Abstract
Phosphoinositols are an important class of phospholipids that are involved in a myriad of cellular processes, from cell signaling to motility and adhesion. Vinculin (Vn) is a major adaptor protein that regulates focal adhesions in conjunction with PIP2 in lipid membranes and other cytoskeletal components. The binding and unbinding transitions of Vn at the membrane interface are an important link to understanding the coordination of cell signaling and motility. Using different biophysical tools, including atomic force microscopy combined with confocal fluorescence microscopy and Fourier transform infrared spectroscopy, we studied the nanoscopic interactions of activated and autoinhibited states of Vn with lipid membranes. We hypothesize that a weak interaction occurs between Vn and lipid membranes, which leads to binding of autoinhibited Vn to supported lipid bilayers, and to unbinding in freestanding lipid vesicles. Likely driving forces may include tethering of the C-terminus to the lipid membrane, as well as hydrophobic helix-membrane interactions. Conversely, activated Vn binds strongly to membranes through specific interactions with clusters of PIP2 embedded in lipid membranes. Activated Vn harbored on PIP2 clusters may form small oligomeric interaction platforms for further interaction partners, which is necessary for the proper function of focal adhesion points.
Introduction
Phosphoinositols play an important functional role in different processes at the membrane-cytosol interface and are important regulators of cell physiology (1). For example, phosphatidylinositol 4,5-bisphosphate (PIP2) recruits and activates proteins related to the actin regulatory machinery at the plasma membrane and consequently affects cell shape and mobility, as well as several other processes (2, 3, 4). Actin-binding proteins that serve to link the actin cytoskeleton with cell-cell and/or cell-matrix adhesion complexes at the plasma membrane also rely on PIP2 for their smooth functioning. The molecular mechanisms underlying the binding and unbinding to PIP2 with various regulator and adaptor proteins at these membrane interfaces still remain to be understood.
Vinculin (Vn) is an adaptor protein and a key regulator of focal adhesions (FAs) (5, 6, 7). Downregulation of Vn causes increased migration rates and smaller and fewer adhesions compared with wild-type cells (8, 9, 10, 11). Structurally, Vn comprises five loosely packed, antiparallel α-helix bundle domains. The head domain consists of four bundles connected to the five-helix bundle Vn tail (Vt) domain by a disordered proline-rich region that functions as a hinge for the molecule (12). An extensive intramolecular interaction between the head and tail domains characterizes the inactive closed state of the molecule. This interaction has been shown to be disrupted by talin, which releases the Vt domain to bind to other ligands, including PIP2 (13, 14) and F-actin (15, 16). Although the ligand-binding sites are structurally distinct, they are conformationally and thus thermodynamically coupled. The activation of Vn seems to be achieved by a combinatorial input of ligands to distinct regions leading to a stable open-state conformer (17). Once activated, Vn is allowed full access and there are direct interactions of the head with talin and α-actinin; the neck with ponsin, vinexin, and Arp2/3; and the Vt domain with F-actin, PIP2, and paxillin (6, 7). Once Vn is recruited to nascent FAs, the interaction with talin causes activation of integrins and enlargement of FAs (18).
PIP2 itself functions at the cell adhesion sites and has been proposed to prevent F-actin binding (19), regulate FA turnover (20), and activate Vn (17). However, recent evidence revealed that a Vn mutant deficient of the PIP2-binding site is still able to localize FAs (20). The crystal structure of Vn revealed that the binding site of PIP2 is occluded by an extended coil of the Vt (21). Further, it has been shown in recent years that the mechanical tension emanating from integrin receptors plays a role in the activation of Vn at FAs through talin activation (22, 23, 24).
Activated Vn (Vac) has also been shown to exist as oligomers at the sites of cellular adhesions, which results from intermolecular tail domain interactions (14, 21, 25, 26). Vt dimerization can be induced by high concentrations as well as by F-actin binding (14, 21, 27). PIP2 binding also causes an enhanced dimer-/oligomerization (28). These Vt dimers/oligomers are mediated by different ligands and have also been shown to be distinct (21, 25).
Currently, we still face a conceptual gap in our understanding of the Vn-phosphoinositol interaction, especially with respect to PIP2 molecules embedded in lipid bilayers. Here, we report the harboring of Vn-binding site 3 activated vinculin, VBS3-Vac, on PIP2 clusters in lipid bilayers using confocal fluorescence microscopy and surface-sensitive, high-resolution techniques, including atomic force microscopy (AFM). Additionally, we report a weak interaction of the C-terminal amino acids of unactivated Vn with lipid bilayers, which is supported by complementary attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy data.
Materials and Methods
Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dioleoyl-sn-glycero- 3-phospho-(10-rac-glycerol) sodium salt (DOPG), 1,2-dipalmitoyl-sn-glycero-3-phospho-(10-rac-glycerol) sodium salt (DPPG), and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) (ammonium salt) (PIP2) were purchased from Avanti Polar Lipids (Alabaster, AL). Cholesterol (Chol) was obtained from Sigma-Aldrich (Deisenhofen, Germany). Stock solutions of lipid (DOPC, DPPC, and Chol) in chloroform (Merck, Darmstadt, Germany) and PIP2 in chloroform/methanol/water (20:9:1, v/v) were prepared. N-(lissamine rhodamine B sulfonyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethylammonium salt (N-Rh-DHPE) was obtained from Molecular Probes/Invitrogen (Eugene, OR).
Vn was obtained as a glycerol stock and lyophilized powder from Hypermol EK (Bielefeld, Germany). A Vt domain fragment and Vn-binding site 3 (VBS3) were obtained from Hypermol EK as lyophilized powder. For measurements, the lyophilized powders were dissolved in 20 mM Tris, 1 mM MgCl2, pH 7.4 buffer solution and used within a week. Vn was activated by addition of VBS3 solution in 100-fold molar excess to obtain the maximum amount of Vac.
Lysine labeling of Vn was carried out using the TORNADO-SMI 550 Protein Labeling Kit (Hypermol EK). The degree of labeling was calculated to be 58%. The excitation wavelength was 550 nm and the emission was ∼565 nm. Cysteine labeling was carried out using the Atto 488 maleimide (Atto-tec, Siegen, Germany) fluorescent dye derivative. Excess dye was removed using a Nap-5 column (GE Healthcare, Little Chalfont, UK) and further dialysis was performed to exchange buffer. The degree of labeling was calculated to be 42.6%.
AFM
Large unilamellar vesicle solutions were prepared as described previously (29). Briefly, the lipid solutions were mixed with the desired composition of DOPC/DPPC/Chol (1:2:1), DOPC, and DOPC/PIP2 (9:1). The majority of the solvent was evaporated with a nitrogen stream and subsequently removed by drying under vacuum overnight. The dry lipid mixture was hydrated with 1 mL of 20 mM Tris, 1 mM MgCl2, pH 7.4. The hydrated lipid mixture was then vortexed, kept in a water bath at 65°C for 15 min, and sonicated for 10 min. After five freeze-thaw-vortex cycles and brief sonication, large multilamellar vesicles were formed and transformed to large unilamellar vesicles of homogeneous size by use of an extruder (Avanti Polar Lipids) with polycarbonate membranes of 100 nm pore size at 65°C. Vesicle fusion on mica was carried out by depositing 30 μL of the extruded lipid vesicle solution together with 40 μL of Tris buffer on freshly cleaved mica and incubating it in a wet chamber at 70°C for 2 h. The supported lipid bilayer (SLB) was then transferred to a measuring cell filled with Tris buffer for AFM imaging (29, 30).
AFM experiments were carried out on a NanoWizard 3 NanoOptics (JPK Instruments AG, Berlin, Germany) set up on an IX71 microscope (Olympus, Tokyo, Japan). Tapping-mode topographic images were taken in the constant-deflection mode, using V-shaped silicon nitride cantilevers (Veeco, Santa Barbara, CA) with a typical spring constant of 0.3 N/m. A constant line rate was maintained at 0.5 Hz. The images were processed using JPK data-processing software.
Simultaneous fluorescence and AFM imaging was facilitated with a combined setup of the NanoWizard 3 NanoOptics (JPK Instruments AG) with a Microtime 200 (Picoquant, Berlin, Germany), a time-resolved confocal microscope that has single-molecule sensitivity and operates in the time-correlated, single-photon counting mode. Pulsed diode lasers (LDH series; Picoquant) were used as the source of the excitation light (488 nm and 560 nm wavelength) for the different fluorescent dyes. The emission light was transmitted through a dichroic mirror (z488/561rpc; AHF, Tübingen, Germany) and passed through a 50 μm pinhole to the single-photon avalanche diode detectors. A water-immersion objective (60×, NA 1.2; Olympus) was used and all measurements were carried out at 20°C.
FTIR spectroscopy
Lyophilized protein was hydrated with buffer containing D2O to completely remove any traces of water. The measurements were performed at 25°C with a Nicolet 5700 FTIR spectrometer equipped with a liquid nitrogen-cooled MCT (HgCdTe) detector and a cell with CaF2 windows that were separated by 50 μm mylar spacers. The spectrometer was purged continuously with dry air to remove water vapor. Typically, FTIR spectra of 128 scans were acquired with a resolution of 2 cm–1 and the corresponding processing was performed using GRAMS software (Thermo Electron, Waltham, MA). After background subtraction, the spectra were baseline corrected and normalized by setting the area between 1700 and 1600 cm–1 to 1 to allow for a quantitative analysis of the time evolution of secondary-structure changes. For analysis of the secondary-structure changes, second derivative and Fourier self-deconvolution were applied to the normalized spectra to identify the components of the amide-I′ band region. These peaks were then fitted to the normalized raw spectra using a Levenberg-Marquardt curve-fitting routine with bands of Gaussian and Voigt line shapes (31). All spectra were fitted with a similar set of peaks and parameters. In ambiguous cases (such as in the overlap region of unordered and α-helical bands), information from NMR and x-ray diffraction was taken into account. The area under each peak represents the fraction of the respective component (assuming similar transition dipole moments for the different conformers) and was finally used to determine the percentages of the secondary-structure component.
ATR-FTIR spectroscopy
DOPC was dissolved in chloroform to obtain the final concentration of 0.5 mg mL−1. The solvent was evaporated under a stream of N2 to obtain a uniform lipid film. Complete removal of the solvent was ensured by vacuum drying overnight. The dried lipid films were hydrated using a D2O-based Tris buffer (20 mM Tris, 1 mM MgCl2, pH 7.4). Large unilamellar vesicles were prepared from these films as described above. ATR-FTIR spectra were recorded using a Nicolet 6700 infrared spectrometer (Thermo Fisher Scientific, Waltham, MA) at a spectral resolution of 2 cm−1. The ATR out-of-compartment accessory consists of a liquid-jacketed ATR flow-through cell with a trapezoidal Ge crystal (80 × 10 × 4 mm, angle of incidence: 45°; Piketech, Madison, WI). The freshly prepared solution of large unilamellar vesicles was injected into the ATR flow cell, which was held at 25°C; this led to the spontaneous formation of SLBs on the surface of the Ge crystal used. Adsorption of the membrane was controlled by following the increase of the -CH2 lipid band intensities over time. After adsorption overnight, the membrane was washed with buffer over a period of 6 h. Protein solution prepared in D2O buffer was added to the bilayers and time-dependent spectra were recorded. The spectra were processed using GRAMS software (Thermo Electron). After subtraction of buffer together with the membrane and noise, the spectra were baseline corrected between 1710 and 1585 cm−1 and normalized to the amide-I′ band area (also between 1710 and 1585 cm−1).
Confocal laser scanning microscopy
Lipids were doped with N-Rh-DHPE to obtain a molar ratio of 500:1. Giant unilamellar vesicles (GUVs) were prepared by electroformation (32) in a temperature-controlled, in-house-made chamber on optically transparent and electrically conductive indium tin oxide-coated glass slides (SPI Supplies, West Chester, PA). Images were recorded by a confocal laser scanning microscope (MRC 1024, extended for multiphoton excitation; Biorad, now Zeiss, Oberkochen, Germany) coupled via a side-port to an inverted microscope (Eclipse TE-300 DV, infinity corrected optics; Nikon, Tokyo, Japan), enabling fluorescence excitation in the focal plane of an objective lens (Nikon Plan Fluor 40×, NA = 0.6, extra-long working distance of 3.7 mm, and Nikon Plan Appochromat VC 60× A WI, NA = 1.2 collar rim corr.). Fluorescence in the green and red PMT channels (emission bandpass filters, 522 nm/FWHM 35 nm and 580 nm/FWHM 40 nm, respectively) was sequentially acquired by alternating the excitation with the 488 and 568 nm lines of a Kr-Ar-laser S11 (Portmann Instruments AG, Biel-Benken, Switzerland). Image acquisition was controlled by LaserSharp2000 software (formerly Biorad, now Zeiss).
Results
To study the nanoscopic interactions of activated and inactivated Vn molecules with neutral model biomembranes, we first prepared SLB systems with a composition of DPPC/DOPC/Chol (1:2:1) and with pure DOPC only. Before conducting measurements, we checked the lipid bilayers for homogeneity and the absence of membrane defects. As is known from literature data, the raft-like DPPC/DOPC/Chol membrane exhibits phase separation into liquid-ordered (Lo) and liquid-disordered (Ld) domains, and thus serves as a well-established model system for heterogeneous natural membranes (29, 33). To confirm the formation of a bilayer in the case of a homogeneous membrane, we recorded force curves, which showed force jumps typical for the presence of a lipid bilayer (Fig. S1 in the Supporting Material).
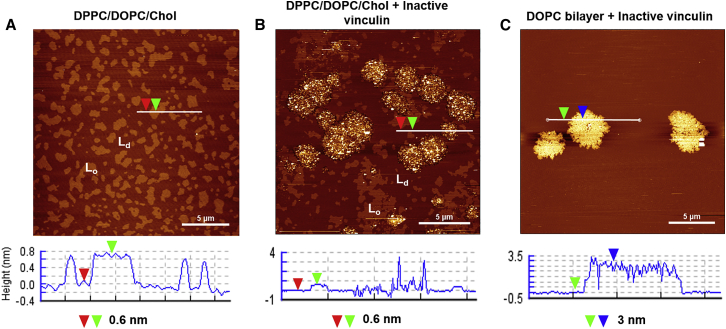
Intriguingly, we found that the inactivated full-length Vn molecule was able to bind to SLBs of zwitterionic lipid composition in a concentration-dependent fashion (Fig. 1) and form clusters. This behavior was observed irrespective of the presence or absence of PIP2 (Fig. S2). To exclude the possibility of an interaction between the protein and the solid support, 40 nM Vn was added to bare mica, and no adsorption was noted. Upon addition of higher concentrations of NaCl, the rate of growth of Vn clusters on the supported membrane decreased (Fig. S3); however, the size of the clusters that formed remained essentially unchanged. This could be due to an altered lipid packing density in the presence of high salt concentrations, leading to a modified binding behavior. Further, the preferential binding of the protein in the Ld phase rather than the Lo phase is clearly visible, indicating at least partial insertion/adsorption of Vn into the bilayer areas of lower packing density and conformational order.
Figure 1.
(A and B) AFM images of a DPPC/DOPC/Chol (1:2:1) lipid bilayer membrane on mica (A) before injection of protein solution and (B) after injection of 40 nM Vn. (C) Addition of 40 nM Vn to a DOPC bilayer. The AFM images are shown with a vertical color scale, with the height increasing from brown to white. (A) The absence of membrane defects and the Lo and Ld domains can be clearly observed with a height difference of 0.6 nm. (B) Addition of 40 nM Vn causes preferential partitioning and clustering of the protein in the Ld phase, resulting in heights of up to 3 nm. (C) Clustering of the protein is also observed in the homogeneous membrane consisting of pure DOPC bilayers. A statistical analysis showing a histogram of the height distribution over the image is presented in Fig. S4. To see this figure in color, go online.
Interestingly, Vn did not show significant binding to freestanding GUVs, which is consistent with various liposome studies that showed that only Vac is able to bind to PIP2-containing lipid vesicles (19, 28). We address this issue regarding different binding behaviors in flat supported membranes and GUVs later in the text.
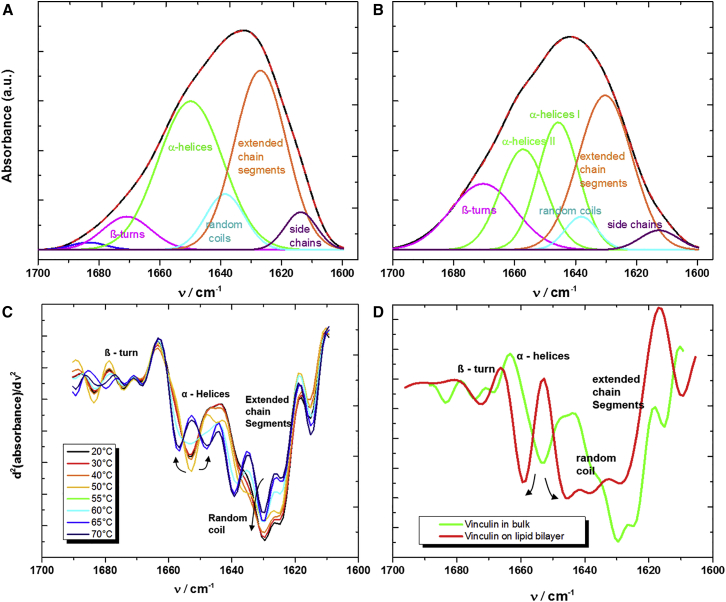
To further characterize the conformational state of Vn achieved upon binding to DOPC lipid bilayers, ATR-FTIR spectroscopy was carried out. The conformational changes that accompany the binding of protein to a lipid bilayer can be evaluated using the second-derivative spectra of the amide-I′ band (31). Changes in the secondary structure can be monitored by deconvoluting the amide-I′ band using a quantitative fit analysis with Gaussian and Lorentzian distribution functions. The resolved subbands can then be assigned to different secondary-structure components, also taking into account the x-ray diffraction and NMR data. As determined by FTIR spectroscopy, native Vn in bulk solution has six subbands (Fig. 2 A) appearing at 1613, 1627, 1638, 1649, 1670, and 1683 cm−1. On the basis of previous studies, we assigned bands at 1670–1683 cm−1 to β-turns, ∼1649 cm−1 to α-helices, ∼1638 cm−1 to random structures, and ∼1613 cm−1 to side chains (34, 35, 36). The band at 1627 cm−1 is generally assigned to intramolecular β-sheets, but here we assigned it to extended chain segments that connect the helical rods (37, 38), in agreement with the x-ray crystal structure of Vn and its domains (12, 17).
Figure 2.
Conformational changes of Vn as monitored by FTIR spectroscopy. (A) FTIR spectra of Vn in bulk solution. (B) ATR-FTIR spectra of Vn bound to the DOPC lipid bilayer. The local minima and maxima from the second derivative and Fourier self-deconvolution, respectively, were used to reveal the underlying amide-I′ subbands, thereby allowing the population of secondary-structure elements to be calculated. Black curves represent the original FTIR spectra and red lines show the fitted curves. (C) Second-derivative amide I′ spectra of Vn in bulk solution after denaturation with increasing temperature. (D) Second-derivative spectra comparing the secondary structure of bulk Vn with Vn bound to the lipid bilayer. To see this figure in color, go online.
Vn adsorbed on the lipid bilayer exhibits major conformational changes compared with Vn in bulk solution. The second-derivative spectra show that the helical band is split into two subbands (peaks at 1645 and 1657 cm−1) upon membrane binding (here denoted as α-helices I and II; Fig. 2, B and D), indicating different helical conformations with different degrees of solvation (31). The band at 1645 cm−1 represents highly solvated helices, whereas the band at 1657 cm−1 represents a helix in much less polar surroundings, perhaps due to partial insertion into the lipid bilayer.
To determine whether the binding of Vn to the SLB caused unfolding-like conformations, we recorded complementary temperature-dependent FTIR data for bulk Vn (Fig. 2 C). As expected, the temperature-induced denaturation of Vn was accompanied by a marked increase in random coil structures, from 9% to 35% (Fig. S5), which was not detected in the case of membrane-bound Vn. Therefore, binding of Vn to supported lipid membranes is likely accompanied by conformational changes in the α-helical tail region only, which could serve as driving force for membrane anchoring and subsequent clustering.
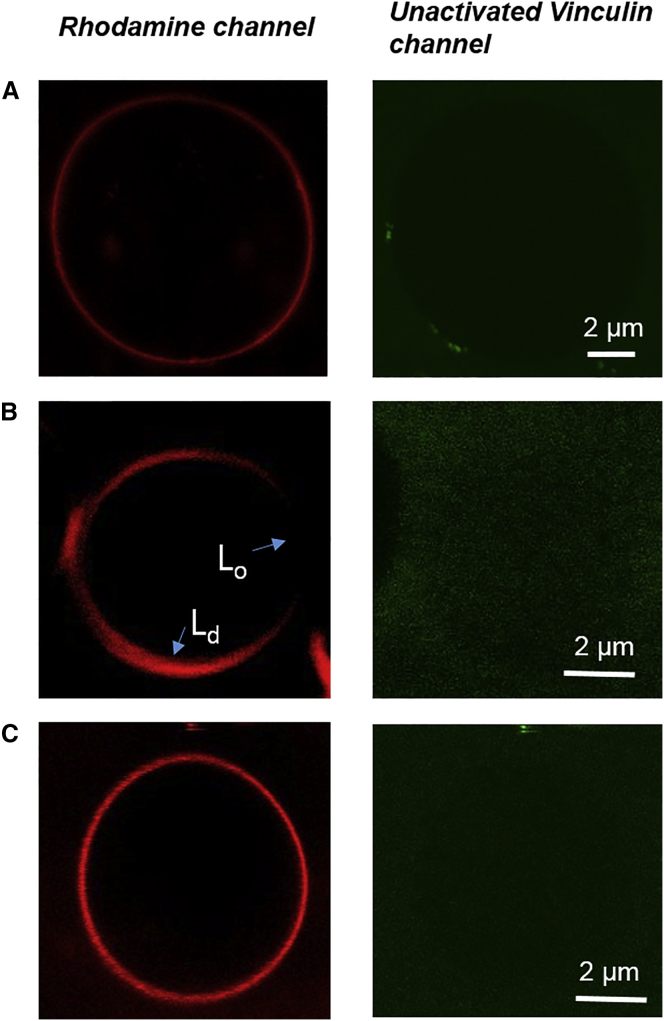
Previous studies have shown that unactivated Vn cannot significantly bind to liposomes. Further, Vac has been shown to specifically bind PIP2-containing liposomes (13, 14). Here, we show that Vn binds to SLBs even in the unactivated state. To examine this issue, we performed protein-binding studies using GUVs. Vn was labeled with Atto-488 via cysteine labeling. As expected, the cysteine-labeled Vn was also found to bind to the SLB, as confirmed by coupled AFM and fluorescence imaging (Fig. S6). Interestingly, we observed no marked binding to GUVs with different lipid compositions, with or without PIP2, as can be seen in Fig. 3. Neither the presence of PIP2 nor lipid phase separation in the vesicles changed the unbinding behavior of labeled full-length Vn with the vesicles, which is also in good agreement with previous studies (28).
Figure 3.
Confocal fluorescence microscopy images of GUVs. The red channel shows a rhodamine-labeled membrane and the green channel shows labeled unactivated Vn. (A) DOPC/PIP2 (9:1). (B) DPPC/DOPC/Chol (1:2:1). (C) DOPC. The lipid/protein ratio is 3000:1. The highest protein concentration for which no binding was observed was 2 μM (lipid/protein ratio 70:1). To see this figure in color, go online.
One way to understand these different interactions between SLBs and freestanding lipid bilayers is to take the bending excitations into account. Studies that examined stacks of membranes revealed an interplay among long-range attractive van der Waals interactions, short-range hydration, and electrostatic repulsive interactions (39, 40, 41), as well as long-range, steric-fluctuation-induced repulsive interactions (42, 43). According to the soft-shell theory, freestanding lipid membranes may exhibit pronounced bending excitations. The repulsive-bending excitations may play a negligible role in binding in the case of intrinsically strong and specific ligand-membrane interactions. However, we propose that for weak interactions, such repulsive-bending excitations may dominate, leading to unbinding transitions (40, 43). Owing to the presence of strong bending excitations in GUVs, weak membrane-Vn C-terminal interactions are largely suppressed. Conversely, a damping of bending excitations may be attained in SLB membranes, resulting in binding of inactive Vn. Thus, the different behaviors observed for Vn binding to GUVs and SLBs may be attributed to weak binding of unactivated Vn to lipid membranes only.
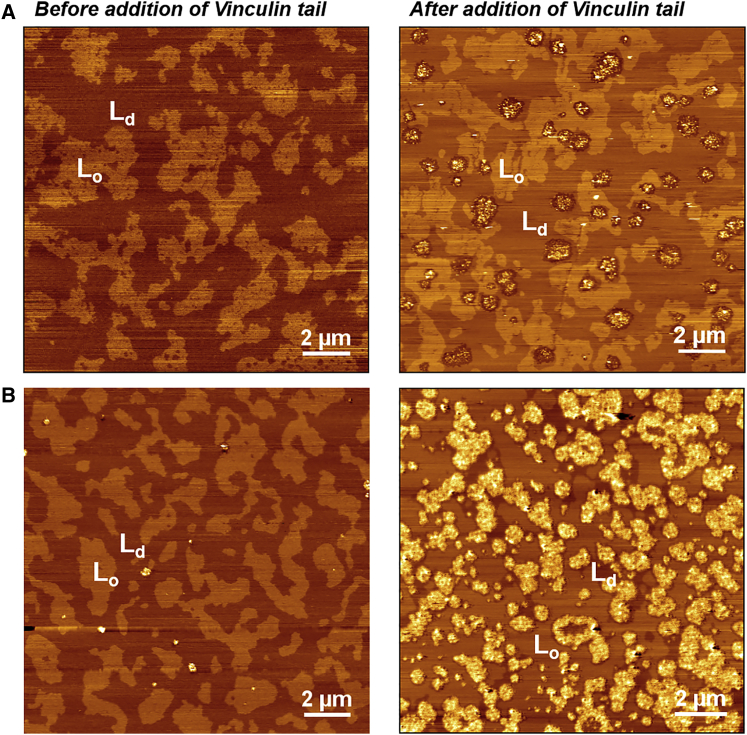
Interestingly, upon lysine labeling, Vn binding to the supported bilayer was significantly inhibited (data not shown). The crystal structure recently presented by Chinthalapudi et al. (28) revealed that Lys-915, Lys-1061, Ser-913, Lys-944, and Arg-945 are mainly associated with PIP2 in the Vn-PIP2 complex. Further, Lys-1061 was revealed to play a major role in binding to the lipid bilayer (28). Lipid-binding-deficient mutants have been shown to suppress the FA turnover rate and decrease cell migration (28). Cell studies have indicated that Lys-944 and Arg-945 mutations cause more immobile fractions than Lys-1061. A study investigating the interaction of peptides 1052–1066 of Vn with liposomes revealed a possible insertion of C-terminal amino acids into a membrane devoid of PIP2 (44). Hence, one possibility is that along with potentially specific binding to PIP2, the C-terminal region interacts weakly with lipid membranes. To examine this issue, we studied the binding of the Vt domain alone with SLBs of different lipid compositions. Interestingly, we observed a behavior similar to that found for the full-length Vn. Irrespective of the presence or absence of PIP2, the Vt domain bound to the SLBs, including neutral lipids (Fig. 4). These partitioning and clustering behaviors are similar to those observed in the full-length inactive Vn, and preferential partitioning into the Ld phase was observed as well. It is well established in the literature that, similarly to full-length Vac, the Vt domain also binds specifically only to PIP2-containing liposomes and micelles (14, 19, 21, 28). The binding behavior of the Vt to SLBs lacking PIP2, akin to the inactivated full-length Vn, implies a conceptual gap in our understanding of the interaction of Vn with the membrane. From the above studies, we can infer that this different binding behavior of full-length Vn to SLBs devoid of PIP2 most likely derives from the Vt domain.
Figure 4.
(A and B) AFM images of SLBs with lipid compositions of (A) DPPC/DOPC/Chol (1:2:1) + 0.8% PIP2 and (B) DPPC/DOPC/Chol (1:2:1). Left: images obtained before addition of the Vt domain show the presence of phase separations marked as Lo or Ld. Right: images of SLBs after addition of the Vt, which specifically partition into the Ld phase. The lipid/protein ratio is 6000:1. To see this figure in color, go online.
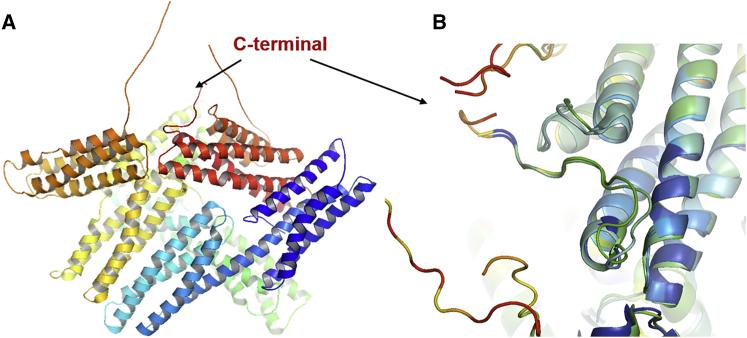
An overlay of the two known crystal structures of full-length Vn (PDB: 1ST6 and 1TR2) shows that unlike the other lysine residues, the C-terminal is located in a relatively unordered region and is solvent exposed (Fig. 5), easily allowing a weak interaction with the lipid bilayer. This also explains the inhibition of membrane binding of Lys-labeled Vn. Hence, eventually in concert with further weak binding motifs, the C-terminus region may play a role in strengthening the membrane-Vn interaction with binding to acidic phospholipids such as PIP2.
Figure 5.
Crystal structure of Vn. (A) A full-length Vn molecule showing the disordered C-terminal region. (B) The C-terminal region and its solvent accessibility. The overlay of two known crystal structures of Vn, PDB: 1ST6 and 1TR2, was constructed using Pymol (http://www.pymol.org). To see this figure in color, go online.
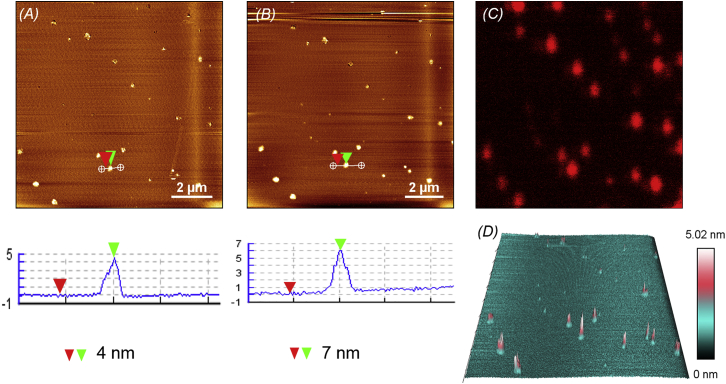
Next, we investigated the binding and oligomerization propensities of Vac in the presence of PIP2 embedded in lipid bilayers. A lipid bilayer membrane with a DOPC/10% PIP2 composition gave rise to significant clustering of PIP2 in the presence of divalent cations (Fig. 6 A). Activation of Lys-labeled Vn was achieved by incubation with VBS3, which has been shown to activate Vn sufficiently when added in a 100-fold molar excess (45, 46).
Figure 6.
AFM images of PIP2 harboring Vn coupled with confocal fluorescence microscopy images. (A) PIP2 clusters that formed in DOPC/10%PIP2 membranes in the presence of divalent cations (heights ≈ 4 nm). (B) Addition of 40 nM Vac, showing PIP2 clusters along with Vac (heights ≈ 7 nm). (C) Fluorescence imaging coupled with the AFM image (B) depicting Lys-labeled Vn. (D) Three-dimensional representation of PIP2 clusters (light blue) harboring Vac molecules (pink). The lipid/protein ratio is 6000:1. To see this figure in color, go online.
Unlike unactivated Vn, VBS3 activated vinculin specifically binds to PIP2 clusters, as is clearly visible in the fluorescence microscopy and AFM images depicted in Figs. 6, B and C. The binding of Vac to the PIP2 clusters is also indicated by a small increase in the height of the structures (∼3 nm). Fluorescence microscopy images combined with AFM images provide substantial evidence of PIP2 clusters harboring Vac molecules. The specificity of binding of VBS3-Vac to PIP2 was checked using other anionic lipids. SLBs with a membrane composition of DPPC/DOPC/DPPG/DOPG/Chol (45:20:5:5:25; 10% negatively charged lipids) did not show significant binding of VBS3-Vac (Fig. S7). The binding of VBS3 alone was also tested and revealed no significant binding (Fig. S8).
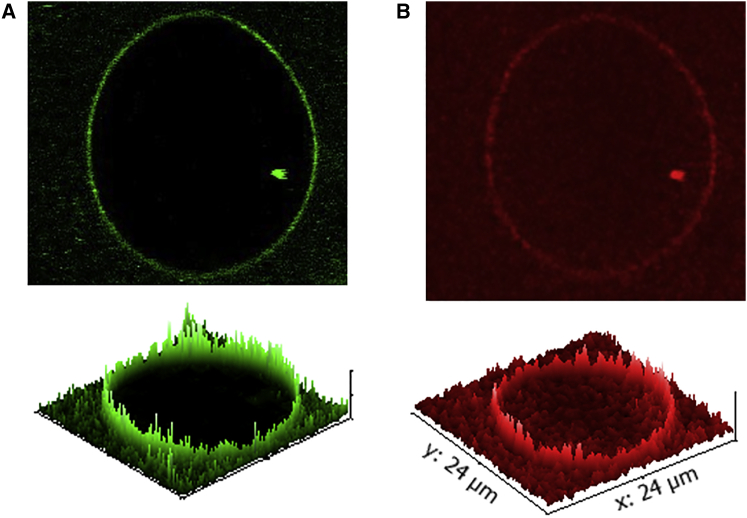
Further, VBS3-Vac can be seen bound to the GUV containing 10% PIP2 labeled with TopFluor PIP2 (Fig. 7), unlike unactivated Vn, which binds to SLBs only. It can also be noted that Vac does not lead to clustering of PIP2 on lipid bilayers. This is consistent with the fact that the PIP2-binding site in Vn does not consist of canonical PIP2-binding motifs or positive amino acid residue sequences that sequester PIP2 (47).
Figure 7.
Confocal fluorescence microscopy images of a GUV with a lipid composition of DOPC/PIP2 (9:1). (A) A 0.1 mol % TopFluor-PIP2-doped lipid membrane. (B) Labeled VBS3-Vac (2 μM; the lipid/protein ratio is 70:1). To see this figure in color, go online.
As discussed above, the driving force for the binding of unactivated Vn to the lipid membrane may arise from an initial tail-domain-mediated interaction, and the subsequent conformational/reorientational changes in Vn, as revealed by ATR-FTIR spectroscopy, may lead to intermolecular (probably hydrophobic) interactions, resulting in clustering. Conversely, in the case of the Vn-PIP2 complex, the dominant force is the specific interaction of Vac with the binding sites of PIP2, and, consistently, this interaction does not lead to the formation of larger clusters or aggregates, in contrast to the scenario observed for unactivated Vn. It is worth noting that unlike PIP2-binding proteins, which have a PIP2-binding motif (e.g., FERM domain proteins), Vac does not sequester PIP2 molecules, since the cluster size observed remains unchanged in SLBs and no clustering is observed in GUVs (3). Notably, the Lys-labeled unactivated Vn, which is rendered incapable of binding to a neutral bilayer membrane, retains its binding affinity for PIP2 molecules on a lipid bilayer when activated.
Discussion
PIP2 binds to numerous adhesion proteins and contributes to the regulation of FAs. However, the precise role and mechanism of control of PIP2 in cell motility and cell adhesion are still hotly debated. Vn is a prominent structural component of adhesion sites, and the activation and function of Vn has been shown to be spatially and temporally regulated at sites of FAs. Also, the interaction of PIP2 with Vn has been shown to play a role in FA turnover (20). Different PIP2-binding sites in Vn have been explored, and higher-order oligomerization was shown to result from Vn-PIP2 complex formation (28). Generally, the binding of a protein to a lipid membrane (apart from specific binding-site interactions) is also subject to contributions from the properties of the lipid membrane itself, such as lipid packing, surface charge density, and dynamic surface roughness, as well as hydrophobic contributions stemming from lipid-protein interactions.
Here, we report for the first time, to our knowledge, the binding of Vac molecules to PIP2 clusters embedded in lipid bilayers. We revealed a different binding behavior of unactivated Vn with supported membranes and freestanding lipid vesicles. Preferential binding of the protein in Ld lipid domains was observed, suggesting partial insertion of the protein into the hydrophobic environment of the membrane. This cannot be conclusively asserted based on these studies, however. Further, the conformational changes that accompanied the binding, as monitored by ATR-FTIR spectroscopy, also hint at partial insertion of a (amphipathic) helical segment of unactivated Vn into the lipid bilayer, which as a result of its further repercussions leads to clustering by hydrophobic and/or lipid-mediated entropic forces. The unbinding transition of the unactivated protein observed with GUVs is in accord with other liposome studies, which demonstrated that only Vac binds markedly to PIP2-containing liposomes. The unbinding transition can be interpreted in terms of stronger repulsive undulation forces present in the freestanding GUV system. This was previously pointed out by Helfrich (42), who demonstrated that entropic thermal fluctuations due to bending excitations in a membrane can contribute significantly to the repulsive forces. The presence of a strong site-specific binding affinity, as in the case of PIP2, renders the attractive forces dominant and the bending excitations negligible, leading to strong binding as observed experimentally in PIP2-containing bilayers. Damping of these thermal fluctuations, such as in supported bilayers or, presumably, generally in bilayers embedded in confined and crowded geometries, allows weak binding even of unactivated Vn.
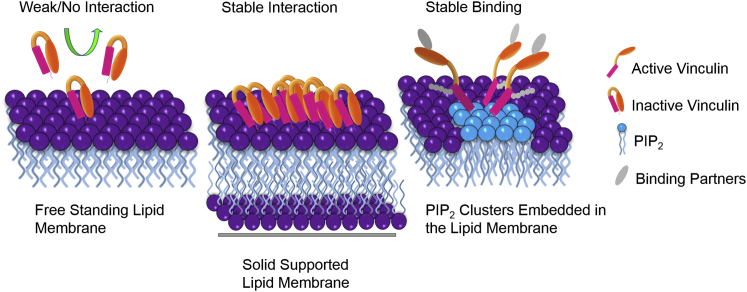
Notably, we also observed that binding of Vn is inhibited by Lys labeling of Vn. Additionally, it was shown that the carboxy terminal of the protein, along with Lys-1061, can potentially insert into lipid membranes (44), and the carboxy terminus, along with its few residues, is fairly flexible and solvent exposed. Therefore, our hypothesis assumes that the weak binding of Vn with the membrane occurs at the carboxy terminus. This weak interaction may open pathways for further binding partners to associate or dissociate (Fig. 8).
Figure 8.
Schematic representation of the interaction of Vn with zwitterionic freestanding and solid SLBs, and stable harboring of Vac on PIP2 clusters embedded in lipid membranes. To see this figure in color, go online.
Conversely, VBS3 activated vinculin shows a distinctly different binding behavior compared with the lipid membrane. PIP2 clusters embedded in the membrane exhibit a small increase in height (∼3 nm) upon binding of Vac. Lys-labeled Vac definitely reveals specific binding to PIP2 clusters in fluorescence microscopy (Fig. 6 D) coupled with AFM. VBS3-Vac is also able to bind to GUVs containing PIP2. This PIP2-specific dominant interaction does not lead to sequestering of PIP2 as observed with canonical PIP2-binding motifs (3). Unlike the case with unactivated Vn, lateral clustering is not observed for VBS3-Vac. Thus, the conformational changes that accompany PIP2 binding via specific residues in Vn do not lead to extended hydrophobic clustering on the membrane. Rather, residue-specific interactions seem to play the sole role in processes leading to the oligomerization of Vac on membranes (Fig. 6) (21, 28). Through such controlled oligomerization, PIP2 is able to direct Vn sequestration at FAs, which is necessary for their proper function.
Author Contributions
M.D. and R.W. designed the research and wrote the manuscript. M.D. performed the research.
Acknowledgments
This research was supported by the Deutsche Forschungsgemeinschaft.
Editor: Claudia Steinem.
Footnotes
Eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30707-X.
Contributor Information
Mridula Dwivedi, Email: mridulavd@gmail.com.
Roland Winter, Email: roland.winter@tu-dortmund.de.
Supporting Material
References
- 1.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Ling K., Doughman R.L., Anderson R.A. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin S., Wang J., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 4.Yin H.L., Janmey P.A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 5.Jockusch B.M., Rüdiger M. Crosstalk between cell adhesion molecules: vinculin as a paradigm for regulation by conformation. Trends Cell Biol. 1996;6:311–315. doi: 10.1016/0962-8924(96)10022-2. [DOI] [PubMed] [Google Scholar]
- 6.Zamir E., Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler W.H., Liddington R.C., Critchley D.R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Coll J.L., Ben-Ze’ev A., Adamson E.D. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. USA. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volberg T., Geiger B., Ben-Ze’ev A. Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J. Cell Sci. 1995;108:2253–2260. doi: 10.1242/jcs.108.6.2253. [DOI] [PubMed] [Google Scholar]
- 10.Xu W., Baribault H., Adamson E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 11.Saunders R.M., Holt M.R., Critchley D.R. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Borgon R.A., Vonrhein C., Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Weekes J., Barry S.T., Critchley D.R. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 1996;314:827–832. doi: 10.1042/bj3140827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer S.M., Playford M.P., Campbell S.L. Lipid binding to the tail domain of vinculin: specificity and the role of the N and C termini. J. Biol. Chem. 2009;284:7223–7231. doi: 10.1074/jbc.M807842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson R.P., Craig S.W. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 16.Peng X., Nelson E.S., DeMali K.A. New insights into vinculin function and regulation. Int. Rev. Cell Mol. Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakolitsa C., Cohen D.M., Liddington R.C. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 18.Niggli V., Sommer L., Burger M.M. Interaction in situ of the cytoskeletal protein vinculin with bilayers studied by introducing a photoactivatable fatty acid into living chicken embryo fibroblasts. Eur. J. Biochem. 1990;187:111–117. doi: 10.1111/j.1432-1033.1990.tb15283.x. [DOI] [PubMed] [Google Scholar]
- 19.Steimle P.A., Hoffert J.D., Craig S.W. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J. Biol. Chem. 1999;274:18414–18420. doi: 10.1074/jbc.274.26.18414. [DOI] [PubMed] [Google Scholar]
- 20.Chandrasekar I., Stradal T.E.B., Ziegler W.H. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- 21.Bakolitsa C., de Pereda J.M., Liddington R.C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 22.del Rio A., Perez-Jimenez R., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao M., Goult B.T., Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galbraith C.G., Yamada K.M., Sheetz M.P. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson R.P., Craig S.W. Actin activates a cryptic dimerization potential of the vinculin tail domain. J. Biol. Chem. 2000;275:95–105. doi: 10.1074/jbc.275.1.95. [DOI] [PubMed] [Google Scholar]
- 26.Molony L., Burridge K. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 1985;29:31–36. doi: 10.1002/jcb.240290104. [DOI] [PubMed] [Google Scholar]
- 27.Hüttelmaier S., Mayboroda O., Rüdiger M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 1998;8:479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- 28.Chinthalapudi K., Rangarajan E.S., Izard T. Lipid binding promotes oligomerization and focal adhesion activity of vinculin. J. Cell Biol. 2014;207:643–656. doi: 10.1083/jcb.201404128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weise K., Kapoor S., Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 30.Weise K., Triola G., Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J. Am. Chem. Soc. 2009;131:1557–1564. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- 31.Kapoor S., Triola G., Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc. Natl. Acad. Sci. USA. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelova M., Dimitrov D. A mechanism of liposome electroformation. Trends Colloid Interface Sci. II. 1988;67:59–67. [Google Scholar]
- 33.van Duyl B.Y., Ganchev D., Killian J.A. Sphingomyelin is much more effective than saturated phosphatidylcholine in excluding unsaturated phosphatidylcholine from domains formed with cholesterol. FEBS Lett. 2003;547:101–106. doi: 10.1016/s0014-5793(03)00678-1. [DOI] [PubMed] [Google Scholar]
- 34.Lenk T.J., Horbett T.A., Ratner B.D., Chittur K.K. Infrared spectroscopic studies of time-dependent changes in fibrinogen adsorbed to polyurethanes. Langmuir. 1991;7:1755–1764. [Google Scholar]
- 35.Wu Y., Murayama K., Ozaki Y. Two-dimensional attenuated total reflection/infrared correlation spectroscopy studies on concentration and heat-induced structural changes of human serum albumin in aqueous solutions. Appl. Spectrosc. 2002;56:1186–1193. [Google Scholar]
- 36.Chittur K.K. FTIR/ATR for protein adsorption to biomaterial surfaces. Biomaterials. 1998;19:357–369. doi: 10.1016/s0142-9612(97)00223-8. [DOI] [PubMed] [Google Scholar]
- 37.Reisdorf W.C., Jr., Krimm S. Infrared amide I′ band of the coiled coil. Biochemistry. 1996;35:1383–1386. doi: 10.1021/bi951589v. [DOI] [PubMed] [Google Scholar]
- 38.Gilmanshin R., Williams S., Dyer R.B. Fast events in protein folding: relaxation dynamics of secondary and tertiary structure in native apomyoglobin. Proc. Natl. Acad. Sci. USA. 1997;94:3709–3713. doi: 10.1073/pnas.94.8.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipowsky R., Leibler S. Unbinding transitions of interacting membranes. Phys. Rev. Lett. 1986;56:2541–2544. doi: 10.1103/PhysRevLett.56.2541. [DOI] [PubMed] [Google Scholar]
- 40.Lipowsky R. The conformation of membranes. Nature. 1991;349:475–481. doi: 10.1038/349475a0. [DOI] [PubMed] [Google Scholar]
- 41.Rand R.P. Interacting phospholipid bilayers: measured forces and induced structural changes. Annu. Rev. Biophys. Bioeng. 1981;10:277–314. doi: 10.1146/annurev.bb.10.060181.001425. [DOI] [PubMed] [Google Scholar]
- 42.Helfrich W. Steric interactions of fluid membranes in multilayer systems. Z. Naturforsch. A. 1978;33:305–315. [Google Scholar]
- 43.Sackmann E., Smith A.-S. Physics of cell adhesion: some lessons from cell-mimetic systems. Soft Matter. 2014;10:1644–1659. doi: 10.1039/c3sm51910d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diez G., List F., Goldmann W.H. Direct evidence of vinculin tail-lipid membrane interaction in beta-sheet conformation. Biochem. Biophys. Res. Commun. 2008;373:69–73. doi: 10.1016/j.bbrc.2008.05.182. [DOI] [PubMed] [Google Scholar]
- 45.Witt S., Zieseniss A., Illenberger S. Comparative biochemical analysis suggests that vinculin and metavinculin cooperate in muscular adhesion sites. J. Biol. Chem. 2004;279:31533–31543. doi: 10.1074/jbc.M314245200. [DOI] [PubMed] [Google Scholar]
- 46.Bass M.D., Patel B., Critchley D.R. Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem. J. 2002;362:761–768. doi: 10.1042/0264-6021:3620761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humphries J.D., Wang P., Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.