de-glycosylation, dehydration, and acetylation. This is the chemical basis of MCGs that are directly related to their pharmacological activities. This study illustrated the differences between MCG and CG, and provided a basis for further MCG research.
4. Conclusion
The generic UPLC/oa-QTOF-MSE sample-profiling strategy allows multiple groups of complex samples to be studied by using the MVA approach. The combination of high-resolution UPLC separation and high-resolution MS detection, coupled with MVA, allowed details of the samples to be profiled, enabling important variance markers to be measured, even at low concentration levels. Our results constitute the first time that differences between CG and MCG have been observed systematically at the chemical-component level.
Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (no. 81073041) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (no. 20100013120010).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2015.11.001.
Contributor Information
Feng Wei, Email: weifeng@nifdc.org.cn.
Xiang-ri Li, Email: lixiangri@sina.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Ang-Lee M.K., Moss J., Yuan C.S. Herbal medicines and perioperative care. J Am Med Assoc. 2001;286:20. doi: 10.1001/jama.286.2.208. [DOI] [PubMed] [Google Scholar]
- 2.Attele A.S., Wu J.A., Yuan C.S. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 3.Lars P.C., Martin J., Ulla K. Simultaneous determination of ginsenosides and polyacetylenes in American ginseng root (Panax quinquefolium L.) by high-performance liquid chromatography. J Agric Food Chem. 2006;54:8995–9003. doi: 10.1021/jf062068p. [DOI] [PubMed] [Google Scholar]
- 4.Zhang D., Yasuda T., Yu Y., Zheng P., Kawabata T., Ma Y., Okada S. Ginseng extract scavenges hydroxyl radical and protects unsaturated fatty acids from decomposition caused by iron-mediated lipid peroxidation. Free Radic Biol Med. 1996;20:145–150. doi: 10.1016/0891-5849(95)02020-9. [DOI] [PubMed] [Google Scholar]
- 5.Yun T.K., Lee Y.S., Lee Y.H., Kim S.I., Yun H. Anti-carcinogenic effect of Panax ginseng C.A. Meyer and identification of active compounds. J Korean Med Sci. 2001;16:S6–S18. doi: 10.3346/jkms.2001.16.S.S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joo S.S., Won T.J., Lee D.I. Reciprocal activity of ginsenosides in the production of proinflammatory repertoire, and their potential roles in neuroprotection in vitro. Planta Med. 2005;71:476–481. doi: 10.1055/s-2005-864145. [DOI] [PubMed] [Google Scholar]
- 7.Jung C.H., Seog H.M., Choi I.W., Choi H.D., Cho H.Y. Effects of wild ginseng (Panax ginseng C.A. Meyer) leaves on lipid peroxidation levels and antioxidant enzyme activities in streptozotocin diabetic rats. J Ethnopharmacol. 2005;98:245–250. doi: 10.1016/j.jep.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine-ginseng (I): etymology, pharmacognosy, phytochemistry, market and regulations. Curr Med Chem. 2009;16:2475. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelova N., Kong H.W., Van Der Heijden R., Yang S.Y., Choi Y.H., Kim H.K., Wang M., Hankemeier T., Van Der Greef J., Xu G. Recent methodology in the phytochemical analysis of ginseng. Phytochem Anal. 2008;19:2–16. doi: 10.1002/pca.1049. [DOI] [PubMed] [Google Scholar]
- 10.Liu C.X., Xiao P.G. Recent advances on ginseng research in China. J Ethnopharmacol. 1992;36:27–38. doi: 10.1016/0378-8741(92)90057-x. [DOI] [PubMed] [Google Scholar]
- 11.Lü J.M., Yao Q., Chen C. Ginseng compounds: an update on their molecular mechanisms and medical applications. Curr Vasc Pharmacol. 2009;7:293. doi: 10.2174/157016109788340767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y., Zhao W., Zhang Q., Zhao Y., Zhang Y. Purification and characterization of a novel and unique ginsenoside Rg1-hydrolyzing β-d-glucosidase from Penicillium sclerotiorum. Acta Biochim Biophys Sin. 2011;43:226–231. doi: 10.1093/abbs/gmr001. [DOI] [PubMed] [Google Scholar]
- 13.Li S.Y., Wang S. Brief introduction to the origin, shape, production area, sort, and differentiating of wild ginseng. Asia-Pac Tradit Med. 2008;4:37–39. [Google Scholar]
- 14.Zhong F.L., Li P.Y. Research progress of ginseng under forest. J Ginseng Res. 2006;2:8–10. [Google Scholar]
- 15.Zhong W.T., Wu X.Z., Yu Z.J., Jin J.R. Definition and identification of wild ginseng, garden ginseng, ginseng under forest. J Ginseng Res. 2006;2:14–15. [Google Scholar]
- 16.Chinese Pharmacopeia Committee . Chinese Medicine Science and Technology; Beijing: 2005. Pharmacopeia of People’s Republic of China. [Google Scholar]
- 17.Liu D., Li Y.G., Xu H., Sun S.Q., Wang Z.T. Differentiation of the root of cultivated ginseng, mountain cultivated ginseng, and mountain wild ginseng using FT-IR and two-dimensional correlation IR spectroscopy. J Mol Struct. 2008;883–884:228–235. [Google Scholar]
- 18.Bu H.B., Wang F., Lin H.Y., Guo Z.Y., Yuan S.X., Pan L.L., Xu X.J., Li X.R., Wang G.L., Lin R.C. Nondestructive recognization of mountain cultivated ginseng and garden cultivated ginseng by FTIR microspectroscopy. Guang Pu Xue Yu Guang Pu Fen Xi. 2013;33:3028–3031. [PubMed] [Google Scholar]
- 19.Liu Z., Ruan C.C., Liu T.Z., Wang L.J., Zheng Y.N., Sun G.Z. Simultaneous determination of 14 kinds of ginsenosides in similar wild ginseng, fresh ginseng, white ginseng, and red ginseng by HPLC. Chin Tradit Herb Drugs. 2012;43:2431–2434. [Google Scholar]
- 20.Kim S.J., Shin S.S., Seo B.I., Jee S.Y. Effect of mountain grown ginseng radix, mountain cultivated ginseng radix, and cultivated ginseng radix on apoptosis of HL-60 cells. Korea J Herbol. 2004;19:41. [Google Scholar]
- 21.Hwang J.W., Oh J.H., Yoo H.S., Lee Y.W., Cho C.K., Kwon K.R., Yoon J.H., Park J., Her S., Lee Z.-W. Mountain ginseng extract exhibits anti-lung cancer activity by inhibiting the nuclear translocation of NF-κB. Am J Chin Med. 2012;40:187–202. doi: 10.1142/S0192415X12500152. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H.M., Li S.L., Zhang H., Wang Y., Zhao Z.L., Chen S.L., Xu H.X. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J Pharm Biomed Anal. 2012;62:258–273. doi: 10.1016/j.jpba.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Kim N., Kim K., Lee D., Shin Y.S., Bang K.H., Cha S.W., Lee J.W., Choi H.K., Hwang B.Y., Lee D. Nontargeted metabolomics approach for age differentiation and structure interpretation of age-dependent key constituents in hairy roots of Panax ginseng. J Nat Prod. 2012;75:1777–1784. doi: 10.1021/np300499p. [DOI] [PubMed] [Google Scholar]
- 24.Song H.H., Moon J.Y., Ryu H.W., Noh B.S., Kim J.H., Lee H.K., Oh S.R. Discrimination of white ginseng origins using multivariate statistical analysis of data sets. J Ginseng Res. 2014;38:187–193. doi: 10.1016/j.jgr.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie G.X., Ni Y., Su M.M., Zhang Y.Y., Zhao A.H., Gao X.F., Liu Z., Xiao P.G., Jia W. Application of ultra-performance LC-TOF MS metabolite profiling techniques to the analysis of medicinal Panax herbs. Metabolomics. 2008;4:248–260. [Google Scholar]
- 26.Wilson I.D., Nicholson J.K., Castro-Perez J., Granger J.H., Johnson K.A., Smith B.W., Plumb R.S. High resolution “ultra performance” liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.