Abstract
While altered TP53 is the most frequent mutation in gastric cancer (GC), its association with molecular or clinical phenotypes (e.g., overall survival, disease-free survival) remains little known. To that end, we can use genome-wide approaches to identify altered genes significantly related to mutated TP53. Here, we identified significant differences in clinical outcomes, as well as in molecular phenotypes, across specific GC tumor subpopulations, when combining TP53 with other signaling networks, including WNT and its related genes NRXN1, CTNNB1, SLITRK5, NCOR2, RYR1, GPR112, MLL3, MTUS2, and MYH6. Moreover, specific GC subpopulations indicated by dual mutation of NRXN1 and TP53 suggest different drug responses, according to the Connectivity Map, a pharmacological drug-gene association tool. Overall, TP53 mutation status in GC is significantly relevant to clinical or molecular categories. Thus, our approach can potentially provide a patient stratification strategy by dissecting previously unknown multiple TP53-mutated patient groups.
TP53 has been recognized as one of the most often mutated genes in cancer1,2,3. Germline TP53 mutations, present in hereditary conditions such as Li-Fraumeni (LFS) and Li-Fraumeni-like (LFL) syndromes3, cause inherited cancer, while somatic TP53 mutations are associated with 5–50% of cases of every known cancer3. In gastric cancer (GC) specifically, a leading cause of cancer deaths in the Asia-Pacific region, ~50% of reported cases show TP53 somatic mutations2. Also, the long history of TP53’s role(s) in responses to DNA-damaging agents in cancer chemotherapy4, is another therapeutically important aspect.
Despite the prevalence of TP53 mutations in cancer, many retrospective studies have failed to identify associations between TP53 abnormalities (e.g., mutations, amplifications) and clinicopathologic phenotypes5, and the lack of well-established clinical significance between patient outcomes and TP53 status has become one of the most controversial topics in cancer research, including GC and colorectal cancer (CRC)1,5,6,7. Discrepancies in reported associations are believed to result primarily from tumor heterogeneity, the complexity of p53 pathways, and defining distinct clinical stages5. Nevertheless, assessments of patient TP53 mutational status, in combination with transcriptional statuses of other genes, have been somewhat beneficial in segregating specific cancer subpopulations5. For example, a patient subpopulation consisting of mutant TP53 and wild-type KRAS in metastatic and chemotherapy-refractory CRC showed better clinical outcomes when treated with the EGFR antibody, cetuximab8, suggesting that the efficiency of molecular targeted therapy (e.g., cetuximab, trastuzumab) depends on TP53 status, in combination with other genetic alterations, even though the mode of action of the targeted therapy is not directly relevant to p53 signals. Consequently, through further combinatorial dissection of TP53 status and other genetic alterations, patient selection (and tailored therapy) may be superior to other therapeutic strategies. Thus, not only is TP53 mutational status significant in and of itself, it also holds clinical significance in combination with other genetic alterations, and thus should be routinely explored.
In this study, we systematically explored TP53 mutations, in combination with other genomic anomalies, in The Cancer Genome Atlas (TCGA)9 GC patient datasets. In GC, we previously established a WNT pathway subnetwork as a new therapeutic target10,11, which we then integrated with TP53 mutation status, and other genetic alterations, to define distinct GC tumor subpopulations. Among these subpopulations, we herein show statistically significant differences in clinical implications, as well as in molecular characteristics, across certain GC subpopulations. In addition, we suggest drug response differences, between cell lines associated with such subpopulations, representing our initial preclinical study of various “tailored” therapeutic interventions for GC.
Results
Patient Grouping Based on Expression Patterns
While TP53 mutation status is important in GC pathogenesis1, GC is a highly heterogeneous disease12, and its clinically significant association with TP53 mutation status remains little explored1,3,13. In fact, GC patient survival analysis in TCGA GC dataset9 showed no significant clinical outcomes in overall survival (OS) or disease-free survival (DFS), based on TP53 status (Supplementary Figure S1).
Here, for finding the significant clinical relevance of TP53 mutation to GC, we reduced the confounding effects of heterogeneity by dividing tumors into subsets, based on the mutational statuses of various genes related to a signaling network. In other words, by dividing GC patients into subpopulations, we subsequently inspected associations between TP53 mutation status and clinical relevance.
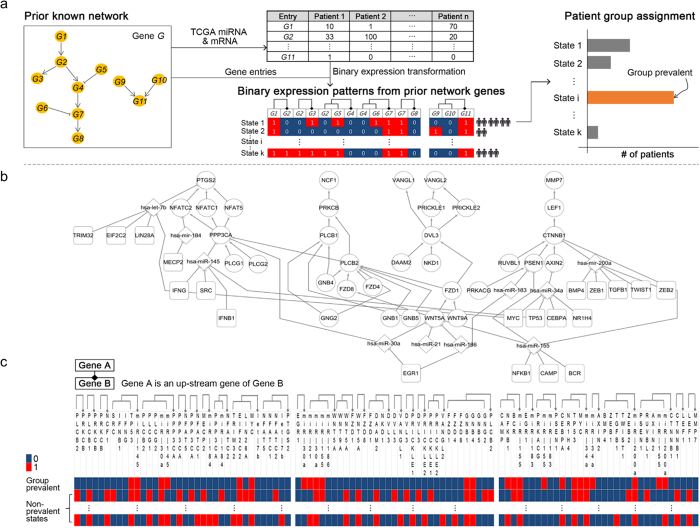
For the patient grouping, we first utilized a previously delineated GC signaling network10 and a GC expression dataset (TCGA)9. Given the network with a smaller number of entries, the network expression patterns for individual samples were divided into several network states by transforming the entries’ expressions into binary values (Fig. 1a). A network state was defined as the set of the binary expressions for the network entries. Then, the patient group with the most prevalent state (henceforth, “Group prevalent”) was identified (Fig. 1a).
Figure 1. Patient grouping: an overview of patterning continuous gene expression to binary gene expression, based on signal pathways and classifying some patient subpopulation with a number of cases in a certain expression pattern.
(a) We set the known network from our previous finding as a prior knowledge. We transformed each gene’s expression values to binary values, 0 s or 1 s. We sorted values of each gene, calculated gradient between two values repeatedly to find the greatest gradient (distance) in certain gene’s expression values. We replaced each expression value to 1 if the continuous value is greater than the threshold value else to 0. We arranged the pathway interaction structure to a flat table similar to computer’s memory. For example, a G2 is downstream of a G1 and upstream of G3 and G4 respectively, so in the table structure, we put repeatedly G2 on the right column of G1 and the left column of G3 and G4. We labeled all combinations of each gene’s value (0 or 1) as State 1, 2, …, k then counted all samples in a certain state. Then we identified a group (Group prevalent) that has the most samples in a certain state. (b) We picked up the WNT-relating signal pathway subset10 with transcription factors (rounded square), miRNAs (diamond shapes), and circle shapes are genes involved the WNT-relating signal pathway. (c) We transformed every expression values (from TCGA GC transcriptome9) to binary values and arranged the WNT-relating signal pathway structure as flattened table structure and counted samples for each states. Then we identified the network entry expression state (in the first row) having the most patient samples, assigning to Group prevalent.
As the previously delineated network, we set our prior result10, demonstrating that WNT pathway context, in combination with miRNAs and transcription factors (TFs), represents a specific GC-associating network (Fig. 1b). In Fig. 1c, the binary expression patterns for the network in Fig. 1b, are shown. Based on the binary expression patterns, we obtained Groups prevalent.
Molecular Phenotypes and Clinical Relevance of Subgroups in Association with TP53
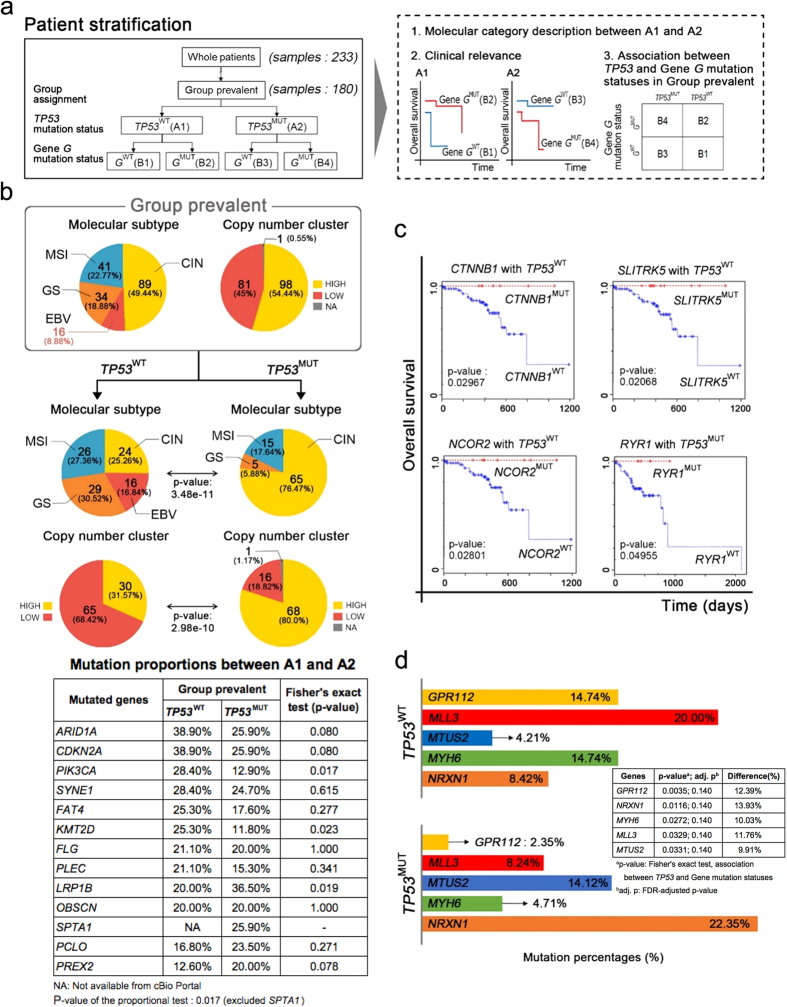
Using the whole 233 TCGA GC samples, we obtained Group prevalent by using the procedures of Fig. 1a. Subsequently, we divided 180 “Group prevalent” patient samples into two groups, TP53WT and TP53MUT, according to TP53 mutation status (Fig. 2a). The two groups are depicted as “A1” and “A2”, respectively (Fig. 2a). Once the groups were defined, we aimed at identifying the clinical relevance of TP53 mutation status by combining it with the mutation statuses of other genes (Fig. 2a). For this purpose, we dissected Groups “prevalent” into further subgroups (Fig. 2a), as examined by statistical tests between the subgroups for identifying significant clinical relevance (Fig. 2a). For simplicity, throughout the manuscript, we henceforth say patients having gene G mutations as GMUT, and those lacking gene G mutations as GWT. For example, TP53MUT patients indicate patient tumors having TP53 mutations.
Figure 2. Molecular Phenotypes and Clinical Relevance of Group Prevalent in Association with TP53 mutation.
(a) In Group prevalent, we checked TP53 mutation status and assigned “A1” for TP53WT, and “A2” for TP53MUT (in the left rectangle). Then, we performed the three analyses (in the right dash rectangle). First, we analyzed the differences of molecular phenotypes between A1 and A2 (depicted in (b)). We analyzed survival (clinical relevance) with every gene’s mutation status, for each A1 and A2 (depicted in (c)). Finally, we performed Fisher’s exact test for each genes’ mutational status to confirm its statistical association with TP53 mutation status (depicted in (d)). (b) Subdivision of molecular subtype and copy number clusters show a different ratio between TP53WT and TP53MUT groups, within the entire population of Group prevalent. Also, the proportion of mutated genes appears differently between TP53WT and TP53MUT groups (in the mutation profile table). We performed the proportional test14 of the table to measure the significance of the proportional difference of overall mutation rates between A1 and A2. It resulted in p-value 0.01779. The mutation proportions were obtained from cBioPortal38. (c) Survival analysis of every gene according to its TP53 co-status in Group prevalent. In the TP53WT group, patients who had mutations of CTNNB1, SLITRK5 or NCOR2 showed better overall survival (OS) than others. In the TP53MUT group, a mutation in RYR1 affects OS. (d) Fisher’s exact tests and FDR multiple comparison corrections identify five genes significantly related to TP53 status in the Group prevalent, GPR112, MLL3, MTUS2, MYH6, and NRXN1, showing significant different proportions between TP53WT and TP53MUT groups. The adjusted p-values of the five genes are 0.140.
While (as mentioned above) we found that the TP53 mutation status among whole TCGA GC populations did not statistically significantly associate with clinical outcomes (e.g., overall survival) (Supplementary Figure S1), it however associated with some molecular categories (Supplementary Figure S2). These findings led to our further dissection of the GC TCGA patient datasets, looking specifically for TP53-significant clinical implications.
These extended assessments (Fig. 2a) showed that TP53 mutation status does indeed associate with clinical outcomes, as well as molecular phenotypes. Consequently, for the most frequent network expression state (i.e., Group prevalent), we combined TP53 mutation status with that of other genes, resulting in a TP53-related population stratification of GC patients.
The patients of Group prevalent (in the rounded rectangle of Fig. 2b) have high (54.44%) versus low (45%) copy number clustering with other genomic anomalies, including 89 patients classified with chromosomal instability (CIN), 41 with microsatellite instability (MSI), 34 with genomically stable (GS), and 16 with Epstein–Barr virus (EBV)-positive. In the further subdivision of Group prevalent according to TP53 status, TP53WT patients in Group prevalent showed a more uniform distribution of all subtypes, as compared to the total patients of Group prevalent (Fig. 2b). Even so, the TP53MUT patients in Group prevalent were more biased toward CIN subtype than to the other subtypes (GS, MSI, EBV): only 5 and 15 patients presented with GS and MSI respectively, and none with EBV (Fig. 2b). With regard to copy number clustering (Fig. 2b), the TP53WT patients in Group prevalent appeared as a ratio of 3 to 7, high and low cluster each. However, 80% (68) of the patients in the TP53MUT group were high in copy number alterations, while 18.82% (16) of patients were low (Fig. 2b and Supplementary Table S1). Moreover, specifically mutated genes appeared differently between the two groups (TP53WT and TP53MUT in Group prevalent), with ARID1A, CDKN2A, SYNE1, FLG, LRP1B, and OBSCN present in both groups, with ≥20%, and PIK3CA, KMT2D and PLEC mutated in ≥20% in the TP53WT group but not in TP53MUT group (12.9%, 11.8%, and 15.3%, respectively, in the TP53MUT group) (in the table of Fig. 2b). PREX2, SPTA1 and PCLO were present in ≥20% of GC patient tumors in the TP53MUT group; however, only 12.6% PREX2 and 16.8% PCLO mutations, with no available data of SPTA1, in the TP53WT group (in the table of Fig. 2b). We performed the proportional test14 of the table to measure the significance of the proportional difference of overall mutation rates between A1 and A2, resulting in p-value 0.01779 although OBSCN has the same mutation rates in two groups, A1 and A2.
We also revealed that TP53 mutation statistically significantly associated with clinical outcomes (e.g., overall survival) across certain subpopulations. For example, in the TP53WT patients within Group prevalent, we noted an overall survival difference between CTNNB1MUT patients and CTNNB1WT patients (in the upper-left panel in Fig. 2c). In Fig. 2c, the mutation status of the additional genes (SLITRK5, NCOR2, RYR1) significantly associated with overall survival in a given TP53 mutation status, within Group prevalent.
To mine genes significantly related to TP53 status in the Group prevalent, we performed Fisher’s exact tests, while also assessing mutation ratios between the two groups (TP53WT and TP53MUT) in the Group prevalent. Subsequently, Benjamini-Hochberg (BH) false discovery rate (FDR)15 multiple comparison corrections were preformed (see “Mutation Proportions between TP53WT and TP53MUT in Group prevalent” in Methods section for the details). We set FDR-adjusted p-value significance cutoff to 0.15, and selected 5 significant genes, GPR112, MLL3, MTUS2, MYH6 and NRXN1 that show their significant differences of their mutation proportions between TP53WT and TP53MUT groups within Group prevalent (Fig. 2d). We then chose NRXN1, since it showed a higher mutation rate (22.35%) in the TP53MUT Group prevalent, demonstrating an 8.42% mutation rate in TP53WT in Group prevalent (Fig. 2d). We divided the TP53MUT in Group prevalent into two groups again, NRXN1WT and NRXN1MUT (for the description in the next section).
Patient Tumors with NRXN1 Mutation, Concurrent with TP53 Mutation, Significantly Associated with Different Drug Responses
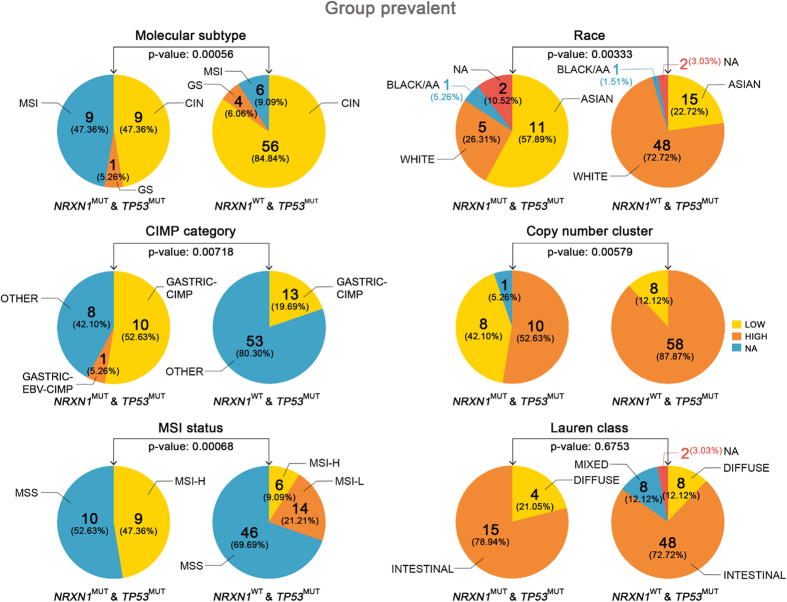
Within the TP53MUT patients in Group prevalent, we inspected several clinical or molecular differences between NRXN1WT (the “B3” group in Fig. 2a) and NRXN1MUT (the “B4” group in Fig. 2a), with Fig. 3 showing that most of the molecular/clinical categories including molecular subtypes, race, CIMP (CpG island methylator phenotype), copy number alterations, and MSI status (except Lauren class) significantly differed between NRXN1WT and NRXN1MUT within the TP53MUT patients in Group prevalent. These significant differences, between the two groups, imply different biological functions, indicating different pharmacological responses.
Figure 3. Two groups (NRXN1MUT with TP53MUT, and NRXN1WT with TP53MUT) in Group prevalent show statistically significant differences in molecular and clinical profiles.
Molecular subtypes, race, CIMP, copy number alterations, and MSI status (except Lauren class) are statistically different by the proportional tests. In the case of NRXN1MUT with TP53MUT group, the ethnic proportion shows ASIAN (57.89%) and WHITE (26.31%), but in the case of NRXN1WT with TP53MUT group, WHITE is 72.72% and ASIAN is 22.72% with p-value, 0.00333. The equivalent table for the figure is in Supplementary Table S5.
In order to show non-randomness (representativeness) of our two subgroups (NRXN1WT (B3) vs. NRXN1MUT (B4) in TP53MUT of Group prevalent) against all GC patients, regarding clinical and molecular features, we performed bootstrapping re-sampling five times (see “Experiment design 1” in Supplementary Method S1 for detailed explanation). In the bootstrapped samples, we did not find significant differences in the majority of clinico-molecular categories (Supplementary Table S6; see also Supplementary Method S1). It suggests that clinical and molecular profiles of our two subgroups are not random. In addition, we performed the same bootstrapping procedures for TP53MUT patients (see “Experiment design 2” in Supplementary Method S1) as well as TP53WT patients (see “Experiment design 3” in Supplementary Method S1) to demonstrate non-randomness of our two subgroups. Analysis (Supplementary Table S7, Supplementary Table S8) also showed that the bootstrapped samples were not significant in the majority of clinico-molecular categories. It complies with non-randomness of our two subgroups (NRXN1WT (B3) vs. NRXN1MUT (B4) in TP53MUT of Group prevalent).
For looking at the different drug responses, we examined differences in drug sensitivity between NRXN1WT and NRXN1MUT, within the TP53MUT patients in Group prevalent. In addition, we inspected clinico-molecular profiles of TP53WT and NRXN1WT (the “B1” group of Fig. 2a) patients in Group prevalent (Supplementary Figure S5).
Before using the Connectivity Map (CMAP) 2.016, the compilation of specific drug-associated gene expression “signatures”, we first aimed at identifying GC cell lines corresponding to the two groups. Previous genomic characterizations17 revealed that SNU-668, NCI-N87, and NUGC-3 cell lines possess TP53MUT and NRXN1MUT (Supplementary Table S2). Golub17 group also reported that other GC cell lines, including MKN74 and SNU-620 (Supplementary Table S2), possessed TP53MUT and NRXN1WT. We next aimed at identifying which cell lines in Supplementary Table S2 are aligned with the “Group prevalent” patients by using the WNT signaling genes of Fig. 1c. Using the correlation classification method18,19 (see details in Method), the GC cell lines were aligned with their GC patient groups (see column “Is it Group prevalent?” in Supplementary Table S2), identifying three cell lines (SNU-16, FU97, and SNU-668 cells) were assigned to Group prevalent of the TCGA GC patients. Through the mutation status of the three cell lines, the two cell lines, SNU-16 and FU97, were considered as the representative cell lines for Group prevalent patients possessing NRXN1WT and TP53MUT (B3). SNU-668 was considered as the representative Group prevalent patients having NRXN1MUT and TP53MUT (B4).
We then used the three-cell line expression as input for CMAP 2.0 (broadinstitute.org/cmap), for extrapolating drug-associated transcriptomes for the two patient groups (NRXN1MUT/TP53MUT vs. NRXN1WT/TP53MUT in Group prevalent). Based on the log2 transformed gene expression of the three cell lines from the CCLE20 (www.broadinstitute.org/ccle), we calculated the fold-changes of NRXN1MUT/TP53MUT cells (SNU-668) over NRXN1WT/TP53MUT cells (SNU-16, FU97). It is noted that, due to the small number of the cells lines of interest, we used the fold-changes instead of p-values and set the fold-change cutoff as 50 (either of greater than 50 or less than 1/50). We obtained the highly or less expressed genes between the two groups. Subsequently, we used the selected genes as input for CMAP 2.016 (broadinstitute.org/cmap), for extrapolating drug-associated transcriptomes for the two patient groups (NRXN1MUT/TP53MUT vs. NRXN1WT/TP53MUT in Group prevalent).
As a result, small compounds that could work differentially in the two groups were identified (Table 1), suggesting drug response differences according to NRXN1 mutation status within TP53 mutation. In Table 1, cardiac-relating agents (vanoxerine, (+)-isoprenaline) as well as antineoplastic agents (exisulind, etoposide) were highly ranked for reversing gene expression of cells possessing TP53MUT and NRXN1WT. But, in TP53MUT/NRXN1MUT cells, an antiviral agent (levcycloserine), an antimalarial agent (chloroquine), and a cholesterol-lowering drug (tetraethylenepentamine) were associated with reversing gene expression of the cells. Considering the different drug preferences of these cell lines, NRXN1 mutation status, within TP53-mutated patients in Group prevalent, demonstrates that our approach may provide different optimal pharmacological options, according to TP53 mutation status-related patient subpopulations in GC, thus conferring individualized patient benefits.
Table 1. Different compound response for TP53 mutant GC cells according to NRXN1 mutation status.
| Mutation status of GC cells | CMAP compounds | Scorea (p-value) | Mutation status of GC cells | CMAP compounds | Scoreb (p-value) |
|---|---|---|---|---|---|
| NRXN1MUT & TP53MUT (SNU-668) | Spaglumic acid | −0.863 (0.038) | NRXN1WT & TP53MUT (SNU-16, FU97) | Exisulind | 0.9 (0.021) |
| 5248896 | −0.855 (0.042) | Sulfaquinoxaline | 0.811 (0.014) | ||
| Levcycloserine | −0.853 (0.00088) | Mebeverine | 0.804 (0.0028) | ||
| Diphemanil metilsulfate | −0.841 (0.00028) | Etoposide | 0.8 (0.003) | ||
| Chloroquine | −0.8 (0.0032) | Protriptyline | 0.771 (0.0053) | ||
| Paroxetine | −0.777 (0.0051) | Vanoxerine | 0.765 (0.0058) | ||
| Ramifenazone | −0.758 (0.007) | (+)-isoprenaline | 0.762 (0.0061) | ||
| Ambroxol | −0.74 (0.0090) | Fendiline | 0.745 (0.032) | ||
| Tetraethylenepentamine | −0.723 (0.00089) | Benzydamine | 0.742 (0.0084) | ||
| Oxybuprocaine | −0.709 (0.015) | Pimethixene | 0.717 (0.044) |
aThis score is the enrichment score and its p-value reported by CMAP 2.0. The negative scores indicate that the compounds could reverse the gene expression profiles of the GC cells with NRXN1MUT & TP53MUT.
bThis score is the enrichment score and its p-value reported by CMAP 2.0. The positive scores indicate that the compounds could reverse the gene expression profiles of the GC cells with NRXN1WT & TP53MUT.
Connectivity Map (CMAP) 2.0 reported differentially preferable compounds (or perturbagen) for reversing the expression profiles for the given GC cells. The list contains the top 10 ranked enrichment score compounds (with the p-value less than 0.05) for the two cell groups (NRXN1MUT & TP53MUT; NRXN1WT & TP53MUT). It is noted that SNU-668, SNU-16, and FU97 cells were assigned to Group prevalent.
Discussion
The NCI-MATCH trial moves toward precision medicine to find a drug-mutation pairing in a certain disease subpopulation21. In precision medicine, genomic alterations (e.g., mutations, copy number, etc.) have been used to serve as biomarkers for “individualizing” treatment of distinct patient subpopulations with specific “targeted” drugs. As TP53 is one of the most prevalent mutations in cancer overall, its roles in cancer biology have been extensively studied to determine its role in the biological importance in cancer development1,2,3. However, it still remains to be answered to find clinical significance for association between TP53 mutations and molecular/clinical categories (e.g., OS, and molecular subtypes)1,2,3. In that line, our approach, utilizing TP53 mutation and network gene expression states in combination with other genes mutation status, one can find TP53-relating significant associations from clinical or molecular categories. It potentially can be used in certain disease subtype identification for molecular targeted therapies.
In our previous report10, actin cytoskeleton signaling (including focal adhesion) and chemokine signaling were revealed to associate with GC. NRXN1 has also been linked with actin cytoskeleton dynamics in neurodevelopment and autism22, although its role(s) in cancer remain unstudied23. Through Ingenuity Pathway Analysis (IPA)24 we detailed a network where TP53 and NRXN1 connect via 64 genes (Supplementary Figure S3), in agreement with experimental evidence-based publications. Also, IPA revealed the genes involved in the pathway to be enriched in the IPA Top Functions & Diseases terms: “Cellular Growth and Proliferation,” “Gene Expression,” and “Cellular Development.” Thus, both TP53 and NRXN1, in this specific network may associate with two of the “hallmarks” of cancer25, namely “sustaining proliferative signaling” and “evading growth suppressors”. It implies that the mutation statuses of TP53 and NRXN1 in Group prevalent need to be experimentally validated with regard to the two cancer hallmark phenotypes. Also, CTNNB1, CEBPA, SRC, PTGS2, PRKCB, PPP3CA, NFKB1, MYC, and LEF1 are all connectors within our WNT-relating network in Fig. 1b, incidentally, in the paths (Supplementary Figure S3) between TP53 and NRXN1. Further experimental validation studies will assess the possible biological effects of these genes on the GC WNT-related network, using the connectors according to the mutation statuses of TP53 and NRXN1.
Since laminin G domains in NRXN1 (1477 amino acids in length) interact with a partner, NLGN1 (neuroligin 1), structural analysis of NRXN1 (neurexin 1) gives clues on structure influence on the interaction by NRXN1 missense mutations in the tumors. The NRXN1 missense mutations (Supplementary Table S3) in the NRXN1MUT and TP53MUT patients within the Group prevalent were correctly aligned to a region corresponding to the first laminin G domain (a.a. 1 to 256 in its protein sequence). Due to no available 3-D structures for the region of the NRXN1 protein sequence, we used a homology modeling26 for 3D structure generation of the domain (details in Supplementary Figure S4). Based on a neuroligin/neurexin-1beta complex structure (PDB ID: 3B3Q), we aligned and superimposed the homology model of NRXN1 for identifying structural effects of the missense mutations. As shown in Supplementary Figure S4, the R124C and D254G mutations are closely localized to the physical interface between NLGN1 and the first laminin G domain of NRXN1, and they could change physico-chemical properties in the interface interaction of the two proteins. Considering the connections (Supplementary Figure S3) between NRXN1 and TP53, through our WNT-relating pathway network, the effect of the NRXN1 mutations needs to be measured in WNT signaling, possibly linking to other signaling subnetworks that underlie the two cancer hallmark phenotypes.
In the study, we utilized a binary based network approach for exploring patient stratification to identify clinical relevance. In general, depending on batches, samples and outliers, a cutoff-based binary transformation of gene expression could be affected27. In the line, our result needs to be further evaluated. Also note that the TCGA Gastric Cancer research group9 concluded that the GC samples did not show major batch effects.
In conclusion, TP53 mutation status can reveal significant relevance in clinical or molecular categories, by utilizing pathway-guided network states and other gene mutation statuses.
Method
Data Collection
To study TCGA human stomach adenocarcinomas, we used TCGA GC patient RNA-Seq/miRNA-Seq expression processed by UCSC Cancer Genomics Browser (CGB) group28. According to UCSC CGB28 data description, the GC patient gene expression was calculated by transforming log2(x + 1), where x is the RPKM value of the level 3 data from TCGA data coordination center. Also, miRNA expression was calculated by transforming log2(x), where x is the RRM (reads per million) value of the level 3 data from the coordination center. We downloaded an RNA-Seq dataset (version: TCGA_STAD_exp_HiSeq-2015-01-28), a miRNA-Seq dataset (version: TCGA_STAD_miRNA_HiSeq-2015-02-24), and a somatic mutation dataset (version: TCGA_STAD_mutation_curated_broad_gene-2015-01-28; curated by Broad Institute Genome Sequencing Center), from the UCSC CGB28. The total number of cancer patients was 376, and their corresponding clinical information was also downloaded from the same web page. Out of 376, we obtained 233 patients of which each has the complete set of mRNA expression, miRNA expression, and mutation. These patients are in our scope for the following analysis.
Data Normalization and Binarization
We performed median normalization on the samples, and we transformed expression values of each gene into binary values by following an edge detection algorithm27. To binarize expression values, given a specific gene, we sorted the gene expression values in an ascending order and calculated all the gradients between the two neighboring expression values. Subsequently, we obtained the greatest gradient (distance) of which the two adjacent expression values (the lower one, and the upper one) were identified. Setting the lower one as the cutoff of the given gene, we transformed the expression values less than or equal to the cutoff to 0; and greater than the cutoff to 1. We repeated the procedures for whole genes in miRNAs and mRNAs.
GC Signaling Network Construction
We constructed the signaling network (of Fig. 1b) with 34 WNT signaling genes identified by manual curation as well as PATHOME10. Our previous studies10,11 demonstrated that the WNT signaling plays an important role in GC tumorigenesis by using computational analysis, in-vitro assay, and xenograft. Also, 10 miRNAs regulating the WNT signaling genes, and 20 upstream regulators (e.g., transcription factors (TFs), signaling molecules) associated with the miRNAs were added to the network, resulting in Fig. 1b (details in Supplementary Table S4). The miRNA-target relations and the miRNA’s upstream regulators were obtained from two literature-evidence based databases: miRTarBase release 4.229, and TransmiR v1.130. Finally, we transformed the network into a tabular representation of the binary expression values of the network entries (Fig. 1c).
Patient Grouping Based on Network Expression States and Mutation Statuses
We next mapped binary expression levels to the “flattened” signal pathway structure (Fig. 1c), and counted tumor database samples for all network expression statuses. We labeled “Group prevalent” in which the network expression status has the majority of samples. We further dissected the Group prevalent according to mutation statuses of TP53 and another genes (Fig. 2a), using Fisher’s exact test for obtaining p-values in the association between TP53 mutation status and another gene (say, Gene G in Fig. 2a) mutation status in Group prevalent, and the log-rank test for possible clinical relevance with TP53MUT or TP53WT within Group prevalent, according to the mutation status of some specific gene G.
Mutation Proportions between TP53 WT and TP53 MUT in Group prevalent
Based on the whole 233 TCGA patients, Group prevalent were identified by the procedures of Fig. 1a. Then, we divided “Group prevalent” (180 patients) into two groups, TP53WT (“A1” in Fig. 2a) and TP53MUT (“A2” in Fig. 2a) according to TP53 mutation status (Fig. 2a). Using the TCGA gastric cancer dataset from cBioPortal31 (cbioportal.org), we obtained all the mutation rates of genes for the two groups. Subsequently, we selected the genes of which mutation rates are greater than or equal to 20% in either of the two groups (TP53WT (A1) in Group prevalent vs. TP53MUT (A2) in Group prevalent). These genes were listed in the table of Fig. 2b. For example, in PLCO, its mutation rate 16.80% in A1 and 23.50% in A2. We calculated the p-values (the last column in the table of Fig. 2b) of the individual genes by using Fisher’s exact tests. For Fisher’s exact test, given a gene G, 2 by 2 contingency table was obtained. In the contingency table, one factor is TP53 mutation status (TP53WT, TP53MUT), and the other factor the G mutation status (GWT, GMUT). In addition, we performed the proportional test14 (prop.test function in R package) for measuring the significant difference of mutational distributions of the listed genes (in the table of Fig. 2b) between A1 and A2.
We narrowed down all the genes in the TCGA GC somatic mutation data through the following three steps before statistical tests: (i) we obtained the mutational proportions of each gene for TP53MUT patients (A2) and TP53WT patients (A1) in the Group prevalent; (ii) for each gene, we calculated difference of the two mutational proportions between TP53MUT (A2) and TP53WT (A1) patient groups in Group prevalent; (iii) setting the difference greater than 5% as cutoff, 537 genes were obtained.
We tested the 537 genes by using the Fisher’s exact tests, resulting in the 537 p-values (equivalently, unadjusted p-values). Subsequently, we performed the FDR multiple comparison corrections15 (using p.adjust() function in R package) for the 537 p-values, resulting in 537 FDR-adjusted p-values. Since consensus of FDR significance cutoff has not been reached so far, we looked into FDR cutoffs of diverse studies32,33,34,35,36,37. FDR cutoff ranging from 0.1 to 0.25 has also been accepted in popular bioinformatics tools (including GSEA32 and DESeq236) as well as scientific publications33,34,35. Considering the FDR cutoff range, we set 0.15 as our FDR-adjusted p-value cutoff, and 263 significant genes out of the 537 were obtained. We selected five significant genes (as shown in the table of Fig. 2d) out of the 263 genes. All the FDR-adjusted p-values of the five genes were 0.140.
Alignment of GC cell lines with Group prevalent patients by the correlation classification method (CCM)
We used an existing algorithm, the correlation classification method (CCM)18,19 package, to match the cell lines to the patient groups (Group prevalent vs. the other group). The package took two gene expression matrices: one for the gene expression matrix of the whole GC patients for the WNT signaling genes; and the other for the gene expression matrix of the GC cell lines (of Supplementary Table S2) for the WNT signaling genes. It is noted that, through the group assignment in Fig. 2a, all the GC patients were already assigned to either Group prevalent or the other group. The CCM package measures the similarity (Spearman’s rank correlation) between the cell lines and the patient groups via the two gene expression matrices and assigns the cell lines to their representative patient groups (either one of the two groups: Group prevalent or the other group).
The gene expression of the GC signaling (the WNT signaling) was inspected in both the whole TCGA GC patients (processed by UCSC CGB28 group) and the GC cell lines (described in Supplementary Table S2). The GC patient gene expression of the UCSC CGB was log2(x + 1), where x is the RPKM value. From the UCSC CGB data, we obtained gene expression matrix (row: genes; column: patients) of the whole GC patients for the WNT signaling. The GC cell line gene expression of the GC signaling (the WNT signaling) was obtained from the Cancer Cell Line Encyclopedia20 (CCLE) data normalized and processed by the cBioPortal31 (cbioportal.org) group. From the CCLE data, we obtained the gene expression matrix (row: genes; column: cell lines) of the GC cell lines for the WNT signaling. By taking these two expression matrices, the CCM package reported that SNU-668, SNU-16, FU97 correspond to Group prevalent patients.
Additional Information
How to cite this article: Park, S. et al. Clinical Relevance and Molecular Phenotypes in Gastric Cancer, of TP53 Mutations and Gene Expressions, in Combination With Other Gene Mutations. Sci. Rep. 6, 34822; doi: 10.1038/srep34822 (2016).
Supplementary Material
Acknowledgments
This research has been performed by a subproject of KISTI’s project No. P16018 (Development of HPC-based Big Data for healthy Aging Society) funded by (Ministry of Science, ICT, and Future Planning) (to S.N.). This work was supported by the Gachon University Gil Medical Center (Grant number: 2016-06) (to S.N.), and by a grant from the KOBIC Research Support Program (to J.L.). This work was supported by the National Research Foundation (NRF-2015R1A2A1A10052661 to Y.H.K.). Authors appreciate Curt Balch for editing the draft.
Footnotes
Y.H.K. is an employee of CrystalGenomics Inc. (Seongnam-si, Korea). This entity has no financial interest in the results of this study or public deposition of any data.
Author Contributions Supervised the study: S.N. Performed the study, and wrote the draft: S.P. and S.N. Performed the structural analysis: J.L. Helped the draft: Y.H.K., J.P., J.-W.S. and J.L. Helped the Figure design: Y.H.K.
References
- Fenoglio-Preiser C. M., Wang J., Stemmermann G. N. & Noffsinger A. TP53 and gastric carcinoma: a review. Hum Mutat 21, 258–270, doi: 10.1002/humu.10180 (2003). [DOI] [PubMed] [Google Scholar]
- Tan P. & Yeoh K. G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 149, 1153–1162, doi: 10.1053/j.gastro.2015.05.059 (2015). [DOI] [PubMed] [Google Scholar]
- Olivier M., Hollstein M. & Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2, a001008, doi: 10.1101/cshperspect.a001008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Ong K., Giaever G. & Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol 20, 648–659, doi: 10.1016/j.chembiol.2013.04.007 (2013). [DOI] [PubMed] [Google Scholar]
- Robles A. I. & Harris C. C. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2, a001016, doi: 10.1101/cshperspect.a001016 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro A. J., Lain S. & Lane D. P. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer 92, 434–444, doi: 10.1038/sj.bjc.6602358 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccarati A. et al. Mutations and polymorphisms in TP53 gene–an overview on the role in colorectal cancer. Mutagenesis 27, 211–218, doi: 10.1093/mutage/ger067 (2012). [DOI] [PubMed] [Google Scholar]
- Oden-Gangloff A. et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer 100, 1330–1335, doi: 10.1038/sj.bjc.6605008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209, doi: 10.1038/nature13480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. et al. PATHOME: an algorithm for accurately detecting differentially expressed subpathways. Oncogene 33, 4941–4951, doi: 10.1038/onc.2014.80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. R. et al. HNF4alpha is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut 65, 19–32, doi: 10.1136/gutjnl-2014-307918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. H. & El-Omar E. M. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 11, 664–674, doi: 10.1038/nrgastro.2014.143 (2014). [DOI] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C. F. & Vousden K. H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16, 393–405, doi: 10.1038/nrm4007 (2015). [DOI] [PubMed] [Google Scholar]
- Newcombe R. G. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med 17, 873–890 (1998). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- Lamb J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935, doi: 10.1126/science.1132939 (2006). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer. Nat Commun 5, 3830, doi: 10.1038/ncomms4830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancik G. M., Ru Y., Owens C. R. & Theodorescu D. A framework to select clinically relevant cancer cell lines for investigation by establishing their molecular similarity with primary human cancers. Cancer Res 71, 7398–7409, doi: 10.1158/0008-5472.CAN-11-2427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing, Vienna (AU): R Development Core Team. Available from http://www.r-project.org (2015). [Google Scholar]
- Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607, doi: 10.1038/nature11003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov 14, 513–515, doi: 10.1038/nrd4694 (2015). [DOI] [PubMed] [Google Scholar]
- Persico A. M. & Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci 29, 349–358, doi: 10.1016/j.tins.2006.05.010 (2006). [DOI] [PubMed] [Google Scholar]
- Schuller H. M. Neurotransmission and cancer: implications for prevention and therapy. Anticancer Drugs 19, 655–671, doi: 10.1097/CAD.0b013e3283025b58 (2008). [DOI] [PubMed] [Google Scholar]
- Kramer A., Green J., Pollard J. Jr. & Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530, doi: 10.1093/bioinformatics/btt703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- Kim T. R. et al. A simplified homology-model builder toward highly protein-like structures: an inspection of restraining potentials. J Comput Chem 33, 1927–1935, doi: 10.1002/jcc.23024 (2012). [DOI] [PubMed] [Google Scholar]
- Shmulevich I. & Zhang W. Binary analysis and optimization-based normalization of gene expression data. Bioinformatics 18, 555–565 (2002). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. The UCSC Cancer Genomics Browser. Nat Methods 6, 239–240, doi: 10.1038/nmeth0409-239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. D. et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39, D163–169, doi: 10.1093/nar/gkq1107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lu M., Qiu C. & Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res 38, D119–122, doi: 10.1093/nar/gkp803 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong E. Y. et al. Local false discovery rate estimation using feature reliability in LC/MS metabolomics data. Sci Rep 5, 17221, doi: 10.1038/srep17221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandam A. et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 19, 619–625, doi: 10.1038/nm.3175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. RhoA determines lineage fate of mesenchymal stem cells by modulating CTGF-VEGF complex in extracellular matrix. Nat Commun 7, 11455, doi: 10.1038/ncomms11455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanli K., Karlsson F. H., Nookaew I. & Nielsen J. FANTOM: Functional and taxonomic analysis of metagenomes. BMC Bioinformatics 14, 38, doi: 10.1186/1471-2105-14-38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.