Abstract
BtuB is a TonB-dependent transporter that permits the high-affinity binding and transport of cobalamin (CBL), or vitamin , across the asymmetric outer membrane (OM) of Gram-negative bacteria. It has been shown that binding is necessary for high-affinity binding of CBL to BtuB, and earlier simulations suggested that calcium ions serve to stabilize key substrate-binding extracellular loops. However, those simulations did not account for the lipopolysaccharides in the OM. To illuminate the roles of both and lipopolysaccharides in protein functionality, we performed simulations of apo and -loaded BtuB in symmetric and asymmetric bilayers. The simulations reveal that the oligosaccharides of LPS stabilize the extracellular loops to some degree, apparently obviating the need for . However, it is shown that ions stabilize a key substrate-binding loop to an even greater degree, as well as reposition specific CBL-binding residues, bringing them closer to the organization found in the CBL-bound structure. These results indicate the importance of including realistic membrane models when simulating outer-membrane proteins.
Introduction
Gram-negative bacteria are characterized by the presence of two membrane bilayers, cytoplasmic and outer, which together provide a significant barrier to antibiotic uptake. The outer membrane (OM) protects against both hydrophobic and hydrophilic molecules due to the intrinsic asymmetry of the bilayer (1, 2, 3). In contrast to the cytoplasmic membrane, which has two symmetric leaflets of phospholipids, the OM has a periplasmic, or inner, leaflet of phospholipids and an extracellular (EC) leaflet of lipopolysaccharides (LPS) (4, 5). Whereas phospholipids characteristically have two aliphatic tails, LPS has a varying number of lipid tails, typically four to six depending on the species, as well as a large, charged oligosaccharide headgroup and long O-antigen chains (2, 5, 6, 7, 8). Furthermore, the LPS oligosaccharide has numerous phosphate groups that result in ionic bridging between LPS oligosaccharides via cations, which is predicted to reduce the lateral mobility of LPS (9).
Although it affords Gram-negative bacteria additional protection, the OM complicates the uptake of nutrients and other molecules needed for cellular function (10). To circumvent this uptake problem, an assortment of transmembrane proteins provide pathways for the passive diffusion or transport of molecules across the OM. Characteristic of most OM proteins is a β-barrel structure spanning the membrane, as well as long and charged EC loops. The orientation of these EC loops often plays a significant role in nutrient uptake across the OM (11, 12, 13, 14, 15).
TonB-dependent transporters, a subset of OM proteins, are 22-stranded, monomeric β-barrels that transport large and rare nutrients, typically ferric siderophores and chelates, by coupling to an energy-transducing, cytoplasmic-membrane-bound TonB complex that spans the periplasm (16, 17, 18, 19). Although there are many similarities among TonB-dependent transporters, BtuB uniquely depends on calcium-ion binding to transport cobalamin (CBL), also known as vitamin , across the OM (20, 21, 22). Crystal structures show that five aspartate residues on EC loops 3/4 and 5/6, constituting the so-called aspartate cage (Asp-cage), provide the binding site for two ions, which are necessary for CBL transport (13, 14). The loop positions are known to be sensitive to solutes (23), and protein dynamics are sensitive to membrane composition and thickness (24), necessitating the use of highly accurate models to capture such sensitivity.
In this work, we first introduce an LPS-containing model of the OM that was constructed in silico to perform molecular-dynamics (MD) simulations of OM proteins in their native environment. We then calculate the area per lipid, hydrophobic thickness, and lateral diffusion rate of the asymmetric bilayer. Next, we use this OM model to study the interactions of BtuB with LPS molecules and compare them with those of BtuB in a symmetric phospholipid bilayer, revealing unique protein-LPS interactions in the asymmetric system. Finally, we use this model of the OM to elucidate the role of binding, which is required for high-affinity binding of CBL (21, 22).
Materials and Methods
Simulation systems
Five systems containing BtuB were built to elucidate the effect of LPS and the role of binding in EC-loop stability. The apo-state structure (PDB: 1NQE) was placed in a symmetric 1-palmitoyl 2-oleoyl phosphatidyl-ethanolamine (POPE) bilayer and two asymmetric LPS (outer leaflet)-POPE (inner leaflet) OMs using VMD (25, 26). The -bound state (PDB: 1NQG) was also placed in two asymmetric model membranes (25). The two asymmetric OMs differ only in the initial arrangement of LPS molecules around the protein. Missing EC loops in the crystallographic structures of BtuB were completed using the TonB-bound structure (PDB: 2GSK) (14, 27), as shown in Fig. S1, A and B, in the Supporting Material.
Each system was solvated and then ionized to a salt concentration of 150 mM NaCl. The apo-symmetric (Apo-sym) system has a total of ∼70,000 atoms, with an outer leaflet composed of 66 POPE lipids and an inner leaflet composed of 61 POPE lipids. The disproportionate number of lipids is due to the cross-sectional area of BtuB being greater on the periplasmic side than on the EC side; therefore, more lipids were needed in the outer leaflet to keep the area per lipid roughly identical between layers. The asymmetric-bilayer systems all have a total of ∼137,000 atoms, with an outer leaflet composed of 50 LPS molecules and an inner leaflet of 157 POPE lipids (details can be found in Table S1).
The initial model used for a single LPS was taken from the crystal structure of LPS in complex with FhuA (28). This LPS from Escherichia coli K-12 is the rough form, also known as the RaLPS chemotype (29), and contains only lipid A along with the 10-saccharide core region (see Fig. S2 for a schematic of the core saccharide) (30). Two phosphate groups were added to heptose I, and one group was added to heptose II. Based on the crystal structure occupancy (31), half of the lipid A moieties have one phosphate on the first glucosamine and the other half have two; all have one phosphate on the second glucosamine. Thus, the net charge on each LPS molecule is either −11e or −12e. LPS molecules were neutralized using mostly (97%) ions, along with a few (3%) ions. These ions were initially placed using the cIonize plugin in VMD, which uses a Poisson-Boltzmann solver to iteratively place ions in electrostatic minima (32). The force-field parameters for LPS were constructed by analogy using the CHARMM36 lipid and carbohydrate force fields, and thus are nearly identical to those developed by Wu et al. (7) (see Supporting Material for details).
MD protocol
NAMD2.9 was used for all MD simulations (33) along with the CHARMM36 all-atom force field for protein, ions, and phospholipids (34, 35). A 2-fs time step was employed. Bonded terms were evaluated every time step, and nonbonded terms and long-range electrostatic interactions were updated every 2 fs and 4 fs, respectively. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method (36). van der Waals interactions were cut off at 12 Å, and a force-based switching function was used beginning at 10 Å. Temperature and pressure were maintained at 310 K and 1 atm by means of Langevin dynamics and a Langevin piston, respectively. All bonds involving hydrogen atoms were constrained using the SHAKE algorithm (37).
System equilibration was carried out over multiple steps. First, all atoms were constrained, with the exception of lipid tails, which were equilibrated for 1 ns, allowing them to melt. Second, water, ions, and lipid headgroups were released and equilibrated for 2 ns in the symmetric bilayer. Due to the large initial spacing of the LPS molecules, which was necessary to avoid clashes during building, a surface tension of −100, −200, −250, and −300 dynes/cm equilibrated at 2 ns each was applied to the asymmetric membrane. This process allowed the LPS to eliminate any gaps introduced in construction. We then released the protein side chains for 1 ns before releasing the protein backbone and equilibrating for 10 ns. Simulations were run in the NPT ensemble for 150 ns for the Apo-sym, and 300 ns for the Apo1-, Apo2-, Ca1-, and Ca2-OM systems after equilibration. Analysis of each system was performed on data collected from the last 200 ns of production for the OM systems and the last 100 ns of production for the symmetric bilayer, as shown in Fig. S3. Langevin dynamics were employed to maintain the temperature at 310 K and a Langevin piston was used to keep the pressure at 1 atm throughout equilibration and production simulations.
For steered MD (SMD), an imaginary particle was affixed to the acetylated N-terminus of BtuB with a spring (k = 5 kcal/mol Å2). This imaginary particle was pulled at constant velocities of 1 Å/ns and 0.25 Å/ns normal to the bilayer plane out of the barrel. Twenty-two atoms in a ring around the barrel, one per β-strand, were fixed normal to the bilayer to prevent system propagation due to luminal domain extraction. The 1 Å/ns SMD runs were performed three times for each Apo system, and the results presented were system and time averaged. Due to the amount of time required, the 0.25 Å/ns SMD runs were only performed once each for the Apo-sym and Apo-OM systems.
Analysis
The membrane thickness was calculated by measuring the average distance between acyl chain C2 atoms in the POPE leaflet and C2 or C4 on each of the LPS tails, as done previously (38, 39). The APL was calculated by determining the total lateral surface area of the system, subtracting the cross-sectional contribution from the protein, and then dividing that area by the number of lipids in each leaflet. The cross-sectional area of BtuB was determined using a double summation technique for a plane width of 1 Å normal to the axis of the protein between the periplasmic turns and EC loops. Lateral diffusion coefficients were obtained by calculating the mean-square displacement of the POPE hydrophilic head center of mass as done elsewhere (40, 41). Similarly, diffusion coefficients for LPS were calculated by measuring the mean-square displacement of the lipid A center of mass. The mean-square displacement was also determined for the LPS center of mass, which resulted in nearly identical diffusion values (data not shown). Root mean-square fluctuation (RMSF) values were calculated only for the atom of each amino acid (42) to remove rotamer conformation bias. A full summary of the aforementioned values for each system can be found in Table S2. Sugar conformations for the LPS core oligosaccharides were tracked for each Apo- and Ca-OM simulation and are plotted in Fig. S4.
Results and Discussion
Bilayer dynamics
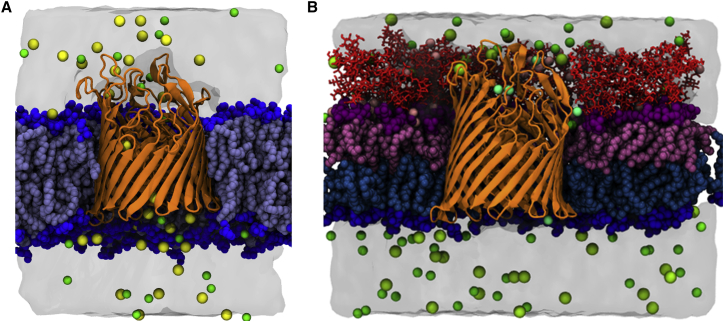
Although the OM typically has an inner-leaflet composition similar to that of the cytoplasmic membrane (43, 44), we simplified the OM model in this study by utilizing an inner-leaflet composition of pure POPE phospholipids. This is an intuitive simplification since the OM inner leaflet consists of ∼80% PE lipids (43). We modeled our OM after K12 E. coli, whose LPS lacks O-antigen chains, possessing only the membrane-forming lipid A and inner- and outer-core oligosaccharides (1, 5, 8). The OM model is shown in Fig. 1 with the symmetric bilayer for comparison. To compare our model system with previous experimental and computational results, we calculated the hydrophobic thickness, lateral diffusion, and area per lipid for the apo BtuB symmetric bilayer system (Apo-sym) (Fig. 1 A), each apo BtuB asymmetric bilayer (Apo1-OM, Apo2-OM), and each -bound BtuB asymmetric bilayer (Ca1-OM and Ca2-OM) system (Fig. 1 B). Furthermore, we simulated the OM model with no protein present.
Figure 1.
(A and B) Snapshot of BtuB in (A) a symmetric bilayer and (B) an asymmetric bilayer. Water is shown as a molecular surface. Ions are shown as yellow for , green for , pink for , and teal for . NaCl concentrations are 150 mM in each system. BtuB is represented as orange ribbons. The phospholipid tails are shown in light blue and headgroups are shown as dark blue spheres. Lipid A of LPS is shown in pink, the corresponding lipid A headgroups are shown as magenta spheres, and the core oligosaccharides are shown in red sticks. To see this figure in color, go online.
To check the validity of the model, we first determined the hydrophobic thickness of each membrane system. As shown in Fig. S5, B–E, each protein-containing OM system, as well as the OM model, has a hydrophobic thickness of ∼25 Å, which matches well with recent results for another OM simulation model (39). When one compares the OM systems with the Apo-sym profile shown in Fig. S5 A, it is clear that the asymmetric OM is much thinner than the symmetric bilayer. The membrane thickness difference is attributed primarily to the tail length difference (LPS has 12–14 carbons per tail, whereas POPE has 16–18), as well as lipid tail interdigitation in the OM systems. Another verification of the hydrophobic thickness is the location of aromatic residues on the protein, specifically phenylalanine, tryptophan, and tyrosine, which are known to reside at amphiphilic interfaces (45, 46). From the density profiles shown in Fig. S5, it is clear that the OM systems match well with the aromatic residue profile of the protein. In contrast, the increased hydrophobic thickness of the Apo-sym system permits PE headgroups to interact with the protein along the β-barrel/EC loop interface, which can be seen in Fig. S6. However, the oligosaccharide number density, as shown in Fig. S5, B–E, indicates that the LPS headgroups align with the EC loops of the protein.
Second, we determined the average lateral diffusion of LPS in the OM systems. It is understood that ionic bridging between core oligosaccharides by divalent cations significantly decreases the lateral mobility of LPS (5). Multiple fluorescence experiments have provided LPS lateral diffusion values ranging from to depending on the LPS variant (9, 47, 48, 49). This is in stark contrast to phospholipid diffusion values, which are on the order of (35). As shown in Fig. S7 and elaborated in Table S2, the diffusion coefficients determined for LPS in the OM systems vary between 6.0 and , and thus are comparable to values found in previous simulations (6, 7). The diffusion coefficients for the inner leaflet of phospholipids are much larger, ranging between 4.3 and , for all protein-membrane systems simulated. This range is only slightly higher than the calculated diffusion coefficient for the Apo-sym bilayer, .
As the final check of the OM model’s validity, we determined the observed area per lipid for each system. The pure OM simulation was performed with 36 LPS and 102 POPE. This ratio was chosen to replicate an experimental value of ∼156 /LPS (50), which was utilized in simulations by Piggot et al. (6). The area per LPS was found to be 179 /LPS for this test system, which was much closer to that reported for a similar variant by Wu et al. (7) (a full comparison can be found in Table S2). This value further contrasts with the measured area per LPS for the protein-containing OM systems, which was ∼10% larger, between 195 and 199 /LPS. Although the area per LPS differs between the pure-OM and protein-OM systems, the large number of LPS-protein interactions (see Tables S3 and S5) and the consistent properties of the LPS leaflet (namely, the thickness and rate of diffusion) suggest that this small difference in density does not lead to a substantive difference in BtuB’s behavior.
A simulation of a pure POPE bilayer with 102 lipids per leaflet was also carried out for 60 ns. After disregarding the first 10 ns, we found that the area per lipid was 57.9 ± 0.9 , which is near the CHARMM36-reported area per lipid for POPE of 57.8 ± 0.1 (51). This value also is only slightly smaller than the measured area per lipid in the Apo-sym case of 58.9 . The area per lipid for the inner leaflet in the OM systems was observed to be 5–10% higher at 61.4–63.7 (see Fig. S8 and Table S2). In light of the results from Wu et al. (7) and the results of the simulations reported here, future systems should be adjusted to target an area per LPS of 180 .
Apo-state comparison
To better understand how LPS affects BtuB’s function, we first compared the interactions of the apo-state BtuB (i.e., with no or CBL) in the Apo-sym and Apo-OM systems described above. The asymmetric distribution of outward-facing charged and polar amino acids on OM proteins (45), the substantial anionic charge of LPS, and the observed alignment of the oligosaccharides with EC loops suggest that LPS should affect protein functionality. This claim is substantiated by observing the stabilizing effect LPS has on the EC loops, the organizational change in the interior luminal domain of BtuB, and the significant difference in force required to partially extract the luminal domain.
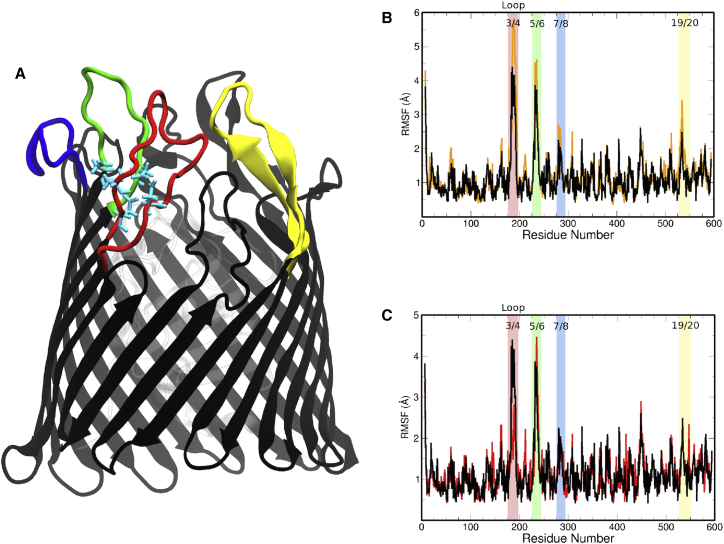
The stabilizing effect of LPS on the protein is exemplified by the RMSF of the protein backbone, particularly the EC loops, as shown in Fig. 2 B. All of the highlighted regions, which correspond to the highlighted loops of BtuB in Fig. 2 A, show a 1–2 Å stabilization when LPS is present. This effect is due to the alignment of the oligosaccharide along the EC loops, as shown in Fig. 3 B, which prevents the loops from folding back over the membrane and thus limits their range of motion, at least on the ∼300-ns timescales investigated here. Further investigation of the oligosaccharide-protein interaction reveals a dense hydrogen-bond network between the LPS and EC loops. These hydrogen bonds occur between the LPS and charged, outward-facing residues in the EC loops, particularly those closer to the EC loop apex, as shown in Fig. S6 and enumerated in Table S3. This contrasts with the membrane-protein interactions observed in the Apo-sym system (Table S4), which occur proximal to the hydrophobic-matching region of the protein along the barrel/EC loop interface (see Fig. 3 A). Although the Asp-cage is exposed, we see limited interactions with ions: an ion binds to Asp195, Asp195, and Asp230 for nearly all but the last 15 ns of the Apo1-OM simulation, and an ion binds to Asp179 for the last 140 ns of the Apo2-OM simulation.
Figure 2.
(A) Ribbon representation of BtuB's β-barrel, with Asp-cage highlighted in cyan sticks. Key EC loops are highlighted in red (loop 3/4), green (loop 5/6), blue (loop 7/8), and yellow (loop 19/20). (B) Plot of RMSFs by residue of the BtuB Apo-sym (orange trace) and Apo1-OM (black trace) bilayers. A substantial RMSF reduction occurs in the highlighted regions, which correspond to the highlighted loops in (A). (C) Plot of RMSFs by residue for the Apo1-OM (black trace) and Ca1-OM (red trace) systems. A slight difference in RMSF is observed in loop 3/4, where binding occurs. RMSF plots for Apo2-OM and Ca2-OM can be found in Fig. S9. To see this figure in color, go online.
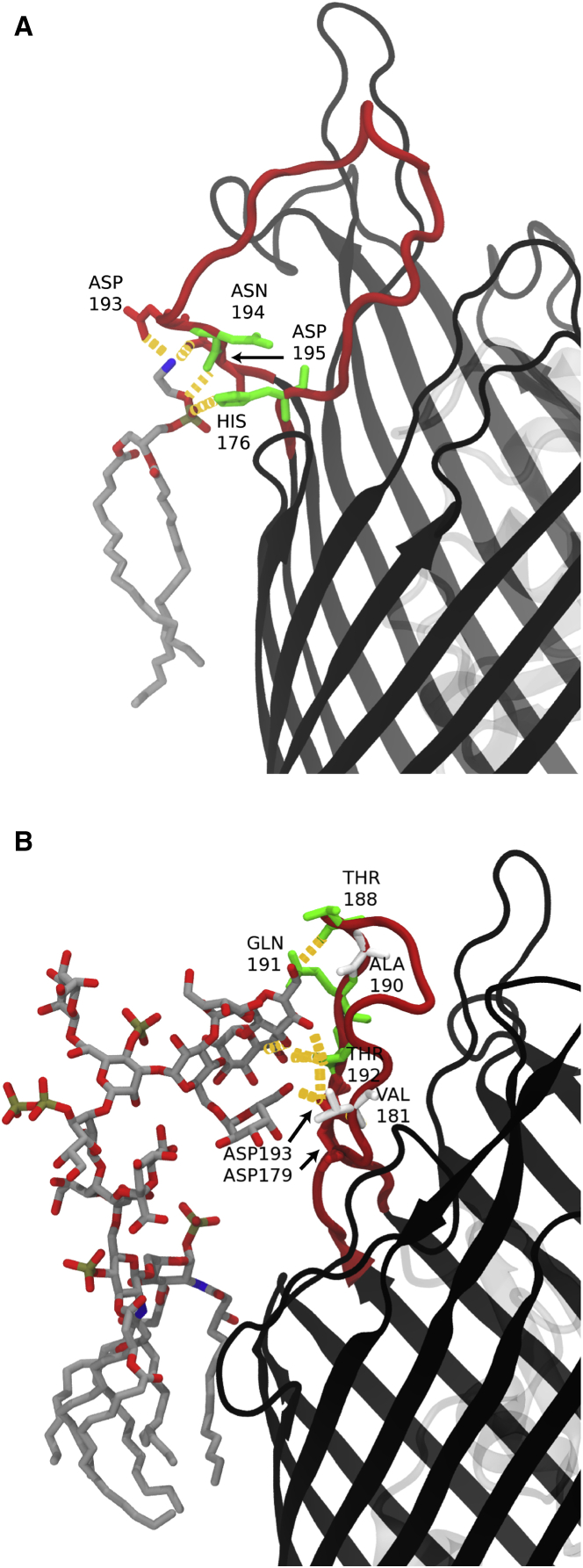
Figure 3.
Hydrogen-bonding interactions between the protein and membrane. Specific protein-lipid hydrogen bonds are indicated as orange dashed lines and BtuB is shown in black ribbon. (A) Apo-sym system, showing a phospholipid (gray sticks) hydrogen bonding (orange dashes) to loop 3/4 of BtuB. Loop 3/4 becomes deformed as Asp193 and Asp195 are attracted toward the zwitterionic POPE headgroup. This interaction causes loop 3/4 to substantially deviate from the conformation observed in the CBL-bound structure. (B) Apo2-OM system, with an LPS molecule (gray sticks) hydrogen bonding (orange dashes) to loop 3/4 of BtuB. The oligosaccharides in the OM systems exhibit hydrogen bonding along the EC loops, including loop 3/4 residues Asp179 and Asp193, in the absence of . Additional hydrogen-bonding interactions can be found in Tables S3–S5. To see this figure in color, go online.
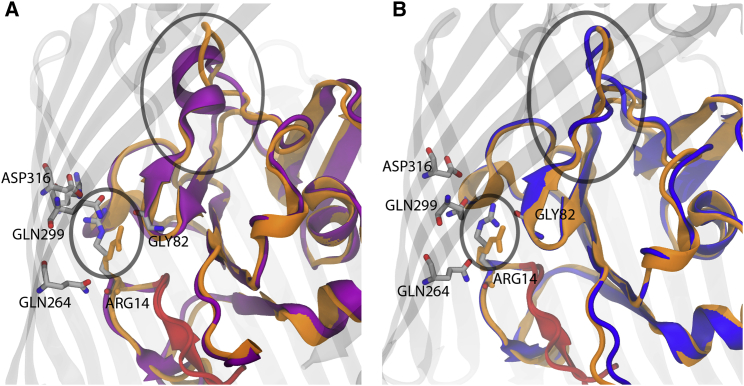
An unexpected effect observed in the Apo1- and Apo2-OM simulations was a conformational change in the luminal domain of BtuB. We first observed a structural difference between the symmetric and OM systems in the region from Gly82 to Ser91. This region forms an α-helix in the Apo-sym simulation within the first 10 ns, whereas in all OM systems this region forms a random coil, as shown in Fig. 4. When this α-helix in the Apo-sym system is compared with the CBL-bound crystal structure (PDB: 1NQH), it can be seen that Ser91, a CBL-binding residue, is farther from its CBL-bound-state position due to this helix. In contrast, the Apo1- and Apo2-OM simulations exhibit excellent alignment with the Gly82 to Ser91 backbone and side-chain orientations found in the CBL-bound crystal structure within the first 20 ns of simulation. These observed structural changes are then maintained throughout the remainder of each simulation (see Fig. S10). Second, we observed a conformational difference in a side-chain rotamer in the Apo-sym and Apo-OM simulations. Arg14, which forms a hydrogen-bonding network with Gln299, Asp316, and Arg358 in the Apo-sym simulation, rotates about its atom in the Apo1-OM toward Gly82. This rotation effectively unlocks the so-called Ton-box, the N-terminal residues that mediate interaction with TonB, from the barrel wall. An unlocked Ton-box is also seen in the CBL-bound crystal structure, although in neither case is full release into the periplasm observed.
Figure 4.
(A and B) Luminal domain organization of the (A) Apo-sym (purple ribbon) and (B) Apo-OM (blue ribbon) systems compared with the CBL-bound crystal structure’s luminal domain (orange ribbon). There is an observed difference in the luminal domain secondary structure (larger oval) of BtuB in the simulated membrane systems. Apo-sym reveals an α-helix between Gly82 and Gly92, whereas each OM system reveals a random coil along the same residues (see also Fig. S10). The random coil conformation brings Ser91, a CBL-interacting residue, closer to the conformation observed in the CBL-bound crystal structure. The smaller oval shows the Arg14 (gray sticks) lock mechanism (A) locked to the barrel wall, as found in the symmetric system, and (B) unlocked, bent toward the barrel interior. The unlocked conformation of Arg14 observed in the simulated OM systems is also found in the CBL-bound crystal structure (orange sticks). See the Supporting Material for movies of lumen extraction using SMD. To see this figure in color, go online.
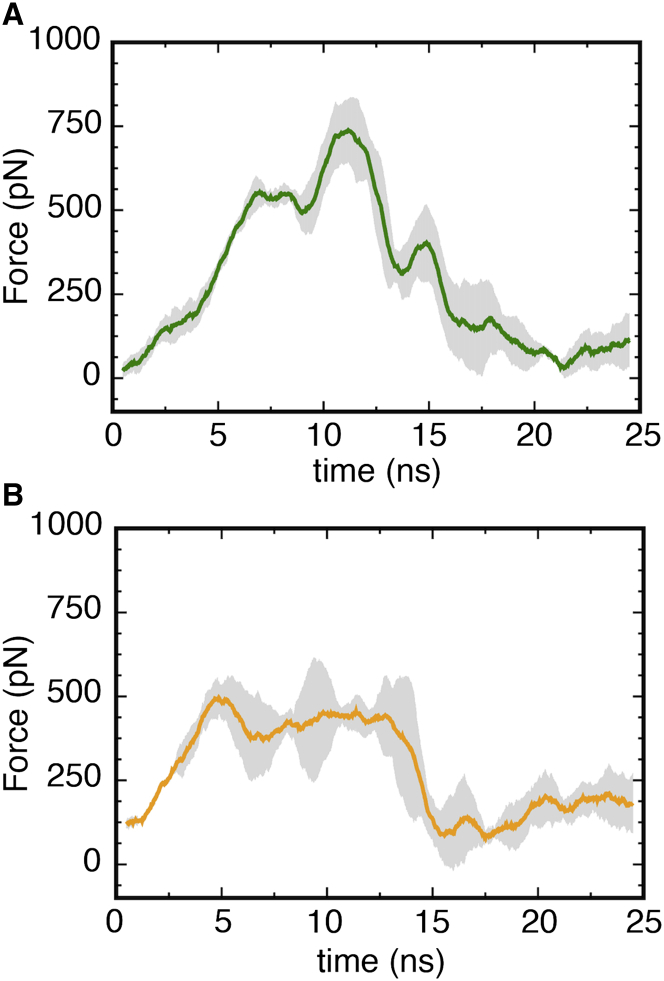
To determine the functional role of the observed structural difference in the luminal domain and the apparent unlocking of Arg14, we performed SMD simulations to extract the Ton-box in the Apo-sym, Apo1-OM, and Apo2-OM systems at a rate of 1 Å/ns, and in the Apo-sym and Apo1-OM systems at 0.25 Å/ns. Although these rates are exceedingly fast compared with the biological process, they are in line with previous simulations (52). The unlocking of Arg14 resulted in an ∼30% lower force required for Ton-box extraction in the Apo-OM systems compared with the Apo-sym system at 1 Å/ns (see Fig. 5). The ∼250-pN higher force peak between 9 and 12 ns in the Apo-sym case is due to Arg14 being electrostatically bound to the barrel wall via the hydrogen-bond network described above. The 500-pN force around 5 ns in both averages correlates with the unbinding of Val10, Thr11, and Ala12 from the luminal domain hydrophobic core. This force would be significantly reduced upon TonB binding due to favorable hydrophobic interactions between the Ton-box and the C-terminus of monomeric TonB (19, 27). The observed difference is further supported by a pulling simulation at a speed of 0.25 Å/ns, which shows an ∼15% lower force required for Ton-box extraction in the OM system compared to the symmetric system (see Fig. S11 C).
Figure 5.
(A and B) Plot of the observed force for Ton-box extraction from the luminal domain in (A) Apo-sym (green) and (B) Apo-OM (orange) systems, with the corresponding standard error shown in gray. Each curve is an average of three time-averaged, constant-velocity SMD runs of 1 Å/ns. The Apo-OM curve is the run average of one Apo2-OM and two Apo1-OM SMD runs. The peak force for Apo-sym occurs around 11 ns and corresponds to unbinding of Arg14 from the hydrogen-bond network, as shown in Fig. 4. The peak force in the Apo-OM systems occurs around 6 ns and corresponds to the hydrophobic interaction of Val10 to Ala12 with the luminal domain. Independent SMD run results can be found in Fig. S11. To see this figure in color, go online.
-bound state comparison
Previous experiments by Bradbeer and Gudmundsdottir (21) and Cadieux et al. (22) indicated that BtuB has a significantly higher binding affinity for CBL in the presence of calcium ions. Furthermore, previous MD simulations by Gumbart et al. (14) showed that calcium binding decreases the fluctuations of the CBL-binding residues compared with the apo state, suggesting that the role of calcium is to stabilize the binding site. However, upon comparing the Apo-sym and Apo-OM systems, we observe that LPS significantly stabilizes many of the CBL-binding residues located on the EC loops, as shown in Fig. 2 B. Thus, the importance of binding in the Asp-cage remains unclear.
Five calcium ions are found in the crystallographic, CBL-bound structure of BtuB, two of which are bound to the Asp-cage located on EC loops 3/4 and 5/6 (13, 14). Earlier mutagenesis experiments on residues in the Asp-cage revealed a dependence on Asp residues in loop 3/4 for effective CBL uptake (22). To evaluate the importance of calcium, we again compared the RMSFs of the Ca-OM systems with those of the Apo-OM systems to illuminate a potential role for binding in the Asp-cage. Figs. 2 C and S9 show a substantial reduction of RMSF in loop 3/4 in the Ca-OM systems (the RMSD is shown in Fig. S12); however, has little to no additional stabilizing effect on the other EC loops. Furthermore, the luminal domain takes on a conformation similar to that observed in the Apo-OM systems, as shown in Fig. S10, indicating that the OM system promotes proper luminal domain organization. Therefore, in combination, the simulation results presented here and previous experimental evidence indicate that binding primarily affects loop 3/4.
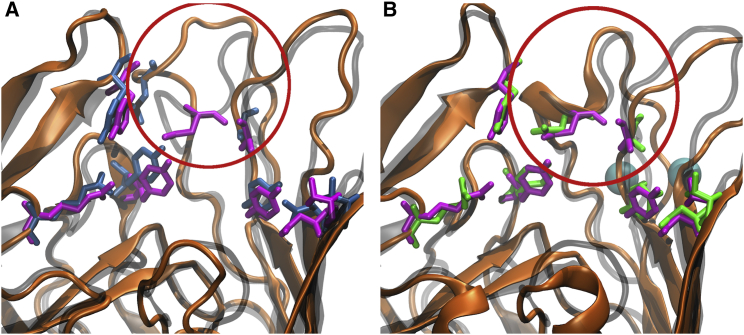
However, since the high-affinity binding of a substrate is not dictated solely by protein fluctuations near the binding site, the stabilization of binding-site loops alone may be insufficient. The presence of calcium in the Asp-cage induces a conformational change in loop 3/4 such that it closely resembles the CBL-bound structure of BtuB, which was not seen in either of the Apo-OM simulations even though they started from very similar conformations (RMSD of 0.8 Å between loop 3/4 of Apo and calcium-bound BtuB). As shown in Fig. 6 A, most of the CBL-interacting residues on the EC loops align with their observed conformation in the CBL-bound crystal structure; however, Asn185 on loop 3/4 is poorly aligned. In contrast, with bound, as shown in Fig. 6 B, there is a nearly perfect alignment of Asn185 to the CBL-bound crystal structure, which after its formation is maintained for the remainder of the simulations (see also Fig. S13). Thus, binding promotes not only the stabilization but also the structural reorganization of loop 3/4.
Figure 6.
(A and B) Comparison of the (A) Apo- and (B) -bound states of BtuB in the simulated OM systems. Orientation differences observed in loop 3/4 are emphasized by the red circle. (A) Apo-state BtuB (orange ribbon) aligned to the CBL-bound crystal structure (transparent gray ribbon) from Chimento et al. (25). Residues within 4 Å of CBL in the CBL-bound crystal structure are highlighted as blue (Apo1-OM) and purple (crystal structure) sticks. (B) -bound BtuB (orange ribbon) aligned to the CBL-bound crystal structure (transparent gray ribbon). Calcium ions are shown as cyan spheres and the green sticks represent the CBL-binding residues in the Ca1-OM system. Replicates from the second OM system can be seen in Fig. S13. To see this figure in color, go online.
The arrangement of residues in the Asp-cage on loop 3/4 due to binding induces a conformational shift of the remaining loop residues. This shift is made apparent by the spontaneous formation of a small α-helix between Tyr183 and Gly187 (Fig. 6 B). The α-helix is first observed 7 ns into the Ca1-OM trajectory and thereafter is maintained for ∼50% of the trajectory. The helix is not formed in the Ca2-OM trajectory; however, loop 3/4 still aligns well with the CBL-bound crystal structure, as shown in Fig. S13. Other recent MD simulations have also revealed spontaneous helix formation for an OM protein, OmpLA, in a native OM bilayer (39).
Conclusions
The OM of Gram-negative bacteria provides an additional barrier to nutrients, antibiotics, pathogens, and other large molecules. This barrier gives these bacteria added protection, but conversely makes the uptake of essential nutrients more difficult. To better understand the local effects of the OM on embedded proteins, we developed an OM model suitable for MD simulations. The hydrophobic thickness and lateral diffusion rates of LPS in this OM model correspond reasonably well with previous simulations and experiments (7, 11, 38, 39, 42); however, the area per lipid/LPS is ∼5–10% higher than that of a similar E. coli OM model (39), suggesting necessary changes for future iterations of our model. Finally, we carried out simulations of an OM transporter, BtuB, in a symmetric phospholipid bilayer and our model OM to characterize novel protein-membrane interactions in the native environment.
Significant differences in protein conformation were observed upon comparison of the apo-state BtuB in the symmetric and model OM systems. Reduced fluctuations of the EC loops in the Apo-OM simulations provide strong evidence that LPS plays an important role in OM-protein dynamics. Whether fluctuations would increase on much longer timescales, beyond the 300 ns simulated here, is uncertain. Additionally, a clear shift in the arrangement of the EC loops was observed between the symmetric and OM systems. For example, in the symmetric system, EC loop 3/4, which is key for substrate binding, becomes greatly displaced from its substrate-bound conformation due to interactions with phospholipids. In all of the OM systems, loop 3/4 maintains an alignment similar to the crystallographic substrate-bound conformation. Surprisingly, the model OM induces an organizational shift in the luminal domain, despite having no direct contact. Lastly, in each OM system, Arg14 becomes unlocked from its interaction with residues in the barrel wall, thereby permitting a lower extraction force for removal of the luminal domain, as observed from SMD simulations.
Previous experimental results indicated that calcium binding is required for CBL binding and transport, although the structural effect of calcium on BtuB was not known. The simulations performed here demonstrate that calcium binding promotes a conformational shift in EC loop 3/4, as well as spontaneous secondary-structure formation in this loop. A comparison of the OM systems reveals that primary loop stabilization occurs as a result of interactions between the protein and LPS, with calcium playing a vital role through CBL-binding-site organization and further loop stabilization. We hypothesize that this function provided by calcium ions is necessary for BtuB alone because CBL is the largest substrate among those transported by TonB-dependent transporters.
Author Contributions
C.B. and J.C.G. designed the study and wrote the manuscript. C.B. acquired and analyzed the data.
Acknowledgments
This work was supported by a National Science Foundation CAREER award to J.C.G. (MCB-1452464). Computational resources were provided by the National Institute for Computational Sciences at Oak Ridge National Laboratory through the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant number OCI-1053575.
Editor: Scott Feller.
Footnotes
Thirteen figures, five tables, two movies, and two data files are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30755-X.
Supporting Material
References
- 1.Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shroll R.M., Straatsma T.P. Molecular structure of the outer bacterial membrane of Pseudomonas aeruginosa via classical simulation. Biopolymers. 2002;65:395–407. doi: 10.1002/bip.10279. [DOI] [PubMed] [Google Scholar]
- 3.Goose J.E., Sansom M.S.P. Reduced lateral mobility of lipids and proteins in crowded membranes. PLOS Comput. Biol. 2013;9:e1003033. doi: 10.1371/journal.pcbi.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piggot T.J., Holdbrook D.A., Khalid S. Electroporation of the E. coli and S. Aureus membranes: molecular dynamics simulations of complex bacterial membranes. J. Phys. Chem. B. 2011;115:13381–13388. doi: 10.1021/jp207013v. [DOI] [PubMed] [Google Scholar]
- 7.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mühlradt P.F., Menzel J., Speth V. Lateral mobility and surface density of lipopolysaccharide in the outer membrane of Salmonella typhimurium. Eur. J. Biochem. 1974;43:533–539. doi: 10.1111/j.1432-1033.1974.tb03440.x. [DOI] [PubMed] [Google Scholar]
- 10.Vollmer W. Bacterial outer membrane evolution via sporulation? Nat. Chem. Biol. 2011;8:14–18. doi: 10.1038/nchembio.748. [DOI] [PubMed] [Google Scholar]
- 11.Piggot T.J., Holdbrook D.A., Khalid S. Conformational dynamics and membrane interactions of the E. coli outer membrane protein FecA: a molecular dynamics simulation study. Biochim. Biophys. Acta. 2013;1828:284–293. doi: 10.1016/j.bbamem.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Faraldo-Gómez J.D., Smith G.R., Sansom M.S. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a comparative study of the ferrichrome-free and bound states. Biophys. J. 2003;85:1406–1420. doi: 10.1016/S0006-3495(03)74573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chimento D.P., Kadner R.J., Wiener M.C. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins. 2005;59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 14.Gumbart J., Wiener M.C., Tajkhorshid E. Coupling of calcium and substrate binding through loop alignment in the outer-membrane transporter BtuB. J. Mol. Biol. 2009;393:1129–1142. doi: 10.1016/j.jmb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eren E., Parkin J., van den Berg B. Toward understanding the outer membrane uptake of small molecules by Pseudomonas aeruginosa. J. Biol. Chem. 2013;288:12042–12053. doi: 10.1074/jbc.M113.463570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds P.R., Mottur G.P., Bradbeer C. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J. Biol. Chem. 1980;255:4313–4319. [PubMed] [Google Scholar]
- 17.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noinaj N., Guillier M., Buchanan S.K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed D.M., Lukasik S.M., Cafiso D.S. Monomeric TonB and the Ton box are required for the formation of a high-affinity transporter-TonB complex. Biochemistry. 2013;52:2638–2648. doi: 10.1021/bi3016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradbeer C., Reynolds P.R., Fernandez M.T. A requirement for calcium in the transport of cobalamin across the outer membrane of Escherichia coli. J. Biol. Chem. 1986;261:2520–2523. [PubMed] [Google Scholar]
- 21.Bradbeer C., Gudmundsdottir A. Interdependence of calcium and cobalamin binding by wild-type and mutant BtuB protein in the outer membrane of Escherichia coli. J. Bacteriol. 1990;172:4919–4926. doi: 10.1128/jb.172.9.4919-4926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cadieux N., Barekzi N., Bradbeer C. Observations on the calcium dependence and reversibility of cobalamin transport across the outer membrane of Escherichia coli. J. Biol. Chem. 2007;282:34921–34928. doi: 10.1074/jbc.M707426200. [DOI] [PubMed] [Google Scholar]
- 23.Kim M., Xu Q., Cafiso D.S. Solutes alter the conformation of the ligand binding loops in outer membrane transporters. Biochemistry. 2008;47:670–679. doi: 10.1021/bi7016415. [DOI] [PubMed] [Google Scholar]
- 24.Xu Q., Kim M., Cafiso D.S. Membrane hydrocarbon thickness modulates the dynamics of a membrane transport protein. Biophys. J. 2008;95:2849–2858. doi: 10.1529/biophysj.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chimento D.P., Mohanty A.K., Wiener M.C. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 27.Shultis D.D., Purdy M.D., Wiener M.C. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson A.D., Hofmann E., Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 29.Raetz C.R.H. Biochemistry of endotoxins. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson A.D., Welte W., Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson A.D., Braun V., Welte W. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 2000;9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone J.E., Phillips J.C., Schulten K. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- 33.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darden T.A., York D.M., Pedersen L.G. Particle mesh Ewald: an N log N method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 37.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 38.Piggot T.J., Piñeiro Á., Khalid S. Molecular dynamics simulations of phosphatidylcholine membranes: a comparative force field study. J. Chem. Theory Comput. 2012;8:4593–4609. doi: 10.1021/ct3003157. [DOI] [PubMed] [Google Scholar]
- 39.Wu E.L., Fleming P.J., Im W. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klauda J.B., Brooks B.R., Pastor R.W. Dynamical motions of lipids and a finite size effect in simulations of bilayers. J. Chem. Phys. 2006;125:144710. doi: 10.1063/1.2354486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulmschneider J.P., Ulmschneider M.B. United atom lipid parameters for combination with the optimized potentials for liquid simulations all-atom force field. J. Chem. Theory Comput. 2009;5:1803–1813. doi: 10.1021/ct900086b. [DOI] [PubMed] [Google Scholar]
- 42.Straatsma T.P., Soares T.A. Characterization of the outer membrane protein OprF of Pseudomonas aeruginosa in a lipopolysaccharide membrane by computer simulation. Proteins. 2009;74:475–488. doi: 10.1002/prot.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delcour A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clifton L.A., Skoda M.W.A., Holt S.A. Asymmetric phospholipid: lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J. R. Soc. Interface. 2013;10:20130810. doi: 10.1098/rsif.2013.0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slusky J.S.G., Dunbrack R.L., Jr. Charge asymmetry in the proteins of the outer membrane. Bioinformatics. 2013;29:2122–2128. doi: 10.1093/bioinformatics/btt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulmschneider M.B., Sansom M.S. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 47.Schindler M., Osborn M.J., Koppel D.E. Lateral diffusion of lipopolysaccharide in the outer membrane of Salmonella typhimurium. Nature. 1980;285:261–263. doi: 10.1038/285261a0. [DOI] [PubMed] [Google Scholar]
- 48.Shawkat S., Karima R., Matsushima K. Visualization of the molecular dynamics of lipopolysaccharide on the plasma membrane of murine macrophages by total internal reflection fluorescence microscopy. J. Biol. Chem. 2008;283:22962–22971. doi: 10.1074/jbc.M801366200. [DOI] [PubMed] [Google Scholar]
- 49.Triantafilou K., Triantafilou M., Cherry R. Fluorescence recovery after photobleaching reveals that LPS rapidly transfers from CD14 to hsp70 and hsp90 on the cell membrane. J. Cell Sci. 2001;114:2535–2545. doi: 10.1242/jcs.114.13.2535. [DOI] [PubMed] [Google Scholar]
- 50.Snyder S., Kim D., McIntosh T.J. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 51.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gumbart J., Wiener M.C., Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys. J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.