Abstract
DNA demethylation plays an essential role in the reactivation of Epstein-Barr virus (EBV) from latency infection. However, it is unclear how epigenetic modification is initiated in responding to stimuli. Here, we demonstrate that ERK/c-Jun signaling is involved in DNA demethylation of EBV immediate early (IE) gene Zta in response to 12-O-Tetradecanoylphorbol-13-acetate (TPA) stimulation. Remarkably, Ser73 phosphorylation of c-Jun facilitates Zta promoter demethylation and EBV reactivation, whereas knockdown of c-Jun attenuates Zta demethylation and viral reactivation. More importantly, we reveal for the first time that c-Jun interacts with DNA dioxygenase Tet1 and facilitates Tet1 to bind to Zta promoter. The binding of c-Jun and Tet1 to Zta enhances promoter demethylation, resulting in the activation of Zta, the stimulation of BHRF1 (a lytic early gene) and gp350/220 (a lytic late gene), and ultimately the reactivation of EBV. Knockdown of Tet1 attenuates TPA-induced Zta demethylation and EBV reactivation. Thus, TPA activates ERK/c-Jun signaling, which subsequently facilitates Tet1 to bind to Zta promoter, leading to DNA demethylation, gene expression, and EBV reactivation. This study reveals important roles of ERK/c-Jun signaling and Tet1 dioxygenase in epigenetic modification, and provides new insights into the mechanism underlying the regulation of virus latent and lytic infection.
Epstein-Barr virus (EBV) is a human herpes virus and is etiologically linked to a number of diseases, including multiple sclerosis, infectious mononucleosis, nasopharyngeal carcinoma, Hodgkin’s lymphoma, Burkitt’s lymphoma and post-transplant lymphoma1,2,3,4,5. EBV belongs to the γ-Herpes virus subfamily and can establish lifelong latency in the host B cells after primary infection, but readily reactivated to produce infectious immortalizing virus in a small percentage of cells6. The switch of EBV from latency to lytic cycle is initiated by the expression of the viral immediate-early gene Zta, also known as BZLF1, ZEBRA and EB17,8. Zta protein is a transcriptional activator that shares partial amino acid homology to a basic leucine zipper family of transcriptional factors, including c-Jun and c-Fos9. The Zta gene expression is regulated at the transcriptional level, which alone leads to trigger EBV reactivation cascade10,11. However, the precise molecular mechanism underlying the regulation of Zta gene is largely unknown.
Extracellular signal-related kinase (ERK) pathway plays an essential role in the regulation of Zta expression and EBV reactivation in response to 12-O-tetradecanoylphorbol-13-acetate (TPA) or other reagents12,13. The c-Jun directly binds on the Zta promoter and activates its activity14,15,16,17,18, and the levels of Zta and phosphorylated c-Jun are much higher in EBV-infected AGS gastric carcinoma cells than in EBV-infected HeLa cells19. It is not clearly whether ERK/c-Jun signaling is required for the epigenetic modification, especially DNA demethylation, to trigger Zta expression and EBV reactivation.
DNA methylation in promoter or enhancer regions normally leads to chromatin compaction that serves as a transcriptional silencer to repress gene expression20,21,22, and conversely, DNA demethylation generates a transcriptional active state. Cytosine 5C position methylation is mediated by several different methyltransferase and usually occurs at CpG dinucleotides23. Recent studies indicate that the ten eleven translocation (Tet) family proteins are capable of oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) to erase existing methylation marks by activating DNA demethylation24,25,26. Furthermore, Tet proteins convert 5mC to another two important cytosine derivatives, 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), in an enzymatic activity-dependent manner27,28. 5caC is specifically recognized and excised by thymine-DNA glycosylase and replaced by unmodified cytosine29. The EBV genome is heavily methylated in latently infected cells and the constitutive activity of lytic viral genes is repressed by methylation, allowing the virus to establish latency infection9,21,30,31,32. In contrast, the viral genome is hypomethylated during the lytic infection2,21,33. However, the mechanism of demethylation and the role of Tet in EBV reactivation remain largely unknown.
In this study, we demonstrated that TPA-induced modulation of ERK signaling in conjunction with Tet1-mediated demethylation of Zta promoter dynamically regulate EBV reactivation. ERK-dependent activation of c-Jun results in the recruitment of Tet1 to the Zta promoter to facilitate Zta promoter hypomethylation, Zta gene expression, and eventually EBV reactivation.
Results
ERK/c-Jun pathway is involved in TPA-stimulated demethylation of Zta promoter
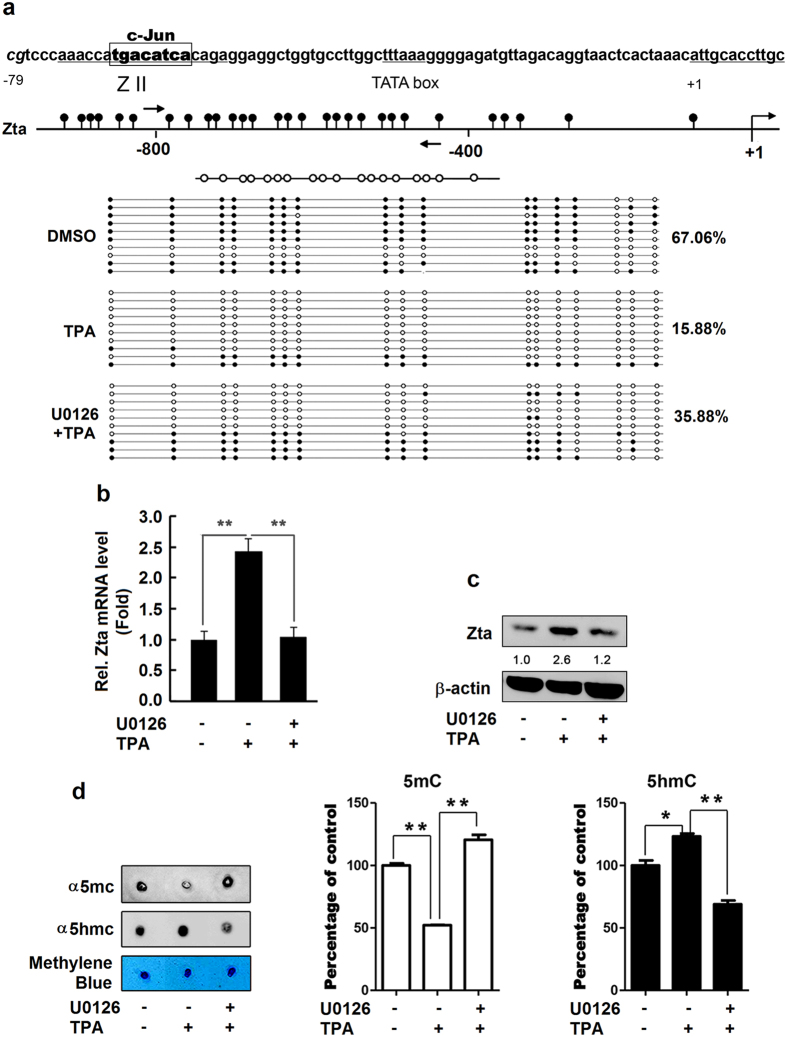
The γ-herpes viruses immediate early (IE) gene promoter contains a CpG island within 1kb of the transcription start site, which is highly methylated in many epithelial, NK- and B-cell original cancer cell lines and induced promoter demethylation following treatment with TPA and DNA methyltransferase inhibitor azacytidine2,34,35,36. However, the mechanism underlying TPA induces epigenetic modification or DNA demethylation is still unknown. As MAPK/ERK signal pathway plays an essential role in expression of Zta and EBV reactivation stimulated by TPA12,13, we speculated that MAPK/ERK signaling may be involvement in the regulation of DNA demethylation. To verify this speculation, B95-8 cells were pretreated with ERK specific inhibitor U0126 and then with TPA and the methylation status of the Zta promoter was assessed by sodium bisulphite sequencing. The Zta promoter methylation was significantly reduced by TPA (67.06% vs. 15.88%), but TPA-mediated repression was largely recovered in the presence of U0126 (15.88% vs. 35.88%) (Fig. 1a), indicating that TPA facilitates Zta promoter demethylation and ERK pathway is involved in such regulation. Furthermore, demethylation was detected within 1 hour after TPA treatment (Fig. S1). The Zta mRNA (Fig. 1b) and Zta protein (Fig. 1c) were stimulated by TPA, but TPA-mediated activations were attenuated by U0126, suggesting that TPA stimulates Zta gene expression through ERK pathway. To further validate the effect of ERK on Zta promoter demethylation, we examined the production of 5mC and 5hmC in the presence of TPA and/or U0126. Dot blot analyses indicated that 5mC was reduced by TPA and the reduction was recovered by U0126, whereas 5hmC was enhanced by TPA and the enhancement was repressed by U0126 (Fig. 1d), implicating that TPA-promoted DNA demethylation is dependent on ERK activation. Taken together, we demonstrated that TPA stimulates Zta promoter demethylation to activate Zta expression through ERK pathway.
Figure 1. ERK/c-Jun pathway is involved in the demethylation of Zta promoter and the expression of Zta gene.
(a–c) B95-8 cells were pretreated with ERK signal pathway specific inhibitor U0126 (20 μM) for 2 h and then treated with phorbol ester 12-O-Tetradecanoylphorbol 13-acetate (TPA) (60 ng/ml) for 24 h. The Zta promoter was analyzed by sodium bisulphite sequencing. The white and black circles indicate unmethylated and methylated CpGs, respectively (a). The level of EBV IE gene Zta mRNA was determined by real-time PCR (b). The level of Zta protein was detected by Western blot analyses (c). Relative protein levels, indicated by numbers below the blots, were determined by densitometry, with internal normalization to β–actin and external normalization to the negative control sample processed in parallel. (d) B95-8 cells were pretreated with U0126 (20 μM) for 2 h and then treated with TPA (60 ng/ml) for 24 h. The levels of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) were determined by dot blot analyses. Bulk genomic DNA was isolated, sonicated, cross-linked to nitrocellulose membrane, and probed with the monoclonal antibody specific for 5hmC or 5mC. The loading control is showing by the methylene blue stain in the bottom panel. The blot was quantitated by densitometry, and a representative experiment is shown from three independent experiments. All data were collected from 3 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Ser73 phosphorylation of c-Jun is essential for ERK-dependent activation of Zta
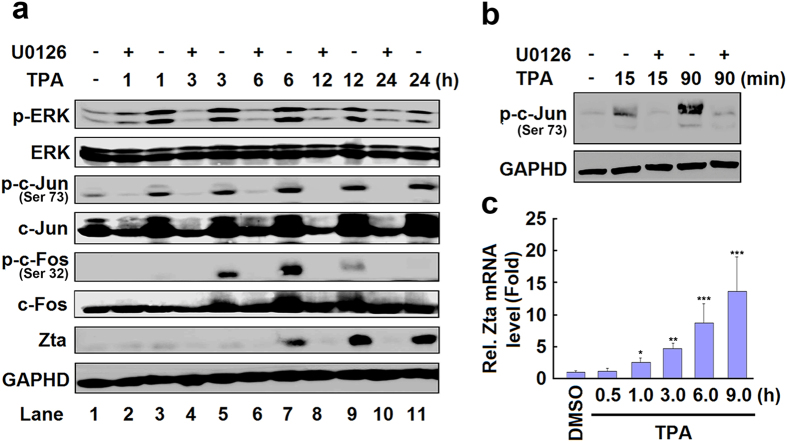
To reveal the mechanism by which ERK signaling stimulates DNA demethylation of Zta promoter, the roles of the downstream targets of ERK in TPA-induced Zta expression was investigated. Since cellular transcription factors including c-Jun and c-Fos are known to activate Zta promoter14,15,16,18,19,37, we examined the involvement of c-Jun and c-Fos in the demethylation of Zta promoter. B95-8 cells were treated with MAPK/ERK inhibitor U0126 for 2 h and then treated with TPA for different times. Zta protein production was stimulated by TPA at 6 h and maintained at a high level until 24 h, ERK phosphorylation was induced by TPA from 1 h to 12 h, c-Jun phosphorylation was induced by TPA at 1 h and increased in a time-dependent manner, whereas c-Fos phosphorylation was up-regulated by TPA from 3 h to 12 h, however, TPA-induced production of Zta and phosphorylation of ERK, c-Jun, and c-Fos were repressed by U0126 (Fig. 2a). In addition, Ser73 phosphorylation of c-Jun was stimulated rapidly by TPA within 15 min and further facilitated at a higher level at 90 min, but such activation was attenuated by U0126 (Fig. 2b). Furthermore, Zta mRNA was induced by TPA, before c-Fos phosphorylation and after c-Jun Ser73 phosphorylation, at 1 h post-treatment in a time-dependent manner (Fig. 2c). These results demonstrated that c-Jun phosphorylation is occurred before Zta expression and suggesting that c-Jun Ser73 phosphorylation is required for Zta expression.
Figure 2. c-Jun Ser73 phosphorylation is essential for ERK-dependent demethylation and activation of Zta promoter.
(a) B95-8 cells were pretreated with U0126 (20 μM) for 2 h and then treated with TPA for 1, 3, 6, 12, and 24 h, respectively. Whole-cell lysates were isolated and the levels of phosphorylated ERK (p-ERK), phosphorylated c-Jun (p-c-Jun), phosphorylated c-Fos (p-c-Fos), ERK, c-Jun, c-Fos, Zta, and GAPDH proteins were determined by Western blotting using corresponding antibodies. (b) B95-8 cells were pretreated with U0126 (20 μM) for 2 h and then treated with TPA for 15 and 90 min. Whole-cell lysates were isolated and the levels of phosphorylated c-Jun (p-c-Jun) and GAPDH proteins were determined by Western blotting. (c) B95-8 cells were treated with TPA for 0.5, 1, 3, 6 and 9 h. Total RNAs were isolated from the cells and the level of Zta mRNA was analyzed by qPCR. *P < 0.05, **P < 0.01 and ***P < 0.001.
c-Jun is required for ERK-dependent DNA demethylation
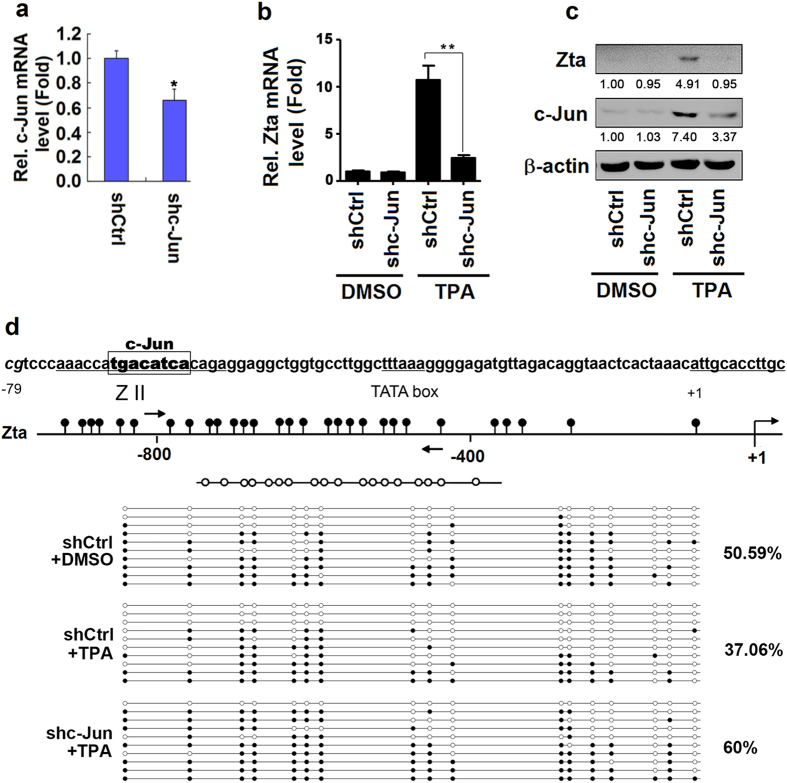
To confirm the requirement of c-Jun for ERK-dependent DNA demethylation induced by TPA, B95-8 cells were transduced with lentivirus expressing shRNA against c-Jun (shc-Jun) for 2 weeks, selected with puromycin, and then treated with TPA. c-Jun mRNA was attenuated in the presence of shc-Jun (Fig. 3a), confirming that shc-Jun is effective. In addition, Zta mRNA (Fig. 3b) and Zta protein (Fig. 3c) were activated by TPA, but the activations were attenuated by shc-Jun, suggesting c-Jun is required for TPA-induced expression of Zta. Moreover, the methylation of Zta promoter was down-regulated by TPA (50.59% vs. 37.06%), whereas this reduction was recovered by shc-Jun (37.06% vs. 60.00%) (Fig. 3d), indicating that c-Jun is involved in TPA-mediated demethylation of Zta promoter. Taken together, we demonstrated that c-Jun is required for the demethylation of Zta promoter induced by TPA.
Figure 3. c-Jun is required for ERK-dependent DNA demethylation of Zta promoter.
(a) B95-8 cells were lentiviruses expressing control shRNA (shCtrl) or shRNA against c-Jun (shc-Jun) for 2 weeks and then selected with puromycin. The total RNAs were isolated and the c-Jun mRNA was analyzed by qPCR (a). (b–d) B95-8 cells were transduced with lentiviruses expressing shCtrl or shc-Jun for 2 weeks, selected with puromycin, and then treated with TPA for 18 h. The total RNAs were isolated form the cells and the level of Zta mRNA was determined by qPCR (b). The cell extracts were prepared and the level of Zta, c-Jun, and β-actin proteins were detected by Western blot analyses (c). The Zta promoter was analyzed by sodium bisulphite sequencing. The white and black circles indicate unmethylated and methylated CpGs, respectively (d). All data were collected from 3 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
c-Jun interacts with Tet1 and recruits Tet1 to the Zta promoter
c-Jun is an important transcription factor involved in cell proliferation, transformation, death, somatic cell reprogramming, and lymphoid leukemia38,39,40,41. However, there is no evidence to support that c-Jun is directly participated in the modification of methylcytosine. Since we showed that 5hmC level was increased when treated with TPA (Fig. 1d), we speculated that c-Jun may promote Zta promoter DNA demethylation through recruiting methylcytosine hydroxylation enzymes (Tet-family proteins).
To test this hypothesis, we initially performed yeast two-hybrid analyses to identify the interaction between c-Jun and Tet1 (a hydroxylase that binds to DNA and modulates gene methylation and transcription via hydroxylation of 5-methylcytosine). Likes BD-p53 interacting with AD-T (as a positive control), a positive interaction was identified between BD-c-Jun and AD-Tet1 CD, but positive interaction was not occurred between BD-c-Jun and AD-vector, between BD-vector and AD-Tet1 CD, or between BD-p53 and AD-Lam (Fig. 4a). To determine whether the interaction of c-Jun with Tet1 is direct, we carried on GST pull-down assays. The results showed that HA-tagged Tet1 CD interacts with GST-c-Jun (Fig. 4b), confirming that c-Jun could bind directly with Tet1. To further examine the interaction of c-Jun and Tet1, co-immunoprecipitation (co-IP) assays were performed, which revealed that GFP-tagged c-Jun, but not GFP-tagged c-Fos, was immunoprecipitated with HA-tagged Tet1 CD (Fig. 4c). Flag-tagged c-Jun was co-immunoprecipitated with HA-tagged Tet1 CD (Fig. 4d, left panel), and reversely HA-tagged Tet1 CD was co-immunoprecipitated with Flag-tagged c-Jun (Fig. 4d, right panel). In addition, the interaction of endogenous expressed c-Jun with HA-tagged Tet1 CD was detected by co-IP assays in HEK293 cells (Fig. 4e). We were also able to coimmunoprecipitate endogenous Tet1 and c-Jun from whole-cell extracts using c-Jun specific antibodies from both control and TPA-treated cells (Fig. S2).
Figure 4. Tet1 binds to Zta promoter through interacting with c-Jun.
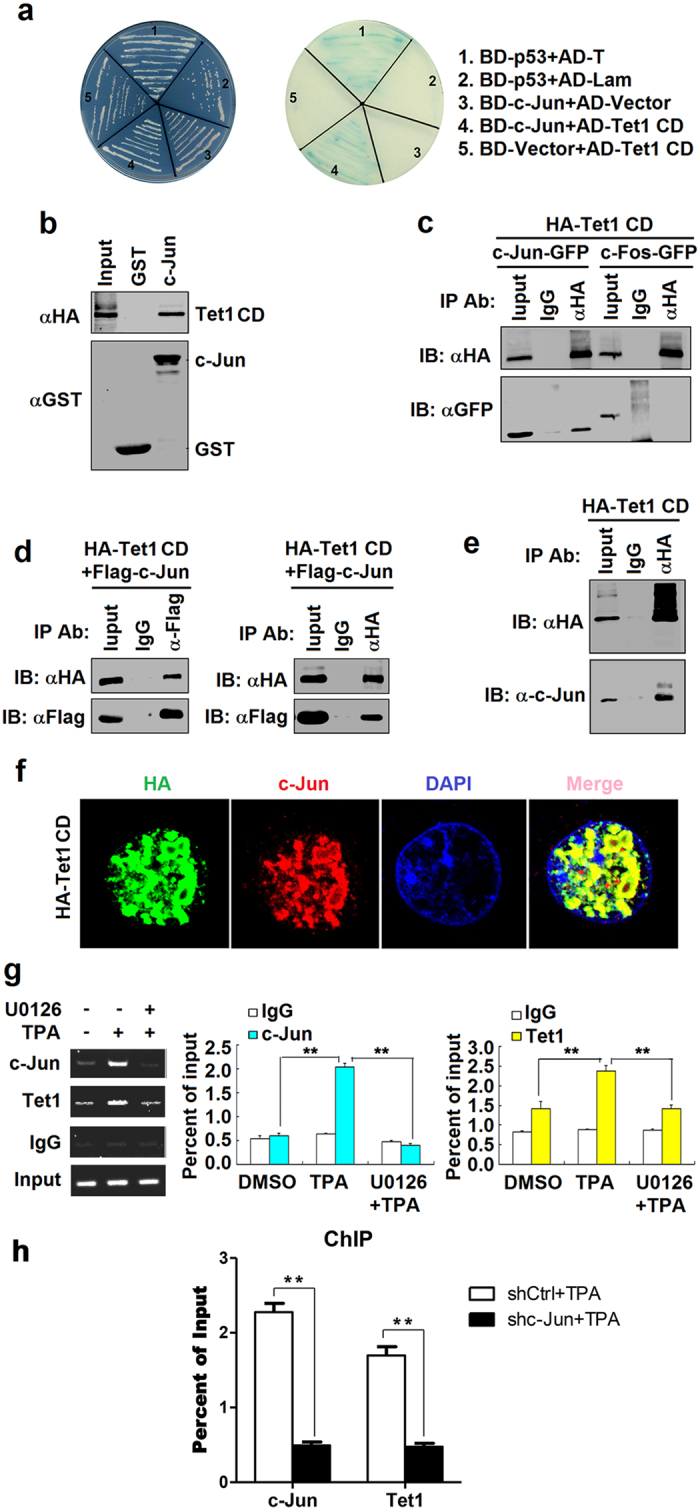
(a) Yeast strain AH109 was co-transformed with the combination of one BD and one AD plasmid, as indicated. Transformed yeast cells containing both plasmids were grown on SD-minus Trp/Leu plates (DDO), and colonies were then replica-plated onto SD-minus Trp/Leu/Ade/His plates (QDO) containing X-α-gal to check for the expression of reporter gene (blue colonies). (b) GST pull-down assay with GST-c-Jun or GST alone incubated with HA-tagged Tet1CD. (c) Co-immunoprecipitation of HA-tagged Tet1 CD and GFP-tagged c-Jun or GFP-tagged c-Fos, followed by western blot analysis with anti-GFP antibody. (d) HA-tagged Tet1 CD and Flag-tagged c-Jun were transfected to HEK293 cells and immunoprecipitated using anti-Flag/HA antibody from cell extracts. Co-immunoprecipitation of HA-Tet1/Flag-c-Jun was detected by western blotting using anti-HA/Flag antibody. (e) HEK293 cells were transfected with pHA-tagged Tet1 CD and immunoprecipitated using anti-HA antibody from cell extracts. Co-immunoprecipitation of HA-Tet1/ c-Jun was detected by western blotting using anti-HA/ c-Jun antibody. (f) HEK293 cells were transfected with HA-Tet1 CD for 24 h. After fixation, the cells were immunostained with anti-HA and anti-c-Jun antibody. The nucleus stained by DAPI and visualized by confocal laser microscopy. (g) For ChIP, B95-8 cells were pretreated with U0126 (20 μM) for 2 h and treated with TPA (60 ng/ml) for 4 h. Cross-linked chromatin was immunoprecipitated with the specific antibodies. (h) For ChIP, c-Jun knockdown and control B95-8 cells were treated with TPA (60 ng/ml) for 4 h. Cross-linked chromatin was immunoprecipitated with the specific antibodies. Error bars indicate standard error of the mean. *P < 0.05, **P < 0.01 and ***P < 0.001.
Moreover, immunofluorescence analysis showed that c-Jun was co-localized with HA-Tet1 CD in the nucleus of HEK293 cells (Fig. 4f).
Finally, to determine whether c-Jun and Tet1 are occupied at coincident Zta promoter in response to TPA stimulation, we performed chromatin immunoprecipitation (ChIP) assays. As shown in Fig. 4g, we observed much more binding of c-Jun to the Zta promoter in response to TPA, more Tet1 was also found to bind to this promoter. Next, we tested whether c-Jun is required for Tet1 binding to the Zta promoter in response to TPA using the Erk signal pathway specific inhibitor U0126 (Fig. 4g) or knocking down c-Jun (Fig. 4h). Both U0126 pretreatment and c-Jun knockdown resulted, as expected, in reduced c-Jun activation and binding. More importantly, the decrease in c-Jun occupancy at the promoter coincides with a significant reduction of Tet1 binding to Zta (Fig. 4h), demonstrating that Tet1 is recruited to the Zta promoter in response to TPA depending on c-Jun. Taken together, we demonstrated that c-Jun interacts with Tet1 and recruits Tet1 to the Zta promoter and suggested that ERK/c-Jun signaling is necessary for the binding of Tet1 to TPA-target Zta promoter.
Tet1 is required for ERK-dependent Zta promoter demethylation and gene expression
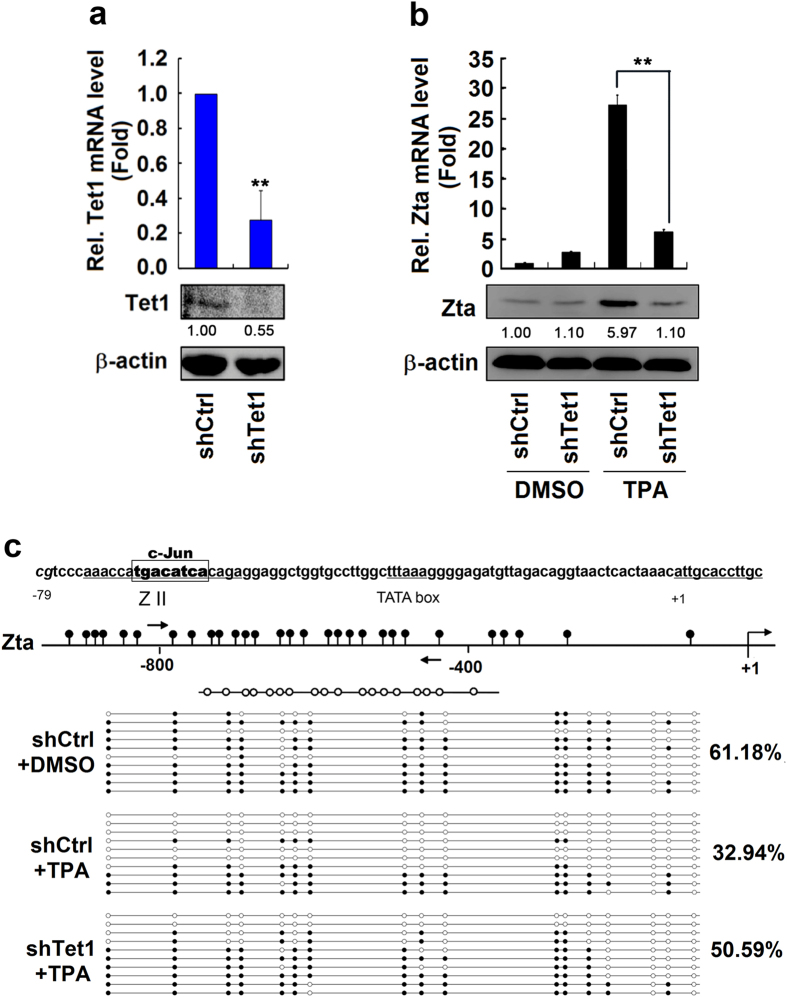
The role of Tet1 in the activation of Zta gene mediated by c-Jun was further evaluated. Tet1 mRNA and Tet1 protein were reduced by lentivirus expressed shRNA against Tet1 (shTet1) in B95-8 cells (Fig. 5a), indicating shTet1 is effective. B95-8 cells were then treated with TPA and transfected with shTet1. The results showed that Zta mRNA and Zta protein were stimulated by TPA, but TPA-mediated activations were significantly repressed by shTet1 (Fig. 5b). The requirement of Tet1 for ERK-dependent demethylation of Zta promoter was then determined. The results revealed that Zta promoter methylation was significantly repressed by TPA (61.18% vs. 32.94%), but TPA-mediated repression was recovered by shTet1 (32.94% vs. 50.59%) (Fig. 5c). These results demonstrated that Tet1 is required for ERK-dependent Zta promoter DNA demethylation and gene expression.
Figure 5. Tet1 is essential for TPA-mediated Zta promoter demethylation and gene expression.
(a) B95-8 cells were transduced with lentiviruses expressing control shRNA (shCtrl) or shRNA against Tet1 (shTet1) for 2 weeks, and selected with puromycin. The total RNAs were isolated form the cells and the level of Tet1 mRNA was determined by qRT-PCR (upper panel). The cell extracts were prepared and the level of Tet1 and β-actin proteins were detected by Western blot analyses (lower panel). (b,c) B95-8 cells were transduced with lentiviruses expressing shCtrl or shTet1 for 2 weeks, selected with puromycin, and then treated with TPA for 18 h. The total RNAs were isolated form the cells and the level of Zta mRNA was determined by qRT-PCR (b, upper panel). The cell extracts were prepared and the level of Zta and β-actin proteins were detected by Western blot analyses (b, lower panel). The Zta gene promoter was analyzed by sodium bisulphite sequencing. White and black circles indicate unmethylated and methylated CpGs, respectively (c). All data were collected from 3 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
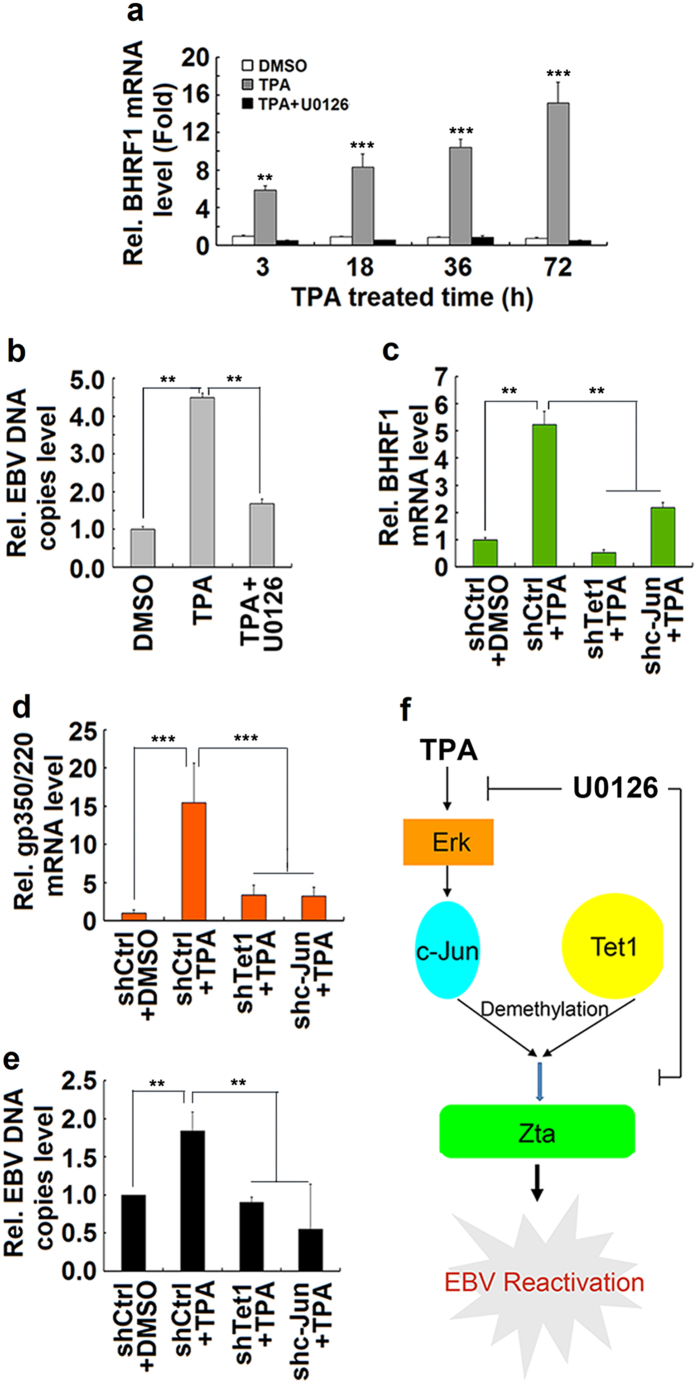
c-Jun and Tet1 are essential for ERK-dependent reactivation of EBV
The role of ERK pathway in the regulation of EBV reactivation was determined. To evaluate the effect of ERK pathway on the regulation of viral lytic gene expression, B95-8 cells were pretreated with U0126 and then treated with TPA for different times. BHRF1 (a key viral lytic early gene) mRNA was stimulated by TPA in a time-dependent manner, but TPA-mediated activation was repressed by U0126 (Fig. 6a). To determine the role of ERK in the regulation of EBV replication, B95-8 cells were pretreated with U0126 and then treated with TPA for 24 h. EBV genomic DNA copy number was activated by TPA in the absence of U0126, but not in the presence of U0126 (Fig. 6b), suggesting that ERK is required for the reactivation of EBV induced by TPA.
Figure 6. c-Jun and Tet1 are essential for ERK-dependent reactivation of EBV.
(a) B95-8 cells were pretreated with U0126 (20 μM) for 2 h and then treated with TPA (60 ng/ml) for different times. RNA was extracted and the BHRF1 gene mRNA was analyzed by qPCR. (b) B95-8 cells were pretreated with U0126 (20 μM) for 2 h and then treated with TPA (60 ng/ml) for 18 h. DNA was prepared from treated cells and the level of EBV genomic DNA was determined by qPCR. Values of viral genome copies are normalized to the control DMSO. (c–e) B95-8 cells were transduced with lentiviruses expressing shCtrl or shTet1 for 2 weeks, selected with puromycin, and then treated with TPA for 18 h. The total RNAs were isolated form the cells and the levels of BHRF1 mRNA (c), pg350/220 mRNA (d), and EBV genomic DNA (e) were determined by qPCR. Values are normalized to the control shCtrl. Error bars represent SD from the mean. All data were collected from 3–6 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Furthermore, the effects of c-Jun and Tet1 on the expression of BHRF1 and gp350/220 (a key viral lytic late gene) and the replication of EBV were determined by using RNAi approach. B95-8 cells were transduced with lentiviruses expressing shCtrl, shc-Jun, and shTet1 for 2 weeks at puromycin selection and then treated with TPA for 18 h. BHRF1 mRNA (Fig. 6c) and gp350/220 mRNA (Fig. 6d) were up-regulated by TPA, but such enhancements were down-regulated by shTet1 or shc-Jun, indicating that Tet1 and c-Jun are required for TPA-stimulated activation of viral lytic early gene and lytic late gene. Finally, EBV genomic DNA copy number was stimulated by TPA, but such activation was attenuated by shTet1 or shc-Jun (Fig. 6e), revealing that Tet1 and c-Jun are required for TPA-stimulated reactivation of EBV. Taken together, we demonstrated that Tet1 and c-Jun are required for ERK-dependent EBV gene expression and reactivation in response to TPA stimulation.
Discussion
Methylation of CpG sites in the vicinity of the herpes virus immediate-early gene promoter usually inhibits gene expression and maintains viral latent infection35,42. In the contrary, DNA demethylation occurs during virus reactivation in vivo or after the treatment with phorbol ester TPA or DNA methyltransferase inhibitor 5-azacytidine in vitro33,35,43. However, the molecular mechanism underlying in the process of DNA demethylation is not understood. In this report, we document evidence that ERK signal pathway directs DNA demethylation stimulated by TPA. U0126, an ERK signaling pathway specific inhibitor, impedes Zta promoter demethylation. Ser73 phosphorylation of c-Jun, an ERK downstream transcription factor, is required for Zta gene transcription. Expectedly, depletion of c-Jun in B95-8 cells abrogates TPA-stimulated DNA demethylation of Zta promoter. More importantly, we demonstrate for the first time that c-Jun (an important transcription factor in the Activator protein-1 family) interacts with Tet1 (an important DNA modifying enzyme in the Ten-eleven translocation hydroxylase family) and recruits Tet1 to the Zta promoter region, leading to the initiation of Zta promoter DNA demethylation, IE gene expression, followed by lytic early and late gene expression, and ultimately EBV reactivation (Fig. 6f).
Previous studies have focused on Zta proximal promoter region (−221 to +12) and identified many important transcription factor binding elements essential for EBV latency or lytic infection, including c-Jun, ATF1, Sp1/3, MEF2D and ZEB115,18,19,37,44. Additionally, there is only one CpG site in the proximal section and different sensitivity of cell types to DNA methyltransferase inhibitor44,45,46, and therefore, the significance of DNA demethylation on regulating Zta gene expression is always undervalued. In this study, we expand the bisulfite genomic sequencing region from −400 to −800 where is abundant with CpG sites. We further demonstrated that Zta gene promoter DNA methylation is significantly decreases in respond to TPA stimulation. The incomplete DNA methylation on Zta gene promoter implicates that the low spontaneous lytic level is permitted. Furthermore, Zta protein preferentially binds to and activates the cytosine-methylated silent viral promoters30,47,48,49,50,51, and also auto-regulates itself promoter52, so Zta may replace Tet1 to promote EBV reactivation without any more DNA demethylation. Therefore, we revel that TPA induces DNA demethylation, but TPA can not present a completely unmethylated status on Zta promoter.
c-Jun is a founding member of the activator protein-1 (AP-1) transcription factor family involved in cell proliferation, transformation, apoptosis, carcinogenesis41,53,54. In p185BCR-ABL-transformed cells, lack of c-Jun leads to methylation within the 5′ region of cell-cycle kinase 6 gene (Cdk6) and down-regulate the production of CDK6 protein38, which implicates that c-Jun may play a key role in maintaining hypomethylation of Cdk6. Recently, the DNA dioxygenase Tet1 has been reported to play an important role in the reprogramming of somatic cells to pluripotency55,56,57,58, and c-Jun impedes somatic cell reprogramming40. Combination with our finding that c-Jun interacts with Tet1 and recruits Tet1 to Zta gene promoter, it is possible that Tet1 is hijacked by c-Jun to regulate the expression of a subset of key reprogramming target genes. However, the assumption still needs further investigation.
In conclusion, we reveal that the activation of ERK/c-Jun signaling is required for the recruiting of Tet1 to the viral immediate-early gene promoter, which leads to the induction of DNA demethylation and viral reactivation. These results would provide new insights into our understanding the role of ERK signaling in the regulation of epigenetic modification changes and also would provide new insights into the mechanism underlying the regulation of EBV reactivation.
Methods
For details of Methods, see Supplemental Methods.
Plasmids, antibodies, and regents
The plasmid HA-Tet1 CD (pAAV-EF1a-HA-hTet1CD-WPRE-PolyA) was a gift from Hongjun Song (Addgene plasmid # 39454)25. We cloned the c-Jun cDNA (isolated from HEK293T cells) into pCMV-Tag 2B (Stratagene) and pGEX-6p-1 (GE Healthcare) vector to create Flag-c-Jun and GST-c-Jun, respectively.
Antibodies against EBV Zta (ZEBRA) (sc-53904, 1:200), c-Jun (sc-1694), c-Fos (sc-52) and ERK (sc-153) were purchased from Santa Cruz Technology Company. Rabbit polyclonal antibodies against p-ERK (p-P44/42 MAPK, # 4370S), p-c-Jun (Ser73, # 3270P) and p-c-Fos (Ser32, # 5348S) were obtained from Cell Signaling Technology. Mouse monoclonal anti-Flag antibody (Cat# F1804) and anti-HA antibody (Cat# H6908) produced in rabbit were purchased from Sigma Aldrich. Polyclonal antibody against Tet1 (Cat# GTX124207) was purchased from Gene Tex. Antibodies against 5-methylcytosine (5mC, MABE146) and 5-Hydroxymethylcytidine (5hmC, Cat# 39769) were obtained from Millipore and Active Motif, respectively.
The TPA (12-O-Tetradecanoylphorbol-13-acetate) (Cat# P8139) and U0126 (Cat# U120) were purchased from Sigma Aldrich.
Additional Information
How to cite this article: Zhang, W. et al. ERK/c-Jun Recruits Tet1 to Induce Zta Expression and Epstein-Barr Virus Reactivation through DNA Demethylation. Sci. Rep. 6, 34543; doi: 10.1038/srep34543 (2016).
Supplementary Material
Acknowledgments
This work was supported by research grants from the Major State Basic Research Development Program of China (973 program) (2012CB518900), the National Natural Science Foundation of China (31230005, 81471942, 31270206, 31200134, and 31170153), and the National Mega Project on Major Infectious Disease Prevention (2012ZX10002006-003 and 2012ZX10004-207).
Footnotes
Author Contributions W.Z. and J.W. conceived the project and designed the experiments; W.Z., D.H., P.W., P.P. and Y.C. performed experiments, and W.Z., Y.L., K.W. and J.W. carried out data analyses; W.Z., K.W. and J.W. wrote the manuscript; J.W. edited the manuscript.
References
- Tse E. & Kwong Y. L. Epstein Barr virus-associated lymphoproliferative diseases: the virus as a therapeutic target. Exp Mol Med 47, e136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. Methylation profiling of Epstein-Barr virus immediate-early gene promoters, BZLF1 and BRLF1 in tumors of epithelial, NK- and B-cell origins. BMC Cancer 12, 125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti M., Giovannoni G. & Aloisi F. Epstein-Barr virus and multiple sclerosis. Curr Opin Neurol 22, 201–206 (2009). [DOI] [PubMed] [Google Scholar]
- Young L. S. & Rickinson A. B. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4, 757–768 (2004). [DOI] [PubMed] [Google Scholar]
- Epstein M. A., Achong B. G. & Barr Y. M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1, 702–703 (1964). [DOI] [PubMed] [Google Scholar]
- Miyashita E. M., Yang B., Lam K. M., Crawford D. H. & Thorley-Lawson D. A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80, 593–601 (1995). [DOI] [PubMed] [Google Scholar]
- Chevallier-Greco A. et al. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J 5, 3243–3249 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. The switch between EBV latency and replication. Yale J Biol Med 62, 205–213 (1989). [PMC free article] [PubMed] [Google Scholar]
- Yu K. P. et al. Latency of Epstein-Barr virus is disrupted by gain-of-function mutant cellular AP-1 proteins that preferentially bind methylated DNA. Proc Natl Acad Sci USA 110, 8176–8181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T., Fujita M. & Kudoh A. Latent and lytic Epstein-Barr virus replication strategies. Rev Med Virol 15, 3–15 (2005). [DOI] [PubMed] [Google Scholar]
- Kenney S. C. & Mertz J. E. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol 26, 60–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton M. & Sinclair A. J. Divergent requirements for the MAPK(ERK) signal transduction pathway during initial virus infection of quiescent primary B cells and disruption of Epstein-Barr virus latency by phorbol esters. J Virol 73, 8913–8916 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi H., Cochet C., Hmama Z., Opolon P. & Joab I. Transforming growth factor beta 1 stimulates expression of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA by an indirect mechanism which requires the MAPK kinase pathway. J Virol 74, 5810–5818 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. L., Chen J. L., Hsu Y. P., Ou J. T. & Chang Y. S. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-beta through cooperativity of Smads and c-Jun/c-Fos proteins. J Biol Chem 277, 23345–23357 (2002). [DOI] [PubMed] [Google Scholar]
- Gao X., Ikuta K., Tajima M. & Sairenji T. 12-O-tetradecanoylphorbol-13-acetate induces Epstein-Barr virus reactivation via NF-kappaB and AP-1 as regulated by protein kinase C and mitogen-activated protein kinase. Virology 286, 91–99 (2001). [DOI] [PubMed] [Google Scholar]
- Borras A. M., Strominger J. L. & Speck S. H. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 gene promoter: role in phorbol ester induction. J Virol 70, 3894–3901 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf I. K. & Rawlins D. R. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J Virol 69, 7648–7657 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington E. & Speck S. H. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol 64, 1217–1226 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W. H. et al. ZEB1 and c-Jun levels contribute to the establishment of highly lytic Epstein-Barr virus infection in gastric AGS cells. J Virol 81, 10113–10122 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. DNA methylation and chromatin - unraveling the tangled web. Oncogene 21, 5361–5379 (2002). [DOI] [PubMed] [Google Scholar]
- Fernandez A. F. et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res 19, 438–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H. The DNA methyltransferases of mammals. Hum Mol Genet 9, 2395–2402 (2000). [DOI] [PubMed] [Google Scholar]
- Tahiliani M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Su Y., Zhong C., Ming G. L. & Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 527, 118–122 (2015). [DOI] [PubMed] [Google Scholar]
- He Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S. et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellmer A., Arteaga-Salas J. M. & Hammerschmidt W. BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression. PLoS Pathog 8, e1002902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. & Ambinder R. F. Methylation of the Epstein-Barr virus genome in normal lymphocytes. Blood 90, 4480–4484 (1997). [PubMed] [Google Scholar]
- Paulson E. J. & Speck S. H. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J Virol 73, 9959–9968 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. T. et al. Azacitidine induces demethylation of the Epstein-Barr virus genome in tumors. J Clin Oncol 22, 1373–1381 (2004). [DOI] [PubMed] [Google Scholar]
- Ben-Sasson S. A. & Klein G. Activation of the Epstein-Barr virus genome by 5-aza-cytidine in latently infected human lymphoid lines. Int J Cancer 28, 131–135 (1981). [DOI] [PubMed] [Google Scholar]
- Chen J. et al. Activation of latent Kaposi’s sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc Natl Acad Sci USA 98, 4119–4124 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K. I. & Ernberg I. Demethylation of the Epstein-barr virus origin of lytic replication and of the immediate early gene BZLF1 is DNA replication independent. Brief report. Arch Virol 144, 2219–2227 (1999). [DOI] [PubMed] [Google Scholar]
- Wang Y. C., Huang J. M. & Montalvo E. A. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology 227, 323–330 (1997). [DOI] [PubMed] [Google Scholar]
- Kollmann K. et al. c-JUN promotes BCR-ABL-induced lymphoid leukemia by inhibiting methylation of the 5′ region of Cdk6. Blood 117, 4065–4075 (2011). [DOI] [PubMed] [Google Scholar]
- Lake R. J. et al. The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure. Plos Genetics 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol 17, 856–867 (2015). [DOI] [PubMed] [Google Scholar]
- Shaulian E. & Karin M. AP-1 as a regulator of cell life and death. Nature Cell Biology 4, E131–E136 (2002). [DOI] [PubMed] [Google Scholar]
- Robertson K. D. The role of DNA methylation in modulating Epstein-Barr virus gene expression. Curr Top Microbiol Immunol 249, 21–34 (2000). [DOI] [PubMed] [Google Scholar]
- Yang Z., Tang H., Huang H. & Deng H. RTA promoter demethylation and histone acetylation regulation of murine gammaherpesvirus 68 reactivation. PLoS One 4, e4556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S. H. & Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8, 100–115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman J. K., Gradoville L. & Miller G. Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J Virol 82, 4706–4719 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T. & Tsurumi T. Epigenetic modification of the Epstein-Barr virus BZLF1 promoter regulates viral reactivation from latency. Front Genet 4, 53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhende P. M., Seaman W. T., Delecluse H. J. & Kenney S. C. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet 36, 1099–1104 (2004). [DOI] [PubMed] [Google Scholar]
- Bergbauer M. et al. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog 6, e1001114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S. J. et al. Methylation-dependent binding of the epstein-barr virus BZLF1 protein to viral promoters. PLoS Pathog 5, e1000356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Q. H., Schelcher C., Verrall E., Petosa C. & Sinclair A. J. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog 4, e1000005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalla M., Schmeinck A., Bergbauer M., Pich D. & Hammerschmidt W. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci USA 107, 850–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. et al. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J Virol 65, 4033–4041 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R. & Wagner E. F. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3, 859–868 (2003). [DOI] [PubMed] [Google Scholar]
- Ozanne B. W., Spence H. J., McGarry L. C. & Hennigan R. F. Transcription factors control invasion: AP-1 the first among equals. Oncogene 26, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- Chen J. et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet 45, 1504–1509 (2013). [DOI] [PubMed] [Google Scholar]
- Costa Y. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 (2013). [DOI] [PubMed] [Google Scholar]
- Chen J. et al. The combination of Tet1 with Oct4 generates high-quality mouse-induced pluripotent stem cells. Stem Cells 33, 686–698 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.