Abstract
Background/Objectives
Pigmentous gallstones occur in South Indians despite significant higher levels of circulating cholesterol. This study was conducted to identify the biochemical and/or genetic causes for the formation of pigmentous gallstones in this ethnic group.
Methods
Plasma lipid profile, bile cholesterol, acids, and phospholipid levels were estimated in patients with gall stone disease and age, sex matched controls using standard protocols. Twenty-seven SNPs related to cholesterol and bilirubin metabolism pathway genes were genotyped in the study population using the Sequenom platform. An equilibrium phase diagram involving bile salt-phospholipid-cholesterol was generated to relate phenotype with the genotype.
Results
There were no significant differences in the lipid profiles between the patients (n = 305) and controls (n = 177). Biliary cholesterol, acids, and phospholipids were significantly different between patients and controls. Single locus analysis revealed association of variants in ABCG6, ABCG8, and UGT1A1 genes with the disease; however when correction was applied as multiple testing was done, only one variant (rs6742078) in UGT1A1 gene was found to be associated with gall stone disease. Equilibrium phase diagram suggested that few samples were in the crystal formation zone. The mutant, but not wild type or heterozygous genotype of SNPs (rs6742078 and rs887829) in UGT1A1 gene, was associated with significantly higher levels of bilirubin.
Conclusions
Higher incidence of pigment stones in South Indians could be due to raised serum bilirubin levels that may be ascribed to variant in the UGT1A1 gene involved in glucuronidation of free bilirubin.
Abbreviations: GSD, gallstone disease; ABCG, 8 ATP-binding cassette, sub-family G (WHITE), member 8; SNPs, single nucleotide polymorphisms; UGT1A1, UDP glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1); ABCG6, ATP-binding cassette protein subfamily G, member 6; LDL, low density lipoprotein; HDL, high density lipoprotein; DNA, deoxyribose nucleic acid; PXR, pregnane C receptor; SD, standard deviation; OR, odds ratio
Keywords: pigmentous gall stones, polymorphisms, UGT1A1 gene, cholesterol gall stones, bilirubin
Introduction
Gallstone disease (GSD) is of global occurrence causing considerable morbidity and mortality apart from being the leading cause of hospitalization and drain on health-care resources.1 Many gallstones are silent, but symptoms and complications ensue in few of the cases, necessitating surgical removal of the gallbladder, usually by laparoscopic cholecystectomy.2, 3 The incidence of GSD in India and rest of the world is on the rise,4 reaching epidemiological proportions in American Indians (60-70%). However, a decrease in its incidence is reported in Hispanics of mixed Indian origin.5
Risk factors associated with GSD include female gender, age, obesity, metabolic syndrome, diabetes, hyperlipidemia, high caloric intake, and family history.6, 7, 8 Identifying risk factors that can be manipulated should provide an opportunity to prevent cholelithiasis.9 Accumulating evidence suggest that the pathogenesis of GSD is multifactorial, with both environmental and genetic factors being involved.10 Importantly, ABCG8 genetic variants confer significant risk for cholesterol gallstone disease.11
Gallstones can be classified into white or yellow colored stones that are rich in cholesterol, whereas pigment gall stones are mixed gallstones that are rich in bilirubin and mixed gallstones. Cholesterol gallstones containing >70% of cholesterol are more prevalent than the other two types. The black or brown colored pigment stones contain lesser cholesterol (<25–30%) and contain majorly insoluble bilirubin and calcium salts that are found in bile, while the cholesterol content in mixed stones is between 30 and 70%.12, 13 The major mechanisms of gall stone formation include biliary cholesterol hyper secretion, super saturation and crystallization, mucus hyper secretion, gel formation and bile stasis. Bile salts attach themselves to cholesterol molecules in the bile to keep them from crystallizing. However, when significant increases in biliary cholesterol contents are unaccompanied by a concomitant increase in bile salts, cholesterol- rich stones can be formed. On the contrary increased bilirubin in the bile leads to formation of pigment stones. Stone formation also occurs if there is an imbalance in the cholesterol transportation process.
Data on biliary cholesterol, bile acids, and phospholipids have earlier been plotted in an equilibrium phase diagram to understand factors leading to stone formation.14 The phase diagram depicts single phase at the bottom (denoting only micelles); two phases on the left (denoting micelles and cholesterol crystals containing zone); two phases on the right (denoting micelles and vesicle containing zone) and three phases at the center (denoting micelles, vesicles and cholesterol crystals containing zone). The relative percentage of cholesterol increases from baseline to top in the phase diagram, indicating progressive tendency of cholesterol crystallization. The relative percentage of phospholipids as compared to the bile salts increases from left to right, indicating more solubilization of cholesterol in vesicles with lower vesicular cholesterol to phospholipid ratios and less cholesterol crystallization. If gallstones are present in the supersaturated bile, competition may occur between the gall stone surface, growth and surrounding bile for available cholesterol molecules.15
Various genetic association studies from India associated polymorphisms in genes namely MspA1 in Cytochrome-17 gene,16 T190C in adrenergic receptor gene (ADRβ3) and C-1291G in ADRA2A gene,17 rs2234693 in ESR1, rs4994 in ADRB3 and rs11887534 in ABCG8,11, 18, 19 Exon4 C>A polymorphism in SLCO1B1,20 −75 G/A in APOA1 gene,21 Intron 5 insertion/deletion polymorphism of RAP gene (LRPAP1).22 Further, UGT1A1 but not HMOX1 gene promoter polymorphisms that confer increased risk of hyperbilirubinemia and gallstones in patients with hereditary spherocytosis.23
Previously we had studied the role of D19H and T400K polymorphisms of ATP-binding cassette, subfamily G, member 8 (ABCG8) gene and identified that D19H, but not SNP T400K, in the ABCG8 gene was significantly associated with Gall stone disease in an Indian population.19 However, individual SNPs have limited predictive value because of their modest effect on risk. Combination of gene variants improves the ability to predict the susceptibility of a population in an efficient manner. Therefore, the current study was envisaged to genotype individuals for SNPs in important pathways related to cholesterol and bilirubin metabolism to identify susceptibility and causative variants for gall stone formation in South Indians. Data generated in this regard were analyzed in combination with estimates of biliary cholesterol, bile salts, pigments and phospholipids, which were used for generating equilibrium phase diagram to understand factors leading to stone formation.
Methods
Study Population
The study group for genotyping comprised patients (N = 305) with gall stone disease as well as age and sex matched controls (N = 177) without the disease. Presence of gallstones was diagnosed by B-mode ultrasound and stone samples were collected after cholecystectomy. Simultaneously bile samples were also collected from patients (n = 45) as well as from controls (n = 39) for estimating biliary cholesterol, bile acids and bilirubin contents. Information concerning demographic details, clinical and family history, and dietary habits was collected from both the study groups using a designed proforma. Written informed consent was obtained from all the individuals, and the study protocol was approved by the institutional research committee.
Serum and Biliary Lipid Analysis
Total cholesterol, low density lipoprotein (LDL), high density lipoprotein (HDL), and triglycerides were estimated in serum using commercially available kits (Randox laboratories, Crumlin, Antrim, UK). Bile was collected from patients with GSD during cholecystectomy for obtaining gallstones and from the controls while they underwent Whipple's procedure for unrelated ailments such as periampullary carcinoma, hepato-jejunostomy. Biliary cholesterol, bile acids, and bilirubin contents were estimated using enzymatic colorimetric methods (Randox laboratories Ltd. Crumlin, UK) and biliary phospholipids were estimated by a colorimetric method (BioAssay Systems, Hayward, USA). Equilibrium phase diagrams were generated as per Venneman, 201014 including data obtained on biliary contents of cholesterol, bile acids, and phospholipids.
Gallstone Analysis
Gallstones collected from patients after cholecystectomies were ground to fine powder in a mortar and pestle. Contents of total cholesterol in the samples were determined24, 25 involving an enzymatic method.
DNA Isolation and Genotyping
DNA was isolated from leukocytes using a commercial kit and as per the manufacturer's protocol (Bioserve Biotechnologies, India). The concentration and integrity of DNA was measured using Nanodrop 1000 spectrophotometer (Thermo Scientific, USA) and agarose gel electrophoresis respectively. DNA with A260/280 ratios between 1.8–2.0 and agarose gel image showing a high molecular weight intact DNA band were included for genotyping analysis. The samples were genotyped for 27 SNPs (Table 1) on the Sequenom platform (Sequenom®, San Diego, CA, USA) using the manufacturer's protocol. The raw data files generated by Mass Array Sequenom were analyzed for the intensity peaks of calibrant to ascertain the quality of the data. An overall call rate of > 95% was maintained. Five percent of the samples were duplicated across the plate and their genotypes were compared, and they had 100% concordance. Negative controls (master mix without DNA) were also included.
Table 1.
Genes Analyzed for Putative Role in the Etiology of Gallstone Disease.
| S. no. | Genes | Chr. locus | SNPs | Metabolism; Function |
|---|---|---|---|---|
| 1 | ABCA1-A | 9q22-q31 | rs2249891 | ABC transporter effluxes cholesterol and phospholipids; HDL metabolism |
| 2 | ABCC6 | 16p13 | rs150468 | HDL metabolism, insulin resistance |
| 3 | ABCC6 | 16p13 | rs212077 | HDL metabolism, insulin resistance |
| 4 | ABCG5 | 2p21 | rs6720173 | Cholesterol metabolism-Cholesterol transportation |
| 5 | ABCG8 | 2p21 | rs11887534 | Cholesterol metabolism-Cholesterol transportation |
| 6 | Apo B | 2p24 | rs520354 | Lipid metabolism pathway |
| 7 | APOA2 | 1q23 | rs3813627 | HDL metabolism, insulin resistance |
| 8 | CAV1 | 7q31 | rs926198 | Regulation of intracellular cholesterol homeostasis |
| 9 | CAV1 | 7q31 | rs3807989 | Regulation of intracellular cholesterol homeostasis |
| 10 | CETP | 16q21 | rs3764261 | Facilitates the exchange of natural lipid among the plasma lipoproteins |
| 11 | CUBN | 10p12 | rs7893395 | HDL metabolism |
| 12 | FABP5 | 8q21 | rs454550 | Free fatty acids carriers in the cytoplasm |
| 13 | LIPC | 15q21-q23 | rs1077834 | Lipid metabolism |
| 14 | LIPC | 15q21-q23 | rs4775065 | Lipid metabolism |
| 15 | LPL | 8p22 | rs6993414 | Encodes the lipoprotein lipase enzyme, have been associated with higher serum LDL levels |
| 16 | MUPCDH | rs3758650 | Mucin | |
| 17 | PNPLA3 | 22q13 | rs738409 | PNPLA3 risk allele is associated with severe forms of hepatic fat accumulation |
| 18 | PXR | rs2461823/A | Mediated lipid accumulation was the result of increased hepatic free fatty acid uptake induced by the activation of the fatty acid transporter | |
| 19 | RXRA | rs11185660 | HDL metabolism, insulin resistance | |
| 20 | SCARB1 (SR-BI) | 12q24 | rs838895 | Impact serum levels of HDL cholesterol and triglycerides |
| 21 | SHBG | 17p13-p12 | rs6259 | Hormone metabolism |
| 22 | SLCO1A2 | 12p12 | rs4149000 | Risk variants present with lower total serum cholesterol concentrations |
| 23 | SLCO1B1 | 12p | rs4149056 | SLCO1B1 encodes a liver-specific member of the organic anion transporter family |
| 24 | SLCO1B1 | 12p | rs11045819 | SLCO1B1 encodes a liver-specific member of the organic anion transporter family |
| 25 | SR-BI | 12q24 | rs2278986 | SR-BI is a plasma membrane protein that binds high density lipoprotein (HDL) with high affinity and mediates selective uptake of cholesterol esters by the liver |
| 26 | UGT1A | 2q37 | rs887829 | Major regulator of bilirubin metabolism |
| 27 | UGT1A1 | 2q37 | rs6742078 | Regulator of bilirubin metabolism |
Statistical Analysis
Data were edited for consistency and was entered into MS-Excel for further analysis. Clinical and biochemical characteristics were compared using Student's t test for continuous variables and proportion test for categorical variables. χ2 test was used on the number of variant carriers in controls and patient groups for identifying SNPs associated with GSD. To correct for multiple comparison testing, the Benjamini-Hochberg false discovery rate correction26 was applied to P values. Multiple logistic regression was used to identify independent predictor variables for GSD. Data was analyzed using Statistical Package for Social Sciences (SPSS Version 17). P value ≤ 0.05 was considered as statistically significant.
Results
Patient Characteristics
A total of 482 subjects including individuals with GSD (n = 305; 43 ± 7.7 years) as well as age and sex matched controls (n= 177; 42 ± 13.5 years) comprised the study group for genotyping. Significant differences between the two groups with regard to demographic and biochemical parameters are as shown in Table 2.
Table 2.
Demographic, Anthropometric and Clinical Details of the Study Group.
| Parameter | Controls (n = 177) | Patient (n = 305) | P value |
|---|---|---|---|
| Age (years) | 42 ± 13.5 | 43 ± 7.69 | 0.36 |
| Sex (Females) | 54 (30.5%) | 178 (58.4%) | 0.001* |
| BMI (kg/m2) | 26.4 ± 6.09 | 26.1 ± 5.05 | 0.58 |
| Diabetes (number) | 21/177 (11.9%) | 51/303(16.8%) | 0.60 |
| Hypertension (number) | 38/177(21.5%) | 78/303(25.7%) | 0.28 |
| Alcohol consumption (number) | 45/177 (25.4%) | 38/302 (12.6%) | 0.001* |
| Laboratory investigations | |||
| Total cholesterol (mg/dL) | 169 ± 24.1 | 171 ± 36.7 | 0.51 |
| HDL (mg/dL) | 39.4 ± 25.8 | 37.6 ± 16.9 | 0.40 |
| LDL (mg/dL) | 100 ± 16.1 | 104 ± 31.8 | 0.10 |
| Triglycerides (mg/dL) | 152 ± 76.6 | 148 ± 70 | 0.56 |
| ALT (IU/L) | 25.43 ± 5.56 | 50.54 ± 72.43 | 0.0002* |
| AST (IU/L) | 25.65 ± 3.59 | 36.43 ± 40.08 | 0.003* |
| WBC (cells/mm3) | 9840 ± 6232 | 9239.29 ± 2178.74 | 0.14 |
| RBC (millions/mm3) | 5.91 ± 8.0 | 4.78 ± 0.52 | 0.01* |
| Hemoglobin (grams/dL) | 12.44 ± 2.87 | 12.87 ± 1.74 | 0.06 |
| Hematocrit (%) | 36.50 ± 7.31 | 39.07 ± 4.59 | 0.0001* |
| Mean corpuscular volume (fl) | 83.27 ± 6.94 | 83.57 ± 5.06 | 0.62 |
| Mean corpuscular hemoglobin (pg) | 27.73 ± 3.03 | 26.71 ± 2.14 | 0.0001* |
| Mean corpuscular hemoglobin concentration (g/dL) | 33.20 ± 1.47 | 31.86 ± 1.68 | 0.0001* |
| Red blood cell distribution width (%) | 14.27 ± 2.15 | 13.86 ± 1.46 | 0.02* |
| Platelets (lakhs/mm3) | 2.62 ± 1.23 | 5.17 ± 5.71 | 0.0001* |
| Polymorphs (%) | 69.17 ± 11.46 | 66.07 ± 9.12 | 0.003* |
| Lymphocytes (%) | 25.10 ± 10.85 | 27.61 ± 7.48 | 0.007* |
| Eosinophils (%) | 2.97 ± 2.03 | 4.50 ± 3.17 | 0.0001* |
| Monocytes (%) | 3.13 ± 0.82 | 2.79 ± 0.79 | 0.0001* |
Unpaired t test.
Statistically significant P value.
Biliary Cholesterol, Bile acid, and Phospholipid Contents
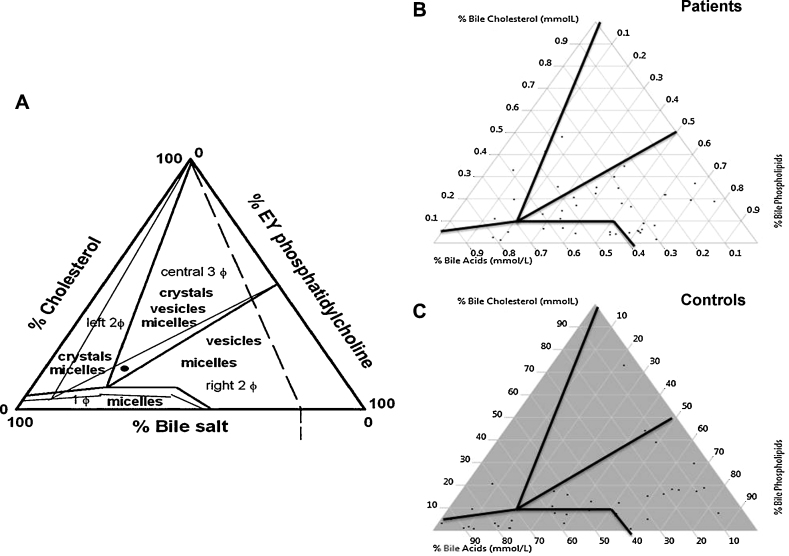
Gall bladder bile could be obtained from controls without GSD (n = 39) and patients with GSD (n= 45) for determining the contents of biliary cholesterol, bile acids and phospholipids (Table 3). Patients with GSD had enhanced levels of biliary cholesterol (P = 0.02), bile acids (P = 0.001) and biliary phospholipids (P = 0.0001) in comparison to the controls. When the data from bile analysis namely bile cholesterol, acids and phospholipids were plotted in a equilibrium diagram, data revealed that four samples from the controls and five samples from patients were in the crystal formation zone. Majority of the samples from the patients and controls were in the vesicles and micelles zone (Figure 1).
Table 3.
Bile Cholesterol, Acids and Phospholipids in the Study Group.
| Controls (n = 39) Mean ± SD–Median |
Patients (n = 45) Mean ± SD–Median |
P value | |
|---|---|---|---|
| Bile cholesterol (mmol/L) | 4.45 ± 5.88–2.44 | 9.40 ± 8.19–7.11 | 0.02* |
| Bile acids (mmol/L) | 16.86 ± 9.75–18.40 | 23.44 ± 8.63–22.80 | 0.001* |
| Bile phospholipids (mmol/L) | 14.04 ± 11.74–10.93 | 26.72 ± 15.45–27.76 | 0.0001* |
Statistically significant P value.
Figure 1.
Equilibrium phase diagram (A) as per reference 14. (B) Patients group. (C) Control group.
Gall Stone Analysis
On the basis of the content of cholesterol and color of the stone, majority (78.65%) of the stones were black or brown type and a few (21.34%) were white or yellow type. Of the 78.65%, 50% of the stones were black, 4.15% were brownish black, 14.23% were brown, 2.17% were brownish yellow, and 8.10% were greenish black. Of the 21.34% of white or yellow stones, 3.16% were greenish white, 5.14% were pale yellow, as well as 3.16% and 9.88% of white and yellow, respectively. Accumulation of biliary bilirubin was significantly high (P = 0.0001) in pigment gallstones (59.37 ± 29.54 mg/dL) compared to yellow colored gallstones (33.0 ± 26.84 mg/dL).
Genotype Association Data
In the single locus analysis, variant SNPs in ABCG8 (rs11887534), ABCC6 (rs150468), UGT1A1 (rs6742078, rs887829) and PXR (rs6771638) were significantly associated with higher risk of gall stone formation. However, as multiple SNPs were evaluated, Benjamini-Hochberg correction26 was applied to the P value from single locus analysis. Only variant in UGT1A1 (rs6742078) was significantly associated with GSD in this multiple SNP analysis.
Association of UGT1A1 SNPs to Mean Serum Bilirubin Levels
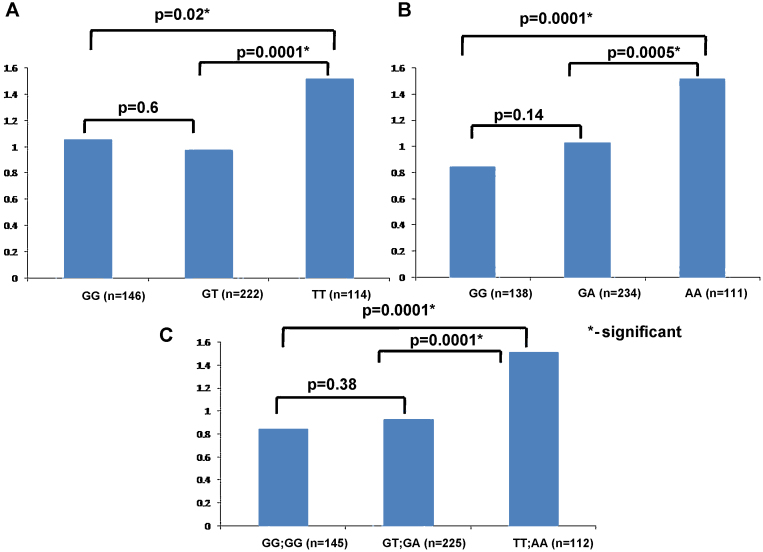
The mean bilirubin levels in sera were compared between wild type, heterozygous and mutant genotypes of the UGT1A1 SNPs (rs6742078 and rs887829). The mean ± SD for rs6742078 SNP genotypes GG, GT and TT were 1.06 ± 2.2, 0.98 ± 1.22 and 1.52 ± 1.46 respectively. The values for rs887829 SNP genotypes GG, GA and AA were 0.85 ± 1.26, 1.03 ± 1.37 and 1.52 ± 1.48 respectively. In both the SNP genotypes, the levels were significantly higher in the mutant genotype as compared to the wild type or to the heterozygous genotype (Figure 2 panel A and B). Combined analysis for both the UGT1A1 SNPs revealed serum bilirubin levels (mean ± SD) to be 0.85 ± 1.26 for individuals with wild type genotypes (GG and GG) as compared to 0.93 ± 0.79 for individuals with compound heterozygous genotype (GT and GA) and 1.52 ± 1.48 for individuals with mutant genotype (TT and AA). There was a significant difference (P = 0.0001) in the mean bilirubin levels between wild and mutant genotypes, compound heterozygous and mutant genotypes (P = 0.0001); however, there was no significant difference in the levels between the wild type and compound heterozygous genotype (P = 0.38) (Figure 2, panel C).
Figure 2.
Mean Bilirubin levels in the study group. (A) Mean bilirubin levels for three genotypes GG, GT, and TT for the SNP rs6742078. (B) Mean bilirubin levels for three genotypes GG, GA, and AA for the SNP rs887829. (C) Mean bilirubin levels for both the SNPs combined.
Multiple Logistic Regression
Age greater than 40 years, female sex and mutant genotype of ABCG8 gene were significant in the multiple logistic regression analysis
Discussion
The present study was conducted with the primary objective of understanding the genetic basis of pigmentous gall stone formation in South Indian patients. Correlative biochemical and genetic analysis of south Indian patients and matched controls revealed (i) increased serum bilirubin contents in patients with GSD and (ii) association of SNP in UGT1A1 (involved in the glucuronidation of bilirubin) gene with GSD.
Even though no significant differences could be noted with respect to the demographic characteristics and circulating lipid profiles between GSD patients and the control group, biliary contents of cholesterol, bile acids, and phospholipids were significantly different between the two groups (Table 3). Increased levels of biliary cholesterol alone do not indicate risk of cholesterol crystallization since it is accompanied with concomitant increase in the levels of bile acids and phospholipids in GSD patients. Such a thought was confirmed upon plotting a equilibrium phase diagram involving biliary contents of cholesterol, bile acids and phospholipids, an approach that resulted earlier in the identification of factors contributing to gall stone formation.14 It may be noted from Figure 1 that few samples from controls and patients were in the crystal formation zone and majority were in the vesicles and micelles zone.
In order to understand the genetic basis for gall stone formation in south Indians, GSD patients from this ethnicity were genotyped for a total of 27 variants predominantly from cholesterol and bilirubin metabolism genes (Table 1). In the present study although in single locus analysis SNPs in ABCG6, ABCG8 and UGT1A1 genes were significantly associated with a higher risk of gall stone formation, upon correction for multiple testing the involvement of UGT1A1 gene (Table 4; P = 0.03; OR = 2.23) in the formation of gall stones was noted. Irrespective of the low P value obtained for this gene, the high OR indicates its importance in the formation of gall stones, an observation akin to those made earlier by Buch et al.27
Table 4.
Genotype Data in the Study Group.
| Name of the Gene – SNP | Risk allele frequency |
P value | Corrected P value | Odds (95%CI) | |
|---|---|---|---|---|---|
| Controls N = 177 | Patients N = 305 | ||||
| LIPC – rs1077834 | 0.51 | 0.52 | 0.87 | 0.97 | 1.03 (0.66–1.61) |
| SLCO1B1 – rs11045819 | 0.75 | 0.63 | 0.35 | 0.68 | 0.58 (0.18–1.85) |
| RXRA – rs11185660 | 0.38 | 0.32 | 0.29 | 0.61 | 0.78 (0.49–1.24) |
| ABCG8 – rs11887534 | 0.05 | 0.13 | 0.02 | 0.14 | 2.52 (1.67–5.95) |
| LIPC – rs12324517 | 0.45 | 0.46 | 0.92 | 0.98 | 1.02(0.65–1.59) |
| ABCC6 – rs150468 | 0.65 | 0.30 | 0.02 | 0.14 | 0.23 (0.05–0.9) |
| ABCC6 – rs212077 | 0.02 | 0.009 | 0.94 | 0.98 | 0.37 (0.06–2.25) |
| ABCA1–A – rs2249891 | 0.11 | 0.11 | 0.98 | 0.98 | 1 (0.47–2015) |
| SR-BI – rs2278986 | 0.28 | 0.25 | 0.62 | 0.90 | 0.84(0.5–1.41) |
| MUPCDH – rs3758650 | 0 | 0.02 | 0.17 | 0.45 | 1.25 (1.35–2.86) |
| CETP – rs3764261 | 0.50 | 0.50 | 0.15 | 0.44 | 1.02 (0.65–1.6) |
| CAV1 – rs3807989 | 0.36 | 0.33 | 0.56 | 0.90 | 0.87 (0.54–1.38) |
| APOA2 – rs3813627 | 0.09 | 0.11 | 0.75 | 0.92 | 1.14 (0.55–2.38) |
| SLCO1A2 – rs4149000 | 0.008 | 0 | 0.76 | 0.92 | 1.52 (0.93–2.48) |
| SLCO1B1 – rs4149056 | 0.08 | 0.04 | 0.67 | 0.92 | 0.56 (0.03–9.03) |
| FABP5 – rs454550 | 0.008 | 0 | 0.73 | 0.92 | 0.18 (1.07–4.64) |
| APOB – rs520354 | 0.06 | 0.10 | 0.81 | 0.94 | 1.69 (0.81–3.51) |
| SHBG – rs6259 | 0.89 | 0.87 | 0.62 | 0.90 | 0.83(0.41–1.69) |
| ABCG5 – rs6720173 | 0.08 | 0.12 | 0.18 | 0.45 | 1.65 (0.77–3.54) |
| UGT1A1 – rs6742078 | 0.17 | 0.31 | 0.003 | 0.03* | 2.23 (1.28–3.87) |
| PXR – rs6771638 | 0.31 | 0.41 | 0.05 | 0.20 | 1.54 (0.99–2.39) |
| LPL – rs6993414 | 0.72 | 0.75 | 0.57 | 0.90 | 1.15 (0.69–1.92) |
| PNPLA3 – rs738409 | 0.56 | 0.62 | 0.29 | 0.61 | 1.27 (0.81–2.01) |
| CUBN – rs7893395 | 0.35 | 0.39 | 0.54 | 0.90 | 1.15 (0.72–1.83) |
| SCARB1 (SR–BI) – rs838895 | 0.04 | 0.07 | 0.13 | 0.41 | 1.45 (0.55–3.86) |
| UGT1A – rs887829 | 0.17 | 0.30 | 0.008 | 0.06 | 2.08 (1.19–3.61) |
| CAV1 – rs926198 | 0.16 | 0.09 | 0.06 | 0.24 | 0.53 (0.27-1.05) |
Statistically significant Odds ratio and Benjamini-Hochberg false discovery rate correction was applied to “P value” to adjust for multiple testing.
In the present study, majority of the stones were of pigment type (black or brown color) with accumulation of biliary bilirubin. Earlier studies suggested that there is a preponderance of pigment stones in South India,28, 29 although there are higher levels of cholesterol in bile. In contrast North Indian gallstones are reported to be predominantly of the cholesterol type.28 However, the genetic basis for such an observation was not explained earlier. Considering the occurrence of pigmentous stones in south India we correlated the mean bilirubin levels to the three genotypes of both the SNPs from UGT1A1 gene. From the obtained results, it was interesting to note that the risk genotype (TT) of rs6742078 had significantly higher mean bilirubin level as compared to the wild type (GG) and heterozygous (GT) genotype, although the levels were within normal limits. Likewise in the other SNP namely rs887829 in the UGT1A1 gene the risk genotype (AA) had significantly higher mean bilirubin level as compared to the wild type (GG) and heterozygous (GA) genotype. Evaluation of data for additive effect of the two SNPs revealed there was none and the compound heterozygotes were not at significant risk of higher bilirubin levels. Only the risk allele carriers (TT and AA) were at a significantly increased risk of higher bilirubin levels. Although both the identified SNPs are in the introns, a recent exome wide association study that assessed the influence of protein coding variants on unconjugated, conjugated and total serum bilirubin levels in well characterized Italian elderly individuals identified low frequency coding variants in first exon of UGT1A1 gene. These coding variants encode for the substrate binding domain. Further it is interesting to note that the coding variants were in strong linkage disequilibrium with 3 intronic variants including rs6742078 and rs887829.30 The present study corroborates these findings including higher gall stone risk and bilirubin levels.
To estimate the strength of the relationship between several independent variables and a continuous dependent variable, multiple logistic regression analysis was carried out with significant parameters from univariate analysis, and it was seen that age (greater than 40 years), sex (female) and variant in ABCG8 gene were significantly associated with GSD, suggesting that these three are independent risk factors to predict gall stone formation in South Indian population (Table 5).
Table 5.
Multiple Logistic Regression Analysis.
| Variable | Risk/non-risk | P value | Odds ratio | 95% CI |
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Age | Greater/less than 40 years | 0.0002 | 2.13 | 1.43 | 3.16 |
| Sex (male) | Female/male | <0.0001 | 0.24 | 0.16 | 0.37 |
| ABCG8 | Mutant/wild | 0.03 | 2.20 | 1.06 | 4.58 |
In conclusion, the present study indicates the importance of significantly higher levels of bilirubin as well as polymorphism in UGT1A1 gene for the genetic susceptibility of south Indians to GSD. Further screening and validation of these SNPs in a larger sample in future could allow the recommendation of including them in the diagnostic workup of South Indian GSD patients.
Conflicts of interest
The authors have none to declare.
Acknowledgements
The authors acknowledge the individuals for consenting to participate in the study. The study was supported by funds from Department of Biotechnology, Government of India (sanction number BT/MB/TF/MED-1/2009) and the funding source had no role in study design, collection, analysis, interpretation of data, writing the report or in the decision to submit the article for publication. None of the authors have any conflict of interest to be disclosed.
References
- 1.Ruhl C.E., Everhart J.E. Gallstone disease is associated with increased mortality in the United States. Gastroenterology. 2011;140:508–516. doi: 10.1053/j.gastro.2010.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Völzke H., Baumeister S.E., Alte D. Independent risk factors for gallstone formation in a region with high cholelithiasis prevalence. Digestion. 2005;71:97–105. doi: 10.1159/000084525. [DOI] [PubMed] [Google Scholar]
- 3.Ransohoff D.F., Gracie W.A. Treatment of gallstones. Ann Intern Med. 1993;119:606–619. doi: 10.7326/0003-4819-119-7_part_1-199310010-00010. [DOI] [PubMed] [Google Scholar]
- 4.Bera S., Bhattacharyya S., Ghose B.C., Bera T., Mukhopadhyay S.K., Saha M. Study of serum lipase, alpha-amylase and pancreatic amylose in gall-stone diseases. J Indian Med Assoc. 2011;109:663–665. [PubMed] [Google Scholar]
- 5.Shaffer E.A. Gallstone disease: epidemiology of gallbladder stone disease. Best Pract Res Clin Gastroenterol. 2006;20:981–996. doi: 10.1016/j.bpg.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Kim S.S., Lee J.G., Kim D.W. Insulin resistance as a risk factor for gallbladder stone formation in Korean postmenopausal women. Korean J Intern Med. 2011;26:285–293. doi: 10.3904/kjim.2011.26.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smelt A.H. Triglycerides and gallstone formation. Clin Chim Acta. 2010;411:1625–1631. doi: 10.1016/j.cca.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Chen L.-Y., Qiao Q.-H., Zhang S.-C., Chen Y.-H., Chao G.-Q., Fang L.-Z. Metabolic syndrome and gallstone disease. World J Gastroenterol. 2012;18:4215–4220. doi: 10.3748/wjg.v18.i31.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinton L.M., Shaffer E.A. Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver. 2012;6:172–187. doi: 10.5009/gnl.2012.6.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D.Q., Cohen D.E., Carey M.C. Biliary lipids and cholesterol gallstone disease. J Lipid Res. 2009;50l:406–411. doi: 10.1194/jlr.R800075-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava A., Mishra A., Singh R., Rai R., Srivastava N., Mittal B. Multi-analytic approach elucidates significant role of hormonal and hepatocanalicular transporter genetic variants in gallstone disease in North Indian population. PLOS ONE. 2013;8:e59173. doi: 10.1371/journal.pone.0059173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trotman B.W., Soloway R.D. Pigment gallstone disease: summary of the National Institutes of Health – international workshop. Hepatology. 1982;2:879–884. doi: 10.1002/hep.1840020624. [DOI] [PubMed] [Google Scholar]
- 13.Kim I.S., Myung S.J., Lee S.S., Lee S.K., Kim M.H. Classification and nomenclature of gallstones revisited. Yonsei Med J. 2003;44:561–570. doi: 10.3349/ymj.2003.44.4.561. [DOI] [PubMed] [Google Scholar]
- 14.Venneman N.G., van Erpecum K.J. Pathogenesis of gall stones. Gastroenterol Clin N Am. 2010;39:171–183. doi: 10.1016/j.gtc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Venneman N.G., van Kammen M., Renooij W., Vanberge-Henegouwen G.P., van Erpecum K.J. Effects of hydrophobic and hydrophilic bile salts on gallstone growth and dissolution in model biles. Biochim Biophys Acta. 2005;1686:209–219. doi: 10.1016/j.bbalip.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Dwivedi S., Agrawal S., Singh S. Association of cytochrome-17 (MspA1) gene polymorphism with risk of gall bladder stones and cancer in North India. Asian Pac J Cancer Prev. 2015;16(13):5557–5563. doi: 10.7314/apjcp.2015.16.13.5557. [DOI] [PubMed] [Google Scholar]
- 17.Rai R., Sharma K.L., Misra S., Kumar A., Mittal B. Association of adrenergic receptor gene polymorphisms in gallbladder cancer susceptibility in a North Indian population. J Cancer Res Clin Oncol. 2014;140(5):725–735. doi: 10.1007/s00432-014-1621-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava A., Srivastava A., Srivastava K., Choudhuri G., Mittal B. Role of ABCG8 D19H (rs11887534) variant in gallstone susceptibility in northern India. J Gastroenterol Hepatol. 2010;25(11):1758–1762. doi: 10.1111/j.1440-1746.2010.06349.x. [DOI] [PubMed] [Google Scholar]
- 19.Siddapuram S.P., Mahurkar S., Duvvuru N.R. Hepatic cholesterol transporter ABCG8 polymorphisms in gallstone disease in an Indian population. J Gastroenterol Hepatol. 2010;25:1093–1098. doi: 10.1111/j.1440-1746.2010.06309.x. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava A., Srivastava A., Srivastava N., Choudhuri G., Mittal B. Organic anion transporter 1B1 (SLCO1B1) polymorphism and gallstone formation: high incidence of Exon4 CA genotype in female patients in North India. Hepatol Res. 2011;41(1):71–78. doi: 10.1111/j.1872-034X.2010.00736.x. [DOI] [PubMed] [Google Scholar]
- 21.Dixit M., Choudhuri G., Saxena R., Mittal B. Association of apolipoprotein A1–C3 gene cluster polymorphisms with gallstone disease. Can J Gastroenterol. 2007;21(9):569–575. doi: 10.1155/2007/329342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixit M., Choudhuri G., Keshri L.J., Mittal B. Association of low density lipoprotein receptor related protein-associated protein (LRPAP1) gene insertion/deletion polymorphism with gallstone disease. J Gastroenterol Hepatol. 2006;21(5):847–849. doi: 10.1111/j.1440-1746.2005.03931.x. [DOI] [PubMed] [Google Scholar]
- 23.Warang P., Devendra R., D'Silva S. Do UGT1A1 and HMOX1 gene promoter polymorphisms increase the risk of hyperbilirubinemia and gallstones in patients with hereditary spherocytosis? Ann Hematol. 2015;94(1):169–171. doi: 10.1007/s00277-014-2123-z. [DOI] [PubMed] [Google Scholar]
- 24.Chandran P., Kuchhal N.K., Garg P., Pundir C.S. An extended chemical analysis of gall stone. Indian J Clin Biochem. 2007;22:145–150. doi: 10.1007/BF02913334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steen G., Blijenberg B.G. Chemical analysis of gallstones. Eur J Clin Chem Clin Biochem. 1991;29:801–804. doi: 10.1515/cclm.1991.29.12.801. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 27.Buch S., Schafmayer C., Völzke H. Loci from a genome-wide analysis of bilirubin levels are associated with gallstone risk and composition. Gastroenterology. 2010;139:1942–1951. doi: 10.1053/j.gastro.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Tandon R. Prevalence and type of biliary stones in India. World J Gastroenterol. 2000;6 tk 4-5. [Google Scholar]
- 29.Jayanthi V., Palanivelu C., Prasanthi R., Methew S., Srinivasan V. Composition of gall stones in Coimbatore district of Tamil Nadu state. Indian J Gastroenterol. 1998;17:134–135. [PubMed] [Google Scholar]
- 30.Oussalah A., Bosco P., Anello G. Exome-wide association study identifies new low-frequency and rare UGT1A1 coding variants and UGT1A6 Coding variants influencing serum bilirubin in elderly subjects: a strobe compliant article. Medicine (Baltimore) 2015;94(22):e925. doi: 10.1097/MD.0000000000000925. [DOI] [PMC free article] [PubMed] [Google Scholar]