Abstract
Background
In this study, we screened and identified an endophyte JG09 having strong biocatalytic activity for ginsenosides from Platycodon grandiflorum, converted ginseng total saponins and ginsenoside monomers, determined the source of minor ginsenosides and the transformation pathways, and calculated the maximum production of minor ginsenosides for the conversion of ginsenoside Rb1 to assess the transformation activity of endophyte JG09.
Methods
The transformation of ginseng total saponins and ginsenoside monomers Rb1, Rb2, Rc, Rd, Rg1 into minor ginsenosides F2, C-K and Rh1 using endophyte JG09 isolated by an organizational separation method and Esculin-R2A agar assay, as well as the identification of transformed products via TLC and HPLC, were evaluated. Endophyte JG09 was identified through DNA sequencing and phylogenetic analysis.
Results
A total of 32 β-glucosidase-producing endophytes were screened out among the isolated 69 endophytes from P. grandiflorum. An endophyte bacteria JG09 identified as Luteibacter sp. effectively converted protopanaxadiol-type ginsenosides Rb1, Rb2, Rc, Rd into minor ginsenosides F2 and C-K, and converted protopanaxatriol-type ginsenoside Rg1 into minor ginsenoside Rh1. The transformation pathways of major ginsenosides by endophyte JG09 were as follows: Rb1→Rd→F2→C-K; Rb2→C-O→C-Y→C-K; Rc→C-Mc1→C-Mc→C-K; Rg1→Rh1. The maximum production rate of ginsenosides F2 and C-K reached 94.53% and 66.34%, respectively.
Conclusion
This is the first report about conversion of major ginsenosides into minor ginsenosides by fermentation with P. grandiflorum endophytes. The results of the study indicate endophyte JG09 would be a potential microbial source for obtaining minor ginsenosides.
Keywords: conversion, endophytes, minor ginsenosides, Platycodon grandiflorum
1. Introduction
Ginseng, the root of Panax ginseng Meyer (Araliaceae), has been a greatly valued herb in Asian countries for over 1,000 yr [1]. It consists of ginsenosides, polysaccharides, polyynes, flavonoids, and volatile oils among other components [2]. Ginsenosides are considered the major active components of ginseng species and are responsible for numerous pharmacological properties, such as antitumor, anti-aging, blood vessel softening, anticancer, anti-inflammatory, and hepatoprotective effect activities [3], [4], [5]. Deglycosylated ginsenosides (minor ginsenosides Rh1, Rh2, F2, Rg3, C-K, etc.) are more pharmaceutically active than major glycosylated ginsenosides (Rb1, Rb2, Rc, Rd, Rg1, etc.), because of their smaller size, higher bioavailability, and better permeability across the cell membrane [6]. However, minor ginsenosides are not present in most natural ginseng plants or are present in very low amounts. Therefore, the study of converting major ginsenosides to minor ginsenosides is of great significance [7]. Various approaches including heating [8], acid treatment [9], and biotransformation [10] have been carried out. Among these methods, biotransformation is shown to be the most effective way to produce minor ginsenosides [9], [11], [12]. Biotransformation is a high specificity, low cost, selective, and environmentally friendly technique, and can be defined as chemical reactions catalyzed by different enzymes of various microbial cells; it can be an alternative for biosynthesis of some important drug metabolites. It also exhibits many advantages over chemical synthesis such as stereo- or region-selectivity, mild reaction conditions, and avoiding complex protection and deprotection steps [13], [14].
Endophytes are considered to be bacterial or fungal microorganisms that colonize healthy plant tissues without causing any apparent symptoms [15]. Recently, the application of endophytes to biotransformation has also attracted people's attention. Due to the endophytes special living environment and long term coexistence with their hosts, the endophytes have developed a specific lifestyle to maintain a stable symbiosis, and they can produce various extracellular enzymes for secondary metabolite biosynthesis. Therefore, endophytes have been used for some complex reactions to obtain more active compounds instead of traditional chemical methods [16]. The application of endophytes to biotransformation of region- and stereo-selective production is a suitable method to obtain more active substances; they have been used in natural compound conversion such as flavans, tetrahydrofuran lignin, and astragalosides [17], [18], [19].
To date, however, no relevant studies have been reported on the conversion of major ginsenosides to minor ginsenosides by endophytes isolated from Platycodon grandiflorum. The present study aimed to enhance the content of minor ginsenosides in ginseng total saponins by endophytes from P. grandiflorum. The optimization of the bioconversion conditions was conducted by single factor tests of the bioconversion procedure.
2. Materials and methods
2.1. Materials
HPLC-grade methanol and acetonitrile (ACN) were supplied by Merck (Darmstadt, Germany). Ultrapure water was prepared by a Milli-Q purification system (Millipore, Bedford, MA, USA). Ginsenosides Rb1, Rb2, Rc, Rd, Rg1, Rg3, Rh1, Rh2, Re, F2, C-K and PPD were purchased from Chengdu Mann Stewart Biological Technology Co., Ltd. (Sichuan, China). The other chemicals used in this study were at least of analytical reagent grade.
2.2. Standard solutions and standard curve preparation
Stock solutions in methanol were prepared for ginsenosides F2 and C-K in concentrations of 2.0 mg/mL and 3.0 mg/mL, respectively. The stock solutions were then diluted with methanol to yield a series of standard solutions with different concentrations for linearity validation and standard curve preparation. Working methanolic solutions were prepared daily from stock solutions.
2.3. Isolation and screening of β-glucosidase-producing endophytes
Plant samples were obtained from a P. grandiflorum field in Yanji, China. The P. grandiflorum was kept refrigerated at 4°C, and endophytes were isolated within 48 h.
The P. grandiflorum samples were washed adequately under running tap water to remove soil, then surface-disinfected through the following sequence of immersions: 1min in 70% (v/v) ethanol, 5–10 min in 5% NaClO, 1 min in 70% (v/v) ethanol, and 1 min in sterile water four times. The outer tissue was removed with a sterile scalpel. Small segments (0.5 cm × 0.5 cm) were cut and pressed on a potato dextrose agar plates (PDA), R2A (Agar medium S) agar plates and Luria-Bertani (LB) agar plates and incubated at 30°C for 5–7 d. Single colonies from these plates were purified by transferring onto new plates.
Esculin-R2A agar was used to isolate β-glucosidase-producing endophytes. The endophytes producing β-glucosidase that hydrolyze esculin appeared as colonies surrounded by a reddish-brown to dark brown zone. Esculin-R2A agar contains (per 1 L): esculin 1 g and ferric citrate 0.5 g with 15.2 g R2A agar, autoclaved at 121°C for 15 min.
2.4. Bioconversion of major ginsenosides in ginseng total saponins
2.4.1. Transformation of ginseng total saponins
The β-glucosidase-producing endophytes were cultured in LL medium consisting of 0.5 g/L NH4Cl, 1.0 g/L K2HPO4, 0.5 g/L KH2PO4, 0.25 g/L MgSO4, and 1.0 g/L yeast extract. The medium was sterilized at 121°C for 15 min, and the initial pH was 7.0. The suspension of the strain was incubated until it reached logarithmic phase growth, then mixed with the same volume of 15 mg/L ginseng total saponins solution and incubated in a shaking incubator (30°C, 150 rpm). The culture was extracted with the same volume of aqueous saturated n-BuOH and concentrated in a vacuum to dryness, then the residue was dissolved in methanol and analysis of the biotransformation ability of ginsenosides was carried out by TLC.
In order to explore the source of minor ginsenosides and the conversion pathway by endophytes, we mixed the suspension culture of endophytes and ginsenosides Rb1, Rb2, Rc, Rd, and Rg1 and confirmed at regular intervals by TLC and HPLC analysis.
2.4.2. Optimization of fermentation conditions
Microbial fermentation requires a suitable fermentation medium, temperature and pH. The optimum fermentation condition is the key to getting a high yield of fermentation products successfully; in order to obtain the best conversion for ginsenosides, single-factor experiments were performed to optimize the medium and pH value. Six culture mediums (H2O, R2A, PDB (Potato Dextrose Broth), LB (Luria-Bertani), NB (Nutrient Broth), LL (Lysogeny Liquid), initial pH of 7.0) were tested one by one. Subsequently, fermentation conditions, on the basis of the optimum medium, at pH values of 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, and 10.0 were investigated.
2.5. Analytic methods
2.5.1. TLC analysis
TLC was carried out using 60F254 silica gel plates (Merck) with CHCl3–CH3OH–H2O (10:5:1, v/v/v, lower phase) as the solvent. The spots on the TLC plates were visualized by spraying with 10% (v/v) H2SO4 solution, followed by heating at 110°C for 5 min.
2.5.2. HPLC analysis
HPLC analysis was performed using an HPLC system with automatic injector and single wavelength UV detector. The separation was carried out on a BDS HYPERSIL C18 (5 μm, 250 mm × 4.6 mm; Agilent 1100, Waltham, USA) with a column temperature of 25°C. The mobile phase was A (water) and B (acetonitrile). Gradient elution started with 77% solvent A and 23% solvent B and was then changed to: A from 77% to 54%, 0–13 min; A from 54% to 32%, 13–33 min; A from 32% to 0%, 45–55 min; A from 0% to 77%, 60–63 min. The flow rate was 1.0 mL/min, and samples were detected by absorption at 203 nm, with an inject volume of 10 μL.
2.6. Phylogenetic analysis
The endophyte genomic DNA was extracted by the method of CTAB (Hexadecyl trimethyl ammonium Bromide) [20]. The 16S ribosomal DNA was amplified using universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [21]. The PCR reaction system was carried out in volumes of 20 μL, containing 2 μL of template DNA, 2 μL of each primer, 2 μL of dNTPs, 2 μL of 10 × PCR buffer (containing Plus Mg2+), 0.4 μL of TaKaRa Taq enzyme, and 10 μL of ddH2O. The amplification program was as follows: an initial denaturation at 95°C for 3 min, 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 90 s, and a final extension at 72°C for 7 min.
The PCR products were purified using a Silica Bead DNA Gel Extraction Kit (Fermentas Life Sciences, Vilnius, Lithuania), in order to obtain more complete sequence information, then ligated into a pGEM-Teasy Vector (Biofine, China), and subsequently used to transform JM109 competent cells. Transformed cells carrying inserts were identified with blue/white colony selection. Recombinant plasmids in transformed cells were cloned and then sequenced (Shanghai Biotechnology Co., Ltd. Megiddo).
The obtained sequencing results were checked for homology using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST), and compared with available data from GenBank sequence databases, getting the closest strain's rDNA sequence. The resulting alignment was used to construct a Neighbor-joining (NJ) phylogenetic tree using MEGA 4.0 software (http://www.megasoftware.net/mega4/mega.html) [22].
3. Results and discussion
3.1. Isolation and phylogenetic analysis of β-glucosidase-producing endophytes
A total of 69 endophytes were isolated from P. grandiflorum by the method of organizational separation. After the Esculin-R2A agar assay, 32 β-glucosidase-producing microorganisms were assayed to verify their activity for converting ginseng total saponins. Among these endophytes, strain JG09 showed the strongest activity for converting ginseng total saponins into minor ginsenosides C-K, F2 and Rh1.
The genomic DNA of strain JG09 was extracted and amplified, recombined and sequenced. Strain JG09 was an endophytic bacteria; after a series of operations with the extracted DNA, such as PCR and recombinant plasmid generation, the resulting gel electrophoresis figure showed the DNA length to be approximately 1,600 bp (Fig. 1). The obtained sequences were compared with those in the GenBank database, and the results are shown in Table 1.
Fig. 1.
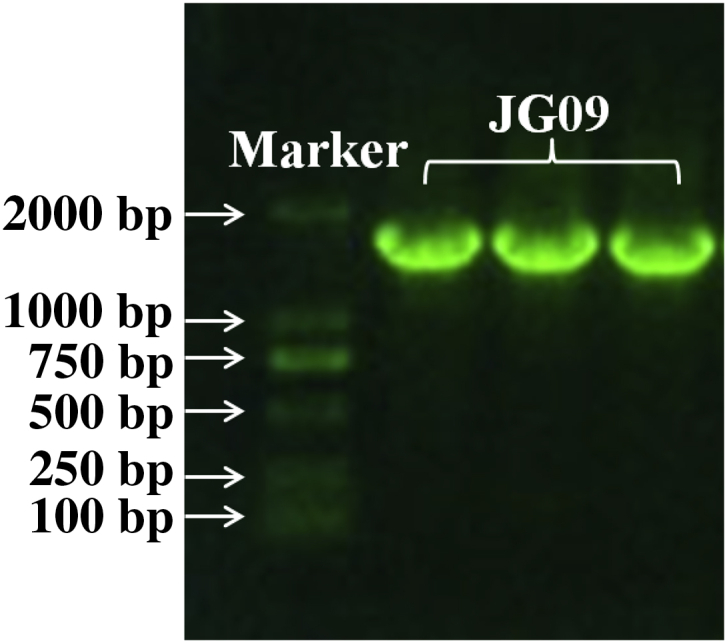
Gel electrophoresis of recombinant plasmid DNA of endophyte JG09.
Table 1.
Similarity between the isolates and closest species in GenBank
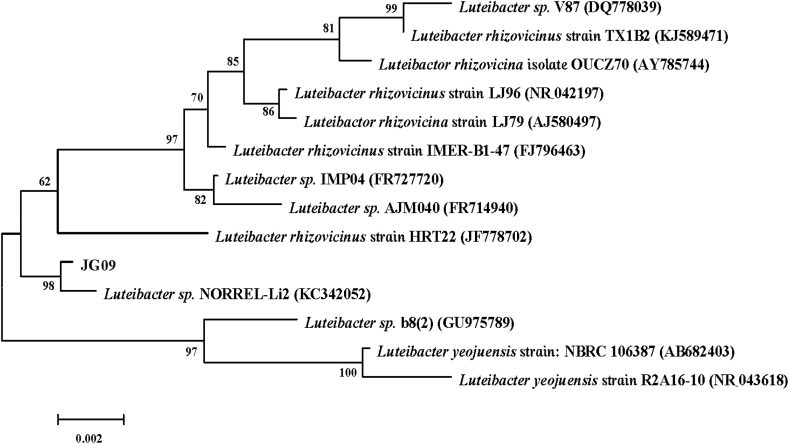
Phylogenetic analysis based on 16S rDNA gene sequences indicated that the strain JG09 belongs to the genus Lucteibacter, and endophyte JG09 is most closely related to Luteibacter sp. NORREL-Li2 (99%), as seen in Fig. 2.
Fig. 2.
A 16S rDNA sequenced-based phylogenetic tree constructed by the Neighbor joining method. The numbers at the nodes are bootstrap values based on 1,000 replications.
3.2. Effect of pH and culture medium on bioconversion of major ginsenosides
The culture medium must meet the demand of microbial cell growth and reproduction, because a large number of cells are needed to generate large amounts of product. However, excessive growth of cells will consume a large amount of nutrients; this sometimes can affect the production capacity of the cell, resulting in a decreased yield of product. So an appropriate media is essential to a microbial strain.
In order to compare the effect of various media, strain JG09 was cultured in the broths of H2O, R2A, PDB, LB, NB and LL, at an initial pH of 7.0, mixing with ginseng total saponins for 7 d. The suspension were analyzed by TLC (Fig. 3). The cultures in PDB broth and LB broth couldn't convert major ginsenosides into C-K, but LL broth fermented by JG09 efficiently converted major ginsenosides into F2 and C-K (Fig. 3), with the maximum degree of conversion of major ginsenosides. This suggested that LL broth was not only suitable for the growth of the strain JG09, but also for the production of F2 and C-K.
Fig. 3.
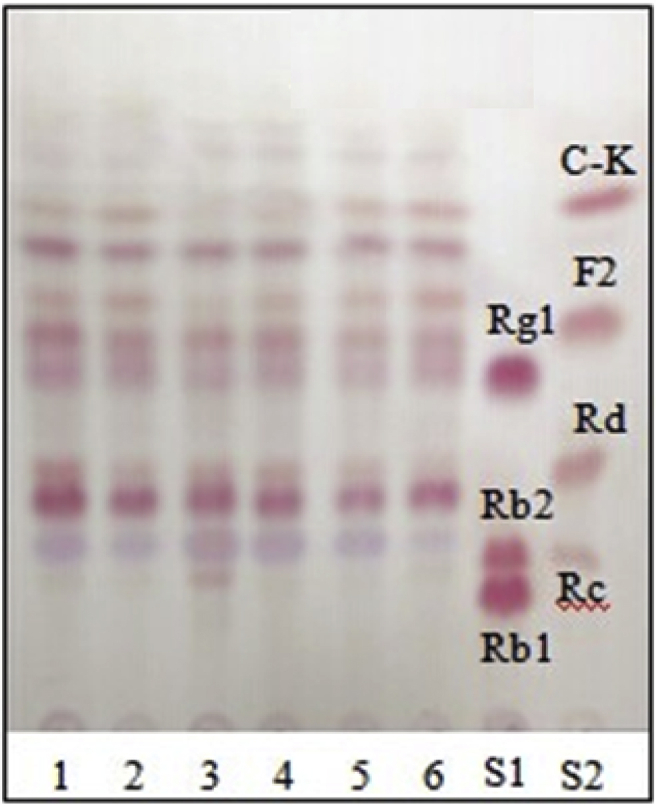
TLC analysis of metabolites of ginseng total saponins by endophyte JG09 cultured in different media: H2O (1), R2A (2), PDB (3), LB (4), NB (5), and LL (6). LB, Luria-Bertani; LL, Lysogeny Liquid; NB, Nutrient Broth; PDB, Potato Dextrose Broth; R2A, Agar medium S; S1 and S2, saponin standards.
The pH value of the culture medium mainly causes the change in the charge of the microorganism's cytomembrane, and impacts the ionization degree of nutrients, thereby affecting absorption of nutrients by the microbial cells. The pH also affects the biologically active substances, such as enzyme activity. Different microbes have different optimum pH values to adapt to the environment. The optimum pH for microbial growth ranges from 4.0 to 9.0 in general. Therefore, fermentation conditions, on the basis of the optimum medium, at pH values of 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, and 10.0 were investigated. The results indicated that the optimum pH of strain JG09 was 4.0 (Fig. 4), which could maximize the content of generated minor ginsenosides F2 and C-K.
Fig. 4.
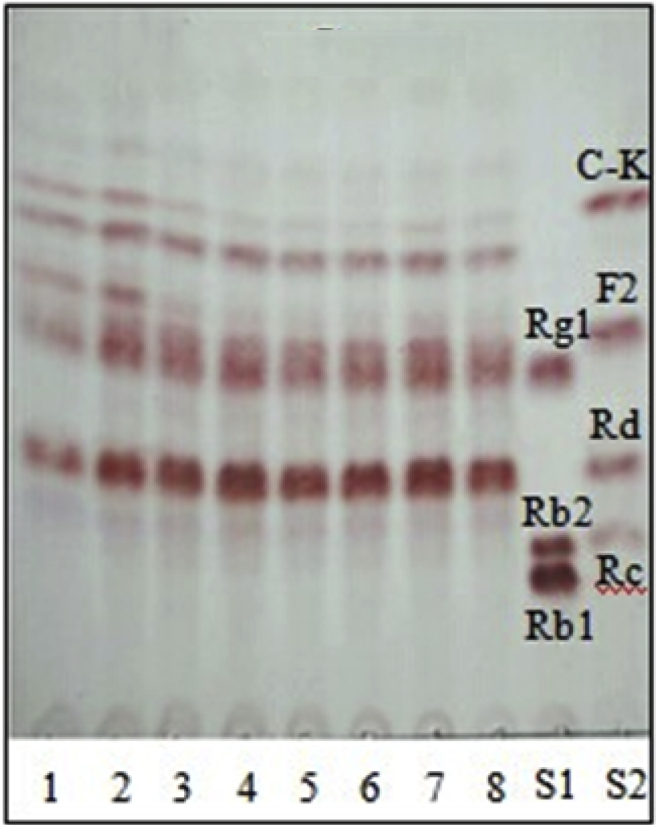
TLC analysis of metabolites of ginseng total saponins by endophyte JG09 cultured at different pH's of 3.0 (1), 4.0 (2), 5.0 (3), 6.0 (4), 7.0 (5), 8.0 (6), 9.0 (7), and 10.0 (8). S1 and S2, saponin standards.
3.3. Bioconversion of major ginsenosides in ginseng total saponins
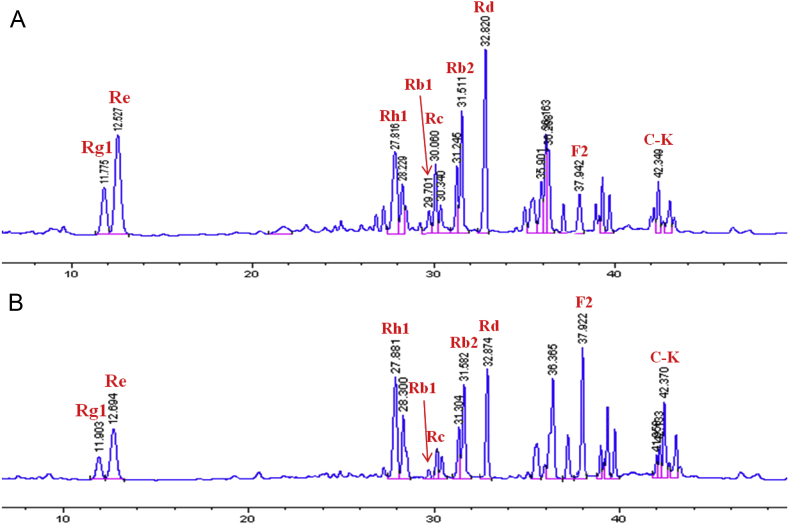
Under optimized fermention conditions, endophyte JG09 was inoculated (3%, v/v) into a 250 mL Erlenmeyer's flask containing 100 mL of sterilized liquid broth and 1.5 g ginseng total saponins for a period of 10 d in a shaking incubator (30°C, 150 rpm). Comparing the change of saponin content before and after fermentation by HPLC analysis, the content of protopanaxadiol-type major saponins (Rb1, Rb2, Rc, and Rd) and protopanaxatriol-type major saponins (Rg1 and Re) was generally decreased, and Rb1 was almost completely transformed after 10 d of incubation. At the same time, a significant increase in the content of minor ginsenosides F2, C-K, and Rh1 was observed (Fig. 5). To further analyze the source of minor ginsenosides F2, C-K, and Rh1, major ginsenosides Rb1, Rb2, Rc, Rd, and Rg1 were incubated with endophyte JG09; these standards of ginsenosides were then injected into HPLC.
Fig. 5.
HPLC analysis of (A) ginseng total saponins and (B) fermented ginseng total saponins. The peaks correspond to ginsenoside Rg1, Re, Rh1, Rb1, Rc, Rb2, Rd, F2, and C-K, respectively.
3.4. Bioconversion pathways of major ginsenosides
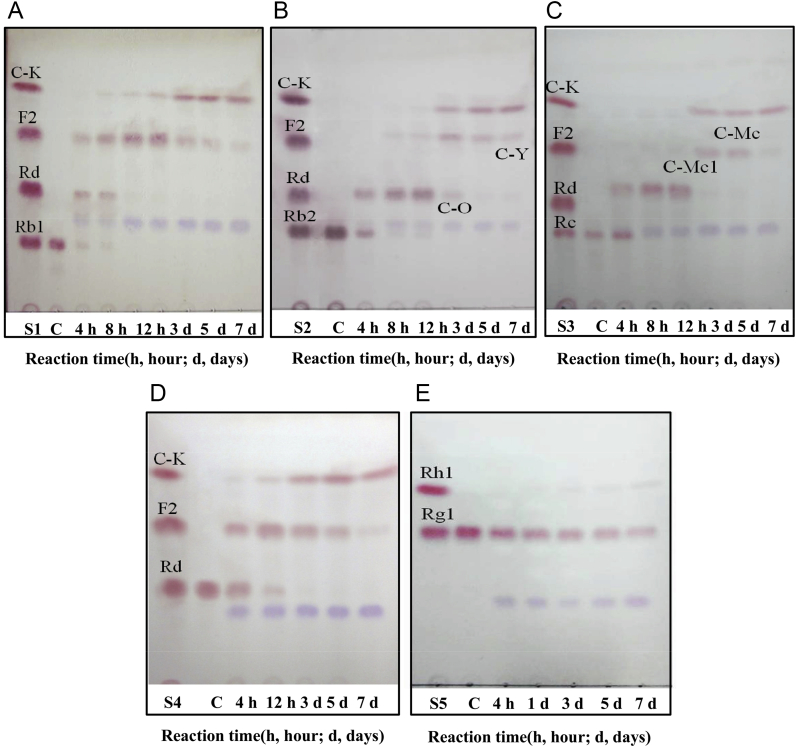
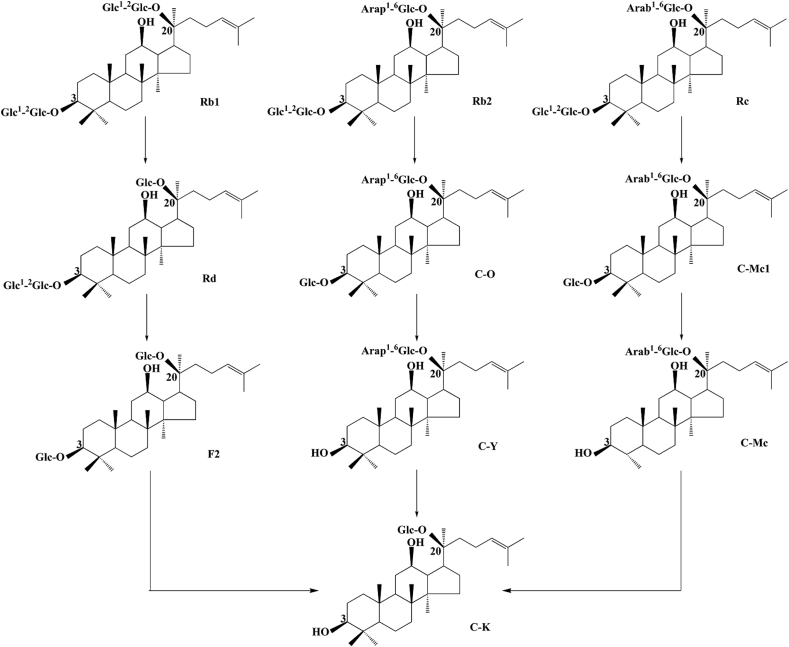
In order to investigate the biotransformation pathway of major ginsenosides into minor ginsenosides by endophyte JG09, the conversion of ginsenosides Rb1, Rb2, Rc, Rd, and Rg1 was carried out by inoculating the ginsenoside into precultured suspensions, followed by 7 d of incubation, during which different individual samples from the same batch at different times were extracted. As shown in Fig. 6, it is clear that endophyte JG09 could convert major ginsenosides Rb1, Rb2, Rc, and Rd into minor ginsenosides F2 and C-K and convert major ginsenoside Rg1 into minor ginsenoside Rh1, based on the Rf values of standards.
Fig. 6.
Time-course TLC analyses of ginsenoside (A) Rb1, (B) Rb2, (C) Rc, (D) Rd, and (E) Rg1 by endophyte JG09. C, blank sample; S1–S5, standard.
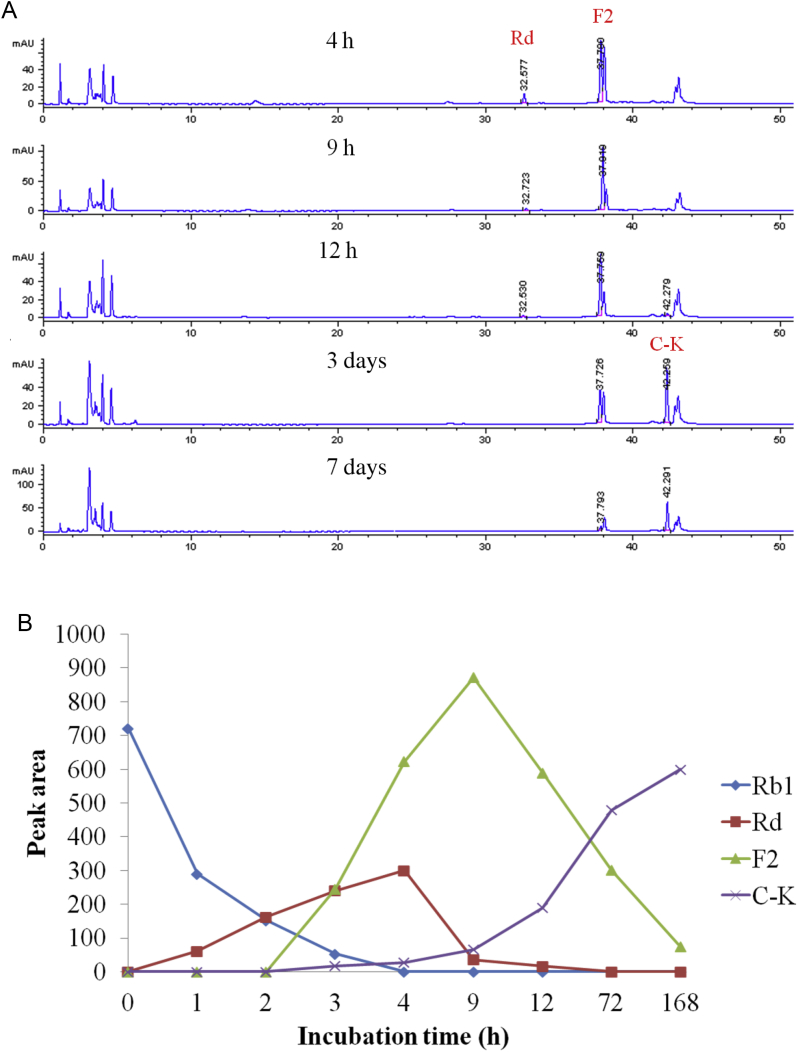
The HPLC profile of the reaction mixture of ginsenoside Rb1 and the endophyte JG09 after incubation is shown in Fig. 7. In the HPLC chromatogram, Rb1 was hydrolyzed by endophyte JG09 to produce ginsenosides Rd, F2, and C-K, following 7 d of incubation. The transformation products of ginsenosides Rb2, Rc, Rd, and Rg1 were also analyzed by HPLC, which yielded the same results to the findings of TLC (Fig. 6).
Fig. 7.
(A) HPLC profile of the metabolites of ginsenoside Rb1 converted by endophyte JG09. (B) The content change of ginsenoside Rb1, Rd, F2, and C-K in the fermentation broth.
In the coculture of endophyte JG09 and ginsenoside Rb1, endophyte JG09 converted ginsenoside Rb1 into C-K as well as Rd and F2; the concentration of Rb1, Rd, F2, and C-K exhibited regular change with reaction time. From 4 h to 9 h, the content of ginsenoside Rb1 and Rd gradually decreased and that of F2 and C-K gradually increased. Ginsenoside Rd was completely converted after 9 h of incubation. It proved that the metabolite Rd was the precursor of metabolite F2 and C-K. Fig. 7 shows that a peak area of ginsenoside F2 increased after 4 h with the generation of ginsenoside Rd, but decreased after 1 d of incubation. In the meantime, the content of ginsenoside C-K significantly increased after 1 d. The peak area of ginsenoside F2 reached the maximum value within 9 h, then the generation of ginsenoside C-K went into the rapidly developing period and this stage should take a longer time. At last, ginsenoside Rb1 was completely transformed and the corresponding peak disappeared (Fig. 6, Fig. 7). This suggested that Rb1 was converted in the following sequence: Rb1→Rd→F2→C-K, as shown in the pathway in Fig. 8, by the different β-glucosidase from endophyte JG09, consecutively hydrolyzing the terminal glucose and inner glucose at the C-3 position, and terminal glucose at the C-20 position.
Fig. 8.
Biotransformation pathway of ginsenosides Rb1, Rb2, Rc, and Rd by endophyte JG09.
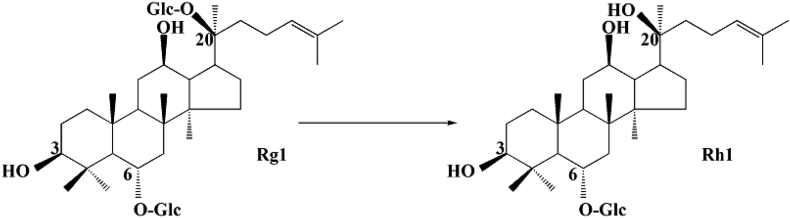
Combining TLC with HPLC results, we also confirmed the conversion pathways of major ginsenosides Rb2, Rc, Rd, and Rg1 by endophyte JG09, the results are as follows: Rb2→C-O→C-Y→C-K; Rc→C-Mc1→C-Mc→C-K; Rd→F2→C-K; Rg1→Rh1, as shown in the pathways in Fig. 8, Fig. 9.
Fig. 9.
Biotransformation pathway of ginsenosides Rg1 by endophyte JG09.
3.5. Standard curve preparation and the maximum production rate calculation for minor ginsenoside F2 and C-K
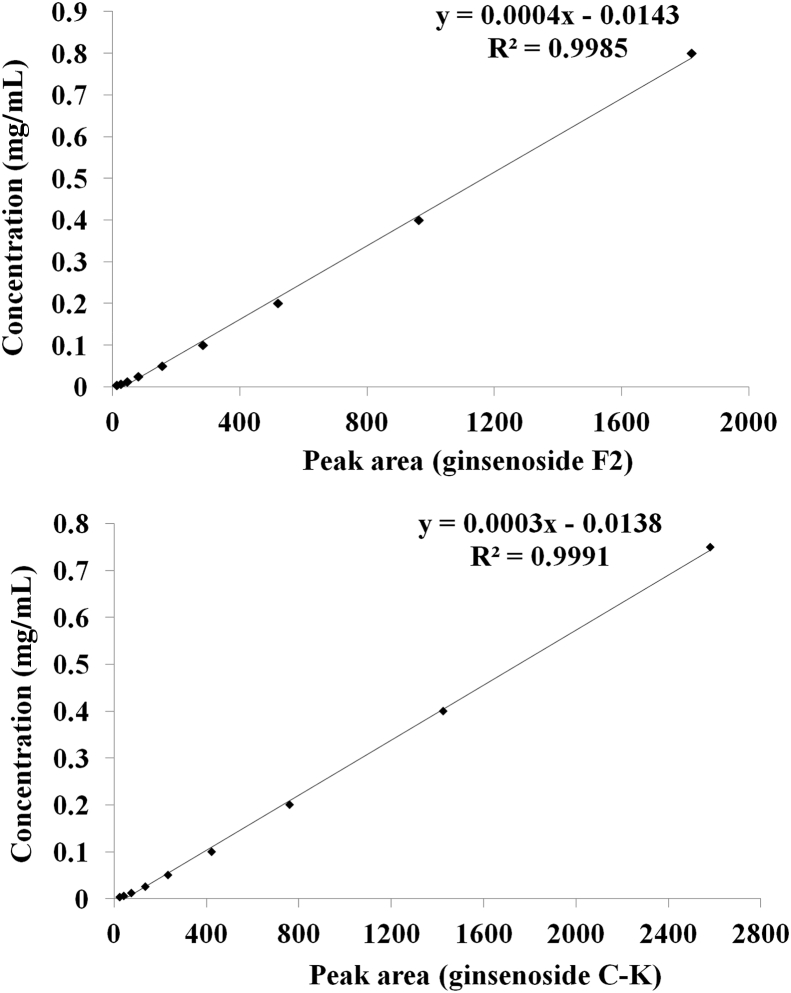
Fig.10 shows the standard curve of ginsenosides F2 and C-K by plotting the area against the concentrations of ginsenosides F2 and C-K. Under these conditions, the full linear range of the assay was extended from 0.003125 mg/mL to 0.75 μg/mL, as indicated in Fig. 10. The linear correlation coefficients R2 are 0.9985 and 0.9991, respectively, so the standard curve can be used to calculate the maximum yield of ginsenosides F2 and C-K during the transformation. By using the molecular weight and the initial concentration of Rb1, the theoretical yield of ginsenosides F2 and C-K were calculated. Under optimum conditions, when Rb1 was added to the culture broth of endophyte JG09, the content of ginsenosides F2 and C-K could reach a maximum value in 7 d. In order to determine the maximum production rate of minor ginsenosides F2 and C-K and assess the transformation effectiveness of endophyte JG09 to ginsenoside Rb1, calculations were carried out using peak area and the standard curve of ginsenosides. The results show that the maximum production yield of ginsenoside F2 at 9 h reaches 94.53%, the maximum production yield of ginsenoside C-K at 7 d reaches 66.34% (Table 2).
Fig. 10.
Standard curve of ginsenoside F2 and C-K.
Table 2.
Actual yield and production rate of ginsenoside F2 and C-K
| Strain | Ginsenoside F2 |
Ginsenoside C-K |
||||
|---|---|---|---|---|---|---|
| Peak area (9 h) |
Actual mass (mg) | Yield (%) |
Peak area (7 d) |
Actual mass (mg) |
Yield (%) |
|
| JG09 | 872.3 | 0.06692 | 94.53 | 599.8 | 0.03323 | 66.34 |
It is well known that P. ginseng has various ingredients including ginsenosides and their derivatives. To date, ginsenosides have been isolated and identified structurally from approximately 60 species of the Panax genus [23]. Major ginsenosides such as Rb1, Rb2, Rc, Rd, and Rg1 are present in greater abundance than any other ginsenosides, however minor ginsenosides Rh1, F2, and C-K have greater pharmacological effects such as tumor-suppressing, antimetastatic, anticarcinogenic, hepatoprotective, neuroprotective, immune-stimulating and vasodilating effects, which are of great significance for human health, though their content is relatively low in ginseng. The anticancer activity of ginsenoside F2 was already applied and noticed [24], [25]. Ginsenoside C-K is the main pharmacologically active metabolite detected in blood after the oral administration of major ginsenosides [26], [27]. It has been shown that ginsenoside C-K possesses various biological activities in vivo or in vitro, such as antigenotoxic activity, antiallergic effect, and antitumor activity [27]. In addition, ginsenoside C-K shows potential hepatoprotective and anti-inflammatory activities and the pharmacological effects of ginsenoside C-K appear to be stronger than those of known minor ginsenosides and some pharmaceutical drugs including diphenyl dimethyl bicarboxylate and indomethacin [4], [5], [28], [29], [30]. There is a study showing that ginsenoside Rb1 is hydrolyzed by intestinal microorganism in the body and is changed to minor ginsenoside F2 [31]. Some studies have looked for suitable microbes or enzymes that can transform major ginsenosides into minor ginsenosides [3], [12], [32], [33]. However, most methods suffer from the fact that they are small scale, difficult, expensive, and low yielding, and there are only a few reports of transforming major ginsenosides into minor ginsenosides by potential resource endophytes.
In this study, we isolated 32 β-glucosidase-producing endophytes from P. grandiflorum and identified endophyte JG09 as Lucteibacter sp., which shows a strong ability to bioconvert major ginsenosides into minor ginsenosides, as confirmed by TLC and HPLC analysis. This is the first report of conversion of major ginsenosides to minor ginsenosides by fermentation with P. grandiflorum endophytes. We found that endophyte JG09 can hydrolyze protopanaxadiol ginsenosides Rb1, Rb2, Rc, and Rd into minor ginsenosides F2 and C-K, and hydrolyze protopanaxatriol ginsenoside Rg1 into minor ginsenoside Rh1 simultaneously. The method was simple, low cost, and high yielding after optimizing the fermentation conditions. This new finding could have great significance in the research of ginsenosides, and this biotransformation has great potential to be applied in the preparation of minor ginsenosides F2 and C-K in the pharmaceutical industry.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of Jilin Province of China (No. 20150101184JC) and the National Natural Science Foundation of China (No. 20862017).
References
- 1.Qi L.W., Wang C.Z., Yuan C.S. Isolation and analysis of ginseng: advances and challenges. Nat Prod Rep. 2011;28:467–495. doi: 10.1039/c0np00057d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine-ginseng (I): etymology, pharmacognosy, phytochemistry, market and regulations. Curr Med Chem. 2009;16:2475–2484. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., Liu Q.M., Sung B.H., An D.S., Lee H.G. Bioconversion of ginsenosides Rb1, Rb2, Rc and Rd by novel β-glucosidase hydrolyzing outer 3-O glycoside from Sphingomonas sp. 2F2: Cloning, expression, and enzyme characterization. J Biotechnol. 2011;156:125–133. doi: 10.1016/j.jbiotec.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Zhang M., Zheng Y.N., Li J., Wang Y.P., Wang Y.J., Gu J., Jin Y. Snailase preparation of ginsenoside M1 from protopanaxadiol-type ginsenoside and their protective effects against CCl4-induced chronic hepatotoxicity in mice. Molecules. 2011;16:10093–10103. doi: 10.3390/molecules161210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Xu Y., Zhu Y., Li X. Anti-cancer effects of ginsenoside compound k on pediatric acute myeloid leukemia cells. Cancer Cell Int. 2013;13:24. doi: 10.1186/1475-2867-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Q.F., Fang X.L., Chen D.F. Pharmacokinetics and bioavailability of ginsenoside Rb1 and Rg1 from Panax notoginseng in rats. J Ethnopharmacol. 2003;84:187–192. doi: 10.1016/s0378-8741(02)00317-3. [DOI] [PubMed] [Google Scholar]
- 7.Shin K.C., Oh D.K. Characterization of a novel recombinant β-glucosidase from Sphingopyxis alaskensis that specifically hydrolyzes the outer glucoseat the C-3 position in protopanaxadiol-type ginsenosides. J Biotechnol. 2014;172:30–37. doi: 10.1016/j.jbiotec.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 8.Kim W.Y., Kim J.M., Han S.B., Lee S.K., Kim N.D., Park M.K., Kim C.K., Park J.H. Steaming of ginseng at high temperature enhances biological activity. J Nat Prod. 2000;63:1702–1704. doi: 10.1021/np990152b. [DOI] [PubMed] [Google Scholar]
- 9.Bae E.A., Han M.J., Kim E.J., Kim D.H. Transformation of ginseng saponins to ginsenoside Rh2 by acids and human intestinal bacteria and biological activities of their transformants. Arch Pharm Res. 2004;27:61–67. doi: 10.1007/BF02980048. [DOI] [PubMed] [Google Scholar]
- 10.Noh K.H., Son J.W., Kim H.J., Oh D.K. Ginsenoside compound K production from ginseng root extract by a thermostable beta-glycosidase from Sulfolobus solfataricus. Biosci Biotechnol Biochem. 2009;73:316–321. doi: 10.1271/bbb.80525. [DOI] [PubMed] [Google Scholar]
- 11.Chi H., Ji G.E. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol Lett. 2005;27:765–771. doi: 10.1007/s10529-005-5632-y. [DOI] [PubMed] [Google Scholar]
- 12.Wu L.P., Jin Y., Yin C.R., Bai L.L. Co-transformation of Panax major ginsenosides Rb1 and Rg1 to minor ginsenosides C-K and F1 by Cladosporium cladosporioides. J Ind Microbiol Biotechnol. 2012;39:521–527. doi: 10.1007/s10295-011-1058-9. [DOI] [PubMed] [Google Scholar]
- 13.Yang X.B., Hou J., Liu D., Deng S., Wang Z.B., Kuang H.X. Biotransformation of isoimperatorin by Cunninghamella blakesleana AS 3.970. J Mol Catal B: Enzym. 2013;88:1–6. [Google Scholar]
- 14.Lv X., Liu D., Hou J., Dong P.P., Zhan L.B., Wang L. Biotransformation of imperatorin by Penicillium janthinellum. Anti-osteoporosis activities of its metabolites. Food Chem. 2013;138:2260–2266. doi: 10.1016/j.foodchem.2012.11.138. [DOI] [PubMed] [Google Scholar]
- 15.Wilson D. Endophyte: the evolution of a term, and clarification of its use and definition. Oikos. 1995;73:274–276. [Google Scholar]
- 16.Wang Y., Dai C.C. Endophytes: a potential resource for biosynthesis, biotransformation and biodegradation. Ann Microbiol. 2011;61:207–215. [Google Scholar]
- 17.Agusta A., Maehara S., Ohashi K., Simanjuntak P., Shibuya H. Stereoselective oxidation at C-4 of flavans by the endophytic fungus Diaporthe sp. isolated from a tea plant. Chem Pharm Bull. 2005;53:1565–1569. doi: 10.1248/cpb.53.1565. [DOI] [PubMed] [Google Scholar]
- 18.Verza M., Arakawa N.S., Lopes N.P., Kato M.J., Pupo M.T., Said S. Biotransformation of a tetrahydrofuran lignin by the endophytic fungus Phomopsis sp. J Braz Chem Soc. 2009;20:195–200. [Google Scholar]
- 19.Yao M.L., Liu J.Z., Jin S., Jiao J., Gai Q.Y., Wei Z.F., Fu Y.J., Zhao J.D. A novel biotransformation of astragalosides to astragaloside IV with the deacetylation of fungal endophyte Penicillium canescens. Process Biochem. 2014;49:807–812. [Google Scholar]
- 20.Porebski S., Bailey L.G., Baum B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. [Google Scholar]
- 21.Kim O.S., Cho Y.J., Lee K., Yoon S.H., Kim M., Na H., Park S.C., Jeon Y.S., Lee J.H., Yi H. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K., Dudley J., Nei M., Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Yang J.L., Gao L.L., Zhu P. Advances in the biosynthesis research of ginsenosides. Acta Pharmaceutica Sinica. 2013;48:170–178. [PubMed] [Google Scholar]
- 24.Mai T.T., Moon J., Song Y., Viet P.Q., Phuc P.V., Lee J.M., Yi T.H., Cho M., Cho S.K. Ginsenoside F2 induces apoptosis accompanied by protective autophagy in breast cancer stem cells. Cancer Lett. 2012;321:144–153. doi: 10.1016/j.canlet.2012.01.045. [DOI] [PubMed] [Google Scholar]
- 25.Shin J.Y., Lee J.M., Shin H.S., Park S.Y., Yang J.E., Cho S.K., Yi T.H. Anti-cancer effect of ginsenoside F2 against glioblastoma in xenocraft model in SD rats. J Ginseng Res. 2012;36:86–92. doi: 10.5142/jgr.2012.36.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa H. Proof of the mysterious efficacy of ginseng: basic and clinical trials: metabolic activation of ginsenoside: deglycosylation by intestinal bacteria and esterification with fatty acid. J Pharmacol Sci. 2004;95:153–157. doi: 10.1254/jphs.fmj04001x4. [DOI] [PubMed] [Google Scholar]
- 27.Bae E.A., Choo M.K., Park E.K., Park S.Y., Shin H.Y., Kim D.H. Metabolism of ginsenoside Rc by human intestinal bacteria and its related antiallergic activity. Biol Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 28.Lee E.S., Choi J.S., Kim M.S., You H.J., Ji G.E., Kang Y.H. Ginsenoside metabolite compound K differentially antagonizing tumor necrosis factor-a induced monocyte-endothelial trafficking. Chem Biol Interact. 2011;194:13–22. doi: 10.1016/j.cbi.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Lee H.U., Bae E.A., Han M.J., Kim D.H. Hepatoprotective effect of ginsenoside Rb1 and compound K on tert-butyl hydroperoxide-induced liver injury. Liver Int. 2005;25:1069–1073. doi: 10.1111/j.1478-3231.2005.01068.x. [DOI] [PubMed] [Google Scholar]
- 30.Park E.K., Shin Y.W., Lee H.U., Kim S.S., Lee Y.C., Lee B.Y., Kim D.H. Inhibitory effect of ginsenoside Rb1 and compound K on NO and prostaglandin E2 biosyntheses of RAW264.7 cells induced by lipopolysaccharide. Biol Pharm Bull. 2005;28:652–656. doi: 10.1248/bpb.28.652. [DOI] [PubMed] [Google Scholar]
- 31.Ko S.R., Suzuki Y., Suzuki K., Choi K.J., Cho B.G. Marked production of ginsenosides Rd, F2, Rg3, and compound K by enzymatic method. Chem Pharm Bull. 2007;55:1522–1527. doi: 10.1248/cpb.55.1522. [DOI] [PubMed] [Google Scholar]
- 32.Cheng L.Q., Kim M., Lee J.W., Lee Y.J., Yang D.C. Conversion of major ginsenoside Rb1 to ginsenoside F2 by Caulobacter leidyia. Biotechnol Lett. 2006;28:1121–1127. doi: 10.1007/s10529-006-9059-x. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y., Yin Z.H., Wu L.P., Yin C.R. Diversity of cultivable β-glycosidase-producing microorganisms isolated from the soil of a ginseng field and their ginsenosides-hydrolysing activity. Lett Appl Microbiol. 2013;58:138–144. doi: 10.1111/lam.12166. [DOI] [PubMed] [Google Scholar]