Abstract
No systems have been reported for genetic manipulation of cold-adapted Archaea. Halorubrum lacusprofundi is an important member of Deep Lake, Antarctica (~10% of the population), and is amendable to laboratory cultivation. Here we report the development of a shuttle-vector and targeted gene-knockout system for this species. To investigate the function of acetamidase/formamidase genes, a class of genes not experimentally studied in Archaea, the acetamidase gene, amd3, was disrupted. The wild-type grew on acetamide as a sole source of carbon and nitrogen, but the mutant did not. Acetamidase/formamidase genes were found to form three distinct clades within a broad distribution of Archaea and Bacteria. Genes were present within lineages characterized by aerobic growth in low nutrient environments (e.g. haloarchaea, Starkeya) but absent from lineages containing anaerobes or facultative anaerobes (e.g. methanogens, Epsilonproteobacteria) or parasites of animals and plants (e.g. Chlamydiae). While acetamide is not a well characterized natural substrate, the build-up of plastic pollutants in the environment provides a potential source of introduced acetamide. In view of the extent and pattern of distribution of acetamidase/formamidase sequences within Archaea and Bacteria, we speculate that acetamide from plastics may promote the selection of amd/fmd genes in an increasing number of environmental microorganisms.
The coldest lake known to support microbial growth is Deep Lake in Antarctica where temperatures drop to −20 °C1. Liquid water remains at these temperatures because the lake is hypersaline (~10× marine salinity). It is a closed, isolated marine-derived system that separated from the Southern Ocean ~3,500 years ago2. Genomic, metagenomic and metaproteomic studies have revealed that the lake community has a number of remarkable features: a low complexity community of haloarchaea that support a high level of community wide, intergenera gene exchange2; genome variation and niche adaptation occurring at the level of genera and strains2,3,4; virus-host interactions involving invasion, evasion and adaptation strategies5. The three most abundant members that represent ~72% of the entire lake community have been cultivated and their genome sequences determined: Halohasta litchfieldiae (~44%), DL31 (an undescribed genus; ~18%) and Halorubrum lacusprofundi (~10%)2. An additional species which represents a minor fraction of the lake community has also been isolated and sequenced: DL1 (Halobacterium sp.; ~0.3%)2. By being able to cultivate the abundant members (representing about three-quarters of the lake’s cellular population), the Deep Lake system is unusual compared to most environmental systems where typically <1% can be isolated and grown as axenic cultures in the laboratory6.
Hrr. lacusprofundi is the most readily isolated species, typically representing the majority of isolates forming colonies on plates from Deep Lake, and was the first psychrophilic member of the Archaea formally described7,8. In addition to the studies described above, Hrr. lacusprofundi has been the subject of genomic9,10,11,12,13, protein14,15,16, membrane lipid17, and physiological18,19,20 studies.
To date, Methanococcoides burtonii (isolated from Ace Lake in the same region of Antarctica)21 has served as the main model for examining cold adaptation of Archaea22,23. However, M. burtonii is not readily cultivated on plates and a genetic system for it has not been developed. The development of genetic manipulation systems for Archaea has greatly facilitated understanding of their molecular biology8,24. Transformation systems for haloarchaea were developed in the late 1980s25, largely built around the well-studied species, Haloferax volcanii and Halobacterium salinarum26,27,28,29, leading to the development of molecular genetic tools including selectable markers27,30,31,32,33, shuttle vectors30,34,35,36,37,38,39,40, reporter constructs41,42,43,44, overexpression systems40, and gene knockout systems28,29,31,32.
Antibiotics and resistance genes for Archaea are different to those for Bacteria but can have parallels with those from Eucarya. Mevinolin, which is derived from the fungus Aspergillus, inhibits 3-hydroxy-3-methylglutaryl coenzyme A reductase (HmgA), which is an essential enzyme in the synthesis of isoprenoid lipids in Archaea. An overexpression mutant of this gene from Hfx. volcanii provides resistance to mevinolin38,45. In humans, cholesterol is produced from the mevalonate pathway and statins that target HmgA are used for controlling cholesterol levels. In haloarchaea, overexpression of the gene that encodes HmgA (hmgA) can also provide resistance to the statins fluvastatin, simvastatin and pravastatin33.
In this study we aimed to develop a system for genetic manipulation of Hrr. lacusprofundi ACAM34. We targeted an acetamidase/formamidase (amd/fmd) gene because they have not been experimentally characterized in Archaea. Hrr. lacusprofundi encodes three amd/fmd genes sharing 29–42% identity, and here we define them as amd1 (Hlac_1866), amd2 (Hlac_2016) and amd3 (Hlac_2285). In recent proteomic studies of Hrr. lacusprofundi ACAM34, amd3 was identified as an abundant protein under a variety of growth conditions (Liao Y and Cavicchioli R, unpublished results). Amd/Fmd enzymes catalyze a single-step reaction (hydrolysis of acetamide or formamide) for which substrate (acetamide or formamide) is readily commercially available. We reasoned that a gene knockout would be unlikely to be lethal as the enzyme does not function in central metabolism, and the gene appears to be mono-cistronic, thereby reducing the likelihood of gene inactivation generating polar effects. Our study describes the development of transformation, construction of a shuttle-vector, disruption and phenotypic characterization of an amd3 mutant, and discusses the ecological and evolutionary significance of the findings.
Results
Plasmid construction
The Hfx. volcanii-Escherichia coli shuttle vector pIDJL4039,40 encodes the ColE1 origin of replication and bla gene for ampicillin selection in E. coli, and the Hfx. volcanii pHV2 origin of replication and pyrE2 for selection in pyrimidine auxotrophs and hdrB gene for selection using thymidine auxotrophy in rich media. It also harbors a soluble-modified red-shifted green fluorescent protein (smRS-GFP) under the control of the tryptophan-inducible promoter from the tnaA gene of Hfx. volcanii (p.tnaA) that is flanked by the Hfx. volcanii L11e ribosomal protein gene terminator (t.L11e) and a synthetic terminator (t.Syn) comprising a T track flanked by G/C-rich sequences. In order to construct a plasmid that conferred resistance to statin drugs, pJWID1 was constructed by cloning the up-regulated mutant of the hmgA gene from Hfx. volcanii33 into pIDJL40 (see Methods and Fig. 1). Strains and plasmids used in this study are listed in Table 1, and PCR primers in Table S1.
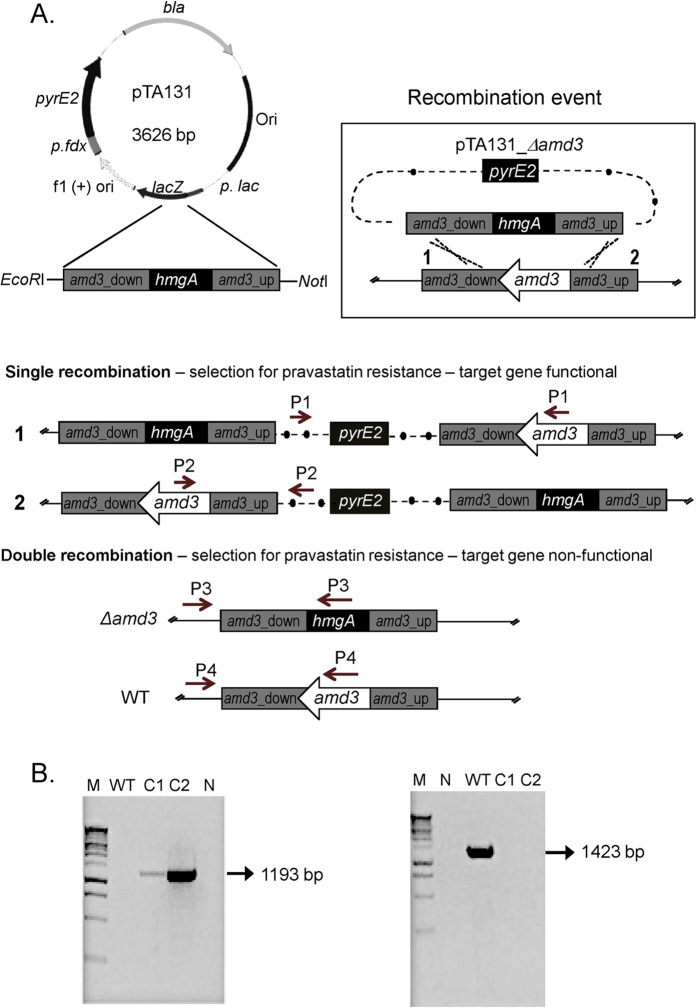
Figure 1. Plasmid maps of pJWID1 and pJWID1_amd3.
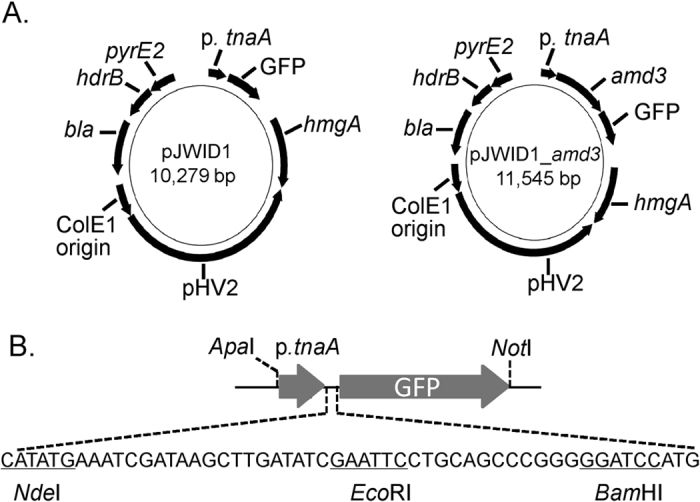
(A) The E. coli-haloarchaea shuttle plasmid pJWID1 is based on pIDJL4039,40 which contains the SmRS-GFP gene under the control of the tryptophan-regulated p.tnaA promoter, and the selection markers bla (ampicillin resistance in E. coli), pyrE2 (for selection in pyrimidine auxotrophs) and hdrB (thymidine auxotrophy in rich media). pJWID1 has the additional selection marker, hmgA (resistance to statins including pravastatin). In pJWID1_amd3 the amd3 gene was inserted at the NdeI and EcoRI sites of pJWID1. (B) Expanded view of the cloning sites in the region containing the p.tnaA promoter and GFP gene.
Table 1. Strains and plasmids.
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| Plasmids | ||
| pTA131 | pBluescript II with BamHI-Xbal I fragment from pGB70 containing pfdx-pyrE2; bla (AmpR) | 31 |
| pIDJL40 | gfp-fusion expression vector derived from pTA962 | 39, 40 |
| pJWID1 | Shuttle vector derived from pTA962 with pyrE2, hdrB, hmgA, bla, SmRS-GFP and pHV2 origin of replication | This study |
| pTA131_Δamd3 | pTA131 with EcoRI-NotI fragment containing hmgA and amd3 flanking regions for gene inactivation | This study |
| pJWID1_amd3 | pJWID1 with NdeI-EcoRI fragment containing amd3 | This study |
| Strains | ||
| E. coli c2925 | dam, dcm strain; used for preparing unmethylated plasmid DNA | New England Biolabs, C2925I |
| Hfx. volcanii DS2 | Source of hmgA gene | 33 |
| Hrr. lacusprofundi ACAM34 | Wild-type | 7 |
| Hrr. lacusprofundi Δamd3 | Hrr. lacusprofundi ACAM34 with amd3 gene disruption | This study |
Development of DNA transformation protocol for Hrr. lacusprofundi
In the initial testing phases for developing transformation, minimal inhibitory concentrations of novobiocin (0.05 μg mL−1), mevinolin (0.01 μg mL−1), simvastatin (0.005 μg mL−1), fluvastatin (0.05 μg mL−1) and pravastatin (1 μg mL−1) were determined and competent cells prepared using a PEG-based procedure26. Initially, the shuttle vectors pJAM20235 and pCBD-sec11b37 were tested, which confer novobiocin resistance from the Haloferax strain Aa2.2 gyrB gene34, but transformants of Hrr. lacusprofundi were not obtained. Success was achieved using PEG-mediated transformation and selection of pJWID1 using 2.5 μg mL−1 pravastatin, with clear differences in resistance observed between transformed (up to 20 μg mL−1) and untransformed cells (Fig. S1). While the hmgA gene can confer resistance to mevinolin, simvastatin and fluvastatin in Hfx. volcanii33, effective resistance was only observed for pravastatin in Hrr. lacusprofundi.
The plasmid was prepared in an E. coli dam dcm strain because plasmid methylation was reported to significantly reduce transformation efficiency in Hfx. volcanii46. Transformation efficiency of Hrr. lacusprofundi increased from 1 ng to 1 μg of DNA with the highest efficiency of ~9 × 107 transformants per μg obtained using 1 μg of pJWID1 (Fig. S2), an efficiency that is similar to transformation of Haloferax strain Aa 2.2 with the novobiocin resistance plasmid, pHK230. Using 10 μg of DNA, the total number of transformants was similar to using 1 μg, translating to ~10-fold decrease in efficiency per μg (Fig. S2). The data indicate there is no benefit to using more than 1 μg of intact plasmid DNA for transforming Hrr. lacusprofundi. Pravastatin resistant colonies were not observed if plasmid DNA was omitted, and the transformation procedure which uses EDTA and PEG600 did not reduce cell viability (data not shown). Plasmid stability was tested by growing transformed cells in liquid medium without antibiotic, plating cells on solid medium in the absence of antibiotic, and assessing the ability of the cells to grow on pravastatin (2.5 μg mL−1) containing plates (Fig. S3). All colonies tested (total 50) were sensitive, indicating the plasmid was readily cured. The relative ease of curing provides potential benefit for experiments requiring plasmid loss. The plasmid was also effectively maintained in strains in the presence of pravastatin (2.5 μg mL−1).
GFP expression from pJWID1
The smRS-GFP gene is under the control of the p.tnaA promoter but the coding sequence is out of frame with the expected start codon (within NdeI) in Hfx. volcanii (Fig. 1)39. The fact that GFP expression occurs (Fig. 2, Fig. S4) demonstrates that the translation machinery in Hrr. lacusprofundi is able to recognize and initiate translation of the GFP ORF in pJWID1. Moreover, expression levels increased with tryptophan concentration (1–3 mM) demonstrating that tryptophan induction also functioned effectively in Hrr. lacusprofundi. This pattern of expression occurred throughout the growth phase from mid-log to mid-stationary phase (data not shown). Expression of GFP was sufficient to readily enable fluorescence microscopy observation of cells (Fig. S4) and quantification of GFP using a fluorescence scanner (Fig. 2). The ability to detect GFP fluorescence in Hrr. lacusprofundi provides the potential for constructing reporter-fusions, tracking plasmid transfer, and performing flow activated cell sorting and GFP-fusion, protein localization experiments (also see Plasmid expression of amd3 below).
Figure 2. Tryptophan induction of GFP expression from pJWID1 in Hrr. lacusprofundi.
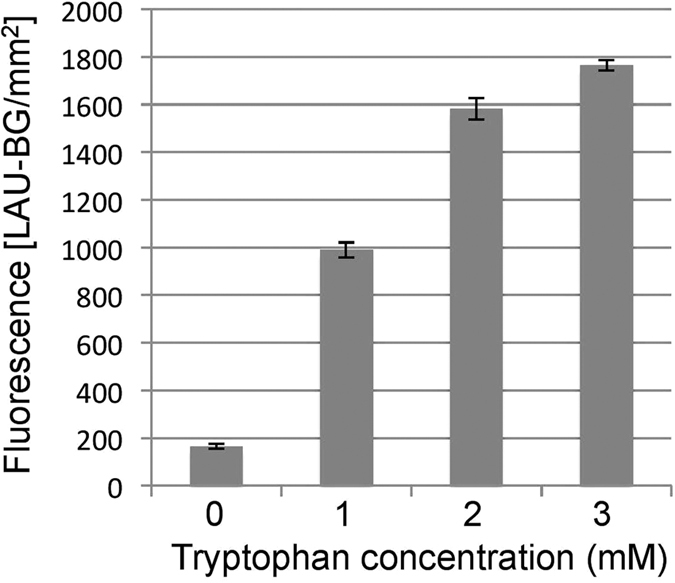
Quantitative measurement of fluorescence emission in response to 0, 1, 2 and 3 mM tryptophan induction. GFP expression levels of Hrr. lacusprofundi harboring plasmid pJWID1 increased with tryptophan concentration. The fluorescence is given in light absorbance units per mm2 [(LAU-BG)/mm2]. Error bars represent standard error of three replicate cultures.
Construction of gene knockouts using plasmid-mediated, gene inactivation
The hmgA gene conferring pravastatin resistance was the only effective antibiotic selection marker we identified (see Development of DNA transformation protocol for Hrr. lacusprofundiabove). To construct a gene knockout, we initially considered developing a pop-in, pop-out approach that uses pyrE auxotrophs32. Hrr. lacusprofundi possesses one orotate phosphoribosyltransferase, pyrE gene (Hlac_0584). However, spontaneous pyrE mutants were not isolated following the passaging of cells through increasing concentrations (200–500 μg mL−1) of 5-fluoroorotic acid (1–2.5-fold above the minimum inhibitory concentration) in the presence of uracil (50 μg mL−1). The approach was therefore abandoned in favor of a strategy that used a suicide plasmid32 and gene exchange with an hmgA inactivated amd3 gene.
To inactivate the amd3 gene, plasmid pTA131_Δamd3 was constructed by cloning the amd3 gene that was inactivated by the insertion of the hmgA gene, into pTA13131(see Methods). Pravastatin resistant (2.5 μg mL−1) colonies of Hrr. lacusprofundi arising from transformation of pTA131_Δamd3 can arise from a single recombination event leading to plasmid integration, or a double recombination event leading to exchange of the wild-type gene for the disrupted gene (Fig. 3A). Genomic DNA extracted from 10 pravastatin resistant colonies was screened by PCR to discriminate between single and double recombination events (see Methods). Two transformants gave a single band using P3 primers, and no product using P1, P2 and P4 primers (Fig. 3B, Fig. S5), which was diagnostic for a double recombination event. The single band (Fig. 3B) matched the size of the product expected for P3 primers (1193 bp), and analysis of the DNA sequence of the PCR product for each of the two transformants confirmed the presence of the hmgA gene within amd3. One of the two strains was designated Hrr. lacusprofundi Δamd3. The frequency of achieving double recombination (2/10 clones) is similar to that previously achieved in Hfx. volcanii for the construction of a pyrE gene disruption (6/16 clones)32.
Figure 3. Construction of amd3 gene disruption.
(A) Pravastatin resistant cells arising from the transformation of pTA131_Δamd3 into wild-type Hrr. lacusprofundi can occur due to a single (plasmid insertion) or double (gene replacement) recombination event. Only the double recombination event produces a defective amd3 gene. Four groups of primers (P1, P2, P3 and P4) were used to determine if single or double recombination events occurred. The region denoted amd3_down corresponds to a 950 bp fragment containing the last 214 bp of amd3 and 736 bp of the downstream sequence, and the region denoted amd3_up corresponds to a 970 bp DNA fragment containing 225 bp of the 5′ coding region of amd3 and 745 bp of the upstream sequence (see Methods). In the section showing double recombination, the gene structure and diagnostic primers are shown for the amd3 disruption (Δamd3) and wild-type strain (WT). (B) A PCR product (1193 bp; left gel image) using the P3 primers was diagnostic of a double recombination event, whereas a PCR product (1423 bp; right gel image) using P4 primers was diagnostic of the wild-type genes being present. (A,B) Lane M, 1 kb DNA ladder; Lane N, no DNA (negative control); Lane WT, untransformed Hrr. lacusprofundi; Lane C1–C2, two strains with double recombination events. C2 was chosen as the Hrr. lacusprofundi Δamd3 strain. The results of other primer sets for C1 and C2 are show in Fig. S5.
Assessment of the phenotype conferred by amd3
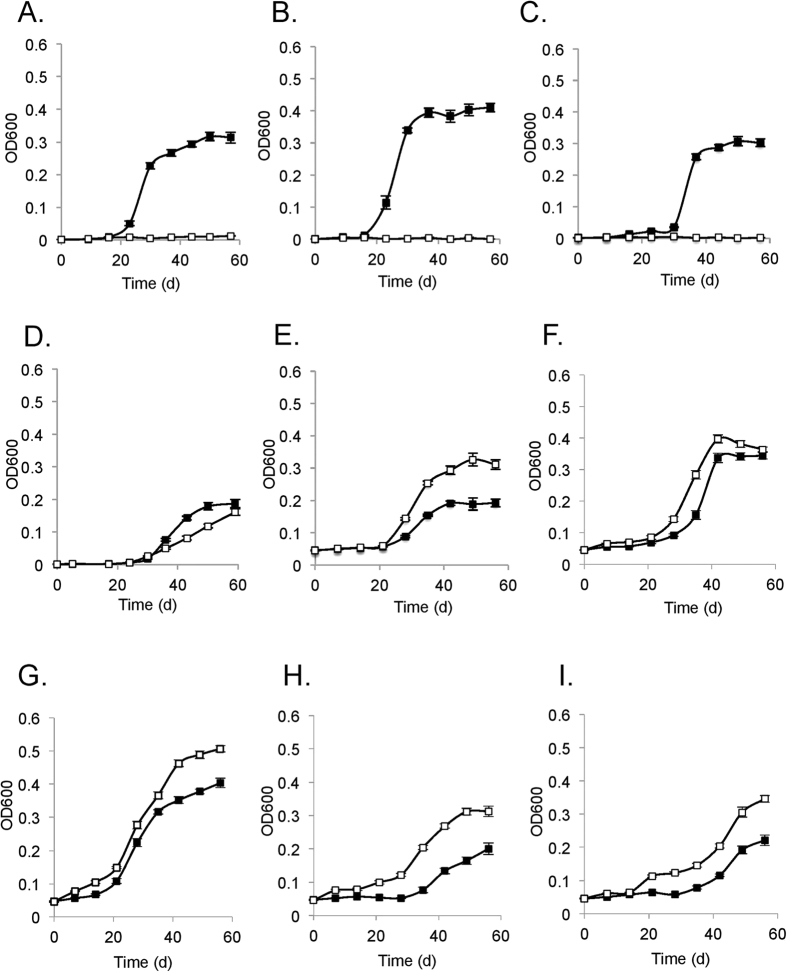
To assess the function of amd3, the wild-type and mutant were grown in media containing various amide substrates (acetamide, formamide, glutamine, asparagine, nicotinamide, urea) and growth assessed using these compounds as a sole carbon, nitrogen, or carbon and nitrogen source in defined media (Fig. 4). The wild-type grew using 10 mM acetamide as a sole source of nitrogen (with 10 mM pyruvate as the carbon source; Fig. 4A), sole source of carbon (with 5 mM ammonium as the nitrogen source; Fig. 4B), or as the sole source of both carbon and nitrogen (10 mM acetamide only; Fig. 4C). In contrast, under the same conditions the mutant was unable to grow (Fig. 4A–C). The phenotypic distinctions between wild-type and mutant were marked, and the results indicate that amd3 is a functional acetamidase gene that enables Hrr. lacusprofundi to grow on acetamide.
Figure 4. Phenotypic characterization of Hrr. lacusprofundi ACAM34 wild-type and Δamd3 mutant strains.
Growth of wild-type (full symbols) and Δamd3 mutant (open symbols) on (A) 10 mM pyruvate and 10 mM acetamide; (B) 5 mM ammonium and 10 mM acetamide; (C) 10 mM acetamide; (D) 10 mM pyruvate and 10 mM formamide; (E) 10 mM pyruvate and 10 mM nicotinamide; (F) 10 mM pyruvate and 10 mM glutamine; (G) 10 mM pyruvate and 10 mM asparagine; (H) 5 mM ammonium and 10 mM glutamine; (I) 10 mM glutamine. Error bars represent standard error of three replicate cultures.
Growth using 10 mM formamide demonstrated formamide could be used by Hrr. lacusprofundi as a sole source of nitrogen (Fig. 4D), but not as a sole source of carbon (data not shown). The Δamd3 mutant was also capable of growth with formamide as a sole source of nitrogen (Fig. 4D). The growth of the mutant lagged behind the wild-type indicating that Amd3 had activity on formamide but Hrr. lacusprofundi possessed other amidases (possibly Amd1 and/or Amd2) that also functioned as a formamidase to enable cells to grow.
Although urea did not support growth of the wild-type or mutant (data not shown), nicotinamide, glutamine or asparagine did support growth as a sole source of nitrogen (Fig. 4E–G). Hrr. lacusprofundi was not capable of growth using nicotinamide or asparagine as a sole source of carbon (data not shown), but both the wild-type and mutant did grow with glutamine as a sole source of carbon, or as a sole source of both carbon and nitrogen (Fig. 4H,I), albeit more slowly than growth on acetamide (Fig. 4C). However, unlike growth on formamide, for nicotinamide, glutamine or asparagine, the growth of the mutant was superior to the wild-type, indicating disruption of amd3 had pleiotropic effects. It is not apparent why inactivation of amd3 would lead to better growth on these substrates. This could possibly occur if Amd3 (in the wild-type strain) produces a metabolite that negatively effects (e.g. allosteric) the regulation of gene expression or activity of the enzymes involved in catabolizing nicotinamide, glutamine, and asparagine.
Plasmid expression of amd3
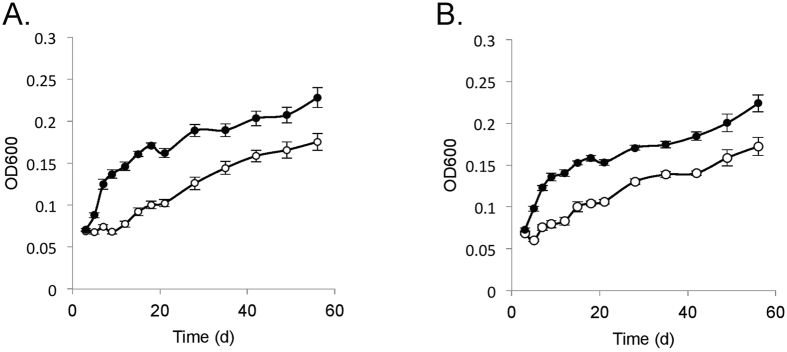
The amd3 gene was cloned in pJWID1 to construct pJWID1_amd3 (Fig. 1). The amd3 gene was cloned from start codon to stop codon downstream of the p.tnaA promoter with the open reading frame terminating prior to the beginning of the GFP open reading frame (i.e. not a translational fusion). Tryptophan (3 mM) was used to induce p.tnaA-mediated amd3 expression in Hrr. lacusprofundi. Tryptophan could not support growth as carbon (with 5 mM ammonium as nitrogen), nitrogen (with 10 mM pyruvate as carbon) or sole source of carbon and nitrogen (data not shown). GFP was expressed from this plasmid although fluorescence levels were lower than for pJWID1 (Fig. S6). Background fluorescence in the absence of tryptophan was also observed for both pJWID1 and pJWID1_amd3, although again it was lower for pJWID1_amd3 (Fig. S6). The data indicate that in the absence of tryptophan, expression from p.tnaA is not completely repressed in Hrr. lacusprofundi (i.e. somewhat leaky expression), and GFP expression can co-occur with expression of cloned genes in pJWID1. To assess the effects of increased gene copies and/or gene expression of amd3 on the ability to utilize acetamide, growth was compared between wild-type Hrr. lacusprofundi harbouring either pJWID1 or pJWID1_amd3 (Fig. 5). Enhanced growth was observed with pJWID1_amd3, particularly when acetamide was the sole carbon or sole carbon and nitrogen source (Fig. 5).
Figure 5. Growth of Hrr. lacusprofundi wild-type harboring pJWID1 or pJWID1_amd3.
Growth of Hrr. lacusprofundi harboring pJWID1 (open symbols) or pJWID1_amd3 (closed symbols) with (A) acetamide as the sole source of carbon; (B) acetamide as the sole source of carbon and nitrogen. Error bars represent the standard error of three replicate cultures.
Acetamidase enzyme activity
Acetamidase enzyme activity was determined (in triplicate) for the wild-type and Δamd3 mutant grown in media containing acetamide plus pyruvate and ammonium (to support growth of the mutant), and wild-type cells harbouring pJWID1 or pJWID1_amd3 grown with acetamide as the sole source of carbon and nitrogen. Activity for the Δamd3 mutant was very low (0.04 ± 0.01 U mg−1) compared to the wild-type (18.1 ± 0.4 U mg−1), and somewhat higher for cells harbouring pJWID1_amd3 (28.6 ± 0.5 U mg−1) compared to pJWID1 (23.8 ± 0.1 U mg−1). These enzyme activity data are consistent with the inability of the Δamd3 mutant to utilize acetamide for growth (Fig. 4A–C), and pJWID1_amd3 enhancing the ability of the wild-type strain to grow on acetamide (Fig. 5).
Characterization of Amd/Fmd sequences in Archaea and Bacteria
To identify Amd/Fmd sequences, UniProtKB was searched using a word search (see Methods). To validate the approach, randomly sampled sequences from the word search were used in a BLAST search of UniprotKB and cross checked to the original set. From a total of 2500 BLAST matches, only two new sequences were obtained, illustrating the word search was effective at retrieving Amd/Fmd sequences. Sequences were clustered using a 90% identity cutoff (OTU0.9) with clusters containing between one and 291 sequences. The 1323 OTU0.9 manually curated clusters consisted of 139 from Archaea and 1184 from Bacteria (Table S2). For Archaea, sequence diversity was highest in Halobacteria (haloarchaea) with 94 clusters and Thermoprotei (14 clusters), and for Bacteria, sequence diversity was highest in Actinobacteria (329), Alphaproteobacteria (245) and Bacilli (172) (Fig. S7).
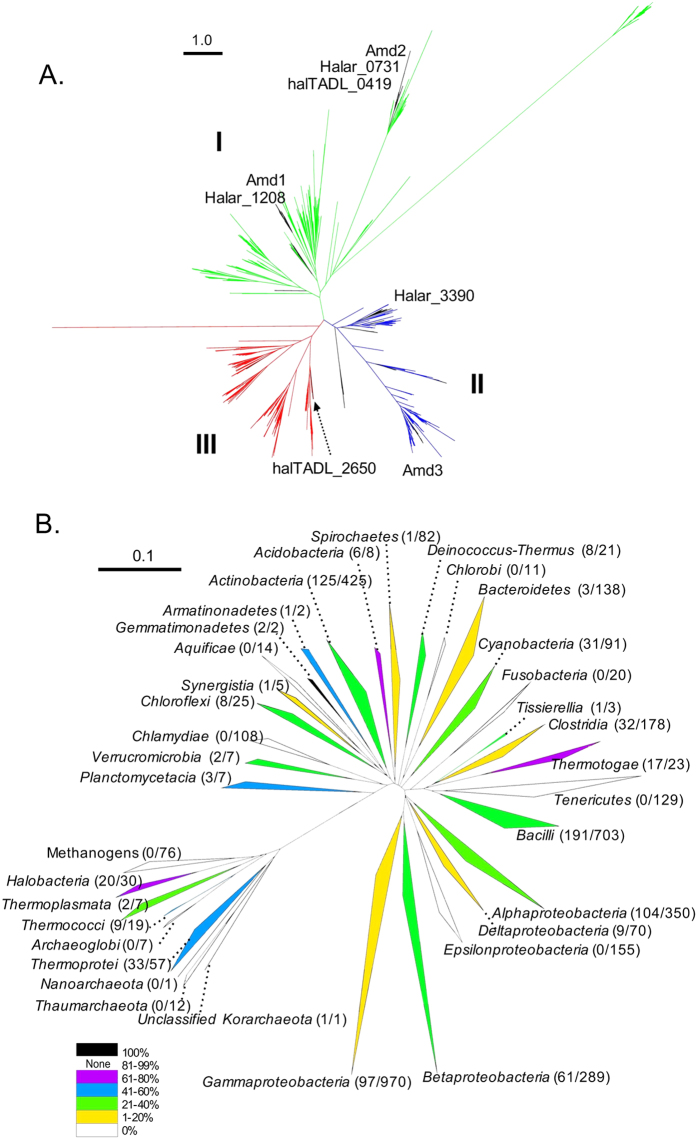
Phylogenetic analyses were performed using one sequence from each cluster. Three distinct Amd/Fmd clades were apparent in trees constructed using all clusters (Fig. 6A, Fig. S8), or a subsample of equal numbers of archaeal and bacterial sequences (Fig. S9). The clade structure was robust (1000 bootstraps) and the three clades rooted (bootstrap 0.996) to a bacterial origin with archaeal sequence ‘islands’ distributed within bacterial clusters. Archaeal sequences were present in each clade, although Clade III consisted almost exclusively of bacterial sequences. Sequences from individual organisms tended to be distributed amongst the clades rather than being confined to a single clade. For the Antarctic haloarchaea, Clade I contained Amd1, Halar_1208, Amd2, Halar_0731 and haltADL_0419; Clade II, Amd3 and Halar_3390; Clade III, haltADL_2650. Amd3 clustered with sequences from an uncultured archaeon and two other haloarchaea (Natronomonas pharaonis and Halopiger xanaduensis) and more distantly to sequences from Clostridia and Bacilli (Fig. S8). Amd1 and Amd2 do not have high sequence identity (42%) and were in subclades of Clade I. The Hht. litchfeldiae Amd/Fmd sequence halTADL_2650 was one of only three archaeal sequences in Clade III, and clustered with sequences from Haloquadratum walsbyi and an unclassified bacterium YEK0303, and more distantly with a Bacillus species (Fig. S8).
Figure 6. Diversity distribution and abundance of Amd/Fmd sequences within Archaea and Bacteria.
(A) Maximum likelihood tree of Amd/Fmd protein sequence clusters (OTU0.9) showing three distinct clades (Clade 1, green; Clade II, blue; Clade III, red) with the root of the tree (bootstrap value, 0.996) being of bacterial origin. Clusters of archaeal sequences are highlighted (black branches). The Deep Lake haloarchaeal Amd/Fmd sequences (Hrr. lacusprofundi: Amd1, Amd2, Amd3; undescribed genus DL31: Halar_0730, Halar_1208, Halar_3390; Hht. litchfeldiae: halTADL_0419, halTADL_2650) were distributed in Clade I (Amd1, Halar_1208, Amd2, Halar_0731 and haltADL_0419), Clade II (Amd3 and Halar_3390) and Clade III (haltADL_2650). Scale bar represents 1 amino acid variation per aligned position. (B) Neighbour-joining tree of 16S rRNA gene sequences for closed genomes from Archaea and Bacteria which possess Amd/Fmd sequences. 16S rRNA gene sequences represented at class or phylum level. Clades are colored according to the proportion of closed genomes that contain Amd/Fmd sequences. The number of Amd/Fmd sequences relative to total number of closed genomes in a clade is shown in parentheses after the name of the clade. Bacterial clades with less than 10 sequences that do not contain Amd/Fmd sequences are not displayed. The scale bar represents 0.1 changes per base position.
Closed genomes were used to quantify the representation (including presence/absence) of Amd/Fmd sequences within taxa (Table S3; Fig. 6B). From 215 archaeal genomes, 65 contained Amd/Fmd sequences at an average 1.28 per genome, with 703 for Bacteria at an average 1.40 per genome (Fig. S10). For Archaea, 20 of 30 closed genomes from haloarchaea contained Amd/Fmd sequences, and for those carrying the genes they had the highest average number (1.75) of genes per genome for Archaea (Fig. S10). The archaeal species which contained the largest number of Amd/Fmd sequences was Haloarcula marismortui which contained four (Table S4). The genomes of Archaeoglobi, Thaumararchaeota Nanoarchaeota and all the methanogens (Methanobacteria, Methanococci, Methanomicrobia, Methanopyri) did not contain Amd/Fmd sequences (Table S4; Fig. 6B). For Bacteria, Amd/Fmd sequences were present in 402 species representing 225 genera (Table S4). The Bacteria that tended to have Amd/Fmd sequences (≥70% of the taxa) were Gemmatimonadetes, Acidobacteria and Thermotogae (Fig. 6B). Starkeya, Modestobacter, Bradyrhizobium, Agromonas and Opitutaceae sp. TAV5 contained ≥5 Amd/Fmd sequences per genome, with the largest number (8) in Starkeya novella (Table S4). Bacterial genomes that lacked Amd/Fmd sequences were from Epsilonproteobacteria, Tenericutes, Chlamydiae, Fusobacteria, Aquificae and Chlorobi (Fig. 6B, Table S2).
Concentration of acetamide in aquatic samples
In order to assess the likelihood of acetamide serving as an environmental growth substrate, the acetamide concentration was determined in water samples from Deep Lake, hypersaline lakes from the Rauer Islands and the Southern Ocean (see Methods). Acetamide was detected at levels similar to the background levels present in Milli-Q water. The presence of 1–2.5 μM acetamide in the controls probably derives from the high-density polyethylene tubes the water samples were stored in. When the standard curve was corrected for background acetamide (Fig. S11), only two ocean samples were above control levels (highest 4.74 μM).
Discussion
Here we report the first procedure for performing gene transfer and gene knockouts for a psychrophilic member of the Archaea. Even for psychrophilic bacteria, few systems are available for genetic manipulation47,48,49,50,51,52. Hrr. lacusprofundi is an important member of the Deep Lake community, representing ~10% of the population throughout the depth of the lake. Because of the relative ease of isolation from environmental samples3, it was the first psychrophilic member of the Archaea formally described7,8. Hrr. lacusprofundi is capable of growth in the laboratory at 4 °C, and very slowly at 0 or −1 °C7,18. Intergenera gene exchange and population level genetic variation is a feature of the Deep Lake community2,3,5. The development of genetic manipulation of strain ACAM34 provides considerable scope for probing mechanisms of adaptation and gene exchange in this species.
Physiological and ecological significance of acetamide
The ability of Hrr. lacusprofundi to utilize acetamide as a sole source of both carbon and nitrogen, but formamide only as a sole source of nitrogen, indicates that cells can utilize ammonium released from either source, but that only acetate and not formate can be used as a carbon source. This is consistent with genomic evidence for Hrr. lacusprofundi, which includes a capacity for acetate assimilation via the glyoxylate cycle3,53, but no evidence for known formate assimilation pathways. This is further corroborated by growth assessments which show that Hrr. lacusprofundi can utilize acetate as a sole source of carbon3,7. The enzyme activity data and pronounced phenotype of the Hrr. lacusprofundi Δamd3 strain and relatively fast growth of the wild-type strain on acetamide illustrates that Amd3 functions effectively in substrate conversion and is unlikely to limit ammonium and acetate utilization pathways. Deamidation of formamide may be calatalyzed by an Amd3 homolog; possibly Amd1 and/or Amd2. The Hrr. lacusprofundi genome encodes genes for a putative asparaginase (COG0252: Hlac_2272) and nicotinamidase (COG1335: Hlac_2101, Hlac_2473) for the deamidation of asparagine and nicotinamide, respectively. A gene for glutaminase is not apparent in the Hrr. lacusprofundi genome; but glutamine can be assimilated (via glutamate) using glutamate synthase (GOGAT). Ammonium liberated by amidases can be assimilated via the glutamine synthetase-GOGAT cycle, which is present in Hrr. lacusprofundi3.
The role of amides such as acetamide and formamide as sources of carbon and/or nitrogen has been examined in fungi54, algae55,56 and Bacteria, including species of Pseudomonas and Burkholderia57,58,59, Alcaligenes60, Nocardia61, and Mycobacterium62. In the marine environment amides have been described as a potential nutrient source, and speculated to be derived from photodegradation of dissolved organic matter, from atmospheric input, or as a byproduct of an unspecified degradative metabolic pathway56. Acetamide is known to be generated through the pyrolytic cleavage of N-acetylated biopolymers such as chitin63 and peptidoglycan64, and constitutes a dominant N-containing product from the fragmentation of soil organic matter, sewage sludge, and chitin-containing biomass63,65,66,67. Acetamide is also a byproduct of the catabolism of nitroimidazole antibiotics by bacteria, through the reductive cleavage of the imidazole ring68,69,70. In the Antarctic environment, acetamide would not be expected to be produced by thermal degradation (such as by fire or volcanic eruptions) of natural biopolymers. Acetamide may be generated endogenously within cells as a transient intermediate from the breakdown of natural imidazoles or other compounds. Our analyses found acetamide in water samples approximated relatively high (μM) background concentrations. If a pool of acetamide is not maintained in cells (and released during cell lysis), whatever acetamide becomes available in the environment is likely to be rapidly metabolized. In the ocean, dissolved organic carbon (e.g. glucose) and dissolved free amino acids are in the ~1–20 nM range and can have a high flux through the labile pool of nutrients71,72, so if acetamide is metabolized as actively, concentrations may not exceed nM levels.
Evolution of the Amd/Fmd sequences in Archaea and Bacteria
Our study appears to be the first to examine the diversity and phylogeny of Amd/Fmd sequences in Archaea and Bacteria. The analyses revealed the presence of three distinct clades of Amd/Fmd sequences which are predicted to originate from Bacteria with subsequent dissemination to Archaea (Fig. 6A). Experimental data is available for very few representatives of the tree. Our data for Amd3 indicate that it effectively converts acetamide. The only other experimentally studied enzyme from Cluster II, Uniprot KB ID Q50228 from Methylophilus methylotrophus, has highest activity on formamide and relatively low activity on acetamide, propionamide, butyramide, and acrylamide73,74. The Cluster I enzyme, UniprotKB ID O25836 from Helicobacter pylori, was reported to only catalyze the conversion of formamide75. While limited, the experimental data (particularly for Cluster II) indicate that functional specialization (substrate preference for acetamide vs formamide) has occurred following lineage-specific gene acquisition.
The highest number of Amd/Fmd sequences occurs in Bacteria, including Modestobacter multiseptatus, Pseudonocardia dioxanivorans, Agromonas oligotrophica, Bradyrhizobium spp., Opitutaceae sp. and Starkeya novella. Bradyrhizobium spp., A. oligotrophica, M. multiseptatus and S. novella are soil bacteria76,77,78,79,80; P. dioxanivorans was isolated from industrial sludge81,82; and the putative methylotroph Opitutaceae sp. TAV5 was isolated from the hindgut of a wood-feeding termite83. Gene duplication and specialization may enhance the competitiveness of these bacterial taxa that are capable of growth by scavenging nutrients in low nutrient environments (especially soil), including growth on single-carbon compounds.
While Amd/Fmd sequence distribution is wide-spread in Archaea and Bacteria, specific taxonomic groups lack them. These tend to be anaerobic or facultative anaerobic microorganisms from aquatic or terrestrial environments (Archaeoglobi, methanogens, Epsilonproteobacteria, Aquificae and Chlorobi) or parasites of animals and plants (Tenericutes, Chlamydiae and Fusobacteria). The only aerobic group lacking Amd/Fmd sequences are Thaumarchaeota which possess an ability to generate energy via ammonia-oxidation84,85. The fact that methanogens lack these genes is notable for several reasons. Firstly, many methanogens are capable of growth on single-carbon compounds (CO2, methanol, formate). Secondly, acetamide is a potential source of acetate for acetoclastic methanogens. Thirdly, 76 methanogen genomes are available that represent species from diverse environments (e.g. deep-sea hydrothermal vents, Antarctic lakes, rice paddies, human and ruminant gastrointestinal tracts), and some are characterized as supporting a high level of horizontal gene transfer86,87,88. The lack of Amd/Fmd sequences in methanogens is suggestive of selection against retention of the genes rather than the existence of a barrier to acquiring the genes.
The Antarctic haloarchaea support a high level of intergenera gene exchange, including long stretches (up to 35kb) of identical DNA2. However, the three dominant genera have distinct metabolisms enabling them to utilize different lake substrates and providing selection for sympatric speciation2,3,4. The clustering of Amd1 with Halar_1208, and Amd2 and Halar_0731 with haltADL_0419 in Clade I may have arisen from intergenera transfer within the community in Deep Lake. However, the presence of Amd3 and Halar_3390 on distinct branches of Clade II, and halTADL_2650 in Clade III which contains very few archaeal sequences, suggests the genes arose from acquisition and selection for specific enzymatic properties as a means of fulfilling specific physiological function(s).
The pattern and extent of distribution of Amd/Fmd sequences within Archaea and Bacteria illustrates the evolutionary significance of the genes. The demonstration that acetamide can sustain microbial growth of Hrr. lacusprofundi as a sole source of carbon and nitrogen, is intriguing, and the sources and concentrations of acetamide in the Antarctic environment need to be accurately determined in order to consider the environmental cues controlling the selection of these genes. In industry, acetamide is used as a plasticizer and solvent89,90, and as a component of pesticides91, thereby providing avenues for acetamide to be introduced into the global environment as an industrial contaminant. Of great concern is the enormous build-up of environmental plastics92,93,94, as these potentially provide a significant anthropocentric source of acetamide. The selection of genetic variants with new capacities to utilize unnatural substrates has been documented for both atrazine pesticides and poly(ethylene terephthalate) plastics95,96,97. Recently, a Rhodococcus sp. was isolated which is capable of utilizing N,N-dimethylacetamide (DMAC) and its degradation product acetamide as sole sources of carbon and nitrogen98. DMAC is an acetamide-based compound that has become an important environmental pollutant that is widely used as an agrochemical and in a wide range of industries98. As the amd/fmd genes are already naturally occurring in a wide range of lineages, enhancing a community’s capacity to breakdown acetamide only requires dissemination and stable inheritance of one gene. As a result, we hypothesize that amd/fmd genes will arise in a greater number of microbial lineages and in a higher proportion of microbial communities that are increasingly exposed to introduced acetamide (e.g. plastics, pesticides, industrial waste).
Methods
Culture conditions, strains, plasmids and PCR primers
Hrr. lacusprofundi ACAM34 was grown in artificial Deep Lake vitamin succinate broth (ADLVSB)7 at 30 °C (see Supplementary information). The phenotype of wild-type and mutant strains was tested using various defined carbon and nitrogen substrates in modified DBCM2 medium99 which had yeast extract and peptone omitted. Acetamide, formamide, glutamine, asparagine, nicotinamide, and urea were used as amide substrates. Pyruvate was used a carbon source and ammonium as a nitrogen source. All carbon substrates were used a 10 mM, nitrogen substrates at 5 mM, and substrates used as both a source of carbon and nitrogen at 10 mM. E. coli strain c2925 (dam, dcm; New England Biolabs) was used to prepare unmethylated plasmid DNA for transformation of Hrr. lacusprofundi, and was grown in Luria-Bertani medium with 100 μg mL−1 ampicillin.
Construction of shuttle vector pJWID1
Plasmid pJWID1 was constructed by cloning the up-regulated mutant of the hmgA gene from Hfx. volcanii33 into the Hfx. volcanii-E. coli shuttle vector pIDJL4039,40. Primers pJ_For and pJ_Rev were initially used to amplify the hmgA region from Hfx. volcanii DS2 and the product was used as template for a second PCR using the same reverse primer, and forward primer pJ_For_M that introduced two point mutations that up-regulate the promoter of hmgA. The resulting fragment was digested with PstI and pIDJL40 was digested with NsiI and alkaline phosphatase to create compatible ends for ligation, and ligation performed to generate plasmid pJWID1 (Fig. 1); the orientation and expected sequence of hmgA was verified by sequencing. A red fluorescence protein (mCherry) version of the plasmid (pJWID4) was also constructed (data not shown).
Transformation, plasmid stability and GFP expression
E. coli was transformed using a standard heat-shock protocol100. For Hrr. lacusprofundi, a polyethylene glycol 600 (PEG600) procedure developed for Hfx. volcanii101 was used with modifications. Compositional changes were made to buffered salt water, regeneration and transformation dilution solutions (the full procedure is in Supplementary information). Cells were harvested for preparation of competent cells after growth in ADLVSB medium reached late-log phase: an optical density (OD600) of 0.8–1.0 (1.3–3.6 × 108 cells mL−1). Transformants were selected on ADLVSB medium supplemented with 0, 0.05, 0.5, 1, 2.5, 5, 7.5, 10, 15 or 20 μg mL−1 of pravastatin (from 5 mg mL−1 stock dissolved in MilliQ-water). Plates were incubated in sealed plastic bags (with wet tissue to maintain moisture) at 30 °C for 15 d. To test transformation efficiency, cells were transformed with 1 ng, 10 ng, 100 ng, 1 μg or 10 μg of plasmid DNA. Plasmid stability was assessed by growing cells (inoculated 1:100) in liquid ADLVSB medium without antibiotic until late log phase, plating cells on solid medium in the absence of antibiotic, and testing the ability of 50 colonies to grow on solid medium containing 2.5 μg mL−1 pravastatin. To assess GFP expression, pJWID1 transformants were grown to mid-log phase in liquid ADLVSB medium supplemented with 2.5 μg mL−1 pravastatin, and 25 mL aliquots supplemented with 0, 1, 2 or 3 mM tryptophan. After further growth, cells were diluted with basal salts (3 M NaCl, 150 mM MgSO4, 40 mM KCl) as required to bring all cultures to OD 600, 0.2. The fluorescence of samples (100 μL) in 96 well plates was quantified using a Fujifilm FLA-5000 Fluorescent Image Analyzer (Fujifilm, Tokyo, Japan) with a 473 nm excitation laser and Fujifilm LPB filter using Fujifilm Science Lab Image Gauge Ver 4.0 software. Basal salt solution was used as a blank and assessments were performed in triplicate, and standard error calculated. Cell fluorescence was also viewed and photographed with a digital microscope (Olympus BX61 microscopy with DP71 camera; Olympus, Tokyo, Japan) using bright-field or fluorescence-field imaging (Olympus WIBA filter).
Construction of the amd3 gene deletion strain
To construct pTA131_Δamd3, an in-fusion high efficiency directional (HD) cloning system (Clontech/Takara Bio, Mountain View, CA, USA) was used for cloning multiple fragments in a single reaction. The plasmid pTA13131 was digested with EcoRI and NotI. The hmgA gene including promoter sequence was amplified by PCR from pJWID1 using primers hmgA_For and hmgA_Rev. A 970 bp DNA fragment containing the 225 bp of the 5′ coding region of amd3 and 745 bp of the upstream sequence was PCR amplified. The primers used, Acet_up_For15 and Acet_up_Rev15_h, contain a 15 bp extension complementary to the NotI end of digested pTA131 and a 15 bp extension complementary to the 3′ end of hmgA, respectively. Similarly, a 950 bp fragment containing the last 214 bp of amd3 and 736 bp of the downstream sequence was amplified by PCR with primers Acet_down_For15_h and Acet_down_Rev15, which contain a 15 bp extension complementary to the 5′ end of hmgA and a 15 bp extension complementary to the EcoRI end of digested pTA131. These three fragments and the linearized vector pTA131 (EcoRI/NotI digestion) were ligated simultaneously according to the manufacturer’s instructions. The pTA131_Δamd3 plasmid was transformed into competent E. coli c2925 and non-methylated plasmid DNA prepared. The plasmid pTA131_Δamd3 was transformed into Hrr. lacusprofundi and plated on ADLVSB medium containing 2.5 μg mL−1 pravastatin, and colonies screened by PCR using primers diagnostic for single or double recombination events (Table S1, Fig. 3B, Fig. S5).
Plasmid expression of amd3
To control the expression of amd3, the gene was cloned to pJWID1 under the control of the p.tnaA promoter39 using the HD cloning system as described above. The amd3 gene was amplified from Hrr. lacusprofundi genomic DNA using primers pJ_2285_FW and pJ_2285_RV, which contain 15 bp extensions complementary to NdeI and EcoRI ends of digested pJWID1. The fragment was ligated into linearized vector pJWID1 (NdeI/EcoRI) to generate pJWID1_amd3. The plasmid was sequenced to confirm the correct insertion event. The plasmid pJWID1_amd3 (or pJWID1 as negative control) extracted from E. coli c2925 was transformed into Hrr. lacusprofundi Δamd3 and plated on ADLVSB medium supplemented with 40, 60, 80, 100, 120 or 150 μg mL−1 pravastatin. The plasmids were also transformed into the wild-type Hrr. lacusprofundi strain with selection on 2.5 μg mL−1 pravastatin.
Acetamidase enzyme activity
Hrr. lacusprofundi harboring pJWID1 or pJWID1_amd3 were inoculated 1:100 grown and grown for 40 d at 30 °C in 50 mL DBCM2 medium supplemented with 10 mM acetamide as the sole source of carbon and nitrogen plus 3 mM tryptophan to induce p. tnaA-mediated amd3 expression. In addition, the wild-type and Δamd3 strain were inoculated 1:100 grown and grown for 40 d at 30 °C in 50 mL DBCM2 medium supplemented with 10 mM pyruvate, 1 mM ammonium and 10 mM acetamide, with the pyruvate and ammonium provided to support growth of the Δamd3 strain. Cells were pelleted by centrifugation for 20 min at 4,500 × g, washed three times in DBCM2 salt solution, and suspended in DBCM2 salt buffer supplemented with 2 mM EDTA (pH 7.2) and 0.4 mM phenylmethanesulphonylfluoride (PMSF). The suspensions were ultrasonically disrupted on ice using a Branson Sonifier 250 (Branson Ultrasonics, Danbury, CT) with the probe output set at 20% amplitude for five periods of 40 s (pulse cycle of 0.5 s on and 0.5 s off), with 40 s cooling on ice between periods of sonication to prevent excessive sample heating. The sonicate was centrifuged at 4,500 × g for 5 min to remove cell debris, and the cell free extract (supernatant) filtered through a 15 mL Amicon centrifugal concentration unit (Millipore, 25 Billerica, MA) with a 3 kDa cutoff by centrifugation at 5,000 × g, with three subsequent buffer exchanges with DBCM2 salt solution to remove EDTA and PMSF and the concentrate (~500 μL) stored at −80 °C until needed. Protein concentration was determined at 562 nm with a microplate reader using Thermo Scientific Pierce BCA Protein Assay Kit (Product No. 23225) according to manufacturer’s instructions. Acetamidase activity was determined by measuring the release of ammonium based on a phenol-hypochlorite ammonia detection protocol102. A standard reaction mixture (100 μL) containing 50 mM KH2PO4–K2HPO4 (pH 7.6), 150 mM NaCl, 10 mM acetamide and 100 μg of crude enzyme (cell free extract) was incubated at 30 °C for 1 h and the reaction terminated by the addition of 350 μL of reagent A (0.59 M phenol, 1 mM sodium nitroprusside). The color was developed by the addition of 100 μL of reagent B (2 M sodium hydroxide, 0.11 M sodium hypochlorite), with the mixture maintained at 30 °C in the dark for 20 min. Absorbance was measured at 600 nm with a microplate reader. The enzyme assays were performed in triplicate and a negative control that contained all reagents but no added cell free extract was included. Enzyme activity was calculated from a standard curve constructed using 0, 0.25, 0.5, 1, 2, 4, 6 and 8 mM NH4Cl. One unit of acetamidase activity was defined as the amount of enzyme that hydrolyzed acetamide to release 1 μM NH3 per minute under assay conditions.
Phylogenetic analysis of Amd/Fmd sequences
Amd/Fmd protein sequences from Archaea and Bacteria were retrieved with a protein name search using “acetamidase”, “formamidase”, “Amd” and “Fmd” from UniProtKB database (18 March, 2016), and sequences recovered clustered with an identity cutoff of 90% (OTU0.9). Representative sequences of each cluster were interrogated and manually filtered to remove irrelevant sequences (e.g. transcriptional regulator of acetamidase genes). Six of the Deep Lake haloarchaeal Amd/Fmd sequences were in the search (Hrr. lacusprofundi: amd1, amd2, amd3; undescribed genus DL31: Halar_0730, Halar_1208, Halar_3390) but the two from Hht. litchfeldiae (halTADL_0419, halTADL_2650) were not and were manually added to the set and aligned against existing clusters using Clustal X 2.0103. Phylogenetic trees were constructed using Fasttree104 using the maximum likelihood method and the robustness of phylogeny tested using 1000 bootstraps. To test the ability of the word search to recover Amd/Fmd sequences, 50 sequences were randomly selected and BLAST used with each sequence against UniProtKB, and the top 50 hits from each BLAST search were cross-checked against the original datasets. To evaluate the effect of bias in the number of bacterial vs archaeal Amd/Fmd sequences, bacterial clusters were randomly subsampled to the same number of archaeal clusters, and the new dataset used for tree construction. To assess Amd/Fmd sequences in closed genomes, the 215 archaeal and 3872 bacterial genomes in Integrated Microbial Genomes (IMG) were searched (22 March, 2016) using the IMG Gene Cassette Search tool for the Pfam 03069 motif (includes acetamidase and formamidase). The 16S rRNA gene sequences corresponding to the archaeal and bacterial genomes were extracted from Silva non-redundant reference SSU database using genome names or retrieved manually from IMG. The 16S rRNA gene sequences were aligned using SINA aligner105 and classified with the least common ancestor method based on the different taxonomies hosted by SILVA. Common gaps in alignments were removed, and phylogenetic trees were constructed using the neighbor joining method in ARB106. The sequences were clustered according to their class or phylum 16S rRNA gene taxonomic classifications.
Acetamide concentration of water samples
Water was collected in acid or ethanol washed, high-density polyethylene bottles from Deep Lake (12/2008; 11/2014; 12/2014), Rauer Islands lakes (01/2015) and the Southern Ocean (10/2008; 12/2008), and cryogenically stored at −80 °C. A total of 12 different samples from these systems, plus controls (100 μL each in duplicate) were dispensed into Pyrex screw cap glass culture tubes (Kimble, ThermoFisher, Sydney, Australia). An acetamide (Sigma, USA) standard curve was prepared in the 0–20 μg mL−1 range. To all samples, controls and standards (for standard curve), 5 μL (0.1 μg μL−1) of stable isotope labeled 13C2, 15N-acetamide internal standard (Medical Isotopes Inc., NH) was added. All samples to be analysed were dried in a vacuum centrifuge at ambient temperature (Savant speedvac, ThermoFisher, Sydney, Australia). To maximize recovery, over-drying was avoided and the hypersaline samples consisted of a moist slurry of crystals. Neat dichloromethane (1 mL) was added to each sample, control and standard, and shaken upright on a mixer platform (Intelli-Mixer, POCD Scientific, Sydney, Australia) for ~2 h at ambient temperature. The liquid extract was transferred to clean glass culture tubes, taking care not to pick up salt crystals, and dried in a vacuum centrifuge (ambient temperature, ~10 min). All samples were resuspended in 100 μL dichloromethane, vortexed briefly and transferred to GC vials with glass inserts. Stable isotope dilution GC/MS quantification of acetamide in lake water was carried out as described previously107 with specific modifications. Analyses were performed on a Hewlett-Packard 6890 plus gas chromatograph interfaced with an Agilent Technologies 5973 mass selective detector. A 4-mm-i.d. straight-walled silanized glass liner containing quartz wool was installed in the injection port and samples injected in the splitless mode (2 μL injection volume). Chromatography of underivatized acetamide was performed on a fused silica capillary column (free fatty acids column; Agilent J&W GC columns: HP-FFAP 50 m length, 0.2 mm id, 0.33 μm film thickness). Helium (BOC Gases, ultra-high purity) was used as the carrier gas at a flow rate of 1.4 mL min−1. The GC/MS conditions were as follows: injector temperature, 230 °C; transfer line, 230 °C; initial oven temperature, 40 °C (for 4 min); then increased to 190 °C at 5 °C min−1; then to 230 °C at 30 °C min−1, with a total run time of 45 min. Mass spectrometry analysis was performed using electron impact ionization mode, and conditions were as follows: electron energy, 70 eV; ion source temperature, 230 °C, MS Quad temperature 150 °C. Single ion monitoring was used to detect the molecular ions of acetamide (59 m/z) and the 13C2, 15N-acetamide internal standard (62 m/z). Peak areas of these ions were integrated and peak area ratios calculated (Chemstation software, RTE integrator, Agilent Technologies Inc, Sydney, Australia).
Additional Information
How to cite this article: Liao, Y. et al. Developing a genetic manipulation system for the Antarctic archaeon, Halorubrum lacusprofundi: investigating acetamidase gene function. Sci. Rep. 6, 34639; doi: 10.1038/srep34639 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council [DP150100244]. YL was funded by the China Scholarship Council (File No. 201206910027). GC-MS results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of the University of New South Wales. This work was undertaken using infrastructure provided by NSW Government co-investment in the National Collaborative Research Infrastructure Scheme. Subsidized access to this facility is gratefully acknowledged. We thank Sarah Payne and Alyce Hancock for collecting samples in Antarctica.
Footnotes
Author Contributions R.C. conceived the research. Y.L. performed all the experiments except J.C.W., I.G.D. and P.M.G.C. designed plasmid pJWID1 and J.C.W. constructed pJWID1, M.J. constructed the phylogenetic tree, and A.P. performed the mass spectrometry. Y.L., R.C. and T.J.W. designed the other experiments performed by Y.L. and analyzed the data and interpreted the findings. R.C., Y.L. and T.J.W. wrote the manuscript. All authors vetted the manuscript and viewed the final version.
References
- Cavicchioli R. Microbial ecology of Antarctic aquatic systems. Nature. Rev. Microbiol. 13, 691–706 (2015). [DOI] [PubMed] [Google Scholar]
- DeMaere M. Z. et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl. Acad. Sci. USA 110, 16939–16944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. J. et al. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant haloarchaea. ISME. J. 8, 1645–1658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschitschko B. et al. Ecophysiological distinctions of haloarchaea from a hypersaline Antarctic lake determined using metaproteomics. Appl. Environ. Microbiol. 82, 3165–3173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschitschko B. et al. Antarctic archaea–virus interactions: metaproteome-led analysis of invasion, evasion and adaptation. ISME. J. 9, 2094–2107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. T. & Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346 (1985). [DOI] [PubMed] [Google Scholar]
- Franzmann P. D. et al. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst. Appl. Microbiol. 11, 20–27 (1988). [Google Scholar]
- Cavicchioli R. Archaea—timeline of the third domain. Nature. Rev. Microbiol. 9, 51–61 (2011). [DOI] [PubMed] [Google Scholar]
- Goo A. Y. et al. Low-pass sequencing for microbial comparative genomics. BMC Genomics. 5, 3 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodt A., Lurie-Weinberger M. N. & Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biol. Direct. 6, 65–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L. K. et al. The immune system of halophilic archaea. Mob. Genet. Elements 2, 228–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L. K. et al. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system IB. RNA Biol. 10, 865–874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C. M. & Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266 (2013). [DOI] [PubMed] [Google Scholar]
- Giaquinto L. et al. Structure and function of cold shock proteins in Archaea. J. Bacteriol. 189, 5738–5748 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S., Capes M. D., Karan R. & DasSarma P. Amino acid substitutions in cold-adapted proteins from Halorubrum lacusprofundi, an extremely halophilic microbe from Antarctica. PLoS One 8, e58587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan R., Capes M. D., DasSarma P. & DasSarma S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 13, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. A. et al. Unsaturated diether lipids in the psychrotrophic archaeon Halorubrum lacusprofundi. Syst. Appl. Microbiol. 28, 19–26 (2005). [DOI] [PubMed] [Google Scholar]
- Reid I. N. et al. Terrestrial models for extraterrestrial life: methanogens and halophiles at Martian temperatures. Int. J. Astrobiology 5, 89–97 (2006). [Google Scholar]
- Fröls S., Dyall-Smith M. & Pfeifer F. Biofilm formation by haloarchaea. Environ. Microbiol. 14, 3159–3174 (2012). [DOI] [PubMed] [Google Scholar]
- Fröls S. Archaeal biofilms: widespread and complex. Biochem. Soc. Trans. 41, 393–398 (2013). [DOI] [PubMed] [Google Scholar]
- Franzmann P. D., Springer N., Ludwig W., De Macario E. C. & Rohde M. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst. Appl. Microbiol. 15, 573–581 (1992). [Google Scholar]
- Cavicchioli R. Cold-adapted archaea. Nature. Rev. Microbiol. 4, 331–343 (2006). [DOI] [PubMed] [Google Scholar]
- Najnin T. et al. Characterization of a temperature-responsive two component regulatory system from the Antarctic archaeon, Methanococcoides burtonii. Sci. Rep. 6, 24278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Albers S. V., Atomi H. & Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608 (2011). [DOI] [PubMed] [Google Scholar]
- Charlebois R. L., Lam W. L., Cline S. W. & Doolittle W. F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 84, 8530–8534 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C. & Doolittle W. F. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35, 148–152 (1989). [DOI] [PubMed] [Google Scholar]
- Peck R. F., DasSarma S. & Krebs M. P. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35, 667–676 (2000). [DOI] [PubMed] [Google Scholar]
- Peck R. F. et al. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. 276, 5739–5744 (2001). [DOI] [PubMed] [Google Scholar]
- Wang G., Kennedy S. P., Fasiludeen S., Rensing C. & DasSarma S. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186, 3187–3194 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M. L. & Dyall-Smith M. L. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172, 756–761 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Ngo H. P., Mevarech M. & Lloyd R. G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan-Banin G., Ortenberg R. & Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185, 772–778 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendoloski D., Ferrer C. & Dyall-Smith M. L. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147, 959–964 (2001). [DOI] [PubMed] [Google Scholar]
- Holmes M. L. & Dyall-Smith M. L. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J. Bacteriol. 173, 642–648 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczowka S. J. & Maupin-Furlow J. A. Subunit topology of two 20S proteasomes from Haloferax volcanii. J. Bacteriol. 185, 165–174 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irihimovitch V. & Eichler J. Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278, 12881–12887 (2003). [DOI] [PubMed] [Google Scholar]
- Fine A., Irihimovitch V., Dahan I., Konrad Z. & Eichler J. Cloning, expression, and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii. J. Bacteriol. 188, 1911–1919 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaseio U. & Pfeifer F. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc. Natl. Acad. Sci. USA 87, 6772–6776 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin I. G. et al. CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Barak S., Liddell S., Wardell K. & Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter C. J. & Maupin-Furlow J. A. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl. Environ. Microbiol. 70, 7530–7538 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I. & Pfeifer F. Use of GFP-GvpE fusions to quantify the GvpD-mediated reduction of the transcriptional activator GvpE in haloarchaea. Arch. Microbiol. 195, 403–412 (2013). [DOI] [PubMed] [Google Scholar]
- Nomura S. & Harada Y. Functional expression of green fluorescent protein derivatives in Halobacterium salinarum. FEMS Microbiol. Lett. 167, 287–293 (1998). [DOI] [PubMed] [Google Scholar]
- Patenge N., Haase A., Bolhuis H. & Oesterhelt D. The gene for a halophilic β-galactosidase (bgaH) of Haloferax alicantei as a reporter gene for promoter analyses in Halobacterium salinarum. Mol. Microbiol. 36, 105–113 (2000). [DOI] [PubMed] [Google Scholar]
- Lam W. L. & Doolittle W. F. Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. J. Biol. Chem. 267, 5829–5834 (1992). [PubMed] [Google Scholar]
- Holmes M. L., Nuttall S. D. & Dyall-Smith M. L. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 173, 3807–3813 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duilio A., Tutino M. L. & Marino G. Recombinant protein production in Antarctic Gram-negative bacteria. Methods Mol. Biol. 267, 225–237 (2004). [DOI] [PubMed] [Google Scholar]
- Miteva V., Lantz S. & Brenchley J. Characterization of a cryptic plasmid from a Greenland ice core Arthrobacter isolate and construction of a shuttle vector that replicates in psychrophilic high G + C Gram-positive recipients. Extremophiles 12, 441–449 (2008). [DOI] [PubMed] [Google Scholar]
- Singh A. K., Pindi P. K., Dube S., Sundareswaran V. R. & Shivaji S. Importance of trmE for growth of the psychrophile Pseudomonas syringae at low temperatures. Appl. Environ. Microbiol. 75, 4419–4426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakermans C., Sloup R. E., Zarka D. G., Tiedje J. M. & Thomashow M. F. Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273-4. Extremophiles 13, 21–30 (2009). [DOI] [PubMed] [Google Scholar]
- Giuliani M. et al. A novel strategy for the construction of genomic mutants of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Methods Mol. Biol. 824, 219–233 (2012). [DOI] [PubMed] [Google Scholar]
- Yu Z. C. et al. Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913. Microb. Cell Fact. 13, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch E. A. et al. Sequencing of seven haloarchaeal genomes reveals patterns of genomic flux. PLoS One 7, e41389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G. A. Regulation of nitrogen metabolism and gene expression in fungi. Microbiol. Rev. 45, 437–461 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson A. H. & Larsson T. The utilization of organic nitrogen for growth of algae: physiological aspects. Physiol. Plant. 48, 542–553 (1980). [Google Scholar]
- Palenik B. & Henson S. E. The use of amides and other organic nitrogen sources by the phytoplankton Emiliania huxleyi. Limnol. Oceanogr. 42, 1544–1551 (1997). [Google Scholar]
- Stanier R. Y., Palleroni N. J. & Doudoroff M. The aerobic Pseudomonads: a taxonomic study. J. Gen. Microbiol. 43, 159–271 (1966). [DOI] [PubMed] [Google Scholar]
- Brown J. E., Brown P. R. & Clarke P. H. Buryramide-utilizing mutants of Pseudomonas aeruginosa 8602 which produce an amidase with altered substrate specificity. Microbiology 57, 273–285 (1969). [DOI] [PubMed] [Google Scholar]
- Oberhofer T. R. & Rowen J. W. Acetamide agar for differentiation of nonfermentative bacteria. Appl. Microbiol. 28, 720–721 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi G. L. Nonfermentative gram-negative bacteria encountered in clinical specimens. Antonie Van Leeuwenhoek 39, 229–242 (1973). [DOI] [PubMed] [Google Scholar]
- Collins P. A. & Knowles C. J. The utilization of nitriles and amides by Nocardia rhodochrous. Microbiology 129, 711–718 (1983). [Google Scholar]
- Draper P. The aliphatic acylamide amidohydrolase of Mycobacterium smegmatis: its inducible nature and relation to acyl-transfer to hydroxylamine. Microbiology 46, 111–123 (1967). [DOI] [PubMed] [Google Scholar]
- Köll P. & Metzger J. Nachweis von acetamid beim thermischen abbau von chitin. Z Lebensm Unters Forsch 113, 111–113 (1979). [DOI] [PubMed] [Google Scholar]
- Morgan S. L., Watt B. E., Ueda K. & Fox A. Pyrolysis GC/MS profiling of chemical markers for microorganisms In Analytical Microbiology Methods: Chromatography and Mass Spectrometry (eds Fox A. et al.) 179–200 (Plenum Press, NY, 1990). [Google Scholar]
- Song X. & Farwell S. O. Pyrolysis gas chromatography atomic emission detection method for determination of N-containing components of humic and fulvic acids. J. Anal. Appl. Pyrolysis. 71, 901–915 (2004). [Google Scholar]
- Cao J. P. et al. Triacetonamine formation in a bio-oil from fast pyrolysis of sewage sludge using acetone as the absorption solvent. Bior. Technol. 101, 4242–4245 (2010). [DOI] [PubMed] [Google Scholar]
- Altarawneh M. et al. Theoretical investigation into competing unimolecular reactions encountered in the pyrolysis of acetamide. J. Phys. Chem. A 115, 14092–14099 (2011). [DOI] [PubMed] [Google Scholar]
- Koch R. L., Chrystal E. J., Beaulieu B. B. & Goldman P. Acetamide—a metabolite of metronidazole formed by the intestinal flora. Biochem. Pharmacol. 28, 3611–3615 (1979). [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Jordan J. C., Vetter W. & Oesterhelt G. Metabolic studies of ornidazole in the rat, in the dog and in man. Xenobiotica 9, 571–581 (1979). [DOI] [PubMed] [Google Scholar]
- Chrystal E. J., Koch R. L. & Goldman P. Metabolites from the reduction of metronizadole by xanthine oxidase. Mol. Pharmacol. 18, 105–111 (1980). [PubMed] [Google Scholar]
- Fuhrman J. A. & Ferguson R. L. Nanomolar concentrations and rapid turnover of dissolved free amino acids in seawater: agreement between chemical and microbiological measurements. Mar. Ecol. Prog. Ser. 33, 237–242 (1986). [Google Scholar]
- Kirchman D. L. et al. Glucose fluxes and concentrations of dissolved combined neutral sugars (polysaccharides) in the Ross Sea and Polar Front Zone, Antarctica. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48, 4179–4197 (2001). [Google Scholar]
- Wyborn N. R., Scherr D. J. & Jones C. W. Purification, properties and heterologous expression of formamidase from Methylophilus methylotrophus. Microbiology 140, 191–195 (1994). [Google Scholar]
- Wyborn N. R., Mills J., Williams S. G. & Jones C. W. Molecular characterisation of formamidase from Methylophilus methylotrophus. Eur. J. Biochem. 240, 314–322 (1996). [DOI] [PubMed] [Google Scholar]
- Skouloubris S., Labigne A. & De Reuse H. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol. Microbiol. 40, 596–609 (2001). [DOI] [PubMed] [Google Scholar]
- Viteri S. E. & Schmidt E. L. Ecology of indigenous soil rhizobia: Response of Bradyrhizobium japonicum to readily available substrates. Appl. Environ. Microbiol. 53, 1872–1875 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. P., McDonald I. R. & Wood A. P. Proposal for the reclassification of Thiobacillus novellus as Starkeya novella gen. nov., comb. nov., in the alpha-subclass of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 50, 1797–1802 (2000). [DOI] [PubMed] [Google Scholar]
- Mevs U., Stackebrandt E., Schumann P., Gallikowski C. A. & Hirsch P. Modestobacter multiseptatus gen. nov., sp. nov., a budding actinomycete from soils of the Asgard Range (Transantarctic Mountains). Int. J. Syst. Evol. Microbiol. 50, 337–346 (2000). [DOI] [PubMed] [Google Scholar]
- Okubo T. et al. Soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica as suggested by genome analysis. Appl. Environ. Microbiol. 79, 2542–2551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler U. & Nouwens A. S. Metabolic adaptation and trophic strategies of soil bacteria—C1-metabolism and sulfur chemolithotrophy in Starkeya novella. Front Microbiol. 4, 304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales R. E., Adamus J. E., White N. & May H. D. Degradation of 1,4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60, 4527–4530 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendra S. & Alvarez-Cohen L. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1,4-dioxane. Int. J. Syst. Evol. Microbiol. 55, 593–598 (2005). [DOI] [PubMed] [Google Scholar]
- Kotak M. et al. Complete genome sequence of the Opitutaceae bacterium strain TAV5, a potential facultative methylotroph of the wood-feeding termite Reticulitermes flavipes. Genome Announc. 3, e00060–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W. & Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- Könneke M. et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. USA 111, 8239–8244 (2014). [DOI] [PMC free article] [PubMed]
- Deppenmeier U. et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461 (2002). [PubMed] [Google Scholar]
- Li J., Wong C. F., Wong M. T., Huang H. & Leung F. C. Modularized evolution in archaeal methanogens phylogenetic forest. Genome Biol. Evol. 6, 3344–3359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garushyants S. K., Kazanov M. D. & Gelfand M. S. Horizontal gene transfer and genome evolution in Methanosarcina. BMC Evol. Biol. 15, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda H. Hand Book on Chemical Industries (Alcohol Based) (ed. Panda H.) 1–374, Asia Pacific Business Press Inc., Delhi, India (2002).
- Cheung H., Tanke R. S. & Torrence G. P. Acetic Acid. Ullmann’s Encyclopedia of Industrial Chemistry. 10.1002/14356007.a01_045.pub2 (2011). [DOI] [Google Scholar]
- Wong C. S. Environmental fate processes and biochemical transformations of chiral emerging organic pollutants. Anal. Bioanal. Chem. 386, 544–558 (2006). [DOI] [PubMed] [Google Scholar]
- Rochman C. M. et al. Classify plastic waste as hazardous. Nature 494, 169–171 (2013). [DOI] [PubMed] [Google Scholar]
- Law K. & Thompson R. Microplastics in the sea. Science 345, 144–145 (2014). [DOI] [PubMed] [Google Scholar]
- van Sebille E. et al. A global inventory of small floating plastic debris. Environ. Res. Lett. 10, 124006 (2015). [Google Scholar]
- Seffernick J. L. et al. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183, 2405–2410 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016). [DOI] [PubMed] [Google Scholar]
- Bornscheuer U. T. Feeding on plastic. Science 351, 1154–1155 (2016). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Biodegradation of N,N-dimethylacetamide by Rhodococcus sp. strain B83 isolated from the rhizosphere of pagoda tree, http://dx.doi.org/10.1016/j.jes.2016.05.013 (2016). [DOI] [PubMed]
- Burns D. G. & Dyall Smith M. Cultivation of haloarchaea. Methods. Microbiol. 35, 535–552 (2006). [Google Scholar]
- Froger A. & Hall J. E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 6, 253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics (ed. Dyall-Smith M.) 43–62 (Martinsried, 2009). [Google Scholar]
- Weatherburn M. W. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39, 971–974 (1967). [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Peplies J. & Glöckner F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westram R. et al. ARB: a software environment for sequence data in Handbook of molecular microbial ecology I: metagenomics and complementary approaches (ed. De Bruijin F. J.) 399–406 (John Wiley and Sons, Hoboken, NJ, 2011).
- Diekmann J., Wittig A. & Stabbert R. Gas chromatographic—mass spectrometric analysis of acrylamide and acetamide in cigarette mainstream smoke after on-column injection. J. Chromatogr. Sci. 46, 659–663 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.