Summary
Background
The Clinical Genome Resource (ClinGen) Electronic Health Record (EHR) Workgroup aims to integrate ClinGen resources with EHRs. A promising option to enable this integration is through the Health Level Seven (HL7) Infobutton Standard. EHR systems that are certified according to the US Meaningful Use program provide HL7-compliant infobutton capabilities, which can be leveraged to support clinical decision-making in genomics.
Objectives
To integrate genomic knowledge resources using the HL7 infobutton standard. Two tactics to achieve this objective were: (1) creating an HL7-compliant search interface for ClinGen, and (2) proposing guidance for genomic resources on achieving HL7 Infobutton standard accessibility and compliance.
Methods
We built a search interface utilizing OpenInfobutton, an open source reference implementation of the HL7 Infobutton standard. ClinGen resources were assessed for readiness towards HL7 compliance. Finally, based upon our experiences we provide recommendations for publishers seeking to achieve HL7 compliance.
Results
Eight genomic resources and two sub-resources were integrated with the ClinGen search engine via OpenInfobutton and the HL7 infobutton standard. Resources we assessed have varying levels of readiness towards HL7-compliance. Furthermore, we found that adoption of standard terminologies used by EHR systems is the main gap to achieve compliance.
Conclusion
Genomic resources can be integrated with EHR systems via the HL7 Infobutton standard using OpenInfobutton. Full compliance of genomic resources with the Infobutton standard would further enhance interoperability with EHR systems.
Keywords: Clinical information systems, standards adoption, HL7, genetics, clinical decision support
1. Background and Significance
Clinical genomics is considered an important, complex, rapidly increasing knowledge domain [1]. Integration of clinical genomics into medical practice is highly desirable, but the need for clinical guidance is significant, as indicated by the US National Human Genome Research Institute (NHGRI) director [2] and the American College of Medical Genetics and Genomics Board of Directors [3]. Further, the President of the United States highlighted the importance of genomics in precision health care (https://www.whitehouse.gov/precision-medicine) in his 2015 State of the Union address. The challenging juxtaposition of complexity, growth and importance in clinical genomics can lead to difficulty in knowledge management, producing gaps in information. When the need for information is not satisfied this can result in medical error or reduced quality of care [4, 5]. Online medical knowledge resources offer a possible solution for satisfying information needs [2, 6]. It has been demonstrated that online resources provide answers for greater than 90% of clinicians’ general patient care questions [7], and there is good evidence that searching electronic resources for information can have positive effects on clinical decision making [8–11].
To support information needs in clinical genomics, the Clinical Genome Resource (ClinGen) was established to “provide high quality, curated information on clinically relevant genes and variants” [12]. The ClinGen Electronic Health Record Work Group (EHR WG) has been tasked with providing integration between EHRs and genomics resources, including future ClinGen resources. As such, the ClinGen EHR WG in conjunction with other ClinGen domain experts has produced a website containing links to several genomics resources thought to be of value in particular contexts. Specifically, the links are organized for different end users (Clinician, Researcher, Laboratory, and Patient) under a clinical activity classification, such as point of care or “just in time” education. Many of these resources are developed by or are accessible through the National Center for Biotechnology Information (NCBI) at the National Institutes of Health [13]. The NCBI offers a rich aggregation of clinical genomics resources, supporting a wide-range of specialization. However, searching many genomic resources and learning how to optimally use each resource is time-consuming and unfeasible for most clinicians. Importantly, in pursuing answers to clinical questions, clinicians note that lack of time and seamless access to resources at the point of care are the main barriers [14]. Thus, a solution is needed to streamline point of care access across the landscape of genomic resources.
As discussed by the eMERGE network in its efforts to aid content authoring [15], and previous implementations within EHR systems [16–18], infobuttons are an information retrieval modality that can address the need for streamlined access to genomic knowledge in the context of patient care. In the clinical setting, infobuttons are used to translate the context of a particular patient encounter into a request for information related specifically to that context from the web’s plethora of electronic resources (e-resources). Infobuttons have been shown to improve the speed with which clinicians find answers to clinical questions [11], producing a positive impact on decision making [11, 19, 20]. Thus, infobuttons offer an attractive tool to improve clinicians’ access to genomic knowledge, even at the point-of-care.
Further motivation to pursue infobuttons for dissemination of genomic knowledge is linked to the US Meaningful Use 2014 EHR certification criteria, which include a requirement [21] for implementation of the HL7 Infobutton standard [22]. The Meaningful Use program also includes financial incentives to providers who adopt certified EHR products. To date, over a thousand EHR products have been certified under the Meaningful Use program[23].
2. Objective
In order to facilitate EHR access to genomic resources, the ClinGen EHR WG decided to develop an infobutton-enabled search interface, compliant with the HL7 Infobutton standard (HL7 IB). The overall goal of the present study is to describe the implementation of the search interface, which leverages an open source, HL7-compliant, infobutton platform called OpenInfobutton. Specifically we aimed to:
develop an OpenInfobutton enabled search engine;
configure OpenInfobutton to access a subset of ClinGen genomic resources;
analyze readiness of this set of genomics resources regarding the HL7 IB standard; and
provide recommendations for publishers to achieve HL7 IB compliance
3. Methods
3.1 HL7 Infobutton standard and OpenInfobutton
An infobutton is a context-aware information retrieval tool that anticipates the information needs of a clinician in a specific clinical context and provides automated links to relevant e-resources [24]. For example, in the context of prescribing clopidogrel in the outpatient setting for a 67 year-old female with a history of stroke, a simple infobutton could provide access to a statement from the American College of Cardiology related to clopidogrel treatment for female patients with a history of stroke.
Infobuttons can be implemented with a web service known as an ‘Infobutton manager’ (IM) [25, 26]. Infobutton managers match the context conveyed in an EHR request to a set of relevant resources; and automatically create infobutton requests for each of the selected e-resources. Importantly, IMs match contextual parameters to specific resources to ensure 1) optimal resources are offered to users, and 2) the order of returned links reflects the relevance of each resource in a particular context. As an example, an infobutton connecting to an IM within the context of computerized provider order entry (CPOE) and containing the term ‘clopidogrel’ could return a link to the FDA approved label for clopidogrel, a link to the common conditions for which clopidogrel may be indicated, and a link to a genomic resource page on testing for pharmacogenetics linked to clopidogrel resistance. Alternatively, an infobutton associated with a ‘history of percutaneous transluminal coronary angioplasty’ (PCI) on the problem list would be unlikely to return a link to FDA labels, although it might have a section on suggested medications, and the response could include information on any genetic variants that have been associated with outcomes of PCI.
The HL7 IB standard is designed to reduce the effort required to integrate e-resources with EHR systems. The standard contains a context information model, a standard set of terminologies to maximize interoperability with EHR systems, and RESTful web-based implementation approaches [27]. The context information model defines a set of context parameters according to four dimensions: the patient, the infobutton user, the care setting (e.g., inpatient, outpatient), and the task undertaken by the user within the EHR task (e.g., a medication order entry, a diagnosis, a laboratory test result). The patient dimension can include a main clinical concept of interest, patient’s age, gender, medications, and additional patient conditions/diagnoses. The user dimension allows the distinction between a healthcare provider and a patient user as well as the preferred language of the target information recipient. Importantly, the HL7 IB standard parameters aids semantic interoperability by providing coded data, and the terminology from which the code is derived. The example shows how a compliant request may appear (► Table 1). Several HL7 IB compliant e-resources have taken advantage of the context within a HL7 IB compliant request to deliver specialized navigation pages that are tuned to support clinical decision making [25].
Table 1.
HL7 IB example request implemented as a URL. The name of the parameter appears after an ampersand (‘&’) and the value after an equals sign (‘=’). The role of each component of the example is explained to the right. Object Identifiers (OIDs), are globally unique identifiers set by the International Organization for Standardization (ISO) and used by HL7 to identify organizations and code systems. Note: more parameters are in the HL7 IB specification.
| Example of Infobutton Request Syntax | Explanation of Component |
|---|---|
| http://clingen.org/tools-resources/web-resources/?searchType=HL7 | Base URL of knowledge resource |
| &representedOrganization.id.root=1.3.6.1.4.1.3768 | OID of requesting organization |
| &patientPerson.administrativeGenderCode.c=F | Patient gender (code) |
| &age.v.v=67 | Patient age (value) |
| &age.v.u=a | Patient age (units) |
| &mainSearchCriteria.v.c=749196 | Search term (code) |
| &mainSearchCriteria.v.cs=2.16.840.1.113883.6.88 | Search term (OID of code system) |
| &mainSearchCriteria.v.dn=Clopidogrel | Search term (display name) |
| &performer=PROV | Role of person performing request (code) |
| &taskContext.c.c=MLREV | Context of the EHR task (code) |
| &informationRecipient=PROV | Role of person consuming results (code) |
| &knowledgeResponseType=application/json | Defines syntax of response |
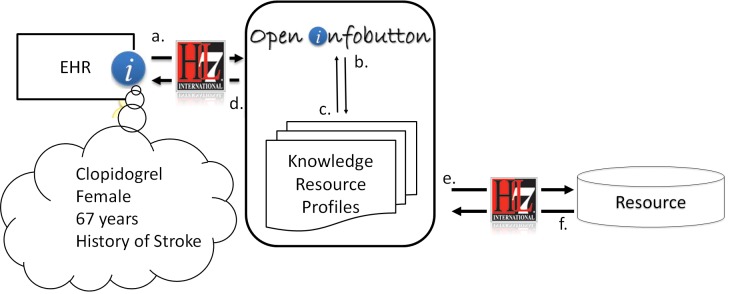
OpenInfobutton is an open source implementation of the HL7 IB standard, and is intended to streamline adoption of infobutton capabilities by healthcare organizations. To date, several large health care organizations, including the Veterans Health Administration [27], have deployed OpenInfobutton. Included in OpenInfobutton is a reference implementation of an IM, and several tools including the resource profile configuration tool known as LITE (Librarian Infobutton Tailoring Environment) [28]. Currently, configured resource profiles allow OpenInfobutton to provide EHRs access to over 40 e-resources. Resource profiles describe the context covered by the resource and, for resources not compliant with HL7, provide mappings to the resource proprietary search engine application program interface (API). ► Figure 1 describes the OpenInfobutton architecture and information flow.
Fig. 1.
OpenInfobutton architecture and information flow from compliant resources. (a) An HL7-compliant URL request that contains context parameters is sent from the EHR to OpenInfobutton. (b) Resource profiles are selected which match the EHR context, and (c) an infobutton response is produced with links to those resources. (d) OpenInfobutton responses can take the form of non-HL7 IB HTML and as HL7 IB XML/JSON. (e) Additionally, if a resource is HL7 IB compliant, OpenInfobutton can send the resource an HL7 IB request and (f) process HL7 IB responses.
The full specification of the parameters available for clinical context can be found in the HL7 IB specification [22]. The parameters we focus on here are the main clinical concept of interest, clinical role and the identifier for the organization making the request, called the organization id. These parameters are included in HL7 IB compliant URLs.
3.2 Selection of genomic resources
A list of resources was compiled by surveying attendees of a ClinGen educational session at the 2015 annual American College of Medical Geneticists meeting (ClinGen web-resources page: https://www.clinicalgenome.org/tools/web-resources/). The resources were organized by the ClinGen EHR WG based upon the role that the resource was expected to satisfy. For example, the patient tab has resources for patient education, but not resources that provide variant level details. In contrast, the clinician tab has resources such as The Pharmacogenomics Knowledgebase (PharmGKB), a clinician-facing pharmacogenomics resource. From the list, resources chosen for the patient and clinician role context were examined for integration using the HL7 IB. Special emphasis was placed on the NCBI resources as they actively maintain an extensive collection of genetic and genomic resources and are closely collaborating with ClinGen.
3.3 OpenInfobutton enabled search engine
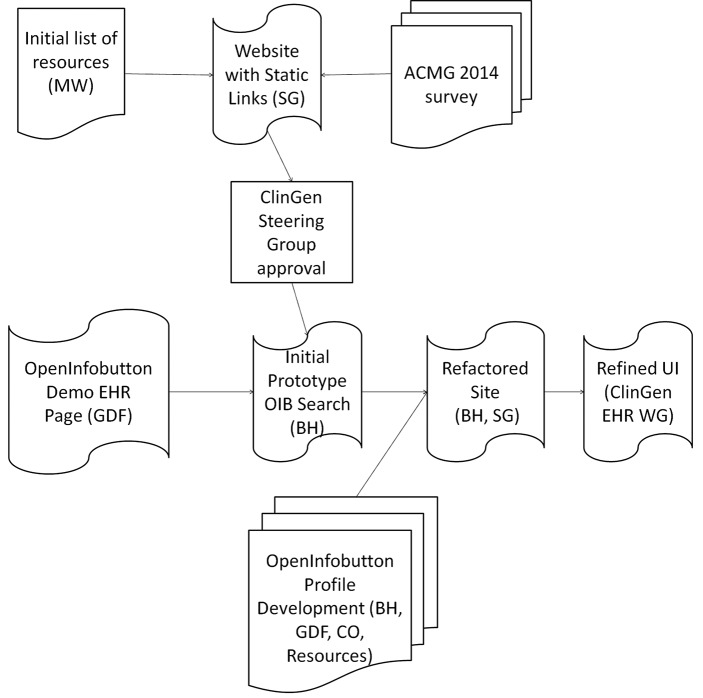
We created a search interface to explore providing genomic resource access to EHRs using the HL7 IB. A diagram of the development process can be seen in ► Figure 2. Though the genomics resources themselves are not compliant with HL7 IB, OpenInfobutton resource profiles allow EHRs to access non-compliant resources through an HL7 IB request. OpenInfobutton accomplishes this through configuring custom API calls.
Fig. 2.
Diagram of website development. Initially, the website contained static links to resources based on an initial list composed by the chair of the ClinGen EHR WG (MW). A survey was performed at the ACMG 2014 conference, after feedback from the ClinGen steering committee and the EHR WG, to gain a broader sense of what resources to include. BH designed/developed the initial prototype for the genomics search interface by combining the OpenInfobutton Demo EHR and the current ClinGen site. SG and BH refactored the site to improve performance, and the UI was further refined with comments from the ClinGen EHR WG. The resource profiles were developed by BH with guidance from GDF. CO aided in gaining feedback from OMIM, PharmGKB and the NCBI. At each stage collaboration between individuals and groups was key.
3.4 OpenInfobutton configuration for genomic resources
OpenInfobutton resource profiles for genomic resources were configured to match the context of the organization making the request (ClinGen), the role of the information recipient (clinician or patient) and the terminology of the search concept (performed by BH with consult of resource representatives). The parameter “requesting organization” provides a means to tailor OpenInfobutton to use only the resources that are available or preferred within a specific health care organization. Health care organizations that wish to access ClinGen resources via infobuttons within EHR systems would need to configure OpenInfobutton to add their organization identifier to ClinGen’s OpenInfobutton profile. We configured Openinfobutton through the LITE application [28].
3.5 Genomics resources and readiness for HL7 IB compliance
According to the HL7 IB specification, compliant resources should provide a RESTful interface that can receive HL7 IB requests and provides HL7 IB responses. The request contains a syntax that presents search context using standard terminologies. Proper semantic understanding of the context requires support of the terminologies used by the request. Further, the HL7 IB specifies syntax for the resource response, including XML and JSON data types, and semantics according to the Atom standard (https://tools.ietf.org/html/rfc4287). We evaluated resource readiness based on the presence of a URL based API, support for HTTP/HTTPS GET and POST protocols, support for standard terminologies, and support for response formats in XML and JSON. The evaluation was carried out by reading the web-accessible documentation for each resource as well as conducting test searches using the resource search API (performed by BH, CO, SG). The results were confirmed by contacting resource representatives (BH performed).
4. Results
4.1 Genomic resources configured for searching
We selected eight genomic resources and two sub-resources for integration by the ClinGen OpenInfobutton search interface. The resource names and urls are found in ► Table 2.
Table 2.
Resource names and base URLs of resources chosen for configuration.
| Resource name* | Resource web-site |
|---|---|
| 1000 Genomes project | http://browser.1000genomes.org/ |
| Clinical Pharmacogenetics Implementation Consortium (CPIC) | Accessed through PharmGKB and NCBI |
| ClinVar | http://www.ncbi.nlm.nih.gov/clinvar/ |
| GeneReviews | http://www.ncbi.nlm.nih.gov/books/NBK1116/ |
| Genetic Practice Guidelines | Accessed through MedGen |
| Genetic Testing Registry (GTR) | http://www.ncbi.nlm.nih.gov/gtr/ |
| Genetics Home Reference (GHR) | http://ghr.nlm.nih.gov |
| MedGen | http://www.ncbi.nlm.nih.gov/medgen/ |
| Online Mendelian Inheritance in Man (OMIM) | http://www.omim.org/ |
| The Pharmacogenomics Knowledgebase (PharmGKB) | https://www.pharmgkb.org/ |
*Genetic Practice Guidelines and CPIC guidelines are examples of specific content accessed as sub-resources of other resources, MedGen and PharmGKB respectively.
4.2 OpenInfobutton enabled search engine
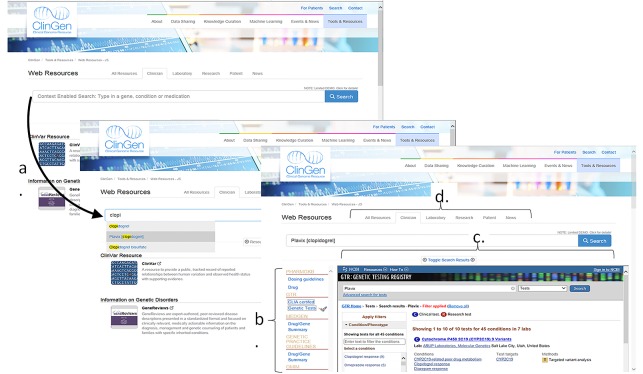
The search user interface consists of a search bar with auto-complete functionality that is populated by a controlled list of relevant genomic terms and concepts (e.g., gene names, genetic conditions) from specific terminologies (► Figure 3). We used the terminology of the search term as an indicator of the genomic domain of inquiry. The three domains used are: Gene (HUGO Gene Symbol), Genetic Disorders/Conditions (OMIM) and Medication (RxNorm). HUGO Gene Symbols is an established international gene name terminology that is required by the HL7 Clinical Genomics Implementation Guide [29]. OMIM was chosen by the ClinGen steering committee to be used to represent genetic disorders within ClinGen, and it is accepted by HL7 as an optional terminology in the HL7 Clinical Genomics Implementation Guide [29]. Further, OMIM currently provides better coverage for rare and genetic diseases than ICD-9, ICD-10 and SNOMED-CT. We use RxNorm to represent drugs given that it is required for EHR certification in the Meaningful Use program [21]. Yet, the use of specific terminologies can be extended according to the HL7 IB specification and feasible within OpenInfobutton’s architecture. For example, OpenInfobutton allows real-time code mappings from HGNC ids and OMIM to EHR terminologies such as SNOMED-CT and ICD10, although we have not taken advantage of this capability.
Fig. 3.
A screenshot of the ClinGen EHR WG OpenInfobutton search interface. (a) As text is typed into the search bar, suggestions for autocompleting are provided from the controlled vocabulary. (b) After search initiation, context dependent links to resources are provided in a menu (c) with the first resource link displayed in a frame. (d) Tabs for clinician or patient provider are found above the search bar.
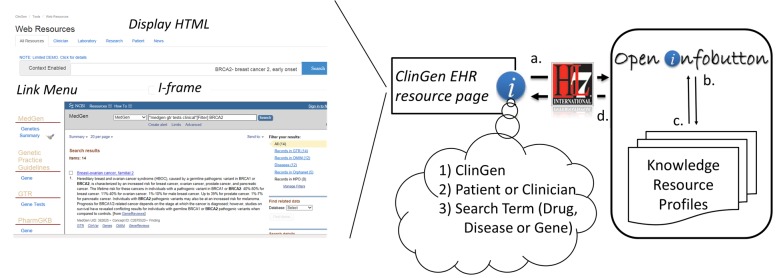
The interaction between the search interface and Openinfobutton (► Figure 4) begins when the ClinGen EHR search interface takes the role of an EHR system and prepares an infobutton request based on search input. The request is sent to OpenInfobutton, which responds using the HL7 Infobutton JSON format. The search interface parses the JSON response, presents the user with a list of indexed links from the matching resources, and loads the first resource into a frame within the ClinGen Web site. The order of the e-resource links is based on the context of the terminology used for the search. For example, when an RxNorm (medication terminology) concept is used it is inferred that a clinician has an interest in pharmacogenomics. PharmGKB is a pharmacogenomics specialized resource, thus links for PharmGKB content appear at the top of the list.
Fig. 4.
Data flow from the ClinGen search interface to OpenInfobutton. (a) When the search button is selected the ClinGen EHR resource page creates an HL7 Infobutton request that sends the context of the search to OpenInfobutton, similarly to an EHR generated infobutton request. The context includes the search term (i. e., main clinical concept of interest, selected from a controlled vocabulary through a drop-down list), the user’s role (clinician, laboratorian, researcher or patient) and the identifier for ClinGen, the organization making the request. (b) Next, the context is matched to the resource profiles within OpenInfobutton. (c) URLs from resources matching the context parameters are collated (d) and sent as a response to the ClinGen EHR Resource. As seen on the left side of the figure, the ClinGen resource page parses the OpenInfobutton response and displays the context sensitive links on a menu to the left of a frame containing the web-site of the first listed resource link.
An important additional context we implemented was the ‘role’ parameter which is set based on the tab selected by the user in the ClinGen EMR page. As an example, when the patient tab is selected in the ClinGen search interface, only GHR is returned for genetic disorder (OMIM) concepts.
4.3 OpenInfobutton configuration for genomic resources
Most resources were configured through OpenInfobutton profiles (example in supplement B) to respond to the context of all three search term domains (Gene, Genetic Disorders/Conditions and Medications). The exception is Gene Reviews, which is retrieved only when the search term is in the Gene domain. For search terms in the Gene and Medications domains, we configured OpenInfobutton to use the text label associated with the search. We followed a similar strategy for the Genetic Disorder domain. However, for OMIM we were able to use the OMIM code (called a MIM number) with some resources. At present only GHR was configured to respond to the patient role.
4.4 Genomics resources and readiness for HL7 IB compliance
► Table 3 describes the readiness of eight genomic resources for compliance with the HL7 IB specification, as of September 14th, 2015. Except for CPIC, all resources provide a URL based API that supports the HTTP/HTTPS GET and POST protocols. CPIC guidelines were made retrievable as a subset of the links returned using the PharmGKB resource. Within PharmGKB, CPIC guidelines are directly accessible only using PharmGKB’s internal proprietary codes. But, in general, PharmGKB supports searches using a proprietary identifier for medications as well as free-text medication terms. Access to concepts defined by MIM numbers was well supported by the NCBI genomic resources (ClinVar, GeneReviews, Genetic Practice Guidelines, Genetic Testing Registry, MedGen), GHR and OMIM itself. Not surprisingly, all resources supported the use of HUGO Gene Nomenclature Committee (HGNC) gene symbols, but not HGNC code based searching. Additionally, OMIM supported retrieval of XML/JSON. Aside from direct URL access, the five NCBI resources can also be accessed through an application program interface (API) called e-utilities [30]. The tool-set provides programmatic access including a URL-based request for XML and JSON. Support for infobutton response using the XML and JSON data types is part of the HL7 IB RESTful specification [22].
Table 3.
Readiness of Genomic Resources regarding the HL7 IB Standard.
| Resource | Response Types | Specific Terminologies used to index the content | Code based searching support for Specific Terminologies |
|---|---|---|---|
| NCBI Resources: ClinVar, GeneReviews, Genetic Practice Guidelines, Genetic Testing Registry, MedGen | HTML/Text XML/JSON (with e-utilities) | OMIM, HGNC Approved Gene Symbol, UMLS, SNOMED-CT | OMIM* |
| GHR (National Library of Medicine) | HTML | OMIM, HGNC Approved Gene Symbol, UMLS, SNOMED-CT | OMIM |
| OMIM | HTML/XML/JSON/JSONP (with Key) | OMIM, HGNC Approved Gene Symbol, UMLS, SNOMED-CT | OMIM |
| PharmGKB | HTML | OMIM, HGNC Approved Gene Symbol, UMLS, SNOMED-CT, NDFRT | None |
| CPIC guidelines | None | None | N/A |
* While code-based searching with OMIM MIM numbers is supported by the www.ncbi.nlm.nih.gov resources, in some cases, we choose to use the search term itself to maximize coverage.
4.5 Search interface go-live and sample HL7 infobutton requests
As of October 2015, the first iteration of the OpenInfobutton search interface was available to the public. Integration with EHRs was enabled with support of HL7 IB requests by the search interface. Additionally, resources within ClinGen such as gene dosage and variant evidence annotations are included in the genomic search and HL7 infobutton response. Response times are usually on the order of seconds. For example, the term BRCA2 retrieves specific, topically organized results from ten resources in less than three seconds. Supplement A contains examples HL7 IB requests for clinical questions regarding Genes, Genetic disorders/conditions, and pharmacogenomics.
5. Discussion
We have successfully enabled infobutton access to eight genomic resources and two sub-resources through OpenInfobutton and the HL7 IB standard. This solution provides access to many EHR systems as compliance with the HL7 IB is required for EHR certification in the US Meaningful Use program. Any EHR system that is HL7 IB compliant can use OpenInfobutton to access the configured genomic resources via context-specific infobuttons located in EHR modules such as problem list, laboratory test results, medication prescriptions, and computerized provider order entry. A next step towards interoperability of genomic resources with EHR systems is for resources themselves to become HL7 IB compliant. HL7 compliance would bring additional benefits, such as the ability to tailor the search results according to the EHR clinical context, more precise information retrieval due to the use of standard terminologies, tuning of resource API results for use by clinicians at the point of care, and the ability to integrate directly with any EHR that is compliant with the HL7 IB standard.
The use of OpenInfobutton has allowed us to configure access to non-HL7 IB compliant genomic resources by EHR systems. However, there are important caveats that resulted in suboptimal results with non-HL7 IB compliant resources. For example, OpenInfobutton access to non-HL7 IB compliant resources will return a URL for the resource even when the URL results in the resources ‘no results found’ page. Furthermore, the resource search URLs used in the resource profiles required manual optimization. After consultation of resource provided documentation, we iteratively optimized the search configurations based on available filters. We chose resource URL search configurations which maximized coverage of the terms and concepts used in the auto-complete functionality of our search interface, but potentially at the expense of precision. Since the IB standard requires EHR systems to send both codes and labels, resource search engines can use a combination of strategies in real time to optimize retrieval. This approach is used by several HL7-compliant resources, such as MedlinePlus and UpToDate. Finally, EHR integration for non-compliant resources is dependent on OpenInfobutton.
5.1 General HL7 IB compliance considerations
We found that many resources required additional work to facilitate HL7 compliance. We therefore provide recommendations to facilitate scalable access to resources using the HL7 IB standard. In general, toward adopting the HL7 IB standard, genomics resources should consider supporting the following requirements in their search APIs.:
Support HTTP/HTTPs POST and GET. (note: HTTPs can provide a layer of security. Also, the method of sending de-identified infobutton requests over HTTPs is accepted under Federal regulations including the Office of the National Coordinator’s EHR Meaningful Use certification criteria.)
Support standard terminologies that are required for EHR certification, such as RxNorm and SNOMED-CT.
-
Support the syntax of standard HL7 IB requests and use context parameters to tailor the search response.
-
i. Example of a request: ‘http://clingen-resource.org/tools-resources/web-resources/?searchType=HL7&representedOrganization.id.root=1.3.6.1.4.1.3768&patientPerson.administrativeGenderCode.c=F&age.v.v=67&age.v.u=a&taskContext.c.c=MLREV&mainSearchCriteria.v.c=749196&mainSearchCriteria.v.cs=2.16.840.1.113883.6.88&mainSearchCriteria.v.dn=Clopidogrel&performer=PROV&informationRecipient=PROV &knowledgeResponseType=application/json’. In the example, the base URL is ‘http://clingen-resource.org/toolsresources/web-resources/?searchType=HL7’ and the text following the base URL contains the contextual parameters. Briefly, the contextual parameters of the example are:
‘representedOrganization.id.root’ is the requesting organization
‘patientPerson.administrativeGenderCode.c’ is the code for patient Gender
‘age.v.v’ and ‘age.v.u’ are the age value and units
‘taskContext.c.c’ has the code for the context of the Task
‘mainSearchCriteria.v.c’, ‘mainSearchCriteria.v.cs’ and ‘mainSearchCriteria.v.dn’ contain the code of the search term, the code system of the search term and a text-version of the search term.
‘performer’ contains a code indicating the role of the person performing the request
‘informationRecipient’ indicates the role of the person the information is intended to be used by.
‘knowledgeResponseType’ indicates the form the client system is expecting the resource response to be in.
-
Support HL7 IB compliant response; including both XML and JSON data types (see the HL7 IB standard specification for examples).
5.2 Recommendations for ClinVar, Gene Reviews, Genetic Practice Guidelines, Genetic Testing Registry, MedGen and GHR
We found that the NCBI resources have many of the foundational components required for HL7 IB compliance, including a URL accessible API that can handle HTTP/HTTPs GET and POST requests, and content indexed with standard terminologies that are required for EHR certification (SNOMED-CT and RxNorm). Although the NCBI APIs do not provide the ability to search using RxNorm and SNOMED-CT codes, NCBI can leverage its ability to search with UMLS codes to provide RxNorm and SNOMED-CT code-based searches. One potential solution is to map incoming requests in RxNorm and SNOMED-CT to UMLS codes and use these codes for searching. Another recommendation regarding terminology support is to support searching with HUGO codes for genes as approved gene symbols change over time.
As mentioned, the NCBI provides a tool called e-utilities for programmatic searches of its databases, with support for XML/JSON responses. E-utilities could be a starting point for an HL7 IB interface to NCBI sites (ClinVar, Gene Reviews, Genetic Practice Guidelines, Genetic Testing Registry, MedGen).
Further, the domain www.ncbi.nlm.nih.gov offers many high-quality genomic resources with specialized content. We suggest that the domain consider enabling a single HL7 IB API for access across all the resources of the domain. The feasibility of this approach is evidenced by the success of our search interface. One request is sent to OpenInfobutton and links to multiple resources are returned, with additional higher level categorization possible using the HL7 IB response format.
Finally, NCBI has the potential to extend accessibility to other e-resources. For example, both the 1000 Genomes resource and the text of CPIC guidelines are currently accessible through NCBI. NCBI adoption of the HL7 IB standard would make, support HL7 IB context aware responses from these resources possible.
5.3 Recommendations for OMIM
OMIM provides two APIs, one requiring an authentication key with the base URL http://www.omim.org/api, which has many of the same foundational components as the NCBI resources. The API accessed through http://www.omim.org/api could be a starting point for an HL7 IB interface. The HL7 IB interface should handle the HL7 IB syntax for responses and requests (as noted in the general considerations section)
5.4 Recommendations for PharmGKB
PharmGKB has implemented some of the foundational features towards HL7 IB compliance, especially a URL-based search mechanism. As a drug reference resource, the most important gap in PharmGKB towards HL7 compliance is support for RxNorm code-based searching. PharmGKB already uses drug names for content indexing. These drug names could be used to automatically identify RxNorm codes via the RxNorm RESTful API provided by the National Library of Medicine [31]. PharmGKB informed the authors that it is extending an API to support JSON/XML data types and code based searching with RxNorm.
5.5 Combining HGNC gene ids and LOINC codes
LOINC codes for genetic tests are another source of context. However, many LOINC codes refer to panels of genes. By using HGNC identifiers one can specify a specific gene. The drawback of using HGNC gene identifiers is that most EHRs are not currently using HGNC gene identifiers. But, with a single LOINC code alone it can be impossible for an EHR to request information for a specific gene result, and knowledge resources would be unable to retrieve gene specific information. Potentially, the use of both a LOINC test code, within the observation parameter, and a HGNC identifier as the main search criteria would add additional layers of context. However, at this time we are not aware of an EHR system that supports this approach.
5.6 Limitations
We did not test implementation of all the possible parameters of the HL7 IB standard and only looked at providing responses for Medications, Genetic Diseases/Conditions and Gene queries. Secondly, we did not consider allelic or genetic sequence-level based searches. Accessing allele-specific or sequence-level information to offer clinical recommendations based on a patient’s specific genotype is a complex but highly important issue for EHR systems not to mention a key goal of the ClinGen project. Efforts to access clinical assertions based on allelic data are frustrated by the absence of widely accepted standards for exchange or storage. As a result, we did not support sequence-level searches other than an attempt using free-text with the 1000 Genomes resource. As a first step toward addressing this issue, the ClinGen Resource is working with key stakeholders to define a common data model to unambiguously describe genetic alleles at the sequence level. ClinGen is also working with NCBI to design and implement a system for assigning unique Allele Identifiers and for providing Registry Web Services (provided by the ClinGen data modeling working-group) that will facilitate linking of genetic test results to HL7 IB compliant resources.
5.7 Future for ClinGen EHR OpenInfobutton enabled search interface and ClinGen Genomics Resource access
The current effort would be enhanced by usability studies to gather and incorporate feedback on the user-interface and content provided by the genomic search, similar to efforts to meet information needs in gene panel reporting [32]. Additionally, it is important to continue to aid the resources mentioned here to improve access to resource content. Further, BH and GDF have conducted a study (manuscript underway) on physicians’ genomics information needs in clinical decision-making. In addition, efforts are underway in HL7 to improve guidance for use of infobuttons in genomic information retrieval.
In the future, the OpenInfobutton-enabled approach will expanded infobutton accessibility to other genomic resources found within ClinGen. This includes clinically actionable data on sequence variant pathogenicity. As standards for representing structured genomic data in the EHR are developed, it will be possible to create links into ClinGen curated variant information from the EHR based on an individual patient’s genotype. ClinGen is actively involved in efforts to establish such standards; the project contributes to the emerging HL7 Fast Healthcare Interoperability Resources FHIR standard for reporting genetic test results, as well as contributing Logical Observation Identifiers Names and Codes (LOINC) codes appropriate for representing genetic test results.
Moreover, Geisinger Health System has recently enabled HL7 infobutton requests within its genomic testing reporting system (MSW personal communication) to utilize the ClinGen/OpenInfobutton enabled access to genomic resources.
6. Conclusions
We have produced the functional architecture needed to perform context-based searching of genomics e-resources using an OpenInfobutton implementation of the HL7 IB standard. OpenInfobutton successfully enabled access to genomic e-resources using HL7 IB standard requests and provided response messages that complied with the standard. Our result demonstrates that OpenInfobutton can reduce the barrier for genomic e-resource providers to utilize the HL7 IB standard for integrating their content with EHR systems. The process of creating the interface also provided an opportunity to evaluate e-resource readiness for HL7 IB standard compliance, and to create recommendations for paths to EHR accessibility and compliance. We found that the genomic resources have many foundational features needed for HL7 IB readiness. To become compliant the resources generally need to adapt their existent interfaces to handle the syntax of an HL7 IB request and return an HL7 IB compliant response. A major feature of handling HL7 requests, that resources should adopt, is support of terminologies used in EHRs, including concept code based searching. To accomplish this, the resources investigated can take advantage of their current usage of codes for other terminologies. Thus, the largest effort will be to determine how the resources wish to use the context provided by EHR systems. Tailored, precise access to genomics information relevant to a specific patient’s context may soon be available at the touch of a button – a crucial function to enhance the value of the anticipated ClinGen resource.
Supplementary Material
Acknowledgements
Representatives of genomics resources: Teri Klein (PharmGKB), Wendy Rubinstein and Donna Maglott (NCBI), Ada Hamosh, and Joanna Amberger (OMIM).
Funding Statement
Funding Bret Heale was supported by a NLM Postdoctoral Training Grant (T15LM007124). ClinGen is funded by the National Human Genome Research Institute
Footnotes
Clinical Relevance Statement
This paper outlines how the Clinical Genomic Resource (ClinGen) made use of the widely-adopted HL7 Infobutton standard and OpenInfobutton to facilitate integration of genomic knowledge into EHRs. The use of the HL7 standard reduces the barrier for EHR systems to support clinical decision making in genomics by providing clinicians with access to precise, context-aware genomics knowledge. Also, guidance is given to aid online knowledge resources in adopting the standard to streamline access to their genomics content.
Conflicts of Interest
The authors declare that they have no conflicts of interest in the research.
Protection of Human and Animal Subjects
Human and/or animal subjects were not included in the project.
References
- 1.Zook J, Salit M. Chapter 23 – Genomic Reference Materials for Clinical Applications. Pfeifer SK, editor. Clinical Genomics [Internet]. Boston: Academic Press; 2015. p. 393–402. Available from: http://www.sciencedirect.com/science/article/pii/B978012404748800023X [Google Scholar]
- 2.Biesecker LG, Green RC. Diagnostic Clinical Genome and Exome Sequencing. N Engl J Med 2014; 370(25):2418–2425. [DOI] [PubMed] [Google Scholar]
- 3.ACMG Board of Directors. Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med 2015; 17(6):505–507. [DOI] [PubMed] [Google Scholar]
- 4.Leape LL, Bates DW, Cullen DJ, Cooper J, Demonaco HJ, Gallivan T, Hallisey R, Ives J, Laird N, Laffel G. Systems analysis of adverse drug events. ADE Prevention Study Group. JAMA 1995; 274(1):35–43. [PubMed] [Google Scholar]
- 5.Institute of Medicine (U.S.), Committee on Quality of Health Care in America. Crossing the quality chasm a new health system for the 21st century [Internet]. Washington, D.C.: National Academy Press; 2001. Available from: http://site.ebrary.com/id/10032412 [Google Scholar]
- 6.Smith R. Strategies for coping with information overload. BMJ 2010; 341(dec15 2): c7126–c7126. [DOI] [PubMed] [Google Scholar]
- 7.Del Fiol G, Workman TE, Gorman PN. Clinical questions raised by clinicians at the point of care: a systematic review. JAMA Intern Med 2014; 174(5):710–718. [DOI] [PubMed] [Google Scholar]
- 8.Pluye P, Grad RM, Dunikowski LG, Stephenson R. Impact of clinical information-retrieval technology on physicians: A literature review of quantitative, qualitative and mixed methods studies. Int J Med Inf 2005; 74(9):745–768. [DOI] [PubMed] [Google Scholar]
- 9.Isaac T, Zheng J, Jha A. Use of UpToDate and outcomes in US hospitals. J Hosp Med 2012; 7(2):85–90. [DOI] [PubMed] [Google Scholar]
- 10.Bonis PA, Pickens GT, Rind DM, Foster DA. Association of a clinical knowledge support system with improved patient safety, reduced complications and shorter length of stay among Medicare beneficiaries in acute care hospitals in the United States. Int J Med Inf 2008; 77(11):745–753. [DOI] [PubMed] [Google Scholar]
- 11.Del Fiol G, Haug PJ, Cimino JJ, Narus SP, Norlin C, Mitchell JA. Effectiveness of Topic-specific Infobuttons: A Randomized Controlled Trial. J Am Med Inform Assoc JAMIA 2008; 15(6):752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, Plon SE, Ramos EM, Sherry ST, Watson MS, ClinGen. ClinGen -the Clinical Genome Resource. N Engl J Med 2015; 372(23):2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2015; 43(Database issue): D6-D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabana MD, Rand CS, Powe NR, Wu AW, Wilson MH, Abboud PA, Rubin HR. Why don’t physicians follow clinical practice guidelines? A framework for improvement. JAMA 1999; 282(15):1458–1465. [DOI] [PubMed] [Google Scholar]
- 15.Overby CL, Rasmussen LV, Hartzler A, Connolly JJ, Peterson JF, Hedberg RE, Freimuth RR, Shirts BH, Denny JC, Larson EB, Chute CG, Jarvik G P, Ralston JD, Shuldiner AR, Starren J, Kullo IJ, Tarczy-Hornoch P, Williams MS. A Template for Authoring and Adapting Genomic Medicine Content in the eMERGE Infobutton Project. AMIA Annu Symp Proc AMIA Symp AMIA Symp 2014; 2014: 944–953. [PMC free article] [PubMed] [Google Scholar]
- 16.Devine EB, Lee C-J, Overby CL, Abernethy N, McCune J, Smith JW, Tarczy-Hornoch P. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: A mixed methods approach. Int J Med Inf 2014; 83(7):473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overby CL, Tarczy-Hornoch P, Kalet IJ, Thummel KE, Smith JW, Fiol GD, Fenstermacher D, Devine EB. Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice. J Pers Med 2012; 2(4):241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Fiol G, Williams MS, Maram N, Rocha RA, Wood GM, Mitchell JA. Integrating genetic information resources with an EHR. AMIA Annu Symp Proc AMIA Symp AMIA Symp 2006; 904. [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino JJ. Use, usability, usefulness, and impact of an infobutton manager. AMIA Annu Symp Proc AMIA Symp AMIA Symp 2006; 151–155. [PMC free article] [PubMed] [Google Scholar]
- 20.Maviglia SM, Yoon CS, Bates DW, Kuperman G. KnowledgeLink: impact of context-sensitive information retrieval on clinicians’ information needs. J Am Med Inform Assoc JAMIA 2006; 13(1):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Health Information Technology: Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology. 2014. Edition, Final Rule, 171, 2012. Available from: http://www.gpo.gov/fdsys/pkg/FR-2012–09–04/pdf/2012–20982.pdf [PubMed]
- 22.HL7 Standards Product Brief – HL7 Version 3 Standard: Context Aware Knowledge Retrieval Application (“Infobutton”), Knowledge Request, Release 2. Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=208
- 23.Certified Health IT Product List (CHPL) | Policy Researchers & Implementers | HealthIT.gov. Available from: http://www.healthit.gov/policy-researchers-implementers/certified-health-it-product-list-chpl
- 24.Cimino JJ, Elhanan G, Zeng Q. Supporting infobuttons with terminological knowledge. Proc Conf Am Med Inform Assoc AMIA Annu Fall Symp AMIA Fall Symp 1997; 528–532. [PMC free article] [PubMed] [Google Scholar]
- 25.Del Fiol G, Huser V, Strasberg HR, Maviglia SM, Curtis C, Cimino JJ. Implementations of the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) Standard: challenges, strengths, limitations, and uptake. J Biomed Inform. 2012. August;45(4):726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimino JJ, Li J, Bakken S, Patel VL. Theoretical, empirical and practical approaches to resolving the unmet information needs of clinical information system users. Proc AMIA Annu Symp AMIA Symp 2002; 170–174. [PMC free article] [PubMed] [Google Scholar]
- 27.Del Fiol G, Curtis C, Cimino JJ, Iskander A, Kalluri ASD, Jing X, Hulse NC, Long J, Overby CL, Schardt C, Douglas DM. Disseminating context-specific access to online knowledge resources within electronic health record systems. Stud Health Technol Inform 2013; 192: 672–676. [PMC free article] [PubMed] [Google Scholar]
- 28.Cimino JJ, Jing X, Del Fiol G. Meeting the electronic health record “meaningful use” criterion for the HL7 infobutton standard using OpenInfobutton and the Librarian Infobutton Tailoring Environment (LITE). AMIA Annu Symp Proc AMIA Symp AMIA Symp 2012; 2012: 112–120. [PMC free article] [PubMed] [Google Scholar]
- 29.HL7 Standards Product Brief – HL7 Version 2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model (US Realm). Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=23
- 30.Sayers E. A General Introduction to the E-utilities. 2010; Available from: http://www.ncbi.nlm.nih.gov/books/NBK25497/ [Google Scholar]
- 31.RxNorm RESTful Web API User’s Guide. Available from: http://rxnav.nlm.nih.gov/RxNormRestAPI.html
- 32.Cutting E, Banchero M, Cimino JJ, Del Fiol G, Hoffman M, Kawamoto K, Pincus H, Suldiner A, Williams MS, Gurses A, Beitelshees A, Pollin T, Overby CL. User-centered design of multi-gene sequencing panel reports for clinicians. J Biomed Inform 2016; 63: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.