Abstract
Aberrant wnt/β-catenin signaling and amplification/overexpression of Myc are associated with hepatoblastoma (HB), the most prevalent type of childhood liver cancer. To address their roles in the pathogenesis of HB, we generated mice in which Myc and mutant β-catenin were targeted to immature cells of the developing mouse liver. Perinatal coexpression of both genes promoted the preferential development of HBs over other tumor types in neonatal mice, all of which bore striking resemblance to their human counterparts. Integrated analysis indicated that tumors emerged as a consequence of Myc-driven alterations in hepatoblast fate in a background of pan-hepatic injury, inflammation, and nuclear factor (erythroid-derived 2)-like 2/Nrf2-dependent antioxidant signaling, which was specifically associated with expression of mutant β-catenin but not Myc. Immunoprofiling of human HBs confirmed that approximately 50% of tumors demonstrated aberrant activation of either Myc or Nfe2l2/Nrf2, while knockdown of Nrf2 in a cell line–derived from a human HB with NFE2L2 gene amplification reduced tumor cell growth and viability. Taken together, these data indicate that β-catenin creates a protumorigenic hepatic environment in part by indirectly activating Nrf2 and implicate oxidative stress as a possible driving force for a subset of β-catenin–driven liver tumors in children.
A mouse model of Wnt/ β-catenin– and Myc-driven hepatoblastoma implicates injury, inflammation, and nrf2 signaling in the pathogenesis of a subset of human hepatoblastomas.
Introduction
Hepatoblastoma (HB) is the most common malignant liver tumor in children under the age of 5 years, with approximately 100 new cases diagnosed annually in the US (incidence ~1.44:1,000,000) (1) and an incidence that is rising at a rate of approximately 4% per year (2). Although the survival rate for HB in North America is approximately 90% for children with low-stage resectable disease, only one-third of HBs are deemed resectable, and children with advanced disease often fail to respond to the standard platinum-based chemotherapy (3). Like other embryonal tumors that develop within immature tissue environments, HBs comprise a heterogeneous group of neoplasms that are thought to originate from hepatoblasts, rapidly dividing undifferentiated cells of the developing liver that give rise to mature hepatocytes and biliary cells (4). Consistent with the concept of “oncogeny as partially blocked ontogeny” (5), pediatric liver tumor development is heavily influenced by developmental context, with histological diversity likely reflecting the uncontrolled expansion of hepatoblasts that have undergone maturational arrest at distinct stages of differentiation during normal liver development. Histologically, HBs are classified as either wholly epithelial (~65%) or mixed epithelial/mesenchymal tumors (~35%), with fetal and mixed fetal/embryonal HBs being the most prevalent epithelial subtypes (6). As histological classification remains the cornerstone of risk stratification and treatment decisions for patients, understanding how genomic, epigenetic, and environmental factors influence or correlate with HB subtype is essential for understanding HB biology and improving outcomes for this rare tumor.
Although most HBs are sporadic, multiple studies have established a linkage between HB and several familial cancer and congenital malformation syndromes (7), most notably familial adenomatous polyposis, a cancer syndrome resulting from mutations in adenomatous polyposis coli, a negative regulator of Wnt signaling. A causative role for Wnt in HB is not only suggested by the high frequency of activating mutations or deletions in CTNNB1 (8) that render the canonical Wnt effector β-catenin constitutively active for Wnt signaling due to impaired degradation of the protein (9), but also by the high frequency of HBs that display Wnt activation via an array of nonmutational mechanisms (10). However, despite an inferred obligatory role for Wnt in HB, forced hepatic expression of mutant β-catenin is not sufficient to induce hepatocarcinogenesis (11, 12), indicating that additional oncogenic “hits” must be required. With the exception of the antioxidant response regulator, nuclear factor (erythroid-derived 2)-like 2 (NFE2L2, also known as Nrf2), the second most frequently mutated gene after CTNNB1, genomic sequencing has produced few candidates reflecting the low rate of mutation in HB (10, 13). However, by virtue of being located on chromosome 8q, a region frequently amplified in human HB (14), and on the basis of being identified as a prominent oncogenic signature in aggressive human HBs (15), Myc has emerged as a potential contributor to HB, yet it is one that has since received little attention. Given the prominence of deregulated Wnt and the relatively unexplored role of Myc in human HB, we set out to determine if Myc cooperated with mutant β-catenin to produce HBs in mice. Using two lines of genetically modified mice, one of which coexpresses a mutant form of β-catenin and Myc and a second that expresses Myc alone, we show that Wnt/β-catenin and Myc efficiently synergize to produce mouse HBs that are histologically and molecularly identical to the most common types of human HBs. Moreover, our results suggest that mouse HBs can develop via two distinct routes: a cell-autonomous route, in which Wnt/β-catenin activation is selected for during Myc-driven HB development in the context of normal liver, and an alternative route, whereby Myc-driven HBs rapidly develop within an hepatic environment characterized by pan-hepatic injury, inflammation, and activation of Nrf2 that is solely associated with expression of mutant β-catenin but not Myc. By also providing evidence for the involvement of Myc and Nrf2 in human HB, we propose that a subset of human HBs may have an etiology rooted in oxidative stress emanating from prior hepatic injury or inflammation and identify Nrf2 as a major component of the protumorigenic field that supports the expansion and neoplastic progression of immature Myc-expressing liver cells.
Results
Neonatal mice coexpressing mutant β-catenin and c-Myc preferentially develop HBs over other hepatocellular tumors.
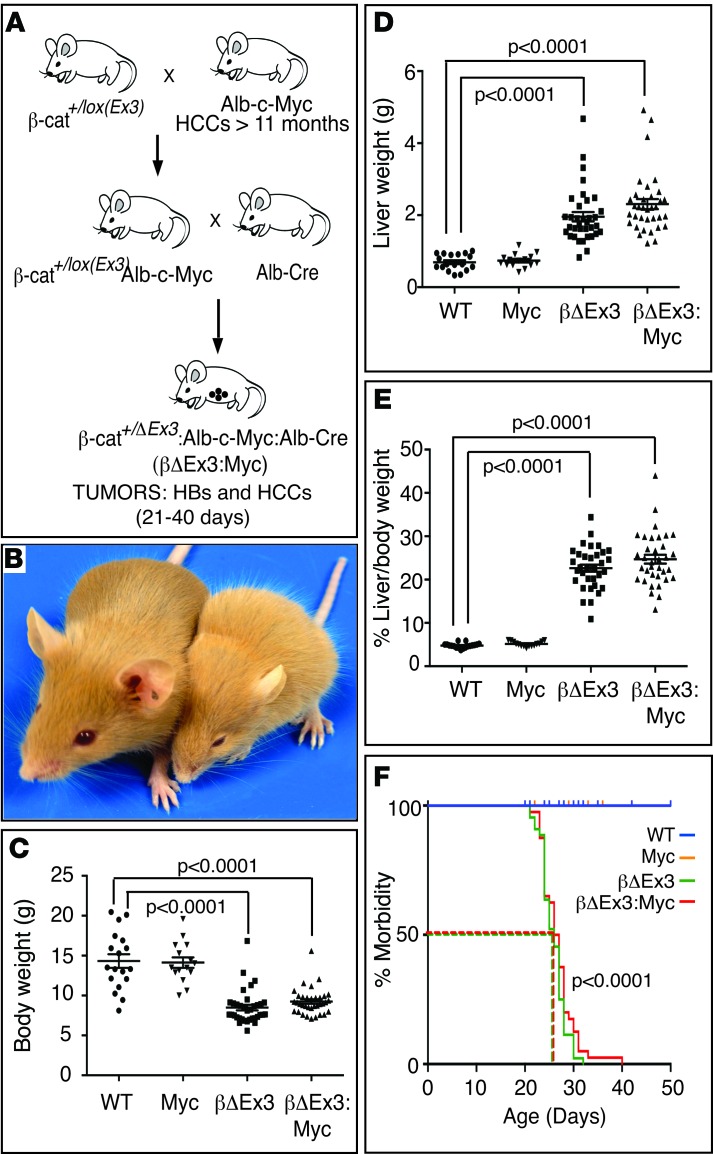
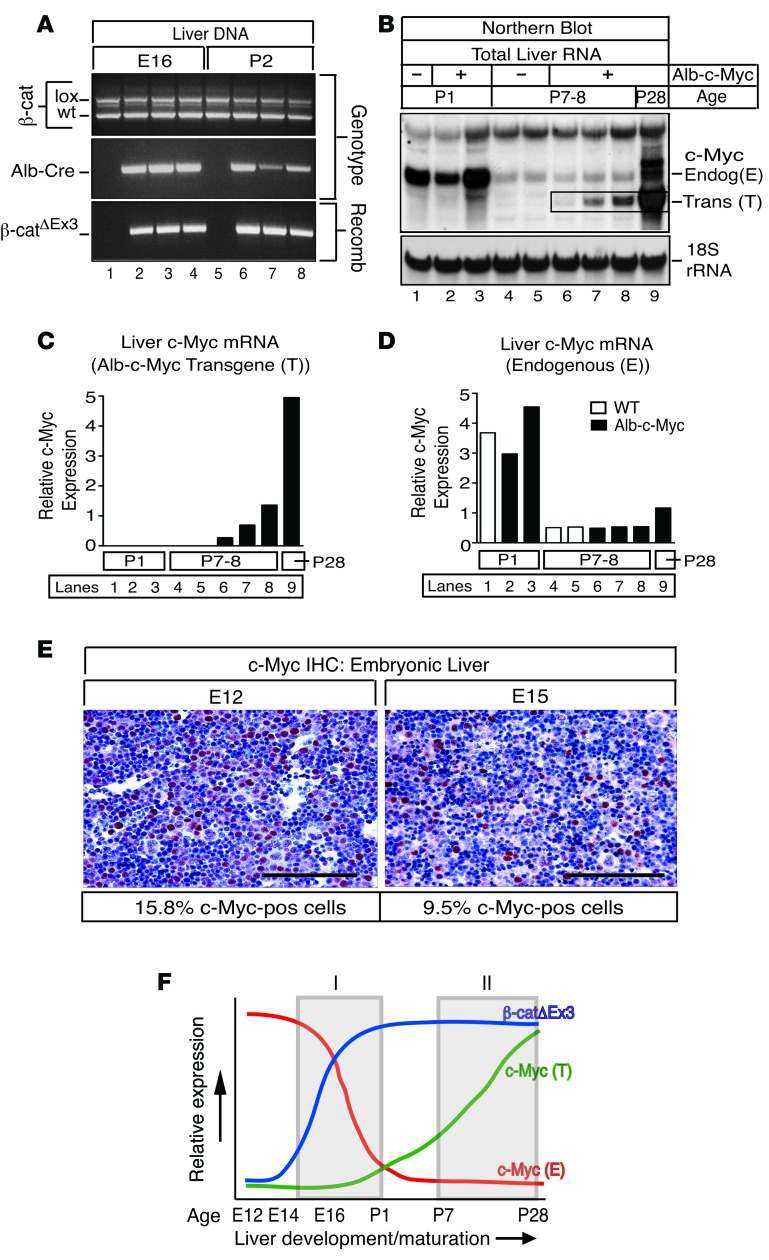
Mice coexpressing c-Myc and one copy of a mutant dominant stable β-catenin Ctnnb1 allele [β-cat+/lox(Ex3)] in immature liver cells (herein referred to as β-catΔEx3:Myc mice) were generated as shown in Figure 1A. Postnatal monitoring and genotyping of progeny born from matings of β-cat+/lox(Ex3):Myc mice to Albumin-Cre mice showed that all possible genotypes were recovered at the expected Mendelian ratio, indicating that hepatic expression of mutant β-catenin, either with or without the Myc transgene, did not cause embryonic lethality. However, a subset of mice became visibly growth retarded beginning at 2 to 3 weeks of age (Figure 1B), a phenotype that segregated with expression of the β-catΔEx3 allele but which was independent of Myc transgene status (Figure 1C). Moreover, in contrast to young mice of the parental Albumin-c-Myc strain (referred to as Alb-Myc mice), which had normal body and liver weights (Figure 1C) and livers that were biochemically and histologically normal, except for mild cellular and nuclear hypertrophy in periportal hepatocytes (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/jci.insight.88549DS1), all mice expressing mutant β-catenin displayed hepatomegaly (Figure 1, D and E) and became moribund, requiring euthanization between 21 and 40 days of age (Figure 1F). Biochemical and histological analysis showed that persistent activation of β-catenin resulted in severe liver dysfunction secondary to extensive hepatocellular damage and overt focal necrosis (Supplemental Figure 1, A and B). Despite the presence of extrabiliary SOX9-positive cells, suggesting a possible regenerative response (Supplemental Figure 1C), and a modest improvement in liver function in β-catenin mice coexpressing the Alb-c-Myc transgene (Supplemental Figure 1B), all β-catenin–expressing mice eventually succumbed to liver failure by 6 weeks of age. That this phenotype was associated with prolonged and persistent expression of mutant β-catenin throughout the perinatal period was inferred by the presence of the recombined β-catenin allele in livers of β-catΔEx3 mice as early as embryonic day 16 (E16) (Figure 2A). In contrast, expression of the Albumin-c-Myc transgene did not become evident until shortly after birth and increased thereafter (Figure 2, B and C, and Supplemental Figure 2). This postnatal increase in transgenic Myc expression contrasted sharply with endogenous Myc, which, despite being abundantly expressed in embryonic and perinatal liver (Figure 2, B, D, and E), declined precipitously after birth (Figure 2, B and D). Expression of mutant β-catenin thus coincided with 2 temporally distinct pools of Myc in livers of β-catΔEx3:Myc mice: an endogenous pool expressed in hepatoblasts from approximately E16–P1 (Figure 2F, gray box I) and a transgene-derived pool expressed in maturing hepatocytes from approximately P7 onward (Figure 2F, gray box II).
Figure 1. Hepatic expression of mutant β-catenin retards growth and cooperates with Myc to promote HBs and HCCs in neonatal mice.
(A) Breeding strategy used to generate β-catΔEx3:Myc (here abbreviated to βΔEx3:Myc) mice coexpressing mutant β-catenin and Myc in the immature liver. (B and C) Hepatic expression of mutant β-catenin stunts postnatal growth. (B) Left, normal sized WT mouse; right, small mouse expressing a single copy of the β-catΔEx3 allele (here abbreviated to βΔEx3) in liver. (C) Body weights. (D and E) Expression of mutant β-catenin with or without the Albumin-c-Myc transgene induces hepatomegaly. (D) Liver weights and (E) percentage of liver/body weight. (F) Kaplan-Meier curve of morbidity. Number of mice analyzed to generate graphs in C–E: WT (n = 18); Myc (n = 15); βΔEx3 (n = 35); and βΔEx3:Myc (n = 35). Number of mice analyzed to generate the graph in F: WT (n = 19); Myc (n = 19); βΔEx3 (n = 44); and βΔEx3:Myc (n = 40). Dashed lines in graph in F indicate the age at which 50% of βΔEx3 and βΔEx3:Myc mice became premorbid, 26 days and 26.5 days, respectively. P values for graphs in C–E were generated using a 2-tailed unpaired t test. P values for graph in F were generated using a log-rank (Mantel-Cox) test.
Figure 2. Expression of mutant β-catenin coincides with 2 temporally distinct pools of Myc in livers of β-catΔEx3 mice.
(A) Cre-mediated recombination of exon 3–deficient β-catenin (β-catΔEx3) is evident in the liver by E16. PCR analysis of DNA isolated from embryonic (E16) and postnatal (P2) livers of β-catΔEx3:Alb-Cre– mice (lanes 1 and 5) (n = 1 each time point) and β-catΔEx3:Alb-Cre+ mice (lanes 2–4 and 6–8) (n = 3 each time point) using genotype- or recombination-specific primers. Recombination is evident only in livers expressing the Alb-Cre transgene (lanes 2–4 and 6–8). (B) Northern blot analysis of endogenous c-Myc mRNA and c-Myc mRNA produced from the Albumin-c-Myc transgene (bounded by black box) in livers of WT and Alb-c-Myc mice. 12 μg of total RNA isolated from livers of WT (lanes 1, 4, and 5) and Alb-c-Myc mice (lanes 2, 3, 6, 7, and 8) at P1 (n = 3), P7–P8 (n = 5), and P28 (n = 1) (lane 9) was gel-fractionated, transferred to a nylon membrane, and hybridized to a radiolabeled c-Myc–specific probe and, thereafter, a radiolabeled oligonucleotide probe specific for 18s rRNA. Endogenous c-Myc mRNA is abundantly expressed at P1 in WT and Alb-c-Myc mice (lanes 1–3) but is negligible by P7–P8 (lanes 4–8). In contrast, the Albumin-c-Myc transgene is expressed by P7–P8 and continues to increase as the liver matures (lanes 6–9, bounded by box, and Supplemental Figure 2). (C) Quantitation of transgenic (T) and (D) endogenous (E) c-Myc mRNA showing relative c-Myc mRNA expression in postnatal liver. Lanes correspond to Northern blot in B. (E) Immunohistochemistry (IHC) of Myc in embryonic liver at E12 and E15. Approximately 16% of cells at E12 and approximately 10% of cells at E15 express Myc. (AEC chromagen [red], hematoxylin counterstain [blue]). Scale bar: 50 μm. (F) Scheme depicting the relative magnitude and temporal-dependent expression of mutant β-catenin relative to endogenous and transgenic c-Myc in β-catΔEx3:Myc mice during liver development and maturation. Mutant β-catenin coincides with a pool of hepatoblast-expressed endogenous c-Myc from approximately E16–P2 (gray box I) and a pool of c-Myc expressed from the Albumin-c-Myc transgene in maturing hepatocytes from approximately P7 onward (gray box II).
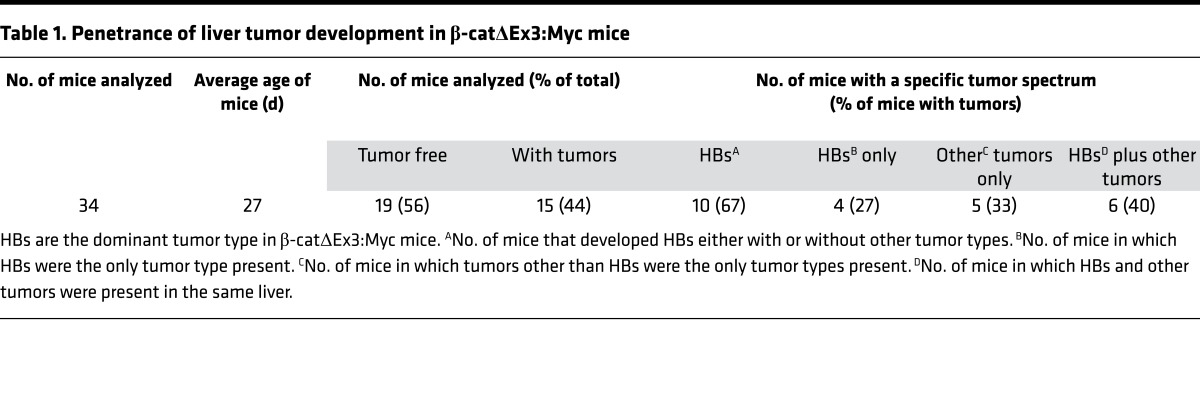
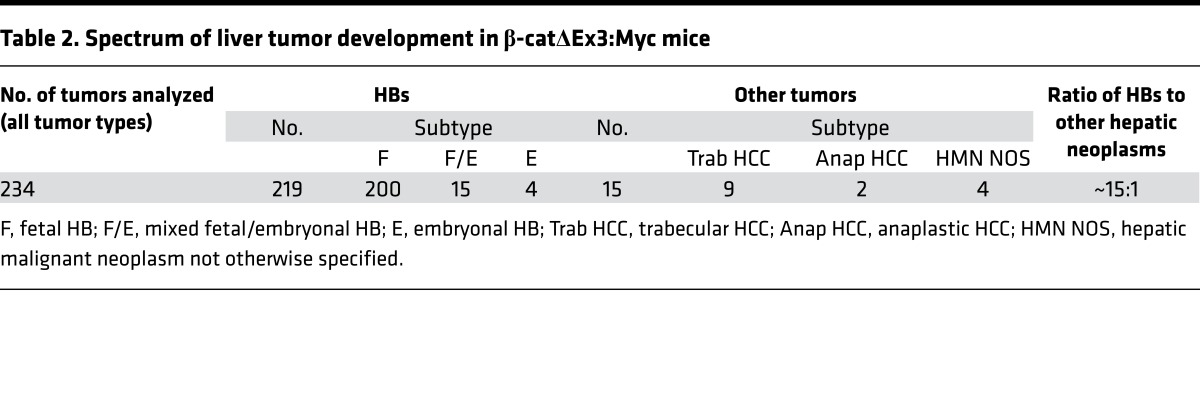
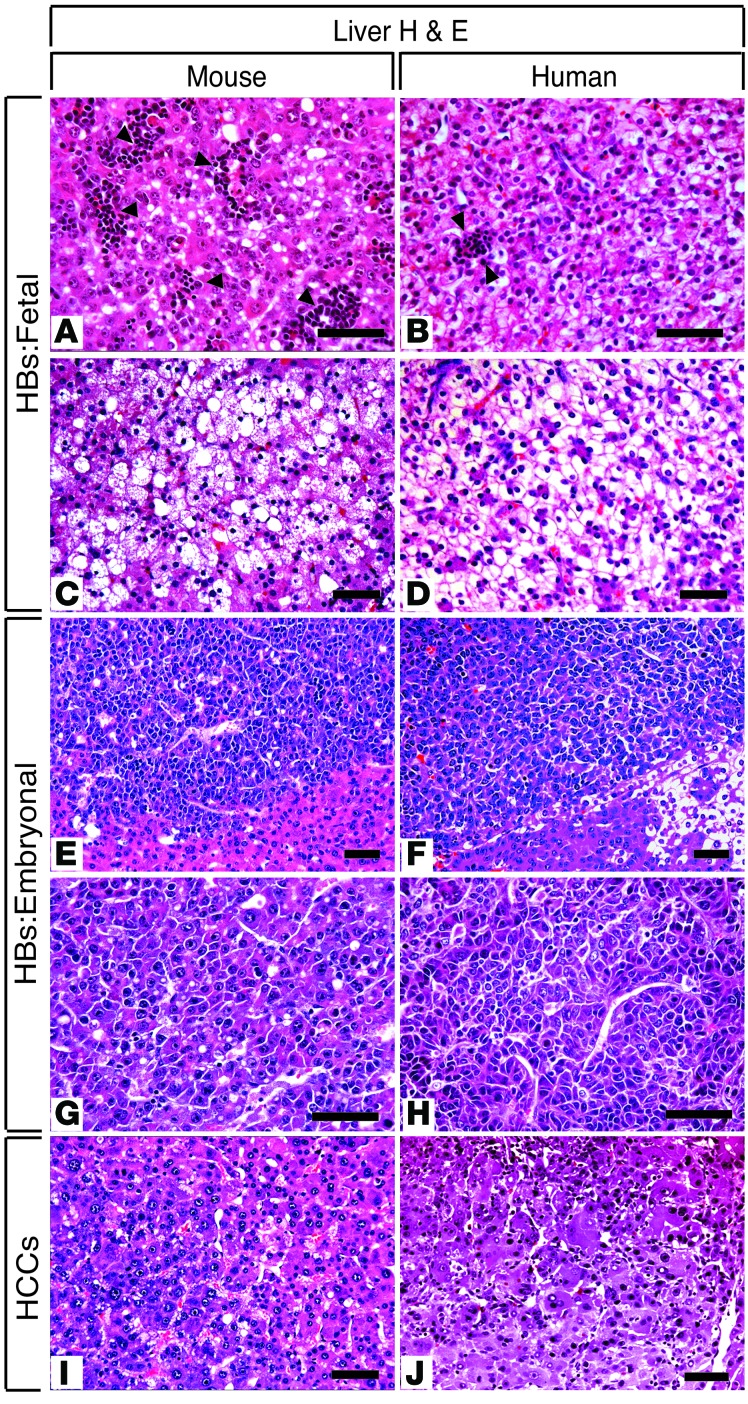
Histological review of enlarged livers from the β-catenin–expressing strains revealed evidence of neoplasia in 15 of 34 (44%) of the β-catΔEx3:Myc mice (Table 1) but not in any of the β-catΔEx3 mice. All tumors were classified as HBs, hepatocellular carcinomas (HCCs), or hepatic malignant neoplasm not otherwise specified (HMN NOS) (Tables 1 and 2), with all bearing striking histological resemblance to their human counterparts (Figure 3). Although tumor number and type varied between animals, analysis of overall tumor incidence by subtype in the tumor-bearing cohort showed that HBs outnumbered HCCs by approximately 15:1 (Table 2). All mouse HBs resembled human epithelial HBs (Figure 3, A–H), approximately 90% of which were designated as well-differentiated fetal HBs (Figure 3, A and C, and Supplemental Figure 3, A–D), with the rest presenting as mixed fetal/embryonal (Supplemental Figure 3, E–G) or moderately differentiated embryonal HBs (Figure 3, E and G, and Supplemental Figure 3, H and I). HMN NOS (Supplemental Figure 3J) and anaplastic HCCs (Supplemental Figure 3K), aggressive tumors that develop in older children and young adults, were rare. Well-to-moderately differentiated HCCs (Figure 3I and Supplemental Figure 3L) resembling human HCCs (Figure 3J) constituted the most prevalent HCC subtype in the mice. Without exception, all tumors developed within livers displaying evidence of substantial injury. Thus, targeted coexpression of mutant β-catenin and Myc to immature hepatic cells in the mouse preferentially drives the development of well-to-moderately differentiated HBs and HCCs in the context of injured liver.
Table 1. Penetrance of liver tumor development in β-catΔEx3:Myc mice.
Table 2. Spectrum of liver tumor development in β-catΔEx3:Myc mice.
Figure 3. Comparative histopathology of mouse and human HBs and HCCs.
Photomicrographs of H&E-stained liver sections showing (A, C, E, G, and I) hepatoblastomas (HBs) and hepatocellular carcinomas (HCCs) that develop in β-catΔEx3:Myc mice, (B, D, F, H, and J) with their human counterparts. In all cases, mouse tumors bear striking resemblance to human HBs. Well-differentiated fetal HBs contained (A and B) eosinophilic or (C and D) clear cells. Extramedullary hematopoiesis is present in (A) mouse and (B) human fetal HBs (arrowheads). Embryonal HBs with a trabecular growth pattern consisting of small angulated cells with a high nuclear/cytoplasmic ratio. Note that, in contrast to the fetal HBs in C and D, (E–H) fat or glycogen is absent. Well-to-moderately differentiated (I) mouse and (J) human microtrabecular HCCs containing hepatoid cells. Nuclear atypia and heterogeneity are evident. Scale bar: 50 μm.
In highlighting the role of Myc in human HB (15), Buendia and colleagues used a genetically engineered mouse strain that develops liver tumors following activation of a tetracycline-regulatable liver-specific Myc transgene (16) (Tg(tet-O-Myc)36aBop, herein referred to as Tet-O-Myc mice). Although originally described as a model of HCC, subsequent analysis showed that HBs and HCCs develop if Myc is induced shortly after birth, however a comprehensive analysis of the overall tumor spectrum has not been reported. We therefore compared HBs that developed in β-catΔEx3:Myc mice with those that developed in Tet-O-Myc mice following induction of c-Myc at approximately 3–4 weeks of age, when hepatocyte differentiation is essentially complete. In contrast to the rapid appearance of HBs in young β-catΔEx3:Myc mice following perinatal activation of Myc and β-catenin, HBs did not appear in Tet-O-Myc mice until approximately 16–20 weeks of age, demonstrating a markedly longer tumor latency without coexpression of the mutant β-catenin allele. Moreover, while well-differentiated HBs arose within injured β-catΔEx3:Myc livers, poorly differentiated HBs with primitive features, including macrotrabecular (Supplemental Figure 3, M–O) or solid (Supplemental Figure 3R) growth patterns and immature tubuloglandular structures (Supplemental Figure 3, P and Q), developed in Tet-O-Myc livers that lacked evidence of injury or lobular disorganization. Thus, while Myc overexpression is sufficient to drive the development of poorly differentiated HBs, forced expression of mutant β-catenin with Myc accelerates tumor development and skews the HB spectrum toward well-differentiated tumors.
As immature cells are more amenable to oncogenic transformation and reprogramming than their mature counterparts (17), temporal differences in Myc activation in livers of β-catΔEx3:Myc and Tet-O-Myc mice could have contributed to the emergence of distinct HB subtypes in each of the strains. However, as cellular responses are also exquisitely sensitive to Myc levels (18), we performed immunohistochemistry (IHC) with a MYC-specific antibody to determine whether differences in the magnitude of transgene-derived MYC could explain the strain-specific HBs. Analysis showed that tumor-specific stabilization of nuclear-localized MYC had occurred in tumors from both strains relative to WT and tumor-adjacent liver (Supplemental Figure 4, A, C, E and G), ruling out major differences in Myc levels as the cause of the different HB subtypes. As stabilizing β-catenin mutations are associated with favorable prognosis in adult HCC (19), we also analyzed the subcellular localization of β-catenin in tumors from both strains to determine if expression of the mutant β-catenin allele in β-catΔEx3:Myc mice, but not Tet-O-Myc mice, could explain the preferential emergence of well-differentiated HBs in β-catΔEx3:Myc mice. IHC showed that, while endogenous β-catenin was correctly localized to the outer cell membrane of hepatocytes and biliary cells in livers of WT mice (Supplemental Figure 4B), mutant β-catenin was abundantly expressed in the cytoplasm of tumor cells in β-catΔEx3:Myc mice (Supplemental Figure 4, D and F), confirming activation of Wnt signaling. However, membranous β-catenin was barely visible in tumors or adjacent liver (Supplemental Figure 4D), with Western blotting confirming that expression of mutant β-catenin had caused an approximately 70% reduction in WT β-catenin (Supplemental Figure 5), likely reflecting disruption of hepatocyte adherens junctions. Parallel analysis of Tet-O-Myc–derived HBs showed that, while membranous β-catenin was visible, so too was abundant cytoplasmic β-catenin (Supplemental Figure 4H), indicating that Wnt activation had also been selected for during Myc-driven tumor progression. The preferential emergence of HBs over HCCs in β-catΔEx3:Myc mice and the selection for Wnt activation in Tet-O-Myc–driven HBs strongly suggest that Myc and activated Wnt/β-catenin are potent, if not “preferred,” cooperative drivers of HB in mice.
Mouse HBs align ontogenically with human HBs.
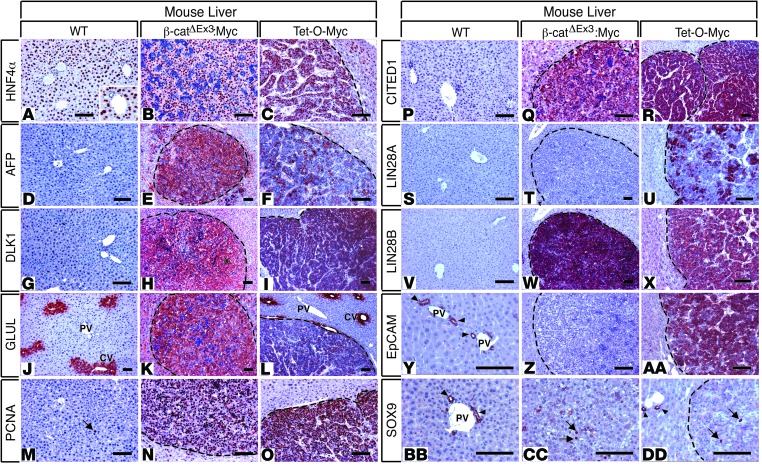
While the young age at HB diagnosis and the resemblance of HB cells to immature liver cells are consistent with the idea that human HBs originate from hepatoblasts, the emergence of HBs in Tet-O-Myc mice following activation of Myc in mature hepatocytes suggests that HBs can also originate from differentiated hepatocytes, presumably via routes involving dedifferentiation or retrograde reprogramming. To determine the extent to which the HBs in β-catΔEx3:Myc and Tet-O-Myc mice reflected distinct ontogenies and aligned with their human counterparts, we performed IHC on tumors from both mouse strains using a panel of antibodies against and oncofetal lineage- and stage-specific liver maturation markers (Figure 4). HBs from both strains expressed markers consistent with a cell identity corresponding to highly proliferative hepatoblasts that had already begun to differentiate toward the hepatocyte lineage and which had lost biliary cell identity (Figure 4). Moreover, despite being histologically distinct, HBs from both strains generally demonstrated quantitative rather than qualitative differences in the expression of most markers, with only EpCAM, GLUL, and focal LIN28A staining providing discrimination between well-differentiated fetal and poorly differentiated embryonal HBs (Figure 4 and Supplemental Figure 6). As EpCAM is primarily expressed in hepatoblasts prior to E14 (20) and GLUL is expressed in hepatoblasts and immature hepatocytes from E15 onwards (21), we concluded that EpCAM–:GLUL+ β-catΔEx3:Myc-driven HBs align with well-differentiated human HBs, while EpCAM+:GLUL– Tet-O-Myc–derived HBs align with poorly differentiated human embryonal HBs, consistent with their histopathological features. Our results suggest that although Myc and activated Wnt/β-catenin drive HB development in both strains, tumors likely develop via distinct routes. While β-catΔEx3:Myc-driven HBs develop via the maturational arrest of hepatoblasts or immature hepatocytes following perinatal activation of Myc in a background of β-catenin–associated injury, Tet-O-Myc–derived HBs likely derive from mature hepatocytes that undergo Myc-driven dedifferentiation or reprogramming, followed by acquisition of a gain of function for Wnt during tumor progression.
Figure 4. Immunohistochemical profiling indicates that HBs from β-catΔEx3:Myc and Tet-O-Myc mice are ontogenically distinct.
Photomicrographs of immunohistochemistry (IHC) of WT livers and tumor-bearing livers of β-catΔEx3:Myc and Tet-O-Myc mice with antibodies specific for (A–C) HNF4α, (D–F) AFP, (G–I) DLK1, (J–L) GLUL, (M–O) PCNA, (P–R) CITED1, (S–U) LIN28A, (V–X) LIN28B, (Y–AA) EpCAM, and (BB–DD) SOX9. Hepatoblastomas (HBs) from both strains express the oncofetal genes AFP, DLK1, CITED1, and LIN28B but lack expression of SOX9 in all but a few tumor cells (arrows in CC and DD). Only embryonal HBs originating in Tet-O-Myc mice express LIN28A and EpCAM. Expression of AFP and GLUL in fetal HBs from β-catΔEx3:Myc mice is higher than in HBs from Tet-O-Myc mice, consistent with a higher degree of tumor cell differentiation in the fetal HBs. Note pericentral-restricted expression of GLUL (glutamine synthetase) in (J) normal and (L) tumor-adjacent liver in Tet-O-Myc mice, indicating maintenance of normal hepatic architecture and zonation in nontumorous liver. The arrow in M highlights a solitary PCNA-positive hepatocyte in liver from a WT mouse, in contrast to an abundance of PCNA-positive cells in HBs (N and O). Arrowheads in Y, BB, and DD indicate EpCAM-positive or SOX9-positive bile ducts in WT liver, respectively. PV, portal vein; CV, central vein. Dashed lines demarcate tumor boundaries. AEC chromagen (red), hematoxylin counterstain (blue). Scale bar: 50 μm. Original magnification for inset in A, ×240.
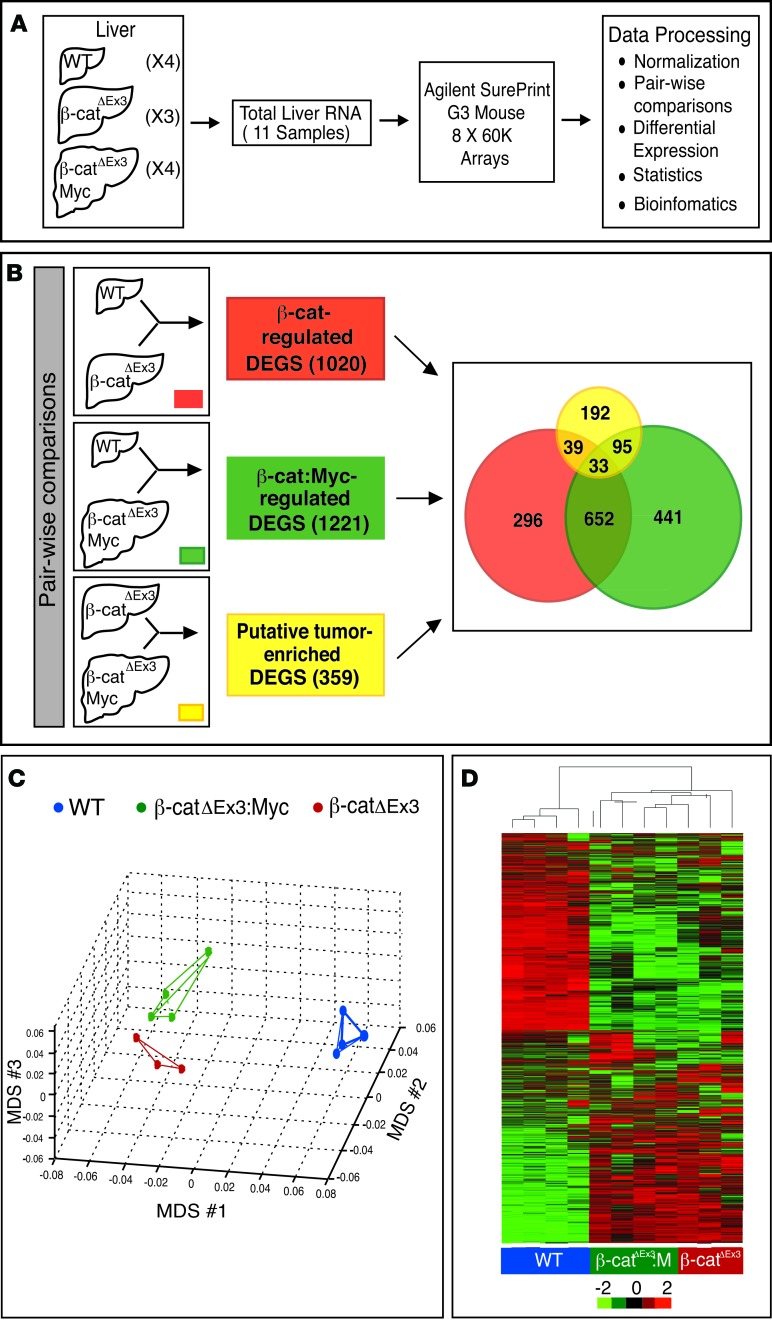
The transcriptome ofβ-catenin/Myc-driven HBs evokes a fetal-like hepatic gene expression program and epigenetic dysregulation. Molecular profiling of human HBs suggests that oncogenic activation and epigenetic derangement against a backdrop of aberrant Wnt signaling drives pediatric liver tumorigenesis (22). To determine the extent to which β-catΔEx3:Myc-driven HBs reflected such etiology, we performed gene expression and bioinformatics analysis. Because tumors were too small to collect for both RNA isolation and histological evaluation, total RNA isolated from whole livers of WT, β-catΔEx3, and β-catΔEx3:Myc mice with the highest incidence of HBs were profiled using gene expression arrays, as outlined in Figure 5A. After obtaining consensus expression profiles for each group, pairwise comparisons were performed against WT liver to generate lists of differentially expressed genes (DEGs) that were either regulated by β-catenin alone (Figure 5B, red box) or coregulated by β-catenin and Myc (Figure 5B, green box) at a level of 4-fold or greater (Supplemental Table 1). To identify candidate tumor-enriched genes, we generated a third list of putative tumor-enriched DEGs (Supplemental Table 1) that comprised mRNAs demonstrating 2-fold differential expression after comparing the list of DEGs obtained from tumor-bearing β-catΔEx3:Myc livers with the list obtained from tumor-naive β-catΔEx3 mice (Figure 5B, yellow box).
Figure 5. Strategy to identify tumor-enriched and β-catenin–regulated hepatic genes.
(A and B) Schematic showing strategy to identify differentially expressed genes (DEGs). (A) Total RNA isolated from livers of 4 WT, 3 β-catΔEx3, and 4 tumor-bearing β-catΔEx3:Myc mice was profiled using Agilent SurePrint mouse arrays. (B) Pairwise comparisons used to generate lists of β-catenin– (red) and β-catenin– and c-Myc–coregulated genes (green). Tumor-enriched DEGs (yellow) were identified by comparing profiles obtained from β-catΔEx3 livers, which never developed tumors, with those from tumor-bearing β-catΔEx3:Myc livers. The Venn diagram shows the number of unique and overlapping mRNAs identified within and between groups. (C) Multidimensional scaling (MDS) plot depicting the relationship among the transcriptional profiles of WT, β-catΔEx3, and β-catΔEx3:Myc livers. Note that β-catΔEx3 and β-catΔEx3:Myc profiles are similar to one another due to common expression of the mutant β-catenin allele. (D) Heatmap depicting hierarchical clustering of DEGs in livers of WT (blue), β-catΔEx3 (red), and β-catΔEx3:Myc (green) mice.
Multidimensional scaling (MDS) (Figure 5C) and hierarchical clustering (Figure 5D) showed that the transcriptomes of β-catΔEx3 and β-catΔEx3:Myc livers differed substantially from WT liver, yet overlapped with each other, reflecting expression of the mutant β-catenin allele in both strains (Figure 5, B–D). Despite this, however, 359 mRNAs demonstrated 2-fold or greater differential expression in tumor-bearing β-catΔEx3:Myc livers relative to those expressing β-catenin alone, representing genes that were most likely to be tumor enriched (Supplemental Table 1). As immunoprofiling had shown high expression of AFP, MYC, DLK1, LIN28B, and CITED1 in HBs relative to surrounding liver (Figure 4), we confirmed the presence of their mRNAs in the tumor-enriched DEGs and used them as “benchmarks” to predict the likelihood that other genes in the list were also tumor enriched, rather than pan-hepatic targets of β-catenin and Myc. Using this strategy, we identified a large number of genes known to be associated with human HBs or HCCs, including Igf2, Igf2as, and H19, as well as platelet-, erythroid-, and neutrophil-specific mRNAs, reflecting HB-associated extramedullary hematopoiesis. Other classes of mRNAs demonstrating enrichment included Wnt pathway genes (Wnt4 and Wif1), metabolic genes (Phgdh, Ldhb, Hif3a), known liver cancer markers (S100a8, S100a9, Tff3, Gpc3, Igdcc4), and several long noncoding RNAs (lncRNAs). mRNAs significantly downregulated in tumor-bearing livers included Ajap1 (P = 0.02), Wnt10a (P = 0.002), and Mettl17 (P = 0.0004).
To validate enrichment of the tumor-specific DEGs and determine if their induction was a function of mutant β-catenin, Myc, or both, we performed semiquantitative (sq) RT-PCR using cDNA prepared from livers of 4- to 6-week-old WT and tumor-bearing β-catΔEx3:Myc mice and each of the parental strains. Analysis confirmed that Dlk1, Igf2, S100a9, Peg3, Phgdh, Afp, Igdcc4 (Nope), Wnt4, and Cited1 were all elevated in tumor-bearing livers coexpressing β-catenin and Myc relative to β-catenin alone (Supplemental Figure 7), confirming broad alignment of the HB transcriptome with that of fetal liver. Unexpectedly, Dlk1, Igf2, S100a9, Peg3, Afp, Igdcc4 (Nope), Phgdh, and Wnt4 were elevated in livers of parental Alb-Myc mice, while Afp and Igdcc4 (Nope) were elevated in livers of β-catΔEx3 mice. Cited1, a highly expressed HB marker (Figure 4), was one mRNA whose expression was markedly elevated in livers coexpressing β-catenin and c-Myc relative to either of the parental strains. Given that 8 of 9 of the most highly expressed tumor-enriched DEGs were already elevated in the livers of tumor-naive Alb-Myc mice, we concluded that Myc sets the stage for tumorigenesis by altering hepatic gene expression and cell fate long before morphological evidence of neoplasia is evident and that Myc is the dominant tumorigenic driver in this strain.
To gain insight into the functional significance of the tumor-enriched DEGs, we performed ingenuity pathway analysis (IPA) and gene set enrichment analysis (GSEA). IPA identified the Wnt/β-catenin signaling pathway as the most significantly altered pathway (Supplemental Table 2), indicating hyperactivation of Wnt signaling in tumors relative to surrounding liver, despite pan-hepatic expression of mutant β-catenin. IPA functions and pathways assigned to the tumor enriched DEGs (Supplemental Table 2) differed substantially from those assigned to the larger list of DEGs co-regulated by mutant β-catenin and Myc (Supplemental Table 3), providing further discrimination between the transcriptomes of tumors and tumor-adjacent liver in β-catΔEx3:Myc mice. Agranulocyte/granulocyte adhesion, diapedesis, and fibrosis were also specified, emphasizing HB development in a background of hepatic injury and inflammation. GSEA highlighted perturbations in mitosis/cell cycle progression; cell communication; p53-, Kras-, and polycomb-regulated gene expression; and the cellular stress response (Supplemental Figure 8A). Given that Myc-associated genes were overrepresented in the tumor-enriched DEGs, we compared and contrasted GSEA-assigned functional classifications of all 3 DEG lists to determine Myc’s contributions to tumor development. Using this strategy, we determined that Myc positively affected chromatin modification, mRNA processing, mitosis, and mitochondrial biogenesis and suppressed the DNA damage response (Supplemental Figure 7B), indicating that Myc promotes HB, in part, by effecting early wholesale changes in hepatic gene expression via epigenetic reprogramming and by promoting mitochondrial biogenesis and genomic instability.
Persistent expression of mutant β-catenin generates a field of pan-hepatic Nrf2-dependent antioxidant signaling.
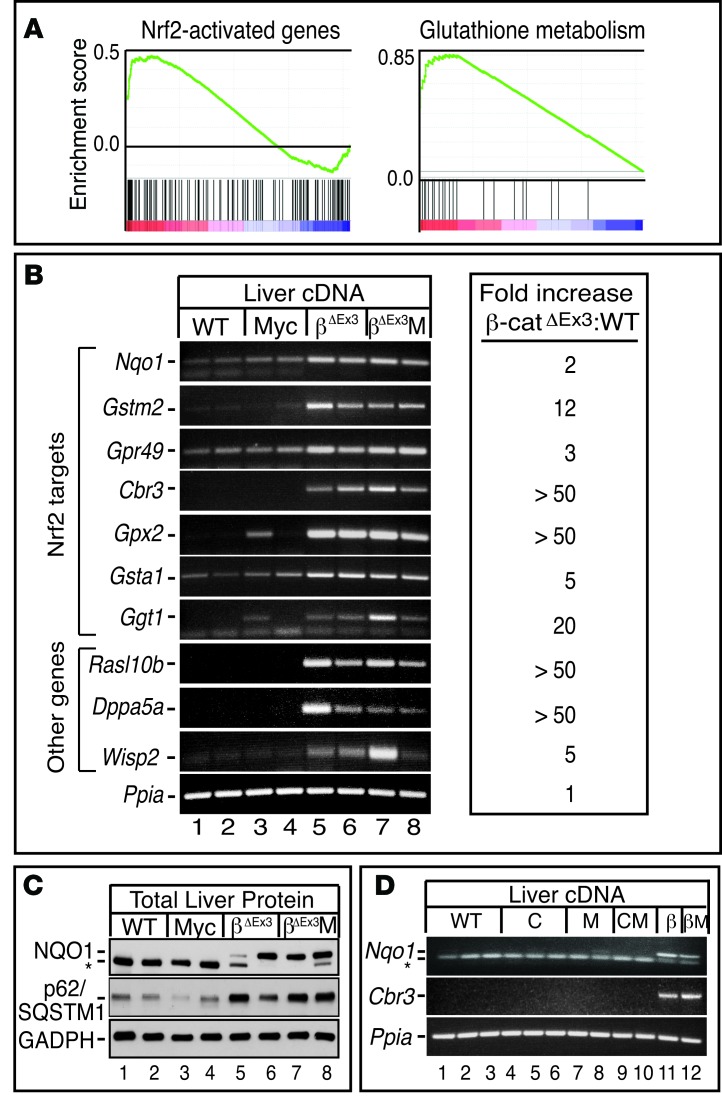
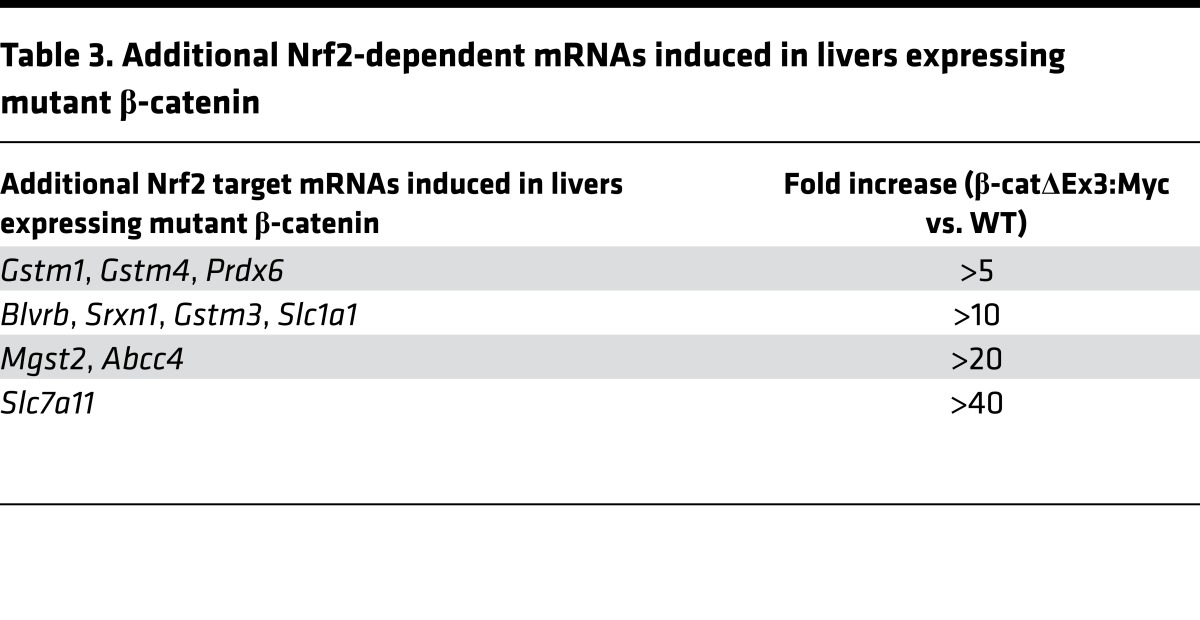
While bioinformatics identified Myc as the dominant driver of HBs in β-catΔEx3:Myc mice, it failed to identify clear protumorigenic functions of mutant β-catenin, forcing us to focus on functional assignments for the β-catenin–regulated genes to gain insight into it’s contributions to tumor development (Supplemental Table 4 and Supplemental Figure 8C). At first glance, we noticed that mitotic and cell cycle–associated signatures that typically accompany hyperplastic liver growth were absent (23), indicating that β-catenin–associated hepatomegaly was unlikely to reflect the direct mitogenic stimulation of hepatocytes. Instead, IPA identified inflammatory-mediated suppression of RXR as the most prominent signature (Supplemental Table 4). GSEA highlighted hepatic immune function and protein oligomerization as functional targets of mutant β-catenin action and, as previously reported (24), identified LEF1- and p53-dependent transcription as positively and negatively enriched oncogenic signatures, respectively (Supplemental Figure 8C). However, most striking of all was that IPA (Supplemental Table 4) and GSEA (Figure 6A and Supplemental Figure 8C) both identified Nfe2l2/Nrf2 antioxidant signaling as a significantly enriched function. Given that Nrf2 is activated in response to different types of cellular stress and is chronically activated in a wide range of human cancers (25), we authenticated the Nrf2 signature by performing sq RT-PCR for Nrf2 and non-Nrf2 targets in the β-catenin–regulated DEG list (Figure 6B and Table 3). In all cases, induction of mRNA was seen in β-catenin–expressing livers relative to WT. Interestingly, although Nrf2 has been reported as a target of Myc (26), Nrf2 target gene induction was not observed in livers of the parental Alb-Myc or Alb-Cre strains or any mice that lacked expression of the mutant β-catenin allele (Figure 6D), confirming that Nrf2’s transcriptional program was uniquely activated in livers expressing mutant β-catenin
Figure 6. Pan-hepatic expression of mutant β-catenin activates Nrf2-dependent antioxidant signaling.
(A) Gene set enrichment analysis (GSEA) of β-catenin–regulated genes using gene sets consisting of previously annotated Nrf2 target genes and genes linked to glutathione metabolism. (B) Semiquantitative (Sq) RT-PCR of β-catenin–regulated Nrf2 target genes and other genes identified by array. mRNAs are elevated in livers expressing β-catenin (lanes 5–8) and are independent of Albumin-c-Myc transgene status. (C) Western blot analysis of NQO1 and p62/SQSTM1 expression in livers of WT (lanes 1 and 2), Alb-c-Myc (lanes 3 and 4), β-catΔEx3 (lanes 5 and 6), and β-catΔEx3:Myc (lanes 7 and 8) mice. In C, the asterisk denotes a lower molecular weight nonspecific band. (D) Sq RT-PCR showing that induction of the Nrf2 mRNA targets Nqo1 and Cbr3 and is solely a function of mutant β-catenin expression (lanes 11 and 12) and that expression of Cre recombinase, either by itself (lanes 4–6) or in combination with the Alb-c-Myc transgene (lanes 9 and 10), does not induce Nrf2 target gene expression. In D, the 85bp Nqo1 amplicon migrates just above the excess primers in the PCR reaction, denoted by the asterisk. C, Alb-Cre mice; M, Alb-c-Myc mice; CM, Alb-Cre:Alb-c-Myc bitransgenic mice; β, β-catΔEx3:Alb-Cre mice; βM, β-catΔEx3:Alb-Cre:Alb-c-Myc mice.
Table 3. Additional Nrf2-dependent mRNAs induced in livers expressing mutant β-catenin.
To pinpoint the exact nature of the cellular stress elicited by mutant β-catenin, we used a commercially available array (RT2 Profiler) to profile gene sets reflecting specific types of cellular stress. Results showed that β-catenin induced a multitude of mRNAs encoding cytoprotective genes involved in glutathione and antioxidant homeostasis (Supplemental Table 5), consistent with IPA and GSEA, which identified a preponderance of processes related to xenobiotic metabolism, glutathione-mediated detoxification, oxidative stress (Supplemental Table 4), and glutathione metabolism (Figure 6A). This was in direct contrast with Myc, which preferentially affected mRNAs involved in the DNA damage response, cell death, and, to a lesser extent, inflammation, most notably Il-6 (Supplemental Table 5). Profiling using a second array enriched for genes specifically involved in counteracting oxidative stress, many of which are known Nrf2 targets, confirmed that mutant β-catenin, but not Myc, activated Nrf2 (Supplemental Table 6). Interestingly, Sqstm1, an autophagy adaptor that is not only a transcriptional target of Nrf2 but which also forms part of a self-amplifying regulatory loop that positively regulates Nrf2 (27), was upregulated in β-catenin livers at the mRNA and protein level (Figure 6C and Supplemental Table 6). Myc and mutant β-catenin thus exert a multilayered level of transcriptional control over hepatic gene expression to promote HB development, wherein Myc alters epigenetic regulation of gene expression and promotes genomic instability within a background of β-catenin–mediated injury and Nrf2/Nqo1-mediated antioxidant signaling.
Myc and Nrf2/Nqo1 are activated in human HB.
Having established the dominant role of Myc in driving mouse HBs and identifying Nrf2 signaling as a secondary consequence of mutant β-catenin expression, we investigated the extent to which human HBs display evidence of Nrf2 activation or overexpress Myc. Using antibodies specific for Myc and NQO1, a known surrogate for Nrf2 activation, we performed IHC on a tissue microarray containing duplicate paraffin cores of 14 clinically annotated human HBs, all but one of which demonstrated evidence of Wnt activation based on cytoplasmic β-catenin localization (data not shown), 3 normal human livers, and a normal human tonsil collected from patients treated at Children’s Medical Center, Dallas, Texas, USA, between 1993 and 2006 (28). IHC for MYC showed that normal liver was either negative or borderline positive for MYC, in contrast to normal tonsil, which expressed moderately high levels of Myc (Supplemental Figure 9 and Supplemental Table 7). Remarkably, half (7 of 14) of the HBs showed moderate or intense nuclear staining for MYC in the tumor epithelium but not in the stroma, while the remainder expressed only negligible MYC. Notably, in MYC-positive HBs containing both fetal and embryonal elements, the trend was for higher MYC expression and a greater number of MYC-positive cells in the embryonal component relative to the fetal component. IHC for NQO1 showed that normal hepatocytes lacked detectable expression, while bile ducts expressed moderate amounts of NQO1 in the cytoplasm (Supplemental Figure 9 and Supplemental Table 7). Of the 14 HBs, 8 (57%) showed elevated expression of NQO1, although expression varied between HBs and was heterogeneous within HBs. Consistent with the association of the Nrf2 signature with β-catenin but not Myc in mouse liver, only 4 tumors stained positive for NQO1 and Myc, indicating that elevated NQO1 expression was unrelated to Myc status in human HB (Pearson correlation coefficient, r = 0.29; P > 0.05).
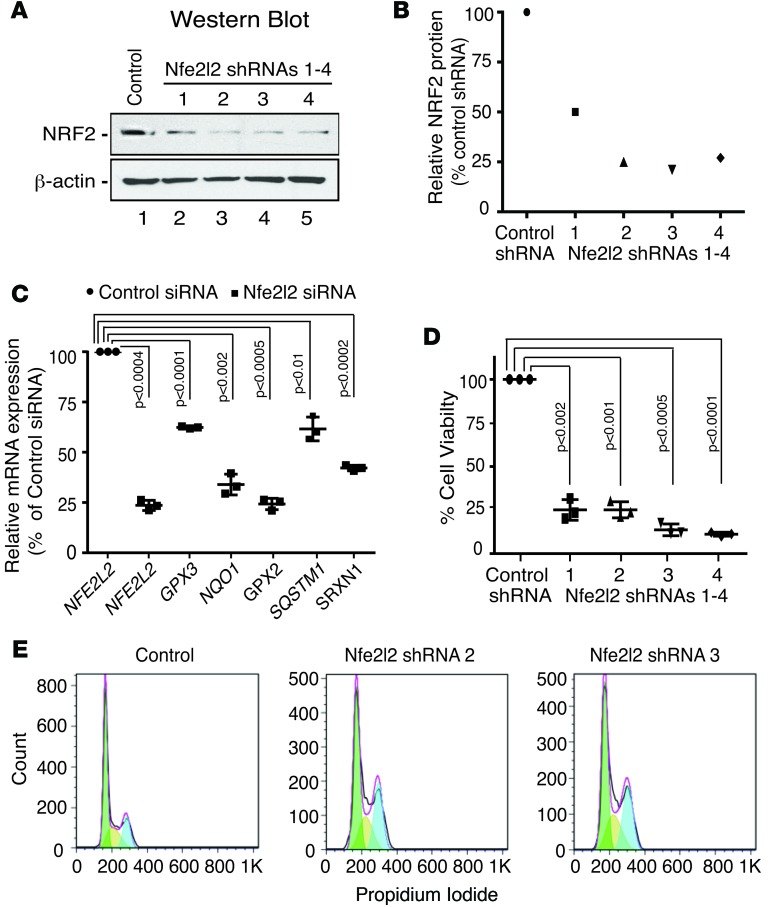
Having found that >50% of the HBs had inferred activation of Nrf2, based on NQO1-positive staining, we assessed the functional significance of Nrf2 for HB growth and viability in vitro by using RNA interference to target Nrf2 in Hep293TT cells, a cell line derived from an aggressive Wnt-activated human HB (29), with elevated Nrf2 expression due to copy number gains in the NFE2L2 gene (Figure 7 and Supplemental Table 8). Knockdown of Nrf2 in Hep293TT cells not only reduced the level of Nrf2 protein and mRNA (Figure 7, A–C) and its target genes (Figure 7C), but also significantly reduced tumor cell viability (Figure 7D) and stalled HB cell proliferation by inducing a G2/M arrest (Figure 7E), indicating that Nrf2 sustains HB cell growth and survival.
Figure 7. Nrf2 maintains HB cell proliferation and survival in vitro.
Hep293TT cells, derived from an aggressive human hepatoblastoma (HB) that expresses high levels of Nrf2, were transfected with control or Nfe2l2-specific shRNAs or siRNAs. (A) Western blot analysis and (B) quantitation showing efficient knockdown of Nrf2 in Hep293TT cells transfected with 4 different Nfe2l2-specific shRNAs. (C) Quantitative RT-PCR of NFE2L2/Nrf2 and Nrf2 target genes in Hep293TT cells transfected with an Nfe2l2-specific siRNA. (D) Suppression of NFE2L2 reduces Hep293TT cell viability. (E) FACS analysis of Hep293TT cells transfected with Nfe2l2-specific shRNAs, as determined by flow cytometry of propidium iodide–stained cells, showing that NFE2L2/Nrf2 knockdown induces a G2/M cell cycle arrest. Colors of the areas under the curves depict cells in G0/G1 (green); S (yellow) or G2/M (blue).Error bars in C and D represent SDs of 3 independent experiments. P values were generated using a 2-tailed unpaired t test.
Discussion
Unlike adult cancers that develop from the acquisition of multiple genomic insults over time, pediatric cancers often display remarkably few mutations (30), a characteristic that is thought to reflect the rapid evolution of childhood cancers from abnormal tissue environments during development. In the case of HB, a tumor with one of the lowest mutation rates of all cancers (13), aberrant activation of Wnt signaling is virtually universal, although generally not sufficient to drive liver tumorigenesis. In this study, we set out to determine if Myc, an oncogenic transcription factor previously implicated in the development of human HBs (15), cooperated with mutant β-catenin to drive HBs when the expression of both genes was targeted to the immature mouse liver. Here, we not only show that Myc indeed efficiently partners with mutant β-catenin to preferentially drive the development of mouse HBs over other liver tumor types, but that the HBs that develop align histologically and molecularly with their human counterparts and emerge before liver development is complete, mirroring onset in children. That Wnt activation is also selected for during Myc-driven hepatocarcinogenesis further suggests that Wnt/β-catenin and Myc are potent, perhaps even “preferred,” collaborators for HB development in mice. In delineating the contributions of both oncogenes to HB development, we identified Myc as the dominant determinant of the HB transcriptome and defined at least one mechanism by which mutant β-catenin likely contributes to liver tumor development, namely by indirectly inducing a protumorigenic field involving pan-hepatic activation of Nrf2/Nqo1 antioxidant signaling.
The rapid onset of tumors in neonates following perinatal activation of β-catenin and Myc, coupled with the fetal liver–like molecular profiles of the HBs in β-catΔEx3:Myc mice, point to hepatoblasts/immature hepatocytes, the postulated cells of origin for human HBs, as being the HB-initiating cells in this strain. In this setting, perinatal activation of the Albumin-Myc transgene likely counteracts the developmentally programmed decline in endogenous Myc to sustain the proliferation and survival of immature mutant β-catenin–expressing hepatic cells that would otherwise arrest or die (31). However, this is unlikely to be the case in Tet-O-Myc mice or other mouse models that develop HBs following oncogene activation in mature hepatocytes (32, 33), raising the question of whether HB transcriptomes and immunoprofiles reflect a hepatoblast cell of origin or the end result of dedifferentiation or reprogramming of mature hepatocytes (34). While an abundance of experimental evidence indicates that aberrant activation of Wnt/β-catenin alone is not sufficient for HB development, this has recently been challenged by the creation of a genetically engineered mouse strain that develops HBs and HCCs when mutant β-catenin is targeted to rare hepatic progenitor cells that express the CBP/p300 transcriptional coactivator protein and Myc target, Cited1 (35, 36). Although the experimental strategy used to target β-catenin to the liver in this strain differs from ours, in both instances, the timing of β-catenin activation overlaps with a period of high fetal liver expression of Myc (37). As HBs derived from mutant β-catenin–expressing CITED1+ cells express high levels of Myc (36) and because β-catΔEx3:Myc-driven HBs expressed high levels of Cited1 (this study), we offer up the possibility that tumors from both strains originate from a subset of mutant β-catenin–expressing CITED1+MYC+ hepatic precursors, a highly specific cellular context in which mutant β-catenin might be sufficient to drive tumor development by acting as a lineage survival oncogene (38). Fate-mapping studies will be required to determine if HBs can develop from cells with widely differing differentiation potentials via maturation arrest, reprogramming, or dedifferentiation.
Our data showing that Nrf2 activation is associated with chronic expression of mutant β-catenin converges with data showing that approximately 10% of human HBs (13) and approximately 6% of β-catenin–mutated human HCCs (39–41) harbor recurrent somatic gain-of-function mutations in NFE2L2, the gene that encodes Nrf2. Although this represents a low percentage of tumors, we reiterate that 57% of the human HBs in our tissue array overexpressed the prototypic Nrf2 target, NQO1, a result that is in agreement with a similar study that investigated Nqo1 mRNA expression in HBs (13). Taken together, these results indicate that Nrf2 activation occurs in a much higher proportion of HBs than is predicted on the basis of NFE2L2 mutational status alone, likely reflecting alternative modes of Nrf2 pathway activation (42).
The link between Nrf2 and oncogenic β-catenin is interesting in light of the fact that evidence of underlying liver disease, a known generator of oxidative stress, is generally absent in HB patients at the time of diagnosis. Nevertheless, a link between oxidative stress and HB has been proposed on the basis of epidemiological studies that show that premature and low birth weight infants, most of which spend extended periods in neonatal intensive care units (NICUs), have an approximately 20-fold elevated risk of HB (43, 44). Routine NICU treatments, including the provision of supplemental oxygen and administration of an array of life-sustaining drugs, place a particularly high burden on the immature liver and have the potential to either induce or exacerbate preexisting oxidative stress (45). Alternatively, collateral damage to the fetal liver caused by intrauterine hypoxia, hazardous environmental exposures, or medications taken by the mother during pregnancy may activate prosurvival pathways to limit damage to the developing liver. As the entire liver is likely to be affected in all of these scenarios, this suggests the possible existence of a “field cancerization” effect in the etiology of HB. Field cancerization is defined as a histologically unremarkable change that predisposes all or part of a tissue to neoplastic progression (46). It is perhaps best exemplified in lung cancer, in which smoking-associated damage in the airway epithelium establishes a preneoplastic field from which tumors later emerge (47). Indeed, recent studies show that half of patients with early-stage non–small-cell lung cancer harbor the same allelic imbalances in histologically normal tumor-adjacent airway cells that are present in tumors (48). Based on our finding that Nrf2 antioxidant signaling was active β-catΔEx3 livers as well as in tumor-naïve and tumor-bearing livers of β-catΔEx3:Myc mice, we propose that Nrf2/nqo1 constitutes a major component of a preneoplastic field orchestrated by mutant β-catenin that drives Myc-driven HB development by promoting the survival of stressed cells. Support for this concept derives from studies showing that hepatic injury provokes activation of β-catenin (49), Myc (50, 51), and Nrf2 (52) and that hepatotoxins accelerate oncogene-driven hepatocarcinogenesis (53). Indeed, very recent data demonstrate that p62/sqstm1, a known Nrf2 target that was elevated 3- to 4-fold in livers expressing β-catenin, constitutes part of such a field that sensitizes livers to diethylnitrosamine-induced HCC (54). Collectively, these data support the idea that a subset of HBs may evolve from hepatic cells that spontaneously acquire oncogenic mutations over time that drive tumor development in a normal liver, while others may have an etiology rooted in oxidative stress emanating from prior hepatic injury or inflammation and develop from cells that survive and outgrow their “less fit” neighbors. In the latter scenario, the preexistence of such a field could accelerate tumorigenesis by lowering the threshold for neoplastic progression in an immature liver; this, perhaps in part, explains why some HBs harbor as few as 2–3 somatic mutations (13).
While the growing association between Nrf2 and β-catenin opens up new therapeutic opportunities for the treatment of Wnt-activated pediatric liver cancers, we emphasize that caution should be taken in light of studies highlighting the complex relationship between Wnt/β-catenin and oxidative stress (55, 56). Moreover, conflicting reports on Nrf2’s role in liver regeneration (57, 58) and the interplay among inflammation, ROS, and cancer (58–61) warrant further studies to determine if Nrf2’s antioxidant program actively drives HB growth or is crucially enacted to limit β-catenin–induced injury and inflammation.
Finally, since being identified as a prominent oncogenic signature associated with aggressive human HBs (15), the concept of Myc as a driver of pediatric liver tumors has received scant attention, despite the fact that liver tumor “addiction” to Myc (16) presents a unique opportunity for therapeutic intervention. While the reasons for this are not entirely clear, the “undruggability” of Myc, coupled with the fact that Myc is neither a transcriptional target of Wnt in the liver (11) nor required for Wnt-dependent liver growth (62) may have diverted interest away from targeting Myc in HB. Moreover, as tumors frequently show hyperstabilization of Myc, mRNA profiling of HBs in the absence of follow up analysis of Myc at the protein level runs the risk of underestimating and diminishing the significance of this oncogenic transcription factor in HB (63, 64). However, in light of the fact that Myc emerged as the dominant driver of HBs in this study and that Myc is overexpressed in chemically induced mouse HBs (65) as well as in HBs and HCCs that develop in mouse models following expression of oncogenes other than Myc (33, 36), we propose that Myc serves as a central signaling hub for an array of other oncogenic HB drivers, making it a potentially important therapeutic target in HB that could benefit from the array of promising new drugs that target Myc and its network (66, 67). Therefore, the feasibility of adopting Myc inhibitory strategies for the treatment of pediatric liver cancers and other embryonal tumors with evidence of Myc amplification/overexpression should be explored.
Methods
Mouse strains, generation of β-catΔEx3:Myc mice, genotyping, and recombination.
Mice coexpressing mutant β-catenin and Myc in the liver (mixed C57BL6J/SJL/129S6 genetic background) were generated by breeding mice harboring a conditional mutant β-catenin (β-cat+/lox(Ex3)) allele (CTNNB1tmMmt) (12) to Alb-c-Myc transgenic mice [Tg(Alb-1, Myc)82Bri] (68). Compound β-cat+/lox(Ex3):Myc mice were then bred to Albumin-Cre transgenic mice [Tg(Alb-1, cre)21Mgn] (69) to generate β-catΔEx3:Myc mice that coexpressed Myc and one copy of the exon-3-deficient β-catenin allele in the liver. Male and female mice between the ages of 21 and 42 days were analyzed. Genotyping of mice and assessment of Cre-mediated recombination were conducted by PCR using primers and conditions listed in Supplemental Table 9.
Analysis of liver function.
Biochemical analysis of liver function was performed on heparin-treated plasma by the University of Texas Southwestern Metabolic Core Facility using a Vitros 250 bioanalyzer. Plasma was isolated from ≥3 individual mice/group.
Tumor incidence and histopathology review.
Tumor incidence was determined by counting the number of tumors and tumor types in 5-μ-thick H&E-stained sections of formalin-fixed liver. In each case, approximately 25% of the liver, consisting of approximately 7 to 9 pieces or 14 to 18 pieces (~1 cm3) in the case of very large livers, was fixed in 10% buffered formalin and embedded in paraffin. Total tumor number was determined by counting the tumors in all of the sectioned liver pieces but was not extrapolated to account for the remaining liver. Histopathology review was conducted independently by MJF, DR, and/or DLT. A final consensus was reached following secondary review by DR and SAC or REH.
IHC.
IHC was performed as described previously (23) using reagents and conditions listed in Supplemental Table 10. Where indicated, microwave antigen retrieval was performed using citrate buffer (pH 6) or 10 mM Tris/1 mM EDTA/0.05% Tween 20 (pH 8).
Scoring of IHC staining for MYC and NQO1 in human HBs, normal human liver, and tonsil.
Each core was assigned overall MYC or NQO1 scores ranging from 0 to 5 based on the relative proportion of positive cells and intensity of protein staining within each core. Threshold criteria used for designating samples as positive or negative for MYC or NQO1 staining are described in Supplemental Table 7.
Preparation of liver proteins, liver RNA, Northern blotting, and Western blotting.
Preparation of liver proteins and total liver RNA and for Northern and Western blotting was performed as previously described (23). Antibodies used for Western blotting are listed in Supplemental Table 10.
Gene expression profiling and determination of DEGs.
Gene expression analysis was conducted using SurePrint G3 Mouse GE 8x60K arrays (Agilent G4852A) containing 39,430 Entrez Gene RNAs and 16,251 lncRNAs using total RNA isolated from the livers of 3 WT mice, 3 mice expressing β-catenin alone, and 4 mice coexpressing β-catenin and c-Myc with multifocal tumor development. RNA integrity and quality were evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies Inc.). Each liver RNA was analyzed on a separate Agilent slide. After processing, microarrays were scanned using an Agilent microarray scanner, and gene expression data were extracted using Agilent Feature Extraction software. Probe annotation of whole genome mouse microarrays was downloaded from the manufacturer’s website (https://earray.chem.agilent.com/earray/). Of the 59,305 probes remaining after removal of control probes, 43,054 represented 24,164 unique genes and 16,251 represented 4,622 unique lncRNAs. Intensity measurements corresponding to gene expression levels were preprocessed with quantile normalization to eliminate array-array variation. Differential expression was detected using Student’s t test at the individual gene level and selected according to the following criteria: (a) fold change of > 4 for DEGS regulated by β-catenin (red box, Figure 5B) or jointly regulated by β-catenin and Myc (green box, Figure 5B) and > 2-fold change for putative tumor enriched DEGs (yellow box, Figure 5B); (b) multiple test adjustment (by Benjamini-Hochberg correction for multiple tests, ref. 70; P < 0.05 for significant DEGs); and (c) average intensity from at least 1 group of greater than 4 (log2). Additional P values for other differential gene expression assessments were also obtained, including rank-sum test and pi value, a unique statistic that combines the t test’s P value and fold change (71). Multi-dimensional scaling (MDS) was performed by applying a MDS algorithm to the gene expression data to visualize the relationship (Pearson coefficient) between samples (72). To visualize gene expression changes, a hierarchical clustering algorithm was applied to the gene expression data by average linkage and Pearson correlation coefficient and used to generate similarity measurements.
Gene set enrichment and other functional bioinformatics analyses.
DEGs obtained from the different liver transcriptome comparisons were as follows: (a) β-catenin (β-catΔEx3) versus WT (red box, Figure 5B): 1434 probes corresponding to 1020 unique genes (> 4-fold change and pi-value > 4). Among these, 415 unique genes were significantly altered with an adjusted P < 0.05. (b) β-catenin and Myc (β-catΔEx3:Myc) versus WT (green box, Figure 5B): 1710 probes corresponding to 1221unique genes, (> 4-fold change and pi-value > 4). Among these, 963 unique genes were significantly altered with an adjusted P < 0.05. (c) β-catenin and Myc versus β-catenin (β-catΔEx3:Myc versus β-catΔEx3) (yellow box, Figure 5B): 454 probes corresponding to 359 unique genes (> 2-fold change and pi-value > 2). Among these 108 unique genes were significantly altered with a raw P < 0.01. Functional analysis was performed using QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity) and Gene Set Enrichment Analysis (GSEA) (http://www.broadinstitute.org/gsea) (73, 74). All 3 gene lists for the putative tumor enriched DEGs (yellow box, Figure 5B), DEGs regulated by β-catenin (red box, Figure 5B) or DEGs co-regulated by β-catenin and Myc (green box, Figure 5B) are available in Supplemental Table 1. Data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE79084.
Post-array validation of mouse liver mRNA expression by sq RT-PCR.
1 μl of cDNA synthesized from DNase 1–treated total mouse liver RNA was used as a template for sq PCR amplification using TAq Polymerase (Qiagen) and primers and conditions listed in Supplemental Table 11. Confirmation of differential expression was obtained by running half of the PCR product on an agarose gel and quantifying band intensity using ImageJ (NIH) software.
Hep293TT cell culture, shRNA- and siRNA-mediated knockdown of NFE2L2, and immunoblotting.
For shRNA-mediated knockdown of NFE2L2, Hep293TT cells were cultured in RPMI 1640 supplemented with 10% FBS, 1 mM sodium pyruvate, 25 mM HEPES buffer, and 1% antibiotics in a humidified incubator at 37°C with 5% CO2. Cells were transiently transfected with nonsilencing shRNA or 4 different shRNAs targeting NFE2L2 using Lipofectamine 2000. Cells were harvested 72 hours after transfection, and Nrf2 protein levels were assessed by Western blotting. Whole-cell lysates were prepared in NENT buffer (100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 20 mM Tris-HCl [pH 8.0]) containing a 2× concentration of protease inhibitor cocktail (Thermo scientific) and cleared by centrifugation at 16,000 g for 30 minutes at 4°C. 30 μg of total protein was subjected to 10% SDS-PAGE gel electrophoresis and transferred to PVDF membranes (Millipore) using a semidry method. Membranes were blocked for 1 hour in 5% nonfat milk/Tris-buffered saline (TBS) containing 0.1% Tween-20 and incubated with primary antibodies (NFE2L2, Abcam; ab62352) and β-actin (Sigma-Aldrich; A5316)) overnight at 4°C. Membranes were washed (3 times for 10 minutes each) in TBS containing 0.3% Tween-20 and incubated with appropriate secondary antibodies for 2 hours at room temperature. Immunoreactive bands were visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore). shRNAs typically resulted in approximately 70%–80% knockdown of endogenous Nrf2 protein. shRNAs were obtained from Open Biosystems.
The following shRNA sequences were used: control nonsilencing shRNA, 5′-CTTACTCTCGCCCAAGCGAGAG-3′; shRNA 1 (V2LHS64258), 5′-GCATTAAAGCAGCGTATC-3′; shRNA 2 (V2LHS 238788), 5′-GCATTAAAGCAGCGTATC-3′; shRNA 3 (V3LHS 306092), 5′-GCATTAAAGCAGCGTATC-3′; and shRNA 4 (V2LHS 239104), 5′-GCATTAAAGCAGCGTATC-3′.
For siRNA-mediated knockdown of NFE2L2, Hep293TT cells (2.5 × 105 cells/well) were seeded into 6-well plates for 24 hours, after which they were transfected with 100 nM of NFE2L2 siRNA (Qiagen) using Lipofectamine RNAiMAX reagent. 72 hours after transfection, RNA was extracted and used to quantify NFE2L2 and target gene mRNA levels by quantitative PCR.
The following siRNA sequences were used: sense strand, 5′-CAUUGAUGUUUCUGAUCUATT-3′, and antisense strand, 5′-uagaucagaaacaucaauggg-3′.
Imaging and quantitation.
Ethidium bromide–stained gels and Northern blot and Western blot films were imaged using ImageJ (NIH) software. mRNA and protein expression were quantified by determining fold changes after first normalizing signal intensities to Ppia (cyclophilinA) for sq RNA analysis of cDNA, 18S rRNA or albumin for Northern blot RNA analysis, and GAPDH or β-actin for Western blot analysis.
Cell viability and cell cycle analysis.
Hep293TT cell proliferation was measured using the CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega) according to the manufacturer’s instructions. Hep293 TT cells (5,000 cells/well) seeded in 96-well plates were transfected with shRNAs. 72 hours after transfection, CellTiter-Glo reagent was added and luminescent intensity was measured using SpectraMax 5 (Molecular Devices). For analysis of cell cycle phase distribution, shRNA-transfected cells were harvested 72 hours after transfection and fixed with ice-cold 70% ethanol. Cells were washed with Dulbecco’s PBS and incubated in buffer containing propidium iodide (50 μg/ml) and RNaseA (20 μg/ml) for 30 minutes at room temperature. Cell cycle analysis was performed using a FACS Calibur Flow cytometer (BD Biosciences).
Quantitative real-time PCR of Hep293TT mRNA expression.
Total RNA was extracted using TRIzol reagent (Invitrogen). Genomic DNA contamination was eliminated by DNase 1 digestion (RNase-Free DNase, Qiagen), and RNA was further purified using an RNeasy MinElute Cleanup kit (Qiagen). Purified RNA was reverse transcribed using an iScript cDNA Synthesis kit (Bio-Rad). For real-time qPCR analysis, 4 μl of 1:20 diluted cDNA was amplified and monitored using the Quantitect SYBR Green PCR Kit (Qiagen) on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Data analysis was conducted using the ΔΔCT method with 18S rRNA as the normalization gene. Dissociation curves confirmed the presence of a single specific PCR product.
The following primer sequences were used: NFE2L2 forward, 5′-GACATGGATTTGATTGACAT-3′; NFE2L2 reverse, 5′-CTGGGAGTAGTTGGCAGATCC-3′; GPX2 forward, 5′-GGTAGATTTCAATACGTTCCGGG-3′; GPX2 reverse, 5′- TGACAGTTCTCCTGATGTCCAAA-3′; GPX3 forward, 5′-AGAGCCGGGGACAAGAGAA-3′; GPX3 reverse, 5′-ATTTGCCAGCATACTGCTTGA-3′; NQO1 forward, 5′-GAAGAGCACTGATCGTACTGGC-3′; NQO1 reverse, 5′-GGATACTGAAAGTTCGCAGGG-3′; SRXN1 forward, 5′-CAGGGAGGTGACTACTTCTACTC-3′; SRXN1 reverse, 5′-CAGGTACACCCTTAGGTCTGA-3′; SQSTM1 forward, 5′-GCACCCCAATGTGATCTGC-3′; SQSTM1 reverse, 5′-CGCTACACAAGTCGTAGTCTGG -3′; 18S rRNA forward, 5′-CCGACCAGAGCGAAAGCAT-3′; and 18S rRNA reverse, 5′-CCTCCGACTTTCGTTCTTGATT-3′.
Microarray data.
The data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE79084.
Statistics.
Statistical significance of results between groups was determined by 2-tailed Student’s t test. Statistical differences were considered significant if P < 0.05. All data are represented as mean ± SD or SEM (as indicated). For graphs, statistical analysis was performed using Prism GraphPad software. Specific P values are included on figures or in figure legends.
Study approval.
All animal experiments were approved by the University of Texas Southwestern Institutional Animal Care and Use Committee.
Author contributions
SAC, REH, and GET designed the research plan. SAC, REH, EAH, and SK conducted all of the mouse experiments. SAC, REH, and YC analyzed data. MJF, DLT, DR, SAC, and REH performed histopathology review. DR provided the HB tissue microarray. HB performed all of the Hep293TT cell culture experiments. KAOD contributed new material. SAC and REH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Mario Villareal for expert animal care, Zhao Lai for microarray processing, and John Shelton and Cameron Perry and all of the staff of the University of Texas Southwestern Molecular Pathology Core for tissue processing. This work was supported by multi-investigator grants from The Cancer Prevention Research Institute of Texas (CPRIT RP101195) (S.A.C, Y.C, D.R, M.J.F, D.L-T. and G.E.T), CPRIT RP120715 (Y.C, G.E.T) and NIH/NCI P30 CA54174 (Y.C, G.E.T), the Anna Schindler Foundation (S.A.C) and the University of Texas Southwestern Excellence in Education Foundation (R. E. H). K. A. O’Donnell is a CPRIT Scholar in Cancer Research and a Sidney Kimmel Foundation Kimmel Scholar. We dedicate this paper to the memory of Anna Schindler.
Footnotes
SK’s present address is: Department of Family Medicine, Baylor Scott and White Memorial Hospital, Temple, Texas, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(16):e88549. doi:10.1172/jci.insight.88549.
Contributor Information
Yidong Chen, Email: ChenY8@uthscsa.edu.
Hima Bansal, Email: BansalH@uthscsa.edu.
Shawn Klapproth, Email: shawn.klapproth@ttuhsc.edu.
Dinesh Rakheja, Email: dinesh.rakheja@utsouthwestern.edu.
Milton J. Finegold, Email: MJFinego@texaschildrens.org.
Dolores Lopez-Terrada, Email: Dhterrad@txch.org.
Robert E. Hammer, Email: Robert.Hammer@UTSouthwestern.edu.
References
- 1.Li J, Thompson TD, Miller JW, Pollack LA, Stewart SL. Cancer incidence among children and adolescents in the United States, 2001-2003. Pediatrics. 2008;121(6):e1470–e1477. doi: 10.1542/peds.2007-2964. [DOI] [PubMed] [Google Scholar]
- 2.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992-2004) Cancer. 2008;112(2):416–432. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 3.Trobaugh-Lotrario AD, Katzenstein HM. Chemotherapeutic approaches for newly diagnosed hepatoblastoma: past, present, and future strategies. Pediatr Blood Cancer. 2012;59(5):809–812. doi: 10.1002/pbc.24219. [DOI] [PubMed] [Google Scholar]
- 4.Shiojiri N. Transient expression of bile-duct-specific cytokeratin in fetal mouse hepatocytes. Cell Tissue Res. 1994;278(1):117–123. doi: 10.1007/BF00305783. [DOI] [PubMed] [Google Scholar]
- 5.Potter VR. Phenotypic diversity in experimental hepatomas: the concept of partially blocked ontogeny. The 10th Walter Hubert Lecture. Br J Cancer. 1978;38(1):1–23. doi: 10.1038/bjc.1978.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. López-Terrada D, et al. Towards an international pediatric liver tumor consensus classification: proceedings of the Los Angeles COG liver tumors symposium. Mod Pathol. 2014;27(3):472–491. [DOI] [PubMed] [Google Scholar]
- 7.Spector LG, Birch J. The epidemiology of hepatoblastoma. Pediatr Blood Cancer. 2012;59(5):776–779. doi: 10.1002/pbc.24215. [DOI] [PubMed] [Google Scholar]
- 8.Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res. 1999;59(2):269–273. [PubMed] [Google Scholar]
- 9.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5(1):e88549. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia D, et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014;60(5):1686–1696. doi: 10.1002/hep.27243. [DOI] [PubMed] [Google Scholar]
- 11.Cadoret A, et al. Hepatomegaly in transgenic mice expressing an oncogenic form of beta-catenin. Cancer Res. 2001;61(8):3245–3249. [PubMed] [Google Scholar]
- 12.Harada N, et al. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 2002;62(7):1971–1977. [PubMed] [Google Scholar]
- 13.Eichenmüller M, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61(6):1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Tomlinson GE, Douglass EC, Pollock BH, Finegold MJ, Schneider NR. Cytogenetic evaluation of a large series of hepatoblastomas: numerical abnormalities with recurring aberrations involving 1q12-q21. Genes Chromosomes Cancer. 2005;44(2):177–184. doi: 10.1002/gcc.20227. [DOI] [PubMed] [Google Scholar]
- 15.Cairo S, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14(6):471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Shachaf CM, et al. Myc inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 17.Beer S, et al. Developmental context determines latency of Myc-induced tumorigenesis. PLoS Biol. 2004;2(11):e88549. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy DJ, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14(6):447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol. 2000;157(3):763–770. doi: 10.1016/S0002-9440(10)64590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, et al. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mech Dev. 2009;126(8-9):665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- 21.Notenboom RG, Moorman AF, Lamers WH. Developmental appearance of ammonia-metabolizing enzymes in prenatal murine liver. Microsc Res Tech. 1997;39(5):413–423. doi: 10.1002/(SICI)1097-0029(19971201)39:5<413::AID-JEMT4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Cairo S, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107(47):20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comerford SA, Schultz N, Hinnant EA, Klapproth S, Hammer RE. Comparative analysis of SV40 17kT and LT function in vivo demonstrates that LT’s C-terminus re-programs hepatic gene expression and is necessary for tumorigenesis in the liver. Oncogenesis. 2012;1:e88549. doi: 10.1038/oncsis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyagishi M, et al. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J Biol Chem. 2000;275(45):35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- 25.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichimura Y, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Litten JB, et al. Activated NOTCH2 is overexpressed in hepatoblastomas: an immunohistochemical study. Pediatr Dev Pathol. 2011;14(5):378–383. doi: 10.2350/10-09-0900-OA.1. [DOI] [PubMed] [Google Scholar]
- 29.Chen TT, et al. Establishment and characterization of a cancer cell line derived from an aggressive childhood liver tumor. Pediatr Blood Cancer. 2009;53(6):1040–1047. doi: 10.1002/pbc.22187. [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decaens T, et al. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology. 2008;47(1):247–258. doi: 10.1002/hep.21952. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen LH, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 2014;26(2):248–261. doi: 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao J, et al. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147(3):690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15(3):244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Méniel V, et al. Cited1 deficiency suppresses intestinal tumorigenesis. PLoS Genet. 2013;9(8):e88549. doi: 10.1371/journal.pgen.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mokkapati S, et al. β-catenin activation in a novel liver progenitor cell type is sufficient to cause hepatocellular carcinoma and hepatoblastoma. Cancer Res. 2014;74(16):4515–4525. doi: 10.1158/0008-5472.CAN-13-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid P, Schulz WA, Hameister H. Dynamic expression pattern of the Myc protooncogene in midgestation mouse embryos. Science. 1989;243(4888):226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- 38.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6(8):593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 39.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46(12):1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 42.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanimura M, et al. Increased risk of hepatoblastoma among immature children with a lower birth weight. Cancer Res. 1998;58(14):3032–3035. [PubMed] [Google Scholar]
- 44.Turcotte LM, et al. Neonatal medical exposures and characteristics of low birth weight hepatoblastoma cases: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2014;61(11):2018–2023. doi: 10.1002/pbc.25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsubasa T, et al. Oxidative stress in very low birth weight infants as measured by urinary 8-OHdG. Free Radic Res. 2002;36(2):189–193. doi: 10.1080/10715760290006510. [DOI] [PubMed] [Google Scholar]
- 46.Rubin H. Fields and field cancerization: the preneoplastic origins of cancer: asymptomatic hyperplastic fields are precursors of neoplasia, and their progression to tumors can be tracked by saturation density in culture. Bioessays. 2011;33(3):224–231. doi: 10.1002/bies.201000067. [DOI] [PubMed] [Google Scholar]
- 47.Kadara H, Wistuba Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc. 2012;9(2):38–42. doi: 10.1513/pats.201201-004MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jakubek Y, et al. Genomic Landscape Established by Allelic Imbalance in the Cancerization Field of a Normal Appearing Airway. Cancer Res. 2016;76(13):3676–3683. doi: 10.1158/0008-5472.CAN-15-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apte U, et al. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol. 2009;175(3):1056–1065. doi: 10.2353/ajpath.2009.080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones BE, Czaja MJ. III. Intracellular signaling in response to toxic liver injury. Am J Physiol. 1998;275(5 Pt 1):G874–G878. doi: 10.1152/ajpgi.1998.275.5.G874. [DOI] [PubMed] [Google Scholar]
- 51.Simile MM, et al. Correlation between S-adenosyl-L-methionine content and production of c-Myc, c-Ha-ras, and c-Ki-ras mRNA transcripts in the early stages of rat liver carcinogenesis. Cancer Lett. 1994;79(1):9–16. doi: 10.1016/0304-3835(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 52.Goldring CE, et al. Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology. 2004;39(5):1267–1276. doi: 10.1002/hep.20183. [DOI] [PubMed] [Google Scholar]
- 53.Beer S, Komatsubara K, Bellovin DI, Kurobe M, Sylvester K, Felsher DW. Hepatotoxin-induced changes in the adult murine liver promote Myc-induced tumorigenesis. PLoS One. 2008;3(6):e88549. doi: 10.1371/journal.pone.0002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umemura A, et al. p62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell. 2016;29(6):935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao GZ, et al. Wnt/β-catenin signaling protects mouse liver against oxidative stress-induced apoptosis through the inhibition of forkhead transcription factor FoxO3. J Biol Chem. 2013;288(24):17214–17224. doi: 10.1074/jbc.M112.445965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang XF, et al. Conditional beta-catenin loss in mice promotes chemical hepatocarcinogenesis: role of oxidative stress and platelet-derived growth factor receptor alpha/phosphoinositide 3-kinase signaling. Hepatology. 2010;52(3):954–965. doi: 10.1002/hep.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beyer TA, et al. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2008;27(1):212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Köhler UA, et al. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology. 2014;60(2):670–678. doi: 10.1002/hep.26964. [DOI] [PubMed] [Google Scholar]
- 59.Beyer TA, Werner S. The cytoprotective Nrf2 transcription factor controls insulin receptor signaling in the regenerating liver. Cell Cycle. 2008;7(7):874–878. doi: 10.4161/cc.7.7.5617. [DOI] [PubMed] [Google Scholar]
- 60.Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med. 2014;6(221):e88549. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 61.Sporn MB, Liby KT. Nrf2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12(8):564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reed KR, et al. B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc Natl Acad Sci U S A. 2008;105(48):18919–18923. doi: 10.1073/pnas.0805778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takayasu H, et al. Frequent deletions and mutations of the beta-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clin Cancer Res. 2001;7(4):901–908. [PubMed] [Google Scholar]
- 64.Turato C, et al. Over-expression of SERPINB3 in hepatoblastoma: a possible insight into the genesis of this tumour? Eur J Cancer. 2012;48(8):1219–1226. doi: 10.1016/j.ejca.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Bhusari S, et al. Genomic profiling reveals unique molecular alterations in hepatoblastomas and adjacent hepatocellular carcinomas in B6C3F1 mice. Toxicol Pathol. 2015;43(8):1114–1126. doi: 10.1177/0192623315599853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dauch D, et al. A Myc-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med. 2016;22(7):744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 67.Posternak V, Cole MD. Strategically targeting Myc in cancer. F1000Res. 2016:e88549. doi: 10.12688/f1000research.7879.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandgren EP, Quaife CJ, Pinkert CA, Palmiter RD, Brinster RL. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4(6):715–724. [PubMed] [Google Scholar]
- 69.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26(2):149–150. doi: 10.1002/(SICI)1526-968X(200002)26:2<149::AID-GENE16>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 71.Xiao Y, et al. A novel significance score for gene selection and ranking. Bioinformatics. 2014;30(6):801–807. doi: 10.1093/bioinformatics/btr671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Meltzer PS. Gene expression analysis via multidimensional scaling. Curr Protoc Bioinformatics. 2005;Chapter 7:e88549. doi: 10.1002/0471250953.bi0711s10. [DOI] [PubMed] [Google Scholar]
- 73.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 74.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.