Abstract
Plant growth promoting rhizobacteria (PGPR) hold promising future for sustainable agriculture. Here, we demonstrate a carotenoid producing halotolerant PGPR Dietzia natronolimnaea STR1 protecting wheat plants from salt stress by modulating the transcriptional machinery responsible for salinity tolerance in plants. The expression studies confirmed the involvement of ABA-signalling cascade, as TaABARE and TaOPR1 were upregulated in PGPR inoculated plants leading to induction of TaMYB and TaWRKY expression followed by stimulation of expression of a plethora of stress related genes. Enhanced expression of TaST, a salt stress-induced gene, associated with promoting salinity tolerance was observed in PGPR inoculated plants in comparison to uninoculated control plants. Expression of SOS pathway related genes (SOS1 and SOS4) was modulated in PGPR-applied wheat shoots and root systems. Tissue-specific responses of ion transporters TaNHX1, TaHAK, and TaHKT1, were observed in PGPR-inoculated plants. The enhanced gene expression of various antioxidant enzymes such as APX, MnSOD, CAT, POD, GPX and GR and higher proline content in PGPR-inoculated wheat plants contributed to increased tolerance to salinity stress. Overall, these results indicate that halotolerant PGPR-mediated salinity tolerance is a complex phenomenon that involves modulation of ABA-signalling, SOS pathway, ion transporters and antioxidant machinery.
The continually growing world population presents a focal challenge for existing agricultural practices to warranty solution to a steadily escalating demand in food production. This increasing want for food is paralleled by striking losses of arable land owing to rising severity of soil annihilation by abiotic environmental conditions. Salinity is a major environmental stress that severely affecting plant productivity worldwide. Exposure of plants to excess salt triggers imbalance of ions, ion toxicity-induced metabolism imbalances and hyperosmotic stress induced water deficit. Plants follow various kinds of salt-tolerant mechanisms like osmolytes and polyamines synthesis, reducing reactive oxygen species level by following antioxidant defense mechanism, and ions transport and their compartmentalization. Plants have stress-specific adaptive responses as well as the response that protects plants against more than one environmental stress1. Numerous abiotic stress-related genes, transcription factors and regulatory elements in promoters, have been characterized2. Plants utilize three strategies to prevent as well as adapt to high Na+ concentrations: (i) active Na+ efflux, (ii) Na+ influx prevention and (iii) Na+ compartmentalization in vacuoles3. Antiporters play a significant role in ion homeostasis in plants. Na+/H+ antiporters (SOS1 and NHX1) minimize the cytotoxicity by maintaining the optimal cytosolic ions concentration. NHX1, located in tonoplast reduces cytosolic Na+ concentration by pumping Na+ in to the vacuole4, whereas plasma membrane localized SOS1 extrudes Na+ in apoplasts5 and both of these process are driven by proton motive force generated by the H+ -ATPase6. Salinity alters the normal homeostasis of the cell and causes an increased production of reactive oxygen species (ROS)7. Under optimal growth conditions, antioxidant defense mechanisms of plants can cope with increased ROS level, whereas, during stress conditions, the antioxidant capacity is overcome by ROS production. Therefore, tight regulation of the steady-state concentrations of ROS by antioxidative mechanism is critical to minimize the oxidative stress-mediated damage in plant cells and consequently allowing ROS to perform useful functions as signal molecules7.
Plant-growth-promoting rhizobacteria (PGPR) are a group of microbes which colonize the plant roots and improve plant growth either directly or indirectly8. Various studies have been already published that explain the effect of PGPR in relieving abiotic stress in different crop plants9,10. PGPR promote plant growth by altering the selectivity of Na+, K+, and Ca2+ and sustain a higher K+/Na+ ratio in plants under salt stress11. With significant improvements in our understanding of PGPR-mediated salinity tolerance in host plant, efforts are being made to understand the tolerance mechanism at the gene expression level. Thus, plant cellular and molecular responses which are generally studied during interactions with various types of symbiotic as well as mutualistic microbes are of great interest, since the responses may be related to an essential process for establishing different relationships between plants and microbes. There are diverse PGPR-induced changes in plants, and growth promotion perhaps results due to a complex combination of various PGPR-induced mechanisms that affect both plant development as well as plant nutrition12. Furthermore, transcriptomic analyses have been conducted for only a few rhizobacterial strains to unravel the changes in molecular mechanisms in plants associated with PGPR-mediated plant growth promotion13,14. Further understanding of the primary mechanisms employed by these bacteria will likely accelerate the recognition of PGPR as appropriate and efficient adjuncts to agricultural practice.
In our earlier experiments, we identified some PGPRs capable of inducing tolerance against NaCl stress in wheat plants (data unpublished); of these Dietzia natronolimnaea strain, STR1 was found to provide maximum protection to the plants under salt stress. D. natronolimnaea strain STR1 was also found to protect Ocimum basilicum plant from salt stress15. With the aim of understanding the possible mechanisms involved in PGPR-mediated salinity tolerance in wheat plants, we studied the expression of different genes involved in salt stress tolerance in plants.
Results
Phylogenetic analysis and functional traits of Dietzia natronolimnaea strain STR1
Phylogenetic analysis confirmed that isolate STR1 (Accession no. KJ413139) has maximum similarity with Dietzia natronolimnaea strain CBS 107.95 (Accession no. NR_026273) (Query coverage 100%, percentage of similarity 99%, E-value 0.0) (Supplementary Fig. S1). Plant growth promoting traits like phosphate solubilisation, indole acetic acid (IAA) production, nitrogen fixation and ACC deaminase activity were found to be negative in D. natronolimnaea STR1. The bacterial strain was also found to be negative for exopolysaccharide production and nifH gene (data not shown). The growth curves at different NaCl concentrations and pH have been shown in Supplementary Fig. S2. D. natronolimnaea STR1 showed the best growth at 0.5 M NaCl and neutral pH (i.e. pH 7).
Colonization of wheat plant with Dietzia natronolimnaea STR1
Scanning electron microscopy confirmed that the untreated control plants did not harbour any bacteria and the roots of D. natronolimnaea STR1-inoculated plants were successfully colonized by D. natronolimnaea STR1 (Supplementary Fig. S3). Scanning electron micrographs confirmed the presence of rhizobacteria on the roots of STR1 inoculated wheat plants (Supplementary Fig. S3).
Effect of Dietzia natronolimnaea strain STR1 inoculation on wheat growth
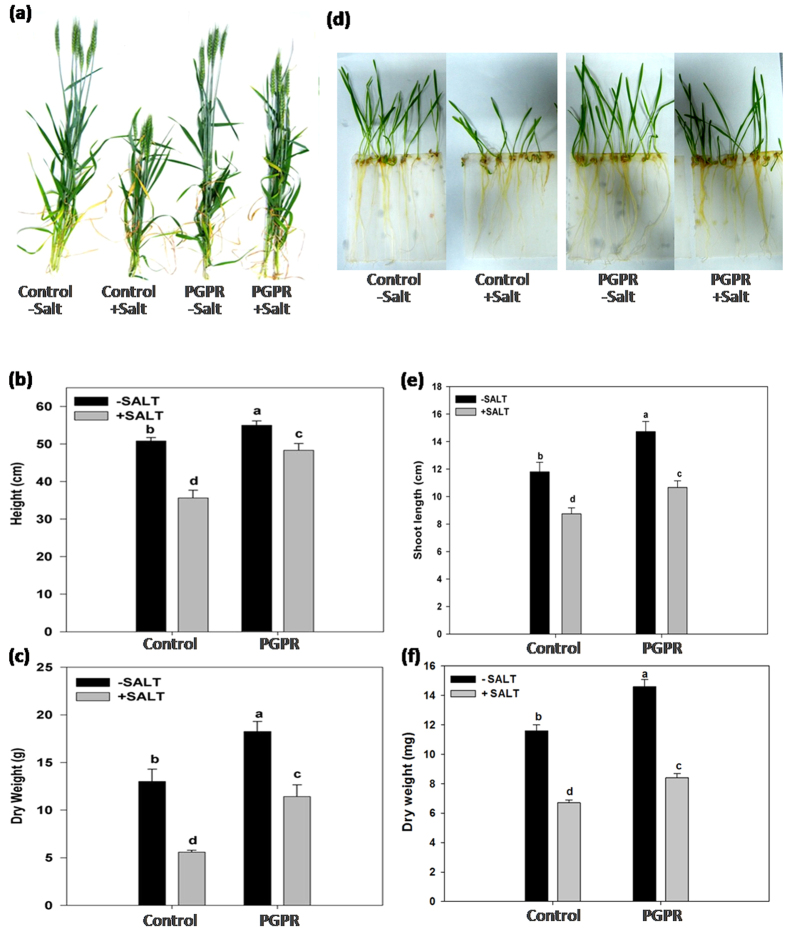
The influence of STR1-inoculation on wheat growth was studied in both saline and non-saline environment. A significant increase in growth of STR1-inoculated wheat was found as compared to uninoculated (control) wheat under both non-saline and saline conditions (Fig. 1). In the case of wheat plants grown in soil, a 57% reduction in dry weight was observed in uninoculated salt-stressed plants in comparison to the non-saline control. PGPR applied plants recorded improved growth in terms of dry weight and plant height (Fig. 1). A marked effect of STR1 inoculation, as well as salinity stress on growth of hydroponically grown wheat plants, was observed (Fig. 1). STR1-inoculated plants recorded higher biomass, shoot and root elongation compare to un-inoculated plants under both non-saline and saline conditions (Fig. 1 and Supplementary Table S1).
Figure 1. Effect of PGPR inoculation on wheat plant growth under non-saline and saline conditions.
(a–c) Plants were grown in soil under glasshouse conditions and saline plants supplemented with 150 mM NaCl via irrigation and harvested 60 d after germination. Dry weight and plant height of wheat shoots were measured. (d–f) 12 d old wheat plants grown hydroponically in Hoagland nutrient solution. NaCl was added to the nutrient solution to obtain a final concentration of 100 mM. Dry weight and height of wheat shoots were measured after 12 d of germination. Control: without bacterial treatment; PGPR: plants inoculated with Dietzia natronolimnaea STR1 strain. Values are mean of five replicates ± standard error of means. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
Effect on photosynthetic pigments and biochemical markers
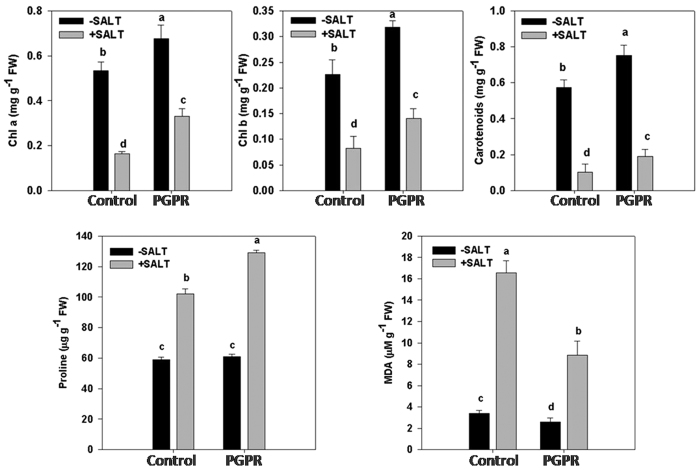
A significant reduction in photosynthetic pigments (chlorophyll a, chlorophyll b and carotenoids) was observed in plants subjected to salt stress irrespective of the bacterial treatments (Fig. 2). PGPR inoculated plants showed increased photosynthetic pigments in comparison to control plants under both saline and non-saline conditions.
Figure 2. Effect of salt stress on photosynthetic pigments, proline and MDA content of PGPR-inoculated wheat plants.
Chlorophyll a, Chlorophyll b, carotenoid, proline and MDA content was measured in leaves of 60 d old wheat plants grown in soil supplemented with 150 mM NaCl and inoculated with STR1. Control: non-inoculated wheat plants, PGPR: plants inoculated with Dietzia natronolimnaea STR1. Values are mean of five replicates ± standard error of means. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
There was no significant change in the proline, and MDA (malondialdehyde) contents among un-inoculated as well as PGPR inoculated plants under non-saline conditions, whereas, an increase was registered when plants were subjected to salt stress (Fig. 2). However, a significant increase was observed in PGPR-inoculated plants compared to un-inoculated control except for the MDA content which was found to reduce in PGPR applied plants (Fig. 2).
Effect on antioxidant enzyme status
The catalase and ascorbate peroxidase levels were similar in both un-inoculated control and rhizobacteria inoculated plants under non-saline conditions (Supplementary Fig. S4). Application of salt stress increased the enzyme levels irrespective of the microbiological treatments; however, the PGPR-inoculated plants recorded significantly higher enzyme content in comparison to control (uninoculated) plants.
Effect on expression of salt stress responsive genes
Hydroponically grown wheat seedlings were used to study the expression of salt stress-responsive genes. A total of 17 genes (Supplementary Table S3) were analyzed for their change in expression under salinity stress and PGPR inoculation in both shoot and root.
Expression of genes related to Abscisic acid metabolism
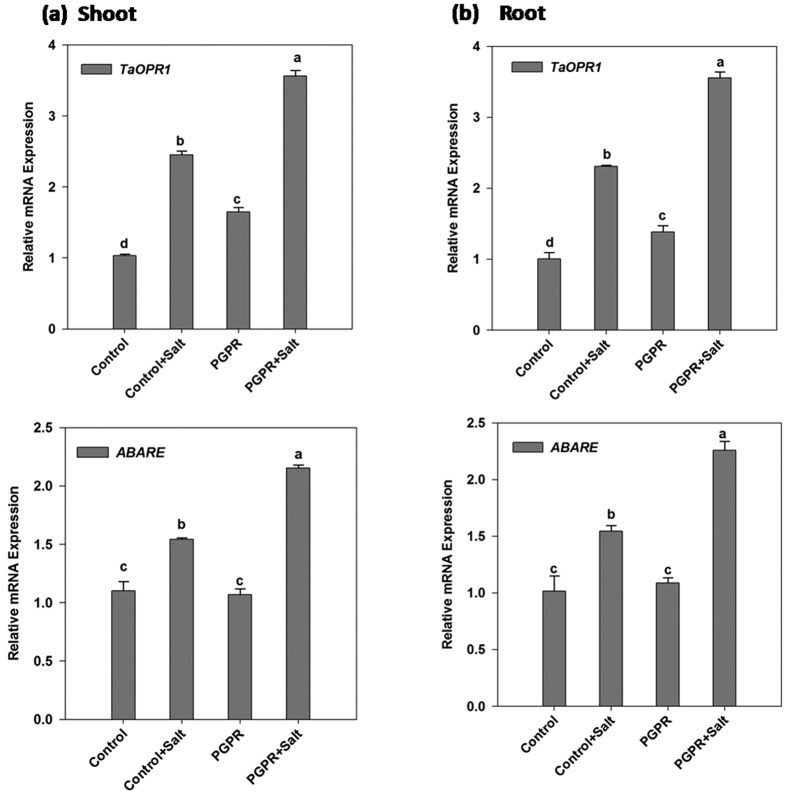
The effect of salt stress and PGPR inoculation in wheat plants on abscisic acid (ABA) metabolism was studied through the change in gene expression patterns of an ABA-responsive gene (ABARE) and TaOPR1 gene (Fig. 3).
Figure 3. Real time expression analysis of TaOPR1 and ABARE in shoot and root of PGPR-inoculated wheat plants subjected to salt stress.
The expression analysis of TaOPR1 and ABARE transcript in (a) shoot and (b) root of PGPR Dietzia natronolimnaea STR1 inoculated 12 d old wheat plants under both non-saline and saline conditions. Un-inoculated wheat plants grown in non-saline condition were used as a control. The data represented means of triplicate biological and experimental repeats; error bars represented SEM. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
The expression pattern of ABA-responsive gene and TaOPR1 in both wheat shoot and root were similar. The expression levels in control and PGPR inoculated plants showed minor differences under non-saline conditions, however, higher expression was observed in salt-stressed plants. PGPR-inoculated plants recorded its maximum increase in both root and shoot in comparison to non-saline control plants.
Expression of genes related to Salt Overly Sensitive (SOS) pathway
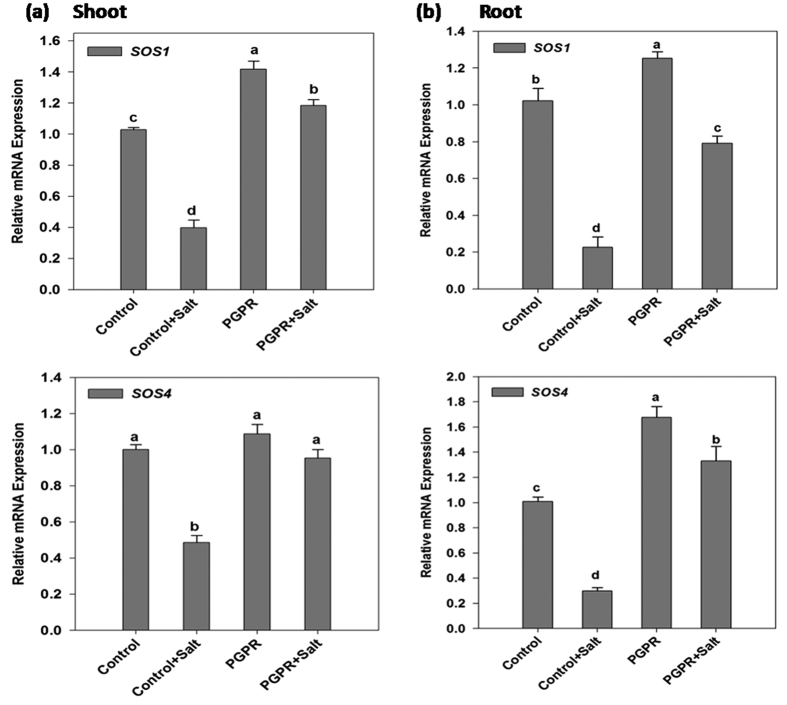
The quantitative expression patterns of SOS1 and SOS4 genes in shoot and root of wheat plants under salinity stress and PGPR inoculation are shown in Fig. 4.
Figure 4. Real time expression analysis of SOS pathway genes viz., SOS1 and SOS4 in shoot and root of PGPR-inoculated wheat plants subjected to salt stress.
The expression analysis of SOS1 and SOS4 transcript in (a) shoot and (b) root of PGPR Dietzia natronolimnaea STR1 inoculated 12 d old wheat plants under both non-saline and saline conditions. Un-inoculated wheat plants grown in non-saline condition were used as a control. The data represented means of triplicate biological and experimental repeats; error bars represented SEM. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
Analysis of variance for SOS pathway genes showed the differential response of shoot under salinity stress as well as PGPR inoculation. PGPR inoculated non-stressed plants recorded a 40% increase in SOS1 expression in comparison to the uninoculated control plants. Application of salinity stress lowered SOS1 gene expression in shoots of both uninoculated and PGPR inoculated plants. However, PGPR-inoculated salt-stressed plants recorded 2 fold higher SOS1 expression in comparison to the uninoculated salt-stressed plants.
The gene expression profile of SOS1 in roots varied with the application of salinity stress and PGPR inoculation. Similar to the shoot response, PGPR inoculated non-stressed plants recorded the highest expression whereas the salt-stressed plants recorded a decrease in the expression levels. However, among the salt-stressed plants, the PGPR inoculated plants showed 2.6 fold higher SOS1 expression level in comparison to the uninoculated plants.
SOS4 expression level in both shoots and roots varied with the treatments. Application of salt stress reduced the expression of SOS4 in shoots of uninoculated control plants. However, salt stress could not affect the expression of SOS4 in PGPR inoculated shoots significantly.
Under non-saline condition, SOS4 expression in roots was higher (65%) in PGPR inoculated plants compared to uninoculated control plants. Salt stress reduced the expression of SOS4 in roots of both uninoculated and PGPR inoculated plants. However PGPR inoculated salt-stressed plants had 3.5 fold higher SOS4 expression than that of uninoculated salt-stressed plants.
Transcription factors
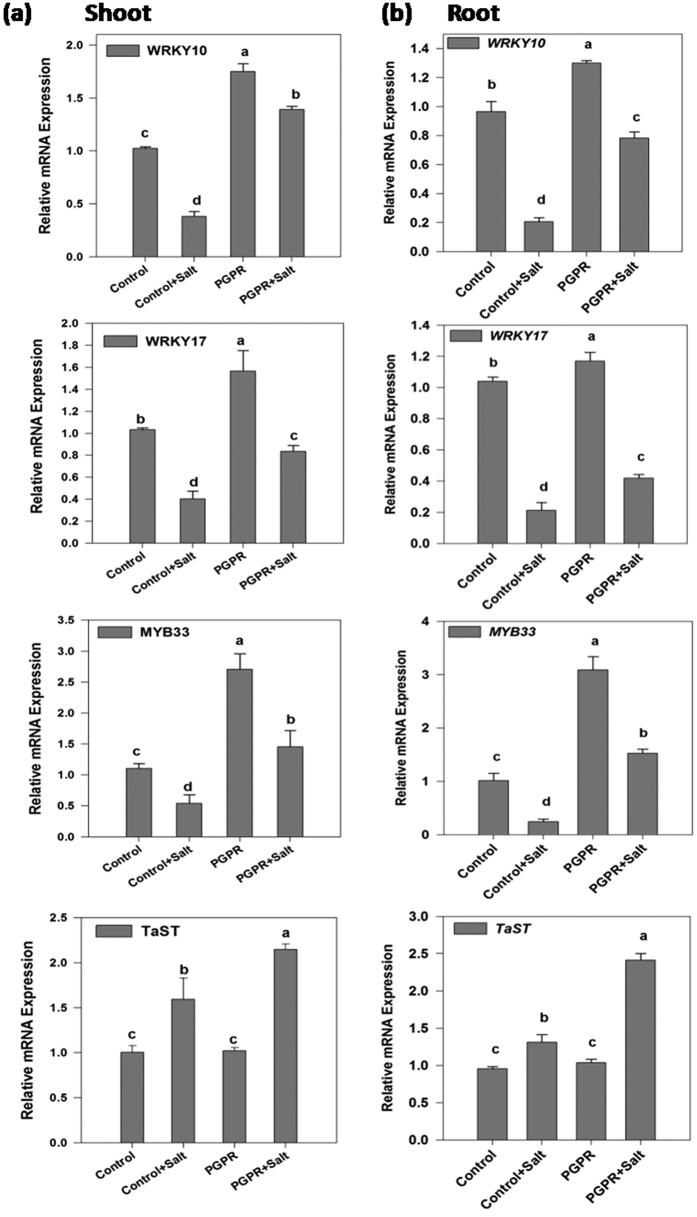
Expression pattern of transcription factors viz., TaWRKY10, TaWRKY17 and TaMYB33 were examined (Fig. 5). TaWRKY10 expression level in the non-stressed wheat shoots and roots escalated in the PGPR applied plants as compared to the uninoculated (control) plants. Salt-stressed uninoculated plants recorded a 60 and 80% decrease in TaWRKY10 expression level in the shoots and roots respectively. However, PGPR-inoculated salt-stressed plants recorded an improvement in the gene expression level as compared to the un-inoculated salt-stressed plants.
Figure 5. Real time expression analysis of WRKY, MYB and TaST in shoot and root of PGPR-inoculated wheat plants subjected to salt stress.
The expression analysis of WRKY10, WRKY17, MYB33 and TaST transcript in (a) shoot and (b) root of 12 d old wheat plants inoculated with PGPR Dietzia natronolimnaea STR1 under both non-saline and saline conditions. Un-inoculated wheat plants grown in non-saline condition were used as a control. The data represented means of triplicate biological and experimental repeats; error bars represented SEM. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
TaWRKY17 expression pattern in shoots and roots was similar to that of TaWRKY10 across the treatments. PGPR-inoculated plants recorded improved TaWRKY17 gene expression in comparison to uninoculated (control) plants under both non-saline and saline conditions.
TaMYB33 expression profile was similar in the wheat shoot and root across the treatments. Among the non-saline plants, the PGPR-inoculated plants recorded approximately 300% (3-fold) increase in the TaMYB33 expression levels in both shoots and roots. Application of salt stress reduced the TaMYB33 expression both shoots (50% reduction) and roots (80% reduction) as compared to their respective control plants. The TaMYB33 expression levels were found to be ameliorated in the PGPR-inoculated salt affected plants in comparison to the control plants.
The expression profile of TaST (T. aestivum salt-tolerant) gene was similar in the roots and shoots of wheat. No significant difference in the TaST expression was observed among the control and PGPR-inoculated non-stressed plants. Salt-stressed control plants recorded 50% higher TaST expression in the shoots in comparison to the non-stressed control plants. In the shoot of PGPR-treated salt-stressed plant TaST expression was almost 1.5 fold higher than that of salt-stressed control plant (Fig. 5).
Ion transporters
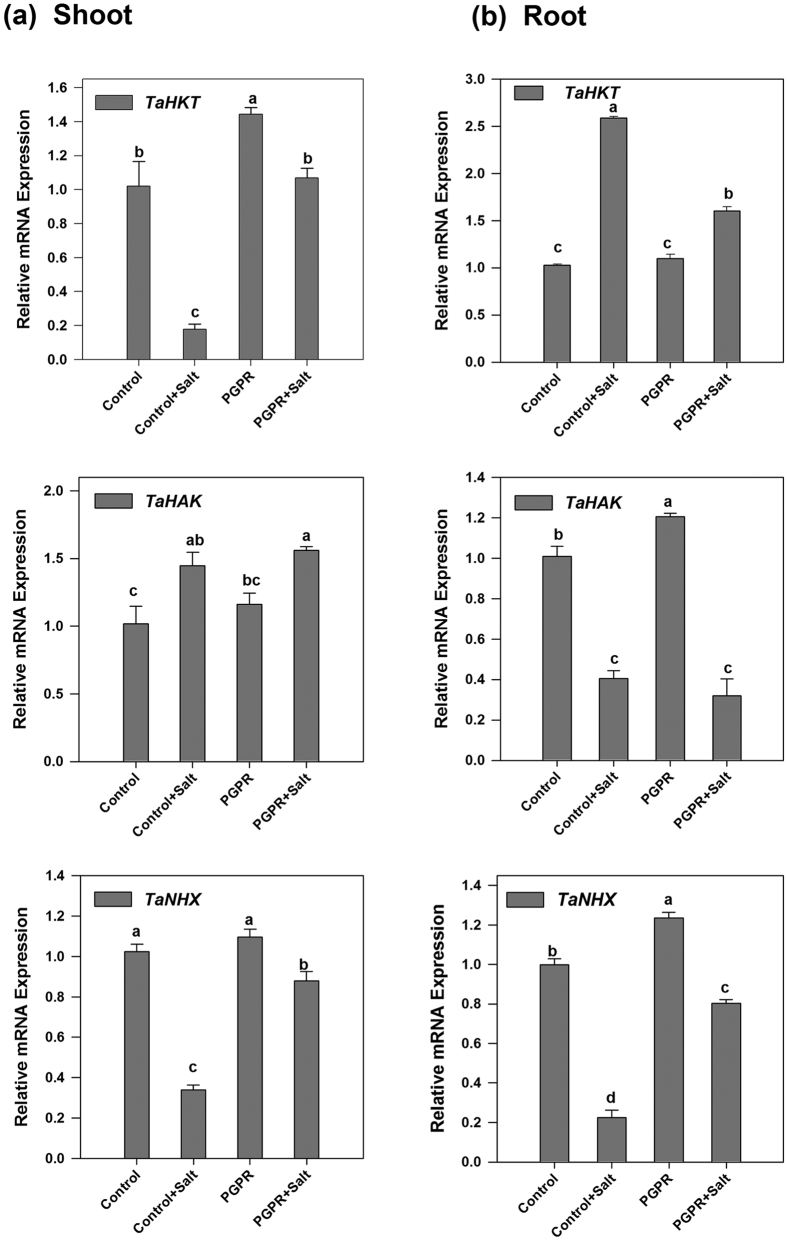
Quantitative RT-PCR analysis was performed to investigate the transcript levels of genes participating in the ion transport (TaHKT, TaNHX, TaHAK).
TaHKT expression showed the tissue-specific response in wheat plants. Under non-saline conditions, no significant difference in transcript levels was recorded between un-inoculated and PGPR inoculated plants (both shoot and root). STR1 exposure up- and downregulates TaHKT gene expression in shoots and roots respectively. PGPR exposure reduced root TaHKT expression by approximately 36% compared with control plants (Fig. 6).
Figure 6. Real time expression analysis of TaHKT, TaHAK and TaNHX in shoot and root of PGPR-inoculated wheat plants subjected to salt stress.
The expression analysis of TaHKT, TaHAK and TaNHX transcript in shoot and root of PGPR Dietzia natronolimnaea STR1 inoculated 12 d old wheat plants under both non-saline and saline conditions. Un-inoculated wheat plants grown in non-saline condition were used as a control. The data represented means of triplicate biological and experimental repeats; error bars represented SEM. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
TaNHX expression was highest in the PGPR-inoculated non-stressed plants in both shoots and roots. Salt-stressed control plants recorded 80% decrease in the expression levels in shoots as well as roots. PGPR-inoculated salt-stressed plants showed enhanced gene expression as compared to that of uninoculated salt-stressed plants (Fig. 6).
Contrary to TaNHX expression profile, TaHAK expression was higher in shoots and lower in roots of salt-stressed wheat plants in comparison to their non-stressed counterparts. Maximum TaHAK expression was observed in shoots of PGPR-inoculated salt-affected plants. The expression levels of TaHAK in roots were similar for uninoculated as well as PGPR-inoculated salt-stressed plants (Fig. 6).
Antioxidant enzymes
To evaluate the effect of salt stress treatments on the antioxidant capacity of PGPR-inoculated plants the gene expression patterns of antioxidant enzymes (POD, CAT, APX, GR, GPX, MnSOD) were analyzed using quantitative RT-PCR (Fig. 7).
Figure 7. Real time expression analysis of POD, APX, GPX, MnSOD, GR and CAT in shoot and root of PGPR-inoculated wheat plants subjected to salt stress.
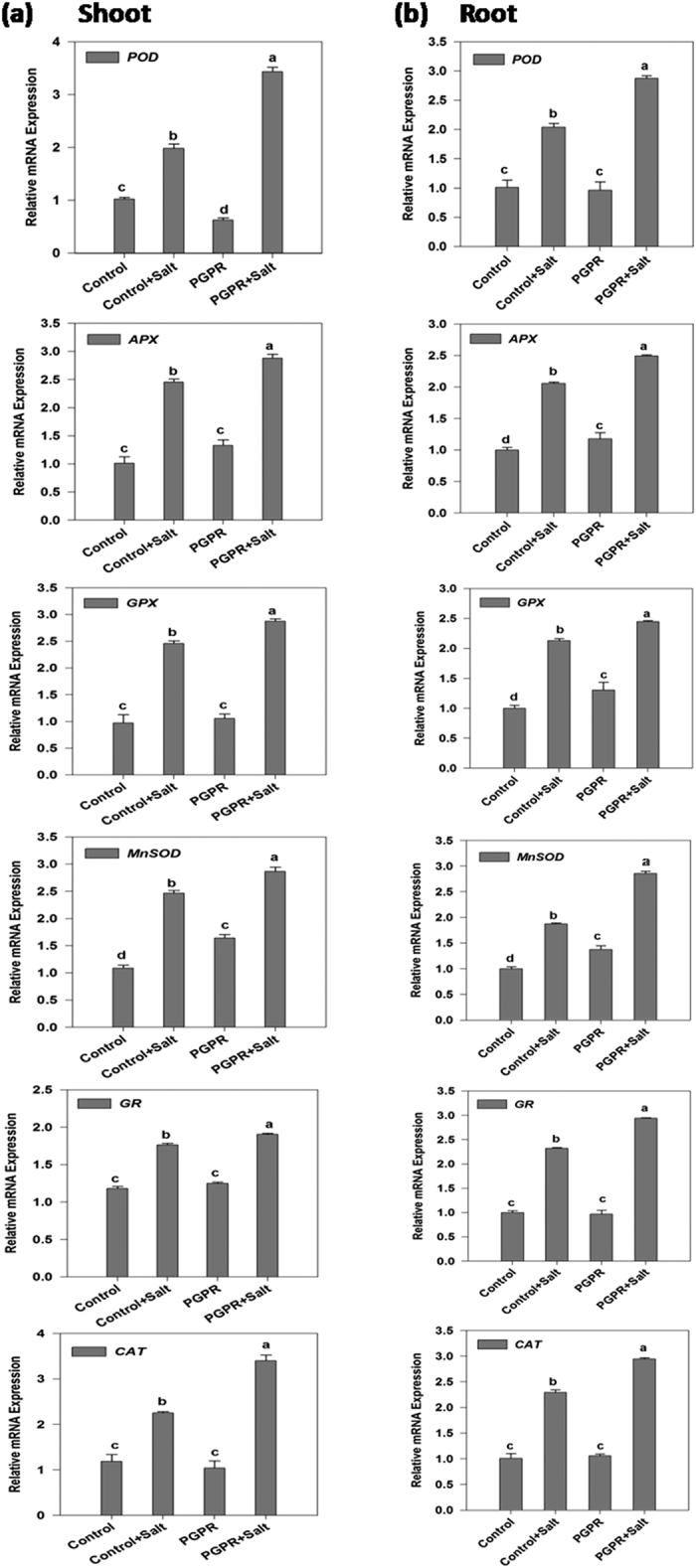
The expression analysis of POD, APX, GPX, MnSOD, GR and CAT transcript in (a) shoot and (b) root of 12 d old wheat plants inoculated with PGPR Dietzia natronolimnaea STR1 under both non-saline and saline conditions. Un-inoculated wheat plants grown in non-saline condition were used as a control. The data represented means of triplicate biological and experimental repeats; error bars represented SEM. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test P < 0.05).
The expression profile of the antioxidant enzymes followed a similar pattern in wheat roots and shoots. The gene expression patterns for all the antioxidants were similar for uninoculated (control) and PGPR-inoculated salt-stressed plants. The gene expression levels were found to be increased (1–2.5 folds) in the control plants when exposed to salinity. The salt-stressed plants applied with the halotolerant PGPR showed an improvement in the gene expression levels as compared to uninoculated salt-stressed plants. Shoot APX, CAT, and POD of PGPR-inoculated salt-stressed plants recorded a 3-fold increase in the gene expression as compared to uninoculated non-stressed plants. Roots of PGPR-inoculated salt-stressed plants recorded a 2–3 fold increase in the gene expression of the antioxidants in comparison with the non-stressed control plants.
Discussion
Rhizobacteria have been well documented to promote plant growth as well as alleviate various abiotic stresses including salinity16. Dietzia natronolimnaea has been extensively exploited for carotenoids production17 and bioremediation of hydrocarbons18. Dietzia species have also been reported to be associated with plants as endophytes19 as well as with plant rhizomes growing in soda lakes20; however, limited information on their role in plant growth promotion exists15. The present study is an effort to identify key genes and transcription factors involved in halotolerant rhizobacteria Dietzia natronolimnaea STR1 mediated salt tolerance of wheat plants. Our results suggested that D. natronolimnaea STR1 promotes the growth of plants in control conditions as well as under salt stress as illustrated by improved dry weight of wheat plants in both glasshouse and hydroponics. STR1 inoculation affected the root architecture of wheat plant as observed by increased root length under both non-saline and saline conditions. Previous reports21 have suggested varying response of rhizobacteria on plant root architecture indicating the specificity of plant-rhizobacterial associations. A recent report22 suggests salt-induced burst in ABA production is correlated with organ-specific carotenoid biosynthesis pathway. The plants inoculated with D. natronolimnaea STR1 recorded higher proline and lower malondialdehyde (MDA) levels under salt-stressed conditions in comparison to the un-inoculated salt affected plants. MDA content, a result of lipid peroxidation, is a marker of the extent of membrane damage owed to oxidative damage due to the salt stress. Reduced MDA content during salt stress in the PGPR inoculated plants matched with the earlier observations correlating reduced lipid peroxidation to better stress tolerance mechanisms10.
The issue of host plant specificity has been well studied in parasitic and mutualistic interactions; however, the question of whether PGPR efficiently interacts only with a specific host remains scantily conversed. Contrary to the popular theory, few reports have suggested strain-specific molecular responses of plants to rhizobacterial inoculation23. Rhizobacteria-mediated plant growth is a multigenic process which may be specific to the participating rhizobacteria and plant species24. The study was hence, conducted to identify the changes in the expression patterns of genes participating in inherent salt tolerance when inoculated with PGPR under non-stressed as well as salt-stressed conditions. The results of the study support the “additive hypothesis”12, suggesting plant growth as affected by bacterial inoculation under salinity stress is a complex phenomenon and involves a cumulative effect of changes in various genes in the global plant metabolic pathways.
Salt stress in plants is a cumulative effect of osmotic and ionic stress which negatively affects the plant growth and yield. Multiple genes are involved in salt tolerance mechanism. A plethora of stress related genes conferring salt stress tolerance in plants have been reported to be involved in signal transduction, ion transporters, transcription regulation and metabolic pathways25,26. Osmotic stress can be alleviated through ABA-dependent and ABA-independent pathways whereas ionic stress is mitigated through the participation of Salt Overly Sensitive (SOS) pathway and ion transporters via ion homeostasis.
A definite signaling pathway required for the control of ionic homeostasis in plants is the SOS signaling pathway which is a key mechanism for exclusion of Na+ and ion homeostasis control at cellular level27. SOS1 is one of the many plasma membrane Na+/H+ antiporters involved in salt tolerance and is crucial for regulating the long distance Na+ transport from root to shoot. SOS4 encodes a pyridoxal kinase, involved in the biosynthesis of pyridoxal-5-phosphate, which acts as an essential cofactor for numerous cellular enzymes. SOS4 regulates Na+ and K+ homeostasis by modulating the activities of ion transporters. It was also suggested that SOS4 plays a precise role in root hair development in Arabidopsis28. The results of the present study indicate a decrease in expression levels of TaSOS1 and TaSOS4 in shoots and roots under salinity stress which is in contrast with its expression pattern in root reported previously29. This could be due to the effect of long-term exposure (10 d) to salt stress in the present experiment whereas Ramezani et al. 201229 reported TaSOS1 and TaSOS4 expression up to the 72 h of salt exposure. The PGPR-inoculated plants always had higher SOS genes expression under salt stress compared to salt stressed un-inoculated plants, suggesting their participation in salt tolerance mechanisms. SOS4 gene is associated with auxin and ethylene mechanisms as well as the development of root hair in Arabidopsis30. The improved SOS4 expression in PGPR inoculated wheat roots can thus be correlated with improvement in root length and overall plant growth in salt-stressed plants.
WRKY TFs play crucial roles in the regulation of many stress induced reactions in plants but until recently unravelling their roles in various abiotic stress responses has lagged behind that of biotic stresses31. TaWRKY10 has been reported to confer salt and drought stress tolerance in transgenic tobacco by regulating the expression of stress-related genes, thus protecting plants from damage32. In the present study, TaWRKY10 expression was found to be reduced in the salt-stressed plants. However, PGPR-inoculated salt-stressed plants recorded increased TaWRKY10 expression. Salinity and drought stress have been reported to upregulate TaWRKY17 expression in plants33. The expression of TaWRKY10, TaWRKY17 was downregulated under the influence of salinity stress, might be due to long term exposure to salt stress. PGPR-inoculated plants had higher TaWRKY10, TaWRKY17 expression in saline condition compare to un-inoculated plants might be a reason to provide tolerance effect under salt stress. The results indicate a positive effect of PGPR inoculation on the expression levels of transcription factors (TFs), hence improved salinity tolerance. WRKY TFs play the important roles in regulation of water/drought-stress by modulating the cellular osmotic balance, ROS scavenging mechanism and expression of different stress-related genes34 as evident by the enhanced and improved growth in PGPR applied plants under glasshouse conditions. Plant growth inhibiting rhizobacteria have been found to induce the response of WRKY18 genes in Arabidopsis demonstrating the direct plant-microbe interaction effects at gene transcript levels35.
MYB proteins are one of the main factors for regulatory networks controlling development, metabolism and responses to abiotic and biotic stresses36. TaMYB33 expression has been found to be upregulated during drought, salt and ABA treatments. The ectopic over-expression of TaMYB33 in Arabidopsis enhanced tolerance to salt and drought stresses with no growth inhibition, indicates that the TaMYB33 is a promising gene candidate for genetic manipulation studies in wheat for providing tolerance to abiotic stress37. In the present study because of the long term exposure to salt stress the TaMYB33 expression was reduced in both root and shoot whereas STR1 inoculated plants had higher TaMYB33 transcript compared to un-inoculated plant during salt stress probably led to increased salt tolerance. Previously reported that overexpression of MYB genes in rice resulted increase in soluble sugars and proline accumulation due to upregulation of proline synthesis and transport related genes, and less accumulation of H2O2 and MDA under salt stress38, suggesting a probable reason for higher proline and reduced MDA levels in rhizobacteria inoculated salt-stressed plants as they had higher TaMYB expression compared to un-inoculated salt stressed plant. It has been found that both the SOS pathway39 and the SOS-independent pathway40 are involved in the perception of the salt stress signal, ionic absorption and intracellular ion compartmentation, however, the involvement of TaMYB33 is a question yet to be answered. A putative MYB like transcription factor (LjMAMI) has been observed to be induced during the mycorrhization process41 indicating the influence of plant growth promoting micro-organism on the gene regulatory system of plants. Hence, it can be concluded that the plant-rhizobacteria interactions are governed by the specificity of interactions at metabolic as well as molecular paradigms.
Ion transporters are known as terminal determinants of ionic homeostasis under salt- stressed conditions. Over-expression of AtNHX1 has been correlated with improved salinity tolerance42. Likewise, in the present study, higher expression of TaNHX in PGPR-inoculated plants was observed in both shoot and roots in comparison to their un-inoculated counterparts suggesting a likely role of halotolerant rhizobacteria in modulating ion transport mechanisms. SOS1 has been associated with Na+/H+ antiporters5 indicating complex and intertwined mechanisms responsible for enhanced salinity tolerance in plants. Involvement of HKT transporters in salt tolerance mechanism has been reported in both dicot and monocot crops43. Hyperaccumulation of Na+ in shoots and lower Na+ in roots have been observed in AtHKT1 mutants indicating a direct role of the gene in Na+ movement across plant tissues. The TaHKT1 expression in the present study has been observed to be considerably higher in PGPR-inoculated plants under both non-saline and saline conditions which could be attributed to improved growth in bacterized plants in comparison to control plants. In congruence with the present study, the tissue-specific response of HKT1 in A. thaliana inoculated with Bacillus subtilis GB03 has been earlier reported to enhance salt tolerance44. HAK type transporters are multigene family members in Arabidopsis, common ice plant and barley, and the various genes show specific type of expression patterns in different tissues of the plants45. Higher TaHAK expression levels can be correlated with improved salinity tolerance in wheat plants. Owing to the tissue-specific response of HAK expression, the transcript levels, unlike other transporters, varied in wheat roots and shoots. TaST is reported to be a salt-inducible gene and reduce intracellular Na+ concentration, increase K+ content, and maintain a high K+/Na+ ratio in the transgenic Arabidopsis46. TaST expression in roots was higher in STR1 treated plants indicating the role of rhizobacteria in triggering the TaST expression hence enhanced salt tolerance. Pathogen invasion, wounding, and oxidative stress and events associated with ROS acceleration have been documented to up-regulate OPR1 genes and have been suggested to be associated with antioxidant activities mediating stress tolerance47. It was observed that TaOPR responds to salinity stress in ABA-dependent manner and its overexpression improve salinity tolerance in Arabidopsis and wheat. Hence, higher TaOPR1 expression levels can be correlated with enhanced antioxidant response.
Application of abiotic stresses causes an increase in cellular level of ROS like superoxide radical (O2−), hydroxyl radicals (OH) and hydrogen peroxide (H2O2), leading to lipid peroxidation of membranes48. Plants have different antioxidant molecules and enzymes that reduce ROS level and alleviate oxidative stress. In the present study, MDA levels reduced in PGPR-inoculated plants, whereas, enzyme activities of CAT and APX in PGPR-inoculated plants growing under NaCl-stress conditions increased significantly when compared with that of the non-inoculated NaCl-stressed plants. The transcript of genes for antioxidative enzymes (APX, CAT, POD, MnSOD, GPX and GR) increased in the PGPR-inoculated plants. Plant tolerance to salinity stress was correlated with the increased expression of CAT, APX, MnSOD, POD, GPX, and GR, suggesting that PGPR triggered the abiotic stress-related defense pathways. Previous reports have demonstrated that plant-associated microorganisms attenuate salt-induced lipid peroxidation as well has higher CAT and APX activities resulting in enhanced salt tolerance49. PGPR have also been known to mediate abiotic stress tolerance in plants through modulation of ROS-scavenging enzyme expression50.
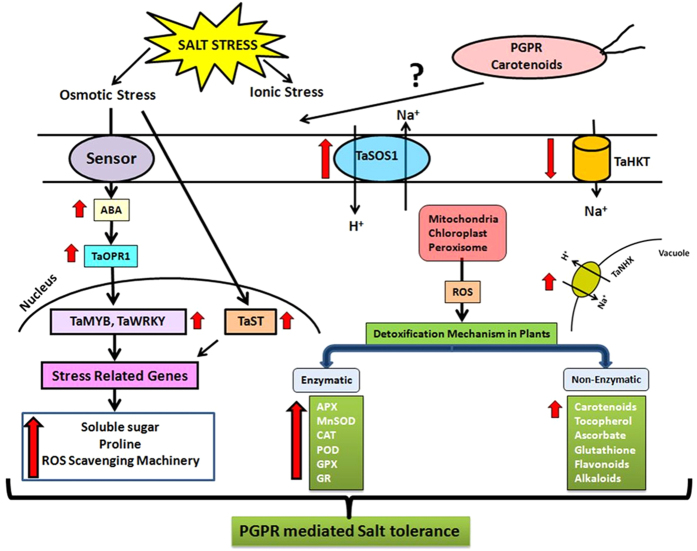
In conclusion, PGPR D. natronolimnaea STR1 promotes growth and protects wheat plants from damage to salt stress. This rhizobacterium appears to confer tolerance to salt stress, at least partly, via modulating the TaMYB and TaWRKY transcription factors. These transcription factors activate adaptive responses by inducing expression of genes encoding ion channels and transporters to eliminate the accumulation of toxic ions in the cytosol, genes involved in the synthesis of osmoprotectants and antioxidant defense mechanism to reduce reactive oxygen species and MDA contents (Fig. 8). However, further evidences are needed to verify the exact role of these transcription factors along with other putative mechanisms participating in PGPR-mediated salinity tolerance in plants.
Figure 8. Overview of modulation of expression of genes involved in PGPR Dietzia natronolimnaea STR1-mediated salinity tolerance in wheat plants.
In the early steps of interaction, Dietzia natronolimnaea STR1 triggered signal transduction which modulated the expression of several genes responsible for salt tolerance. Based on the results we propose that carotenoid producing D. natronolimnaea STR1 participates in salt tolerance via both ABA-mediated and SOS-mediated pathways by up-regulating the expression of ABA-signalling cascade genes (TaABARE and TaOPR1), leading to induction of TaMYB and TaWRKY expression followed by stimulation of expression of a plethora of stress related genes including TaST, a salt stress-induced gene, associated with promoting salinity tolerance. Modulation of SOS pathway related genes (SOS1 and SOS4) and tissue specific responses of ion transporters TaNHX1, TaHAK and TaHKT1, were observed in PGPR applied plants. The enhanced gene expression of various antioxidant enzymes such as APX, MnSOD, CAT, POD, GPX and GR and higher proline content in PGPR-treated wheat plants contributed to increased plant tolerance to salinity stress. The red coloured bold arrows indicate the up-regulation or down-regulation of genes.
Methods
Bacterial strain
The bacterial strain Dietzia natronolimnaea STR1 (Accession no. KJ413139)15 was obtained from the culture collection of Microbial Technology Department, CSIR-CIMAP, Lucknow, India.
Molecular characterization and phylogenetic analysis of bacteria
Bacterial genomic DNA was isolated using the standard procedures of Chachaty and Saulnier 200051. The extracted DNA was quantified using NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Primers used for PCR amplification of the 16S rRNA are described in Supplementary Table S2. For 16S rRNA gene amplification, ~25 ng of genomic DNA and 10 μM of forward (fD1) and reverse (rP2) primers, 0.2 mM of each dNTPs, 0.6 U of Taq DNA polymerase (Bangalore Genei, India), 1X PCR buffer were used for amplification in a Master cycler gradient PCR system (Eppendorf). The thermocycling condition was denaturation at 94 °C for 5 min; 34 cycles of 94 oC for 1 min, 57.4 oC for 1 min, 72 oC for 2 min and a final extension step at 72 oC for 10 min. PCR products were visualized on 1.2% agarose gels and purified using PCR Cleanup Gel Extraction kit (Nucleo-pore, Genetix Biotech Asia Pvt. Ltd. India) following the manufacturer’s protocol. Sequencing was performed on a 3130xl Genetic Analyzer (Applied Biosystems, USA). Sequence analysis was carried out using the NCBI BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) and nearest neighbor sequences were identified and downloaded from the NCBI database. The 16S rRNA gene sequence of STR1 isolate was aligned with those of 29 other bacteria using ClustalW alignment tool (MEGA5)52. A phylogenetic tree was prepared using the bootstrapped neighbor-joining tree method (MEGA5), showed maximum similarity of STR1 with Dietzia natronolimnaea.
Functional traits of STR1 strain
Various plant growth promoting properties were tested in STR1 bacterial strain. The phosphate solubilization test was done on Pikovskaya (PVK) medium53. ACC deaminase activities and indole acetic acid (IAA) production were tested using Dworkin and Foster (DF) minimal salts medium54 and Salkowsky’s reagent in tryptophan amended medium55 respectively. The presence of exopolysaccharides was tested according to the method of Siddikee et al.56. The nifH gene was amplified from the bacterial DNA using primers described in Supplementary Table S2. PCR amplification was performed in 50 μL reaction volume containing 50 ng DNA, 20 μM each of forward (Pol F) and reverse (Pol R) primers, 0.2 mM of each dNTPs (Sigma, USA), Taq polymerase buffer (1X) and 0.6 U of Taq polymerase enzyme (Sigma, USA). PCR conditions consisted of an initial denaturation at 94 °C for 4 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 2 min and a final extension step at 72 °C for 5 min with Master cycler gradient PCR system (Eppendorf). PCR products were analyzed in 1.5% (w/v) agarose (Sigma, USA) gels stained with ethidium bromide and visualized under UV light.
Plant material, growth conditions, and salt stress treatment
Wheat (Triticum aestivum L.) cv. HD 2285, was used in the present study. Seeds of HD 2285 were obtained from Indian Agricultural Research Institute (IARI), Pusa, New Delhi, India. The experiments were performed following completely randomized block design in pots (22 cm top diameter x 12 cm bottom diameter x 17 cm height) containing soil, soil supplemented with NaCl and soil supplemented with NaCl and inoculated with halo-tolerant PGPR (D. natronolimnaea STR1). The experiments were conducted with three replicates in a glasshouse and the potential of STR1 to alleviate salinity stress in wheat was evaluated. Seeds were surface sterilized by gently shaking in 3% sodium hypochlorite solution containing 0.1% Triton X-100 for 5 min and rinsed four times for 5 min in sterile deionized water. Sterilized seeds were soaked in sterile distilled water for 2 h before sowing in pots (five seeds per pot). The potting mixture consisted of sterilized soil [sandy loam (Ustifluvent)] having pH 7.16, EC 0.46 dS m−1, 4.64 g kg−1 organic carbon, 127 kg ha−1 available N (alkaline permanganate extractable), 10.7 kg ha−1 available P (0.50 M NaHCO3 extractable), and 97.5 kg ha−1 available K (1 N NH4OAc extractable). Soil sterilization was performed by autoclaving at 15 lbs/121 °C for 3 h. Pots were watered with sterile water on a daily basis for the first 2 weeks after seed germination and later NaCl stress was applied by supplementing with NaCl solution with concentration gradually increasing to 150 mM. The NaCl concentrations were applied incrementally by 50 mM every week until final concentrations (150 mM) were reached and maintained till plant harvesting. Non-saline plants were irrigated only with sterile water. These plants (60 d after germination) were used for estimation of growth parameters, photosynthetic pigments, total proline content, lipid peroxidation, and activity of catalase and peroxidase.
Gene expression study was performed on hydroponically grown wheat plants. Seeds were surface sterilized in 3% sodium hypochlorite solution containing 0.1% Triton X-100 for 5 min followed by four times washing with sterile distilled water. After extensive washing, seeds were germinated and grown hydroponically on germination sheet rolls placed in 250 mL glass beakers filled with the one-half strength of Hoagland nutrient solution under cool-white fluorescent light (100 μmoles photon m−2 s−1) in 14 h light and 10 h dark photoperiod at 24 oC room temperature. Saline stress was applied from the day of germination and up to 12 d. Hoagland nutrient solution was exchanged every day. Sodium chloride was added to the Hoagland nutrient solution to obtain a final concentration of 100 mM.
Bacterial inoculation
Bacterial inoculation to the wheat plants grown under glasshouse conditions was done prior to the NaCl stress application. Bacterial inoculum was prepared by growing STR1 strains in nutrient broth with 5% (w/v) NaCl at 28 °C for 24 h in an orbital shaking incubator at 100 rpm till late exponential growth phase. Optical density was measured to attain the uniform population of bacteria [~108 colony forming units (CFU) mL−1] in the culture prior to inoculation. Culture was centrifuged at 8,000 × g for 10 min. The pellet obtained was washed with sterile distilled water and then re-suspended in 0.85% saline solution. The bacterial suspension was adjusted to A600nm = 1.0. For control plants only sterilized 0.85% saline was applied57.
For hydroponics experiments, bacterial culture was harvested by centrifugation (8,000 × g for 10 min, 10 °C), washed once with 0.85% NaCl and re-suspended in 0.01 M MgSO4 (~105 C.F.U. mL−1). One mL of bacterial suspension was mixed in 49 mL of plant growth medium (half-strength Hoagland nutrient solution). For mock inoculation, 1 mL of 0.01 M MgSO4 solution was added into the plant growth medium. Colonization of bacteria in root of wheat plant was determined by re-isolating the inoculated bacteria (STR1) from washed roots using glass beads followed by plating out serial dilutions on NaCl amended nutrient agar plates.
Measurement of growth parameters
The soil grown plants were harvested after 60 d of germination, and their plant height (cm) and dry weight were measured. Hydroponically grown wheat seedlings were harvested after 12 d of seed germination and their height and dry weight were measured. For dry weight measurement harvested shoots were dried at 70 °C for 5 d, and their final dry weights were measured.
Measurement of photosynthetic pigments
Chlorophyll a, chlorophyll b and carotenoid content were measured in leaves of soil grown wheat plant. Photosynthetic pigments were extracted from fresh leaf samples in 80% acetone following the procedures described previously10 and the quantification was done following the procedure described by Porra et al.58.
Total proline content
The total proline content in leaves was measured following the procedure described previously59 with slight modifications. Leaves (100 mg) were homogenized in 1.5 mL of 3% sulphosalicylic acid, and centrifuged at 10,000 × g for 10 min in a refrigerated centrifuge. 100 μL of supernatant was reacted with 2 mL glacial acetic acid and 2 mL acid ninhydrin for 1 h at 100 °C and then kept on ice to terminate the reaction. The reaction mixture was extracted with toluene (1 mL) and the absorbance of the extract was recorded at 520 nm. Proline content was determined by using the proline standard curve.
Lipid peroxidation
Amount of lipid peroxidation was determined by estimating the malondialdehyde (MDA) produced by the thiobarbituric acid (TBA) reaction60. Fresh leaf tissues were homogenized in a mortar and pestle with ice-cold extraction buffer (1.6% Na2HPO4. 12 H2O and 0.7% of NaH2PO4. 2 H2O) and centrifuged at 14,000 × g for 30 min. The supernatant was used for the estimation of MDA. 4 mL of 0.5% (w/v) TBA solution containing 20% (w/v) TBA was added to 1 mL of supernatant. The mixture was heated at 95 °C for 30 min and then immediately cooled on ice. The absorbance of the mixture was measured at 532 nm and the correction for non-specific absorbance was done by subtracting the absorbance at 600 nm. MDA concentration was determined by its molar extinction coefficient (155 mM−1 cm−1).
Quantitative real-time PCR analysis
Total RNA was extracted from the shoots and roots of hydroponically grown 12 d old wheat seedlings treated with 100 mM NaCl and PGPR using TRI-reagent (Sigma-Aldrich Inc. MO, USA). DNA contamination from total RNA preparation was removed by using RNase-free DNase I (TaKaRa Bio Dalian CO., Ltd). cDNA was synthesized from approximately 3 μg of total RNA using a first strand cDNA synthesis kit (Thermo Scientific). PCR conditions were 10 min at 95 oC, followed by 40 cycles of denaturation at 95 oC for 15 sec each and annealing/extension at 60 oC for 1 min each. Primers used for the relative quantification of biosynthetic gene transcripts are described in Supplementary Table S3. The threshold (Ct) value for each gene was normalized against the Ct for actin from wheat which was used as the constitutive reference transcript. Uninoculated plants growing in non-saline condition were used as the calibrator. Fluorescent signal intensities were recorded and analyzed on a StepOnePlusTM (Applied Biosystems) Real-Time PCR System.
Scanning electron microscopy
Twelve days old wheat plantlets, grown in Hoagland’s hydroponic system, bacterial cells (D. natronolimnaea STR1) collected from mid-log phase and wheat plantlets inoculated with bacteria STR1, were washed with 0.2 M Sorensen’s phosphate buffer two times. Furthermore, each sample was fixed with fixing solution (prepared in 0.2 M Sorensen’s phosphate buffer) containing 4% (v/v) formaldehyde and 2.5% (v/v) glutaraldehyde for 4 h at room temperature. Fixed samples were washed with 0.2 M Sorensen’s phosphate buffer three times (each wash was given for 15 min). In addition, each sample was dehydrated through a graded series of absolute ethanol (20, 40, 60, 80 and 100% respectively). Each sample was dried in a critical point dryer in a CO2 atmosphere and examined at 20 kv in a JEOL JXA 8100 scanning electron microscope.
Statistical analysis
Statistical analysis of data were carried out by applying ANOVA, suitable to completely randomized design (CRD) using the software PASW Statistics 18. Significant differences among different treatments were carried out using Duncan’s multiple range test at a significance level of P ≤ 0.05. Two trials were conducted for the experiment and both had a similar variance value; hence, the data of both experiments were combined for further analysis. The gene expression data were analyzed using Duncan’s Test (P < 0.05).
Additional Information
How to cite this article: Bharti, N. et al. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6, 34768; doi: 10.1038/srep34768 (2016).
Supplementary Material
Acknowledgments
The authors wish to thank the Director, CSIR- Central Institute of Medicinal and Aromatic Plants, Lucknow, India, for providing necessary facilities and encouragement during the course of investigation and the Council of Scientific and Industrial Research (CSIR), for funding the projects CSIR-NWP BSC109 and CSIR-NWP BSC0117 and University Grants Commission (UGC), New Delhi, India, for providing the financial support in form of fellowships to N.B., V.K.P., S.S.P. and D.B. respectively. Authors are thankful to National Centre of Experimental Mineralogy and Petrology (N.C.E.M.P), University of Allahabad, Allahabad for providing Scanning Electron Microscopy facility. Authors also acknowledge the help of Dr. Rakesh K Shukla of CSIR-CIMAP for critically going through the manuscript.
Footnotes
Author Contributions N.B., S.S.P. and A.K. conceived and designed research. N.B., S.S.P. and D.B. conducted experiments and analysed data. V.K.P. contributed in generation of SEM micrographs. N.B., S.S.P. and A.K. wrote the manuscript. All authors read and approved the manuscript.
References
- Huang G. T. et al. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987 (2012). [DOI] [PubMed] [Google Scholar]
- Agarwal P. K. & Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plantarum. 54, 201–212 (2010). [Google Scholar]
- Rajendran K., Tester M. & Roy S. J. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 32, 237–249 (2009). [DOI] [PubMed] [Google Scholar]
- Gaxiola R. A. et al. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification. Proc. Natl. Acad. Sci. USA 96, 1480–1485 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C. & Zhu J. K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434 (2000). [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S. & Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Env. 33, 453–467 (2010). [DOI] [PubMed] [Google Scholar]
- Lugtenberg B. & Kamilova F. Plant-growth-promoting rhizobacteria. Ann. Rev. Microbiol. 63, 541–556 (2009). [DOI] [PubMed] [Google Scholar]
- Barnawal D., Bharti N., Maji D., Chanotiya C. S. & Kalra A. 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol. Biochem. 58, 227–235 (2012). [DOI] [PubMed] [Google Scholar]
- Bharti N., Barnawal D., Awasthi A., Yadav A. & Kalra A. Plant growth promoting rhizobacteria alleviate salinity induced negative effects on growth, oil content and physiological status in Mentha arvensis. Acta. Physiol. Plant. 36, 45–60 (2014). [Google Scholar]
- Hamdia A. B. E., Shaddad M. A. K. & Doaa M. M. Mechanisms of salt tolerance and interactive effects of Azospirillum brasiliense inoculation on maize cultivars grown under salt stress conditions. Plant Growth Regul. 44, 165–174 (2004). [Google Scholar]
- Bashan Y., Holguin G. & de-Bashan L. E. Azospirillum–plant relationships: physiological, molecular, agricultural, and environmental advances. (1997– 2003), Can. J. Microbiol. 50, 521–577 (2004). [DOI] [PubMed] [Google Scholar]
- Cartieaux F. et al. Transcriptome analysis of Arabidopsis colonized by a plant‐growth promoting rhizobacterium reveals a general effect on disease resistance. The Plant J. 36, 177–188 (2003). [DOI] [PubMed] [Google Scholar]
- Cartieaux F. et al. Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol. Plant Microbe In. 21, 244–259 (2008). [DOI] [PubMed] [Google Scholar]
- Bharti N., Barnawal D., Wasnik K., Tewari S. K. & Kalra A. Co-inoculation of Dietzia natronolimnaea and Glomus intraradices with vermicompost positively influences Ocimum basilicum growth and resident microbial community structure in salt affected low fertility soils. Applied Soil Ecology 100, 211–225 (2016). [Google Scholar]
- Dimkpa C., Weinand T. & Ash F. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694 (2009). [DOI] [PubMed] [Google Scholar]
- Gharibzahedi S. M. T., Razavi S. H. & Mousavi M. Feeding strategies for the improved biosynthesis of canthaxanthin from enzymatic hydrolyzed molasses in the fed-batch fermentation of Dietzia natronolimnaea HS-1. Biores. Technol. 154, 51–58 (2014). [DOI] [PubMed] [Google Scholar]
- Jin Q. et al. Biodegradation of aniline in an alkaline environment by a novel strain of the halophilic bacterium, Dietzia natronolimnaea JQ-AN. Biores. Technol. 117, 148–154 (2012). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Dietzia schimae sp. nov. and Dietzia cercidiphylli sp. nov., from surface-sterilized plant tissues. Int. J. Syst. Evol. Microbiol. 58, 2549–2554 (2008). [DOI] [PubMed] [Google Scholar]
- Micsinai A. et al. Rhizome-associated bacterial communities of healthy and declining reed stands in Lake Velencei, Hungary, Hydrobiologia 506, 707–713 (2003). [Google Scholar]
- Spaepen S., Bossuyt S., Engelen K., Marchal K. & Vanderleyden J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861 (2014). [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola M. Á., Arbona V., Gómez-Cadenas A., Rodríguez-Concepción M. & Rodríguez-Villalón. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS ONE 9, e90765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogue B., Doré H., Borland S., Wisniewski-Dyé F. & Prigent-Combaret C. Which specificity in cooperation between phytostimulating rhizobacteria and plants ? Res. Microbiol. 163, 500–510 (2012). [DOI] [PubMed] [Google Scholar]
- Srivastava S. et al. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 7, 235–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S. et al. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genomics 9, 109–123 (2009). [DOI] [PubMed] [Google Scholar]
- Deinlein U. et al. Plant salt-tolerance mechanisms. Trends Plant Sc. 19, 371–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lining X., Becky S., Tiegang L. & Zhu J. K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14, 575–588 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A. et al. Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress. Mol. Biotechnol. 53, 189–197 (2013). [DOI] [PubMed] [Google Scholar]
- Shi H. & Zhu J. K. SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol. 129, 585–593 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D. L. et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11 (2012). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One 8, e65120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C. F. et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170 (2012). [DOI] [PubMed] [Google Scholar]
- Agarwal P., Reddy M. P. & Chikara J. WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 38, 3883–3896 (2011). [DOI] [PubMed] [Google Scholar]
- Wenke K. et al. Volatiles of two growth-inhibiting rhizobacteria commonly engage AtWRKY18 function. The Plant J. 70 445–459 (2012). [DOI] [PubMed] [Google Scholar]
- Dubos C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010). [DOI] [PubMed] [Google Scholar]
- Qin Y. et al. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 39, 7183–7192 (2012). [DOI] [PubMed] [Google Scholar]
- Yang A., Dai X. & Zhang W. H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. & Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 (2006). [DOI] [PubMed] [Google Scholar]
- Choi J. Y., Seo Y. S., Kim S. J., Kim W. T. & Shin J. S. Constitutive expression of CaXTH3, a hot pepper xyloglucanendotransglucosylase/ hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 30, 867–877 (2011). [DOI] [PubMed] [Google Scholar]
- Volpe V. et al. An AM-induced, MYB-family gene of Lotus japonicus (LjMAMI) affects root growth in an AM-independent manner. The Plant J. 73, 442–455 (2013). [DOI] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A. & Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 (1999). [DOI] [PubMed] [Google Scholar]
- Hauser F. & Horie T. A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 33, 552–565 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant Microbe In. 21, 737–744 (2008). [DOI] [PubMed] [Google Scholar]
- Su H., Golldack D., Zhao C. & Bohnert H. J. The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 129, 1482–1493 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wang G., Shen Y. & Huang Z. The wheat gene TaST can increase the salt tolerance of transgenic Arabidopsis. Plant Cell Rep. 31, 339–347 (2012). [DOI] [PubMed] [Google Scholar]
- Dong W. et al. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol. 161, 1217–1228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgherri C. L. M., Maffei M. & Navari-Izzo F. Antioxidative enzymes in wheat subjected to increasing water deficit and rewatering. J. Plant Physiol. 157, 273–279 (2000). [Google Scholar]
- Baltruschat H. et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 180, 501–510 (2008). [DOI] [PubMed] [Google Scholar]
- Gururani M. A. et al. Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth. Regul. 32, 245–258 (2013). [Google Scholar]
- Chachaty E. & Saulnier P. Isolating chromosomal DNA from bacteria. In The nucleic acid protocols handbook. Humana Press, 29–32 (2000). [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikovskaya R. I. Mobilization of phosphorous in soil in connection with the vital activity of some microbial species. Microbiologiya 17, 362–370 (1948). [Google Scholar]
- Piromyou P. et al. Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand, European J. Soil Biol. 47, 44–54 (2011). [Google Scholar]
- Tang Y. W. & Bonner J. The enzymatic inactivation of indoleacetic acid; some characteristics of the enzyme contained in pea seedlings. Arch. Biochem. 13, 11–25 (1947). [PubMed] [Google Scholar]
- Siddikee M. A., Glick B. R., Chauhan P. S., Yim W. J. & Sa T. Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane- 1-carboxylic acid deaminase activity. Plant Physiol. Biochem. 49, 427–434 (2011). [DOI] [PubMed] [Google Scholar]
- Bharti N., Yadav D., Barnawal D., Maji D. & Kalra A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 29, 379–387 (2013). [DOI] [PubMed] [Google Scholar]
- Porra R. J., Thompson W. A. & Kriedmann P. A. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta. 975, 384–394 (1989). [Google Scholar]
- Bates L. S., Waldren R. D. & Teare I. D. Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207 (1973). [Google Scholar]
- Heath R. L. & Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198 (1968). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.