Abstract
The intestinal microbiota is a large and diverse microbial community that inhabits the intestinal tract, containing about 100 trillion bacteria from 500–1000 distinct species that, collectively, provide multiple benefits to the host. The gut microbiota contributes to nutrient absorption and maturation of the immune system, and also plays a central role in protection of the host from enteric bacterial infection. On the other hand, many enteric pathogens have developed strategies in order to be able to outcompete the intestinal community, leading to infection and/or chronic diseases. This review will summarize findings describing the complex relationship occurring between the intestinal microbiota and enteric pathogens, as well as how future therapies can ultimately benefit from such discoveries.
This article is part of the themed issue ‘The new bacteriology’.
Keywords: intestinal microbiota, inflammation, nutrient, pathogenic bacteria, virulence
1. Introduction
The intestinal microbiota is the collective term describing the large and diverse microbial community that inhabits our intestine. In humans, the microbiota contains about 100 trillion bacteria from 500–1000 distinct species that provide multiple benefits to the host. Among those beneficial functions, the intestinal microbiota plays a central role in (i) shaping the intestinal immune system [1] by contributing to immune system development and maturation, and (ii) nutrient acquisition, by greatly enhancing the metabolic capacity of the gut, thus providing a range of essential nutrients for the host [2]. Another important benefit conferred by the intestinal microbiota to the host intestine is the protection from colonization by exogenous pathogens—a phenomenon nowadays named colonization resistance—and from overgrowth of indigenous pathobionts (potential pathogenic symbionts of the microbiota) [3–5]. The colonization resistance, termed the ‘microbial barrier’ in the early 1980s [6], is the mechanism whereby the intestinal bacteria form a barrier to prevent incursion by new bacteria of other species or other strains of the same species. This notion is well exemplified by the range of infections resulting from the use of antibiotics, such as Clostridium difficile infection [7], as well as by the observation that many enteric pathogens induce stronger disease in mice under germ-free conditions (in the absence of an intestinal microbiota) or following antibiotic treatments [8–12] (figure 1). Mechanisms that regulate the ability of the microbiota to restrain pathogen growth are complex and include competitive metabolic interactions, localization to intestinal niches and induction of host immune responses [4]. Pathogens, in turn, have developed strategies in order to escape from colonization resistance conferred by the commensal community. Unexpectedly, the intestinal microbiota can also play a role in providing nutrients to some intestinal pathogens, or may play a direct role in activating virulence of pathogenic bacteria that will otherwise stay avirulent. In addition, in some particular conditions, the intestinal microbiota may actually drive disease, as is for example the case for inflammatory bowel diseases. This review will summarize these concepts and will describe the most recent findings elucidating the intriguing relationship between the intestinal microbiota and enteric pathogens. We will discuss how the understanding of microbiota–pathogen interactions may ultimately lead to new therapeutic approaches in order to treat infectious diseases.
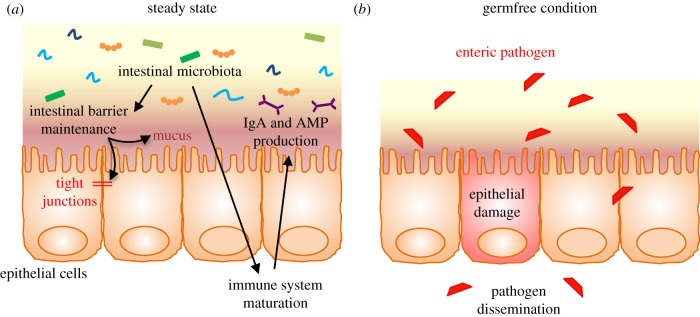
Figure 1.
Microbiota/host homeostasis in the intestine. (a) The intestinal microbiota plays a central role in intestinal barrier maintenance (mucus production and intestinal tight junctions maintenance) and immune system maturation (lymphocytes development, production of IgA and antimicrobial peptides). (b) In the absence of an intestinal microbiota, enteric pathogens can induce epithelial damage and have the potential to disseminate. AMP, antimicrobial peptide; IgA, immunoglobulin A.
2. Role of the intestinal microbiota in colonization resistance
The intestinal microbiota is playing a central role, through multiple mechanisms, in protecting the host intestine from pathogen colonization (figures 1 and 2).
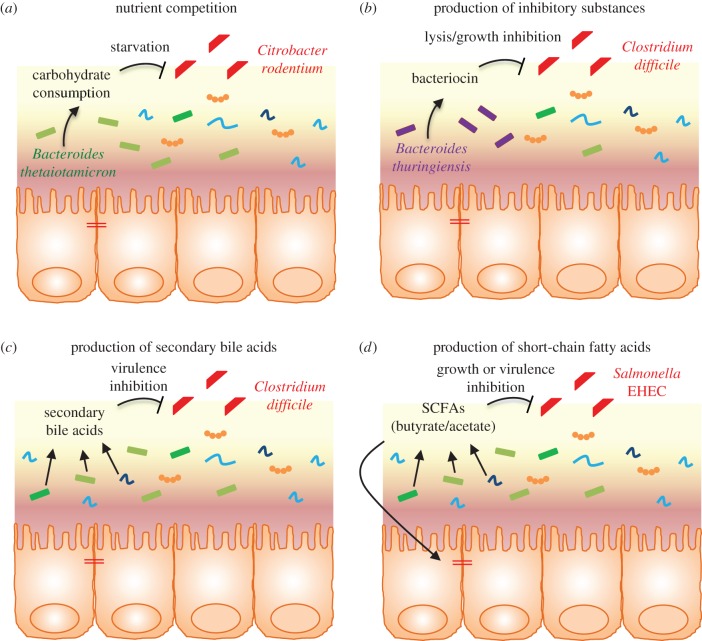
Figure 2.
Intestinal microbiota-mediated colonization resistance. Examples of microbiota-mediated direct inhibition of intestinal colonization by enteric pathogens. Intestinal microbiota prevents colonization by enteric pathogens by competing for nutrients (a) or producing inhibitory substances such as bacteriocin (b), secondary bile acids (c) and short-chain fatty acids (d). EHEC, enterohaemorrhagic Escherichia coli; SCFAs, short-chain fatty acids.
(a). Direct inhibition of colonization by enteric pathogens
The concept of protection of the host intestine from pathogens by commensal bacteria, also called colonization resistance, was first described to be the result of microorganism-mediated direct inhibition [13]. Indeed, many bacteria directly inhibit intestinal pathogens by competing for nutrients or by inducing the production of inhibitory substances. One example highlighting the former is the finding that the commensal Bacteroidetes thetaiotaomicron consumes carbohydrates used by the pathogen Citrobacter rodentium, thus leading to competitive exclusion of the pathogen from the intestine [14] (figure 2). By consuming common limited resources, the gut microbiota induces the starvation of competing pathogens [4]. Through the production of specific metabolites, the intestinal microbiota can also modify the host environmental conditions, then compromising pathogen growth and/or virulence. Butyrate, a short-chain fatty acid (SCFA) produced by the intestinal microbiota, can downregulate the expression of several virulence genes of Salmonella enterica serovar Enteritidis (S. Enteritidis) and Typhimurium (S. Typhimurium) [15] and has been shown to inhibit the growth of enterohaemorrhagic Escherichia coli (EHEC) [16]. Some Bifidobacteria strains can also protect from EHEC infection through the production of acetate [10].
The intestinal microbiota community is also able to produce a large number of broadly bioactive small molecules that act toward other members of the intestinal microbiota and/or toward enteric pathogens. Bacteriocins, for example, are antimicrobial peptides that can have narrow to broad activity spectrums and can selectively kill and/or inhibit the growth of competing bacteria [17]. As an example, Bacteroides thuringiensis is able to secrete a bacteriocin (thuricin CD) that directly targets spore-forming Bacilli and Clostridia, including C. difficile [18]. Similarly, some strains of E. coli are able to produce bacteriocin that can directly inhibit the growth of the EHEC enteric pathogen [19], and anti-Listeria bacteriocins were found to be expressed by Enterococcus faecium and Pediococcus pentosaceus [20,21]. Recently, a computational prediction of biosynthetic gene clusters that encode small molecules in the human gut microbiota identified almost 600 candidate clusters, including many newly annotated antimicrobial peptides, suggesting that these small molecules might mediate commensal–commensal and commensal–pathogen interactions [22]. Another in silico study identified 74 putative-encoding bacteriocin clusters in the gastrointestinal tract, based on the human microbiome project's reference genome database [23,24].
Another somewhat direct inhibitory effect of the intestinal microbiota toward pathogens is through a mechanism involving bile acids. Produced in the liver and delivered into the duodenum, bile acids are subsequently modified by the gut microbiota into a myriad of secondary bile acids that can act as anti-bacterial factors. Highlighting the role of the intestinal microbiota in secondary bile acid production is the observation of very low or undetectable levels in germ-free animals [25] and a dramatically reduced production following antibiotic treatment [26]. The best example of a protective mechanism conferred by microbiota-derived secondary bile acids is the observation that the depletion of microbial members involved in converting primary bile acids into secondary bile acids favours the colonization of the gastrointestinal tract by C. difficile, the most prominent pathogen exploiting antibiotic-mediated alteration of the microbiota [7,26–30]. Importantly, the administration of Clostridium scindens (a bile acid 7α-dehydroxylating bacterium) is sufficient to confer resistance to C. difficile infection in a secondary bile acid-dependent manner. Recent works have described that the intestinal microbiota can also indirectly control enteric pathogens by other mechanisms, such as by shaping the immune system and the inflammatory response (immune-mediated colonization resistance).
(b). Immune system maturation and inflammation process
The microbiota plays a primordial role in the maturation of the intestinal immune system, as demonstrated by the observation that germ-free mice are heavily immuno-depressed. Indeed, the intestine of germ-free animals lack Peyer's patches, and have a reduced expression of both antimicrobial peptides and immunoglobulin A (IgA), molecules implicated in intestinal immunity [31–36]. An example of a member of the microbiota that contributes to the immune system development is Bacteroides fragilis, with the demonstration that monocolonization of germ-free mice with this bacterium is sufficient to promote the development of CD4T lymphocytes [37]. Similarly, segmented filamentous bacteria have been described to be sufficient to drive the differentiation of CD4T cells into Th17 cells, important for protection against the intestinal pathogen C. rodentium [38–41].
The intestinal microbiota is also an inexhaustible source of ligands for the innate immune system. Antibiotic treatment is sufficient to increase susceptibility of mice to dextran sodium sulfate (DSS)-mediated colitis, a phenomenon that can be rescued by the administration of Toll-like receptor ligands [42]. Such a finding initiated the concept that pattern recognition receptor signalling originating from the intestinal microbiota is necessary for the steady-state protection of the intestine [42,43]. This has also been illustrated by the discovery that bacterial flagellin from the intestinal microbiota is able to prevent and cure rotavirus infection though a mechanism requiring flagellin receptors Toll-like receptor 5 (TLR5) and NOD-like receptor C4 (NLRC4) and involving interleukin-22 and interleukin-18 production [44]. Bacteria belonging to the Clostridium group cluster XIVa are able to induce the development of anti-inflammatory T regulatory cells [45], and SCFAs derived from the intestinal microbiota play a central role in maintaining the balance of inflammatory and anti-inflammatory T cell subsets [46–49]. Importantly, some TLR ligands, such as LPS, are modified by the host in order to be less bioactive, indicating the existence of mechanisms to detoxify such pro-inflammatory molecules, thus avoiding over-activation of the intestinal immune system in response to the commensal bacterial population [50–52]. All these reports are good examples of the roles played by the microbiota in the maturation of the intestinal immune system, where the intestinal bacterial population can generate pro-inflammatory or anti-inflammatory responses, both central for avoiding over-activation of the intestinal immune system but conferring protection against enteric pathogens.
(c). Intestinal barrier maintenance
Another mechanism by which the intestinal microbiota protects the intestine against enteric pathogens infection is by acting on the ‘strength’ of the intestinal barrier, such as the thickness and composition of the mucus layer, as well as the maintenance of the intestinal tight junctions. It is indeed primordial for the host to keep the intestinal microbiota at a safe distance from the intestinal epithelium, in order to minimize the appearance of tissue damage and inflammation [53]. The intestinal microbiota is restricted to the intestinal lumen by a mucus layer that overlays the epithelium. Treatment of mice with antibiotics reduces the mucus layer thickness and results in an increased contact of the bacteria with the intestinal epithelium monolayer that can be highly deleterious in the context of an enteric pathogen infection, as exemplified with C. rodentium [54]. In addition, the intestinal microbiota maintains the intestinal barrier though the production of SCFAs, which are a primary nutrient for the colonic epithelium and contribute to the control of mucin production [55,56]. Thus, a decrease in production of SCFAs may result in the reduction of mucus thickness/degradation of the mucus barrier.
3. When the intestinal microbiota leads to enteric pathogen virulence
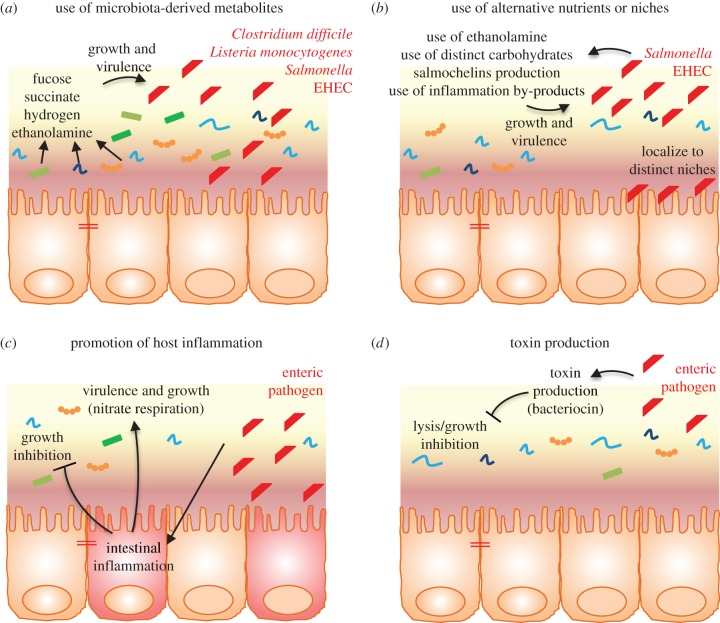
While all the mechanisms described above are central to protect the intestine against bacterial infection and pathogens, the microbiota can also trigger bacterial virulence. For example, the intestinal microbiota can also produce metabolites that might unexpectedly enhance pathogen virulence expression and colonization in the gut [57–61] (figure 3a). Bacteroidetes thetaiotaomicron, found to lead to a competitive exclusion of C. rodentium through the consumption of similar carbohydrates [14], can also cleave sialic acid moieties from mucin and produce high levels of succinate that can lead to an enhanced colonization by C. difficile [58,59]. The production of fucose or succinate from the host mucin by commensal bacteria can also modulate the expression of the virulence factor ler, a master regulator of the locus of enterocyte effacement (LEE) genes in EHEC [60,62], thus contributing to EHEC virulence. Other examples of pathogens that utilize the intestinal microbiota to facilitate their own infection include C. difficile whose spores require by-products from the microbiota, such as bile salts, to germinate [63] and S. Typhimurium that utilizes di-hydrogen generated by the microbiota for its luminal growth [64]. Moreover, microbiota-produced ethanolamine is used as a nitrogen source and a regulator of virulence genes by EHEC, S. Typhimurium and Listeria monocytogenes [65–68]. Akkermansia muciniphila, a mucin degrading commensal bacterium that resides in the mucus layer and that can confer protection against obesity and metabolic disorders [69,70], is also able to exacerbate S. Typhimurium-induced intestinal inflammation by its ability to disturb host mucus homeostasis [71].
Figure 3.
Pathogen strategies to overcome microbiota-mediated colonization resistance. (a) The production of metabolites by commensal bacteria can modulate the growth and expression of virulence genes by enteric pathogens. (b) Enteric pathogens can compete with the microbiota by using alternative nutrients and/or niches. (c) Promotion of intestinal inflammation by enteric pathogens inhibits commensal bacteria growth, conferring an advantage to enteric pathogens. (d) Enteric pathogens produce toxins that can directly and specifically target commensals. EHEC, enterohaemorrhagic Escherichia coli.
4. When intestinal microbiota alteration by a pathogen leads to chronic diseases
While previous examples illustrated how the intestinal microbiota can promote infection by enteric pathogens, some findings also revealed that intestinal microbiota alteration by a pathogen or a pathobiont can lead to chronic diseases. As an example, it was shown that colonization of adherent-invasive E. coli (AIEC, a pathovar of E. coli involved in Crohn's disease pathogenesis) during microbiota acquisition drove chronic colitis in mice lacking the flagellin receptor TLR5 [72]. The observation that such colitis persisted well beyond AIEC clearance leads to the conclusion that AIEC bacteria instigate chronic inflammation by altering the intestinal microbiota composition in a way that increases its pro-inflammatory potential [73]. These data suggest that AIEC, and perhaps other pathobionts, may instigate chronic inflammation in susceptible hosts by altering the gut microbiota composition so as to give it an inherently greater ability to activate innate immunity/pro-inflammatory gene expression [73], leading to the concept of pathobiome [74]. Similarly, Yersinia enterolitica infection of mice lacking the receptor TLR1 leads to an alteration of the microbiota composition and to the generation of anti-commensal immunity, that ultimately leads to the development of chronic intestinal inflammation [75]. Thus, these two findings describe that an acute infection can drive long-term immune and microbiota alterations, leading to chronic inflammatory disease in a genetically predisposed host.
5. How bacterial pathogens emancipate themselves from the intestinal microbiota
As discussed above, the intestinal microbiota is playing a central role, through multiple mechanisms, in pathogen colonization resistance. However, in turn, pathogens have evolved strategies to escape some of those mechanisms (figure 3b–d).
(a). The use of alternative nutrients or niches
Even if the intestinal environment is qualitatively and quantitatively rich in nutrients, the large load of bacteria ultimately leads to competition. One mechanism by which an enteric pathogen can compete with the microbiota is by using a distinct metabolic repertoire. Ethanolamine, which is released into the intestine during epithelial cell turnover, is for example used by some pathogens [76], and genes involved in the use of ethanolamine are preferentially found in the genomes of enteric pathogens [77]. The foodborne illness pathogen EHEC is able to utilize galactose, hexuronate, mannose and ribose as carbon sources, while commensal E. coli cannot use such sugars [78,79]. In addition, some pathogens more efficiently utilize common resources, such as iron. Iron is essential for bacterial growth, and many bacteria produce siderophores in order to acquire ferric iron [80]. As a protective mechanism, host cells secrete lipocalin-2, which is able to block the siderophore enterobactin in E. coli, preventing iron acquisition and proliferation of commensal E. coli in the gut [81,82]. However, Salmonella and some pathogenic E. coli express modified enterobactins, named salmochelins, which are lipocalin-2 resistant, providing an important advantage of pathogens over commensals [4,81,82]. Pathogens can also reside in a distinct niche from the microbiota. Pathogenic E. coli can, for example, localize close to the intestinal epithelial surface, normally devoid of commensal microbiota, through the expression of molecules such as intimin, a LEE-encoded adhesion molecule [14].
Chemotaxis and mobility conferred by flagella enable Salmonella to identify and swim to nutritionally beneficial niches. Indeed, the methyl-accepting chemotaxis receptors Aer and Tsr were observed to respond in vivo to tetrathionate or nitrate, respectively, in order to confer a fitness advantage upon S. Typhimurium during inflammation by enabling the bacteria to seek out favourable spatial niches containing host-derived electron acceptors that boost its luminal growth [83]. Pathogenic bacteria may also benefit from intestinal inflammation and/or by-products of the inflammatory response. For example, reactive oxygen species produced by neutrophils during inflammation react with luminal sulfur compounds (thiosulfate, 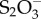 ) to form a new respiratory electron acceptor, tetrationate (
) to form a new respiratory electron acceptor, tetrationate (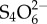 ). Unlike commensals, Salmonella contains the operon ttrSR ttrBCA that allows for the use of
). Unlike commensals, Salmonella contains the operon ttrSR ttrBCA that allows for the use of 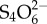 , providing a growth advantage to Salmonella over commensal microbes in an inflamed environment [84]. Similarly, nitrate generated as a by-product of the inflammatory response conferred a growth advantage to the commensal bacterium E. coli over Firmicutes and Bacteroidetes in the large intestine of mice [85]. Although this mechanism of E. coli overgrowth in the inflamed gut involved commensal–commensal competition, pathogenic E. coli strains, which have nitrate reductase genes such as NarZ in their genome, may use a similar mechanism to acquire a growth advantage over the competitive commensal community.
, providing a growth advantage to Salmonella over commensal microbes in an inflamed environment [84]. Similarly, nitrate generated as a by-product of the inflammatory response conferred a growth advantage to the commensal bacterium E. coli over Firmicutes and Bacteroidetes in the large intestine of mice [85]. Although this mechanism of E. coli overgrowth in the inflamed gut involved commensal–commensal competition, pathogenic E. coli strains, which have nitrate reductase genes such as NarZ in their genome, may use a similar mechanism to acquire a growth advantage over the competitive commensal community.
(b). Promotion of host inflammation
Another mechanism by which intestinal pathogens acquire a growth advantage compared to the commensal population is through the promotion of intestinal inflammation, that alters commensal survival. Indeed, the expression of virulence factors by pathogenic bacteria, such as toxins, leads to intestinal inflammation that, in turn, dramatically alters the gut microbiota composition and richness [86]. For example, during intestinal inflammation, neutrophils and macrophages expressing inducible nitric oxide synthetase (iNOS) are recruited, leading to an increased concentration of nitrate in the gut that confers an advantage to Enterobacteriaceae compared to obligate anaerobes, such as Bacteroidetes or Firmicutes [85]. Many E. coli pathovars are indeed able to utilize nitrate as an electron acceptor, such as AIEC associated with inflammatory bowel disease, and can profit from such growth advantage compared to the commensal population to more efficiently colonize the intestine [87–90].
In addition, the host inflammatory environment can enhance the expression of virulence factors, promoting the growth of pathogenic bacteria in host tissues. For example, interferon-ɤ increased the expression of type I lectin by Pseudomonas aeruginosa, allowing adhesion of the bacteria to lung epithelial cells [91]. Hence, the gut inflammatory environment may also promote virulence factor expression by enteric bacteria.
(c). Toxin production
Pathogens are able to produce inhibitory substances/toxins that can directly target the gut microbiota. Through its type VI secretion system, Vibrio cholerae is able to deliver toxic effectors directly to E. coli [92,93]. In addition, bacteriocin production has been reported for Salmonella [94] and pathogenic Shigella strains [95–99], but its role in virulence or as a mechanism to outcompete microbiota are not yet known.
Taking into account that bacteriocins play an important role in bacterial relationships, it is very likely that they can contribute to the successful colonization of the intestine by pathogenic bacteria, by targeting specific commensals and therefore modifying the barrier maintenance, altering the immune surveillance and/or the gut metabolism to promote their colonization. Studying the role of bacteriocins produced by pathogenic strains on the microbiota and on virulence might reveal new mechanisms of pathogenicity and will help to decipher the complex relationship between the intestinal microbiota and some enteric pathogens.
6. Conclusion and perspectives
With the recent appreciation of the important roles played by the intestinal microbiota in health and diseases, a number of studies have highlighted its specific role in protection against enteric pathogen infection. It is important to note here that most mechanisms presented in this review have been discovered from animal models and/or in vitro works. Extrapolation to the human situation has to be considered with caution in a context of different dietary habits, intestinal architecture, microbiota composition, environment, immune system and genetic background. However, new therapeutic approaches may ultimately benefit from understanding the important inhibitory role of the intestinal microbiota against pathogen virulence [100], as exemplified by the recent use of fecal microbiota transplant for the treatment of recurrent C. difficile infection, which has more than 90% effectiveness compared with only 30% when using antibiotic treatment [101]. Targeted manipulation of the intestinal microbiota by bacteriocins and/or other antimicrobials has the potential to be a therapeutic tool for the prevention or treatment of dysbiosis-associated diseases [102]. Based on their very high specificity (at least for some of them), bacteriocins might represent ideal candidates with respect to the targeting of only undesirable populations. Importantly, there are already some proof of concept studies, such as the use of thuricin CD to specifically inhibit C. difficile in a distal colon model [103]. Similarly, bacteriocin production by the probiotic Lactobacillus salivarius UCC118 was shown to significantly protect mice against L. monocytogenes [104]. Moreover, it was recently shown that intestinal colonization with a bacteriocin-producing Enteroccocus faecalis results in the clearance of vancomycin-resistant enterococci, strengthening the concept that bacteriocins may be an effective therapeutic approach to specifically eliminate intestinal colonization by multiple-resistant bacteria without a profound disruption of the commensal population [104]. However, when identifying or choosing a bacteriocin as a therapeutic approach, the putative broad impact on the gut microbiota should be taken in account, even if less drastic than antibiotic use. For example, thuricin CD was also found to strongly inhibit Lactobacillus fermentum [18].
In addition, phage therapy can potentially have beneficial impact on human microbiota and associated host health [105]. Bacteriophages are virus particles that naturally infect bacteria with a high specificity and phage therapy consists of using these bacteriophages as antimicrobial agents [106,107]. Some groups are currently investigating their suitability as therapeutic strategy against some enteric pathogens, for example against AIEC associated with inflammatory bowel disease [108].
Finally, bacteria (both non-pathogenic and pathogenic) synthesize small diffuse signal molecules, called auto-inducers, in order for them to coordinately control the gene expression of the entire community in response to changes in cell density [109]. This process, termed quorum sensing, can be universal or highly specific, enabling bacteria to communicate within and between species. Hence, quorum sensing can have a major impact on the composition of microbial communities, and is also involved in the regulation of virulence gene expression by many pathogenic bacteria [110]. Therefore, the identification of chemical signals, receptors and targeted genes will be essential for our understanding of how bacteria–bacteria communication may be used in preventing colonization by pathogenic bacteria [100].
Additional studies are needed to decipher the complex relationship occurring between the intestinal microbiota and pathogenic bacteria that will ultimately help to define, and maybe engineer, a ‘healthy’ microbiome.
Acknowledgements
This paper is dedicated to our esteemed mentor, Arlette Darfeuille-Michaud, who launched both our scientific careers. We are grateful to Andrew Gewirtz and Pascale Cossart for critical review of the manuscript.
Authors' contributions
N.R. and B.C. wrote the manuscript.
Competing interests
We have no competing interests.
Funding
B.C. is a recipient of the Career Development award from the Crohn's and Colitis Foundation of America (CCFA).
References
- 1.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. ( 10.1038/nri2515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589. ( 10.1038/nrgastro.2012.156) [DOI] [PubMed] [Google Scholar]
- 3.McKenney PT, Pamer EG. 2015. From hype to hope: the gut microbiota in enteric infectious disease. Cell 163, 1326–1332. ( 10.1016/j.cell.2015.11.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. ( 10.1038/ni.2608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow J, Tang H, Mazmanian SK. 2011. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23, 473–480. ( 10.1016/j.coi.2011.07.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducluzeau R, Railbaud P. 1989. Bacterial interactions within the digestive tract. Rev. Sci. Tech. Off. Int. Epiz. 8, 313–332. ( 10.20506/rst.8.2.410) [DOI] [PubMed] [Google Scholar]
- 7.Theriot CM, Young VB. 2015. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 69, 445–461. ( 10.1146/annurev-micro-091014-104115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osawa N, Mitsuhashi S. 1964. Infection of germeree mice with Shigella Flexneri 3a. Jpn. J. Exp. Med. 34, 77–80. [PubMed] [Google Scholar]
- 9.Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. 2012. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes 3, 449–454. ( 10.4161/gmic.21214) [DOI] [PubMed] [Google Scholar]
- 10.Fukuda S, et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. ( 10.1038/nature09646) [DOI] [PubMed] [Google Scholar]
- 11.Bohnhoff M, Drake BL, Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med. 86, 132–137. ( 10.3181/00379727-86-21030) [DOI] [PubMed] [Google Scholar]
- 12.Lawley TD, et al. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77, 3661–3669. ( 10.1128/IAI.00558-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. ( 10.1038/nri3535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. ( 10.1126/science.1222195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72, 946–949. ( 10.1128/AEM.72.1.946-949.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin R, Suzuki M, Morishita Y. 2002. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J. Med. Microbiol. 51, 201–206. ( 10.1099/0022-1317-51-3-201) [DOI] [PubMed] [Google Scholar]
- 17.Kommineni S, et al. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722. ( 10.1038/nature15524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl Acad. Sci. USA 107, 9352–9357. ( 10.1073/pnas.0913554107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schamberger GP, Diez-Gonzalez F. 2002. Selection of recently isolated colicinogenic Escherichia coli strains inhibitory to Escherichia coli O157:H7. J. Food Prot. 65, 1381–1387. [DOI] [PubMed] [Google Scholar]
- 20.Pinto AL, Fernandes M, Pinto C, Albano H, Castilho F, Teixeira P, Gibbs PA. 2009. Characterization of anti-Listeria bacteriocins isolated from shellfish: potential antimicrobials to control non-fermented seafood. Int. J. Food Microbiol. 129, 50–58. ( 10.1016/j.ijfoodmicro.2008.11.005) [DOI] [PubMed] [Google Scholar]
- 21.Chanos P, Williams DR. 2011. Anti-Listeria bacteriocin-producing bacteria from raw ewe's milk in northern Greece. J. Appl. Microbiol. 110, 757–768. ( 10.1111/j.1365-2672.2010.04932.x) [DOI] [PubMed] [Google Scholar]
- 22.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414. ( 10.1016/j.cell.2014.08.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh CJ, Guinane CM, Hill C, Ross RP, O'Toole PW, Cotter PD. 2015. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project's reference genome database. BMC Microbiol. 15, 183 ( 10.1186/s12866-015-0515-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Human Microbiome Project C. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. ( 10.1038/nature11234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. 2015. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96. ( 10.1038/nature14232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1, e00045-15. ( 10.1128/mSphere.00045-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britton RA, Young VB. 2014. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146, 1547–1553. ( 10.1053/j.gastro.2014.01.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990. ( 10.1128/JB.00610-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buffie CG, et al. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. ( 10.1038/nature13828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. 2014. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G310–G319. ( 10.1152/ajpgi.00282.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M. 2008. Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol. 9, 37 ( 10.1186/1471-2172-9-37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rios D, Wood MB, Li J, Chassaing B, Gewirtz AT, Williams IR. 2015. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 9, 907–916. ( 10.1038/mi.2015.121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. ( 10.1038/nrmicro2546) [DOI] [PubMed] [Google Scholar]
- 34.Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130. ( 10.1126/science.1127119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fransen F, et al. 2015. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 43, 527–540. ( 10.1016/j.immuni.2015.08.011) [DOI] [PubMed] [Google Scholar]
- 36.Dolle L, Tran HQ, Etienne-Mesmin L, Chassaing B. 2016. Policing of gut microbiota by the adaptive immune system. BMC Med. 14, 27 ( 10.1186/s12916-016-0573-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. ( 10.1016/j.cell.2005.05.007) [DOI] [PubMed] [Google Scholar]
- 38.Ivanov II, Frutos RD, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. ( 10.1016/j.chom.2008.09.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov II, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. ( 10.1016/j.cell.2009.09.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaboriau-Routhiau V, et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. ( 10.1016/j.immuni.2009.08.020) [DOI] [PubMed] [Google Scholar]
- 41.Lecuyer E, et al. 2014. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620. ( 10.1016/j.immuni.2014.03.009) [DOI] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. ( 10.1016/j.cell.2004.07.002) [DOI] [PubMed] [Google Scholar]
- 43.Araki A, et al. 2005. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J. Gastroenterol. 40, 16–23. ( 10.1007/s00535-004-1492-9) [DOI] [PubMed] [Google Scholar]
- 44.Zhang B, et al. 2014. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346, 861–865. ( 10.1126/science.1256999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atarashi K, et al. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. ( 10.1038/nature12331) [DOI] [PubMed] [Google Scholar]
- 46.Arpaia N, et al. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. ( 10.1038/nature12726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim CH, Park J, Kim M. 2014. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Network 14, 277–288. ( 10.4110/in.2014.14.6.277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furusawa Y, et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. ( 10.1038/nature12721) [DOI] [PubMed] [Google Scholar]
- 49.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. ( 10.1126/science.1241165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bates JM, Akerlund J, Mittge E, Guillemin K. 2007. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382. ( 10.1016/j.chom.2007.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beumer C, Wulferink M, Raaben W, Fiechter D, Brands R, Seinen W. 2003. Calf intestinal alkaline phosphatase, a novel therapeutic drug for lipopolysaccharide (LPS)-mediated diseases, attenuates LPS toxicity in mice and piglets. J. Pharmacol. Exp. Ther. 307, 737–744. ( 10.1124/jpet.103.056606) [DOI] [PubMed] [Google Scholar]
- 52.Koyama I, Matsunaga T, Harada T, Hokari S, Komoda T. 2002. Alkaline phosphatases reduce toxicity of lipopolysaccharides in vivo and in vitro through dephosphorylation. Clin. Biochem. 35, 455–461. ( 10.1016/S0009-9120(02)00330-2) [DOI] [PubMed] [Google Scholar]
- 53.Chassaing B, Gewirtz AT. 2016. Has provoking microbiota aggression driven the obesity epidemic? Bioessays 38, 122–128. ( 10.1002/bies.201500116) [DOI] [PubMed] [Google Scholar]
- 54.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79, 1536–1545. ( 10.1128/IAI.01104-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. 2003. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 52, 1442–1447. ( 10.1136/gut.52.10.1442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimotoyodome A, Meguro S, Hase T, Tokimitsu I, Sakata T. 2000. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 125, 525–531. ( 10.1016/S1095-6433(00)00183-5) [DOI] [PubMed] [Google Scholar]
- 57.Cameron EA, Sperandio V. 2015. Frenemies: signaling and nutritional integration in pathogen-microbiota-host interactions. Cell Host Microbe 18, 275–284. ( 10.1016/j.chom.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng KM, et al. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. ( 10.1038/nature12503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. 2014. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777. ( 10.1016/j.chom.2014.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769. ( 10.1016/j.chom.2014.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pham NT, Lawley TD. 2014. Pathogens’ exploitation of the intestinal food web. Cell Host Microbe 16, 703–705. ( 10.1016/j.chom.2014.11.012) [DOI] [PubMed] [Google Scholar]
- 62.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. ( 10.1038/nature11623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE 5, e8740 ( 10.1371/journal.pone.0008740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maier L, Barthel M, Stecher B, Maier RJ, Gunn JS, Hardt WD. 2014. Salmonella Typhimurium strain ATCC14028 requires H2-hydrogenases for growth in the gut, but not at systemic sites. PLoS ONE 9, e110187 ( 10.1371/journal.pone.0110187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377. ( 10.1111/j.1462-2920.2010.02334.x) [DOI] [PubMed] [Google Scholar]
- 66.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3, e00050-12. ( 10.1128/mBio.00050-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiennimitr P, et al. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17 480–17 485. ( 10.1073/pnas.1107857108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568. ( 10.1128/JB.188.2.556-568.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Everard A, et al. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071. ( 10.1073/pnas.1219451110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneeberger M, Everard A, Gomez-Valades AG, Matamoros S, Ramirez S, Delzenne NM, Gomis R, Claret M, Cani PD. 2015. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5, 16643 ( 10.1038/srep16643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganesh BP, Klopfleisch R, Loh G, Blaut M. 2013. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 8, e74963 ( 10.1371/journal.pone.0074963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvalho FA, et al. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12, 139–152. ( 10.1016/j.chom.2012.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. 2014. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut 63, 1069–1080. ( 10.1136/gutjnl-2013-304909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vayssier-Taussat M, et al. 2014. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front. Cell Infect. Microbiol. 4, 29 ( 10.3389/fcimb.2014.00029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamdar K, et al. 2016. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe 19, 21–31. ( 10.1016/j.chom.2015.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8, 290–295. ( 10.1038/nrmicro2334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S, Hooper SD, Andrade MA, Bork P. 2005. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 3, e134 ( 10.1371/journal.pbio.0030134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fabich AJ, et al. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152. ( 10.1128/IAI.01386-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Bouguenec C, Schouler C. 2011. Sugar metabolism, an additional virulence factor in enterobacteria. Int. J. Med. Microbiol. 301, 1–6. ( 10.1016/j.ijmm.2010.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saha R, Saha N, Donofrio RS, Bestervelt LL. 2013. Microbial siderophores: a mini review. J. Basic Microbiol. 53, 303–317. ( 10.1002/jobm.201100552) [DOI] [PubMed] [Google Scholar]
- 81.Fischbach MA, Lin H, Liu DR, Walsh CT. 2006. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138. ( 10.1038/nchembio771) [DOI] [PubMed] [Google Scholar]
- 82.Singh V, Yeoh BS, Xiao X, Kumar M, Bachman M, Borregaard N, Joe B, Vijay-Kumar M. 2015. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat. Commun. 6, 7113 ( 10.1038/ncomms8113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivera-Chavez F, et al. 2013. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 9, e1003267 ( 10.1371/journal.ppat.1003267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Winter SE, et al. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. ( 10.1038/nature09415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winter SE, et al. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. ( 10.1126/science.1232467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 ( 10.1016/j.chom.2007.08.002) [DOI] [PubMed] [Google Scholar]
- 87.Darfeuille-Michaud A, et al. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421. ( 10.1053/j.gastro.2004.04.061) [DOI] [PubMed] [Google Scholar]
- 88.Chassaing B, Darfeuille-Michaud A. 2011. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140, 1720–1728. ( 10.1053/j.gastro.2011.01.054) [DOI] [PubMed] [Google Scholar]
- 89.Nash JH, et al. 2010. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics 11, 667 ( 10.1186/1471-2164-11-667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miquel S, et al. 2010. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS ONE 5, e12714 ( 10.1371/journal.pone.0012714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu L, et al. 2005. Recognition of host immune activation by Pseudomonas aeruginosa. Science 309, 774–777. ( 10.1126/science.1112422) [DOI] [PubMed] [Google Scholar]
- 92.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 5, 3549 ( 10.1038/ncomms4549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl Acad. Sci. USA 107, 19 520–19 524. ( 10.1073/pnas.1012931107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patankar CV, Joshi LM. 1985. Bacteriocin production in Salmonella. J. Postgrad. Med. 31, 46–51. [PubMed] [Google Scholar]
- 95.Sousa MA, Mendes EN, Apolonio AC, Farias Lde M, Magalhaes PP. 2010. Bacteriocin production by Shigella sonnei isolated from faeces of children with acute diarrhoea. APMIS 118, 125–135. ( 10.1111/j.1600-0463.2009.02570.x) [DOI] [PubMed] [Google Scholar]
- 96.Horak V. 1994. Seventy colicin types of Shigella sonnei and an indicator system for their determination. Zentralbl. Bakteriol. 281, 24–29. ( 10.1016/S0934-8840(11)80633-X) [DOI] [PubMed] [Google Scholar]
- 97.Kaewklom S, Samosornsuk S, Pipatsatitpong D, Aunpad R. 2013. Colicin type 7 produced by majority of Shigella sonnei isolated from Thai patients with diarrhoea. Braz. J. Microbiol. 44, 731–736. ( 10.1590/S1517-83822013000300010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smajs D, Pilsl H, Braun V. 1997. Colicin U, a novel colicin produced by Shigella boydii. J. Bacteriol. 179, 4919–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Padilla C, Lobos O, Brevis P, Abaca P, Hubert E. 2006. Plasmid-mediated bacteriocin production by Shigella flexneri isolated from dysenteric diarrhoea and their transformation into Escherichia coli. Lett. Appl. Microbiol. 42, 300–303. ( 10.1111/j.1472-765X.2005.01829.x) [DOI] [PubMed] [Google Scholar]
- 100.Belizario JE, Napolitano M. 2015. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 6, 1050 ( 10.3389/fmicb.2015.01050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Nood E, et al. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. ( 10.1056/NEJMoa1205037) [DOI] [PubMed] [Google Scholar]
- 102.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. ( 10.1038/nrmicro2937) [DOI] [PubMed] [Google Scholar]
- 103.Rea MC, et al. 2011. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl Acad. Sci. USA 108(Suppl 1), 4639–4644. ( 10.1073/pnas.1001224107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl Acad. Sci. USA 104, 7617–7621. ( 10.1073/pnas.0700440104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823. ( 10.3390/v5030806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. ( 10.1128/AAC.45.3.649-659.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abedon ST. 2014. Phage therapy: eco-physiological pharmacology. Scientifica 2014, 581639 ( 10.1155/2014/581639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsui J, Jacobs J, Braun J. 2014. Bacteriophages as a therapeutic strategy to target adherent, invasive Escherichia coli associated with inflammatory bowel disease. SACNAS National Conference, 16–18 October, Los Angeles, CA.
- 109.Schauder S, Bassler BL. 2001. The languages of bacteria. Genes Dev. 15, 1468–1480. ( 10.1101/gad.899601) [DOI] [PubMed] [Google Scholar]
- 110.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 ( 10.1101/cshperspect.a012427) [DOI] [PMC free article] [PubMed] [Google Scholar]