A high-resolution crystal structure of an unknown glycoside hydrolase enzyme from the rumen of B. taurus is presented. Unique dual carbohydrate-binding domains are revealed.
Keywords: GH29, glycoside hydrolase, α-l-fucosidase, carbohydrate-binding domain, CBM32, PDB5K9H
Abstract
Glycoside hydrolase (GH) family 29 consists solely of α-l-fucosidases. These enzymes catalyse the hydrolysis of glycosidic bonds. Here, the structure of GH29_0940, a protein cloned from metagenomic DNA from the rumen of a cow, has been solved, which reveals a multi-domain arrangement that has only recently been identified in bacterial GH29 enzymes. The microbial species that provided the source of this enzyme is unknown. This enzyme contains a second carbohydrate-binding domain at its C-terminal end in addition to the typical N-terminal catalytic domain and carbohydrate-binding domain arrangement of GH29-family proteins. GH29_0940 is a monomer and its overall structure consists of an N-terminal TIM-barrel-like domain, a central β-sandwich domain and a C-terminal β-sandwich domain. The TIM-barrel-like catalytic domain exhibits a (β/α)8/7 arrangement in the core instead of the typical (β/α)8 topology, with the ‘missing’ α-helix replaced by a long meandering loop that ‘closes’ the barrel structure and suggests a high degree of structural flexibility in the catalytic core. This feature was also noted in all six other structures of GH29 enzymes that have been deposited in the PDB. Based on sequence and structural similarity, the residues Asp162 and Glu220 are proposed to serve as the catalytic nucleophile and the proton donor, respectively. Like other GH29 enzymes, the GH29_0940 structure shows five strictly conserved residues in the catalytic pocket. The structure shows two glycerol molecules in the active site, which have also been observed in other GH29 structures, suggesting that the enzyme catalyses the hydrolysis of small carbohydrates. The two binding domains are classed as family 32 carbohydrate-binding modules (CBM32). These domains have residues involved in ligand binding in the loop regions at the edge of the β-sandwich. The predicted substrate-binding residues differ between the modules, suggesting that different modules bind to different groups on the substrate(s). Enzymes that possess multiple copies of CBMs are thought to have a complex mechanism of ligand recognition. Defined electron density identifying a long 20-amino-acid hydrophilic loop separating the two CBMs was observed. This suggests that the additional C-terminal domain may have a dynamic range of movement enabled by the loop, allowing a unique mode of action for a GH29 enzyme that has not been identified previously.
1. Introduction
Glycoside hydrolases (GHs; EC 3.2.1.–) form a widespread group of enzymes that catalyse the hydrolysis of glycosidic bonds between two or more carbohydrate moieties or between a carbohydrate moiety and a noncarbohydrate moiety. Glycoside hydrolases can be classified according to their catalytic function with a numerical classification (EC number) or a sequence-based classification. The CAZy database uses a sequence-based classification to subdivide these enzymes into more than 130 families (http://www.cazy.org; Lombard et al., 2014 ▸). In addition, two main mechanisms of glycosidic bond hydrolysis of GHs have been characterized, involving either inversion or retention of the anomeric configuration (Rye & Withers, 2000 ▸). Enzymes classed as α-l-fucosidases are currently found in families GH29 and GH95 in the CAZy classification. The best studied family are the GH29 enzymes, which consist of retaining α-l-fucosidases that have been identified in a variety of organisms, including bacteria, plants, insects and animals (Lombard et al., 2014 ▸). Only six X-ray structures of bacterial GH29 enzymes have been deposited in the Protein Data Bank (PDB). These include three proteins from Bacteroides thetaiotaomicron [BT2970 (PDB entry 2wvs), BT2192 (PDB entry 3eyp) and BT3798 (PDB entry 3gza); Lammerts van Bueren et al., 2010 ▸; Guillotin et al., 2014 ▸], one protein from Bifidobacterium longum subsp. infantis (Blon_2336; PDB entry 3ues; Sakurama et al., 2012 ▸), one protein from Thermotoga maritima (TM0306; PDB entry 1hl8; Sulzenbacher et al., 2004 ▸) and one protein from Bacteroides ovatus ATCC8483 (BACOVA_04357; PDB entry 4zrx; Joint Center for Structural Genomics, unpublished work).
The GH29 family can be further divided into two subfamilies by phylogenetic analysis and this partition possibly correlates with a difference in substrate specificity between the two groups (Ashida et al., 2009 ▸; Sakurama et al., 2012 ▸). The group A subfamily (GH29-A) comprises enzymes that show little substrate specificity and act efficiently on model chromogenic substrates such as p-nitrophenyl fucopyranoside (α-l-fucosidases; EC 3.2.1.51). The GH29-family enzymes BT2970 from B. thetaiotaomicron (PDB entry 2wvs) and TM0306 from T. maritima (PDB entry 1hl8) belong to the GH29-A subfamily with verified catalytic activity (Lammerts van Bueren et al., 2010 ▸; Sulzenbacher et al., 2004 ▸). In contrast, the group B subfamily (GH29-B) specifically hydrolyse terminal α(1–3/4)-fucosidic linkages and are poor catalysts for p-nitrophenyl fucopyranoside substrates (α-1,3/1,4-l-fucosidases; EC 3.2.1.111). The GH29-family enzymes BT2192 from B. thetaiotaomicron (PDB entry 3eyp) and Blon_2336 from B. longum subsp. infantis (PDB entry 3ues) have been characterized as belonging to the GH29-B subfamily (Guillotin et al., 2014 ▸; Sakurama et al., 2012 ▸). There are currently no published data characterizing the activity of either the enzyme BT3798 from B. thetaiotaomicron (PDB entry 3gza) or the enzyme BACOVA_04357 from B. ovatus (PDB entry 4zrx). There does not appear to be any clear structural distinction between the two subfamilies. The current crystal structures of GH29 enzymes all appear very similar and show a two-domain configuration with an N-terminal catalytic (TIM-barrel) domain and a C-terminal (β-sandwich) carbohydrate-binding domain. The catalytic residues of GH29 enzymes are well defined from functional studies and comprise an aspartate and glutamate as the catalytic nucleophile and the proton donor, respectively.
We have expressed, crystallized and determined the crystal structure of a predicted GH29 α-l-fucosidase cloned from metagenomic DNA isolated from the rumen of a grass-fed Friesian cow. The bacterial species from which this GH29 enzyme is derived is unknown. The structure reveals three domains with an N-terminal TIM-barrel-like domain followed by two β-sandwich carbohydrate-binding domains. This domain arrangement was unexpected as all available structures at the time comprised two domains only. The structure was solved using molecular replacement with a closely related existing structure (PDB entry 3eyp) which contained only two domains. The third domain was built manually. Recently, a three-domain bacterial GH29 structure has been deposited in the PDB (PDB entry 4zrx). Its second carbohydrate-binding domain is positioned differently to that of GH29_0940, supporting our hypothesis of a dynamic mode of action. Attempts to determine the substrate specificity of this enzyme using p-nitrophenyl (pNP)-conjugated carbohydrates were unsuccessful; however, this result may indicate that this enzyme belongs to the GH29-B subfamily. The multimodular structure of this enzyme is postulated to provide a unique mechanism of substrate recognition in the GH29 family that has not been identified previously.
2. Materials and methods
2.1. Cloning, expression and purification
The open reading frame (ORF) GH29_0940 originated from a bovine rumen microbial metagenomic fosmid clone, Ad-384-0049-J15, which exhibited activities on the substrates 4-methylumbelliferyl β-d-glucopyranoside and 4-methylumbelliferyl β-d-xylopyranoside. The ORF was examined for potential signal peptide, transmembrane sequence and regions of disorder, prior to determining the cloning methodology, using the XtalPred server and the TMHHM and RPSP programs (Slabinski et al., 2007 ▸; Krogh et al., 2001 ▸; Plewczynski et al., 2007 ▸). The gene GH29_0940 coded for a protein of 562 amino acids in length. The gene was synthesized to include attB1/attB2 Gateway cloning sites. Gateway cloning was performed into the destination vector pDEST17 (Life Technologies, USA). Cloning success was confirmed by DNA sequencing prior to vector transformation into electrocompetent Escherichia coli BL21 cells. Macromolecule-production information is summarized in Table 1 ▸. The E. coli transformant was grown in 1 l Luria–Bertani (LB) medium containing ampicillin (100 µg ml−1) and incubated on a rotary shaker (200 rev min−1, 37°C) until the optical density OD600 reached between 0.4 and 0.6. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM to induce expression and the culture was grown at 30°C for a further 20 h. The E. coli culture was centrifuged at 6000 rev min−1 for 20 min at 4°C, resuspended in 40 ml column buffer A (50 mM Tris pH 7.5, 100 mM NaCl, 20 mM imidazole) with the addition of a cOmplete Mini, EDTA-free protease-inhibitor cocktail tablet (Roche, Germany) and was disrupted by sonication. The lysate was centrifuged at 13 000 rev min−1 for 20 min at 4°C, followed by filtration through 1.2, 0.45 and 0.20 µm sterile syringe filters (Sartorius, Germany). The lysate was applied onto a HiTrap Chelating Column (GE Healthcare, Germany) that was charged with nickel and pre-equilibrated with buffer A. The column was washed with buffer A and the recombinant protein was eluted with a gradient of buffer B (50 mM Tris pH 7.5, 100 mM NaCl, 500 mM imidazole). The protein was concentrated (at room temperature) using a 10 kDa molecular-weight cutoff ultrafiltration centrifugation column (Sartorius, Germany), further purified by gel filtration on a S200 16/60 size-exclusion column (GE Healthcare, Germany) in 50 mM Tris pH 7.5, 100 mM NaCl buffer and concentrated.
Table 1. Macromolecule-production information.
| Source organism | Unknown |
| DNA source | Unknown |
| Forward primer† | Gene-specific attB1 primer as designed by GeneArt for Gateway cloning |
| Reverse primer | Gene-specific attB2 primer as designed by GeneArt for Gateway cloning |
| Cloning vector | pENTR221 |
| Expression vector | pDEST17 |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MSYYHHHHHHLESTSLYKKAGFENLYFQSAQVEPCGPVPTENQLRWQDMEMYAFIHYSLNTYTDEEWGYGNEDPQLFNPSSLDCRQWARVCKQAGMRGIIFTAKHHCGFCMWPSAYTEYSVKNSPWKNGKGDVVRELADACREEGLKFAVYLSPWDRNHPAYGQPAYVAYFRNQLRELLTNYGEIFEVWFDGANGGDGWYGGANETRKIDRTTYYQWPETYKMIRQLQPNCLIWNDGSDRGDLRWVGTEAGNVGETNWSLLNHDGEVEWHMLHYGLENGDSWVPGETNTSIRPGWFYHDTENEHVKSLSKLMDTYYKSVGRNSTLLLNFPIAPNGRIHPNDSLRGIAFKKMIGEVFRKNLAEKARTQTKGDETVIDFGKPTTFNRFLAEEDIRYGQRVKKFLLEAEINGQWQQLKDALVENGDGLTTIGHRRIICFPTVNATKLRFTVVNTKCEPFIKKLGVYLAPELTADIPDAGEKKSSNLHLFFSSPTQMMIDWETEQTITSFRYLPPQESKDGTVTHYTLWASTDWSNWTKLASGEFSNVVNNPIWQTIKFQPVRAKILKLDADRLATGNRMAYGDVEVNLKLNIEN |
The attB1 forward primer contained an upstream rTEV cleavage site (underlined)
2.2. Crystallization
Initial crystallization screening was performed using a Mosquito Crystal liquid-handling robot (TTP Labtech, England) with Crystal Screen HT and Index reagent screens (Hampton Research, USA) by vapour diffusion in sitting drops in low-profile 96-well Intelli-Plates (Art Robbins Instruments, USA). Condition D3 [0.1 M HEPES pH 7.0, 30%(v/v) Jeffamine ED-2001 pH 7.0] from the Index regent screen gave needle-shaped crystals in a bundle after 8 d. These conditions were further optimized by manually varying the precipitant and pH in hanging drops in 24-well plates (Hampton Research, USA). Further trials were performed using Additive Screen (Hampton Research) in sitting drops in low-profile 96-well Intelli-Plates. The additive screen yielded needle-shaped crystals in a number of conditions. Table 2 ▸ describes the crystallization conditions of the largest, singular needle, which was flash-cooled in liquid nitrogen for data collection.
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | Art Robbins Instruments Intelli-Plate 96-2 low profile |
| Temperature (K) | 291 |
| Protein concentration (mg ml−1) | 11.0 |
| Buffer composition of protein solution | 50 mM Tris pH 7.5, 100 mM NaCl |
| Composition of reservoir solution | 0.1 M HEPES pH 7.5, 35%(v/v) Jeffamine ED-2001 pH 7.0, 2.0 M sodium thiocyanate |
| Volume and ratio of drop | 1 µl protein solution + 1 µl reservoir solution |
| Volume of reservoir (µl) | 100 |
| Cryoprotection solution | 30%(v/v) glycerol, 0.1 M HEPES pH 7.5, 35%(v/v) Jeffamine ED-2001 pH 7.0 |
2.3. Data collection, processing and structure determination
Two data sets were collected on the macromolecular crystallography beamline (MX1) at the Australian Synchrotron, Melbourne, Australia. Data collection, integration and scaling were performed within Blu-Ice (McPhillips et al., 2002 ▸), XDS (Kabsch, 2010 ▸) and AIMLESS (from the CCP4 suite of crystallographic software; Evans, 2011 ▸; Winn et al., 2011 ▸). Data were collected to 2.47 and 2.03 Å resolution and these resolution limits were determined by the edge of the detectors. Data-collection statistics are given in Table 3 ▸. There was one molecule in the asymmetric unit, with a corresponding Matthews coefficient of 2.54 Å3 Da−1 and a solvent content of 51.7%.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | 3BM1 dipole MX1 beamline, Australian Synchrotron |
| Wavelength (Å) | 0.9537 |
| Temperature (K) | 100.0 |
| Detector | ADSC Quantum 210r |
| Crystal-to-detector distance (mm) | 180 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 1 |
| Space group | P212121 |
| a, b, c (Å) | 65.67, 78.31, 134.53 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.6 |
| Resolution range (Å) | 47.13–2.03 (2.08–2.03) |
| Total No. of reflections | 332797 |
| No. of unique reflections | 45691 |
| Completeness (%) | 99.9 (98.9) |
| Multiplicity | 7.3 (7.0) |
| 〈I/σ(I)〉 | 7.9 (2.1) |
| R r.i.m. † (%) | 10.95 (48.53) |
| Overall B factor from Wilson plot (Å2) | 20.9 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity. R merge = 18.9% (83.2% for the outer shell).
The structure was determined to 2.03 Å resolution. Details of the structure solution and refinement are given in Table 4 ▸. The first and second domains (catalytic and carbohydrate-binding domains) of this protein were solved by molecular replacement using PHENIX (Adams et al., 2011 ▸) and the structure with PDB code 3eyp as a search model. The structure of the third domain (the carbohydrate-binding domain) was built using Buccaneer (from the CCP4 suite of crystallographic software; Cowtan, 2006 ▸; Winn et al., 2011 ▸). This was followed by iterative cycles of manual model building and refinement using Coot (Emsley et al., 2010 ▸) and PHENIX (Adams et al., 2011 ▸). The single polypeptide chain of GH29_0940 is visible from residues 4 to 557, with the N-terminal histidine tag, the first three residues and the last five residues not visible in the electron density. Electron density in the active site indicates that two glycerol molecules are bound. Glycerol was used as the cryoprotectant, which explains its presence.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 46.99–2.03 (2.08–2.03) |
| Completeness (%) | 99.7 (98.9) |
| No. of reflections, working set | 45545 |
| No. of reflections, test set | 2297 |
| Final R cryst | 0.191 (0.288) |
| Final R free | 0.229 (0.348) |
| No. of non-H atoms | |
| Protein | 4499 |
| Ion | 62 |
| Ligand | 48 |
| Water | 635 |
| Total | 5196 |
| R.m.s. deviations | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.225 |
| Average B factors (Å2) | |
| Protein | 19.7 |
| Ion | 52.6 |
| Ligand | 27.7 |
| Water | 25.7 |
| Ramachandran plot | |
| Most favoured (%) | 94.6 |
| Allowed (%) | 4.5 |
The structure of GH29_0940 was compared with all available protein structures using PDBeFold (European Molecular Biology Laboratory–European Bioinformatics Institute; EMBL–EBI). Structural superpositions were calculated using Coot (Emsley et al., 2010 ▸) for the first and second domains or PyMOL (v.1.3; Schrödinger) for the third domain. The solvent-accessible surface area and potential interface area were calculated using PDBePISA (EMBL–EBI). Molecular and surface illustrations were prepared in PyMOL (v.1.3). Coordinate files and structure factors for the orthorhombic crystal form have been deposited in the Protein Data Bank with PDB code 5k9h.
2.4. Sequence alignment
Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) was used to compare the GH29_0940 protein sequence with deposited structures in the Protein Data Bank (PDB). PSI-BLAST is a search method that builds upon protein sequence alignments generated by a run of the BLASTp program, a sequence-similarity search method (the query protein sequence is compared with protein sequences in a target database) which identifies regions of local alignment and reports those above a specified threshold score. We report information on ‘query coverage’, which is defined as the percentage of the query sequence (i.e. GH29_0940) that overlaps the subject sequence.
3. Results and discussion
3.1. Sequence analysis
The ORF GH29_0940 was identified as gene number 940 from within a (bovine rumen microbial metagenomic) fosmid clone which contained a large number of genes identified as carbohydrate-acting enzymes. An annotation of the domain architecture by a database for carbohydrate-active enzymes annotation (dbCAN; Yin et al., 2012 ▸) describes the first domain as a GH29 domain, and the second and third domains as carbohydrate-binding module 32 (CBM32) domains. All data in dbCAN are generated based on the family classifications from the CAZy database (http://www.cazy.org; Lombard et al., 2014 ▸). The glycoside hydrolase class of enzymes is divided into 133 families, with the GH29 family having known α-l-fucosidase (EC 3.2.1.51) and α-1,3/1,4-l-fucosidase (EC 3.2.1.111) enzyme activities. The amino-acid residues involved in the catalytic mechanism are aspartate and glutamate, and there are currently six structures of microbial GH29 enzymes in the database. The CBM32 domain activity is wide-ranging and has been described for only three bacterial species (Micromonospora viridifaciens sialidase, a Yersinia member and Clostridium perfringens).
The DNA sequence of GH29_0940 codes for a mature protein of 562 amino-acid residues with a predicted molecular weight of 65 kDa and a theoretical pI of 5.9 (ProtParam; Gasteiger et al., 2005 ▸). GH29 is predicted to contain three putative conserved domains, comprising an N-terminal α-l-fucosidase domain, a central f5/f8-type C domain and a C-terminal f5/f8-type C domain (PSI-BLAST; Altschul et al., 1997 ▸).
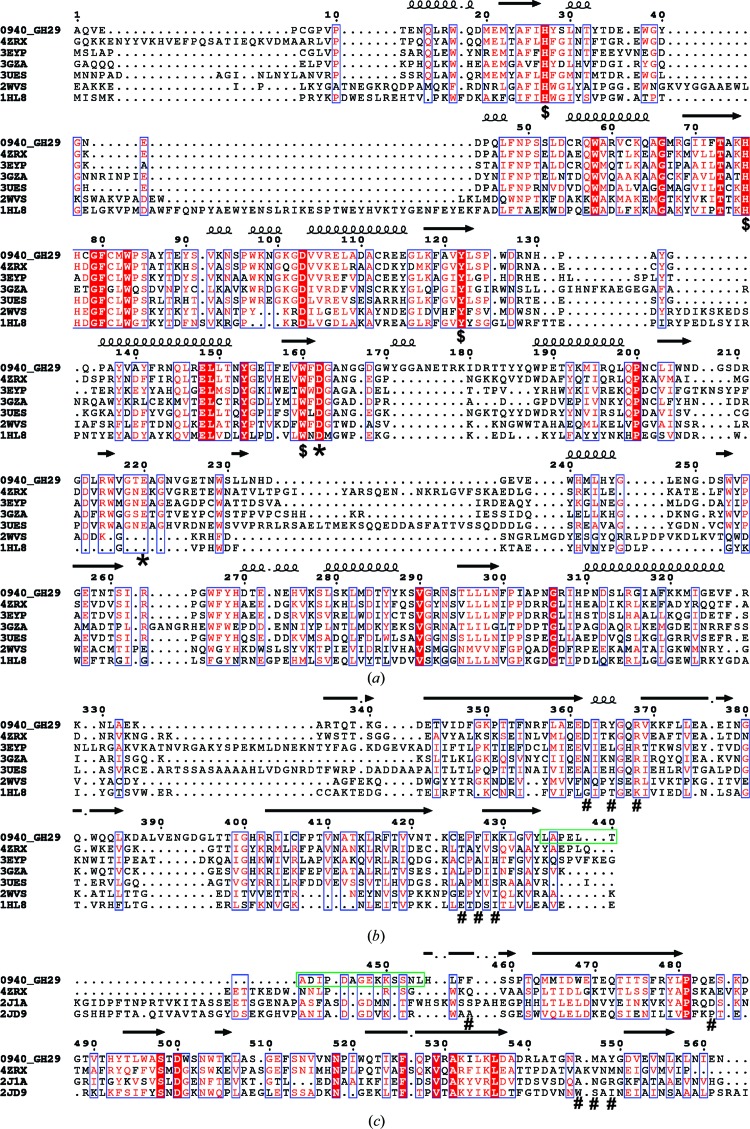
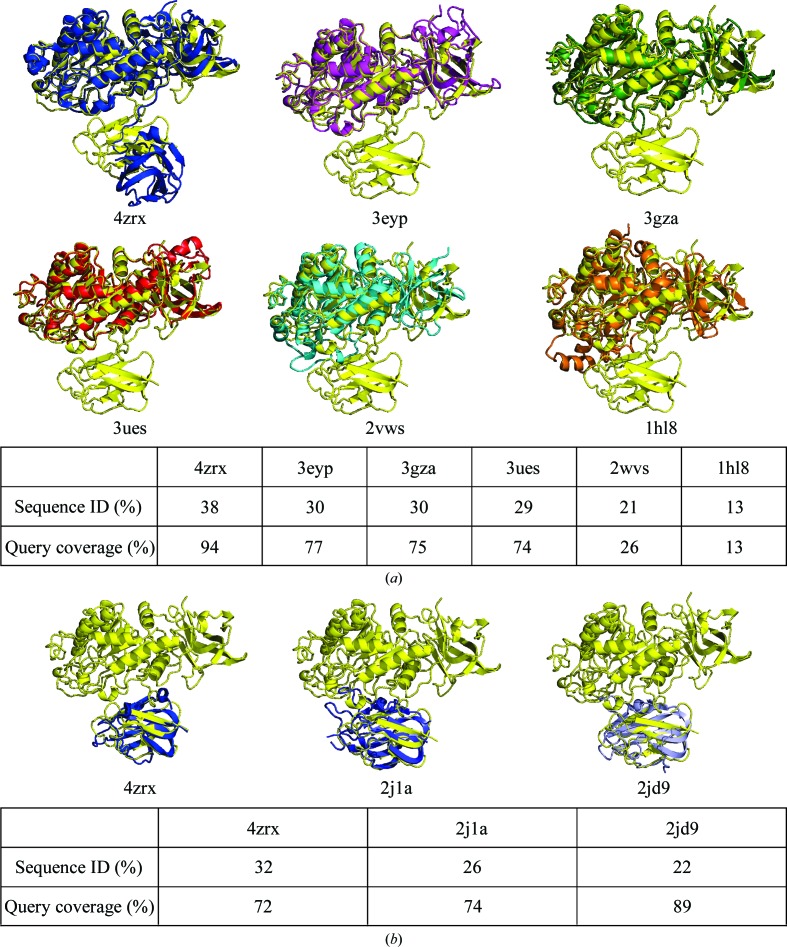
Results from a search using PSI-BLAST against the Protein Data Bank (PDB) show that the amino-acid sequence of GH29_0940 has a high query coverage when compared with the structures of putative α-l-fucosidases from B. ovatus (94%; PDB entry 4zrx) and B. thetaiotaomicron (77%; PDB entry 3eyp). GH29_0940 also has 75 and 74% query coverage when compared with a putative α-l-fucosidase from B. thetaiotaomicron (PDB entry 3gza) and an α-1,3/1,4-l-fucosidase from B. longum subsp. infantis (PDB entry 3ues). Only one of these related PDB structures (PDB entry 4zrx) has any coverage over the second C-terminal carbohydrate-binding domain of GH29_0940. A small number of additional structures show similarity to the second C-terminal carbohydrate-binding domain only. Two examples include a structure of the CBM32 of the β-N-acetylhexosaminidase GH84 protein from C. perfringens (PDB entry 2j1a; Ficko-Blean et al., 2008 ▸) and the structure of a pectin-binding CBM from Yersinia enterocolitica (PDB entry 2jd9; Abbott et al., 2007 ▸). An amino-acid sequence alignment of the related structures mentioned above with GH29_0940 is shown in Fig. 1 ▸. Each domain was aligned independently so as to more accurately illustrate the sequence similarity between the ‘functional’ regions of the proteins.
Figure 1.
Amino-acid sequence alignment of GH family 29 proteins. (a) The catalytic domain, (b) the first carbohydrate-binding domain (CBD) and (c) the second CBD. Identical residues are shown in white with a red background and conservative changes are shown in red with a white background. Secondary structure is depicted with coils for α-helices and black arrows for β-strands. Asterisks denote the two catalytic residues, dollar signs denote catalytic pocket conserved residues, hash signs denote potential residues for carbohydrate binding and the green box indicates the 20-amino-acid loop that connects the first and second CBD. Sequences were aligned using T-Coffee Expresso (Notredame et al., 2000 ▸) and the figure was produced with ESPript 3 (Robert & Gouet, 2014 ▸) with manual modifications.
3.2. Structure and function
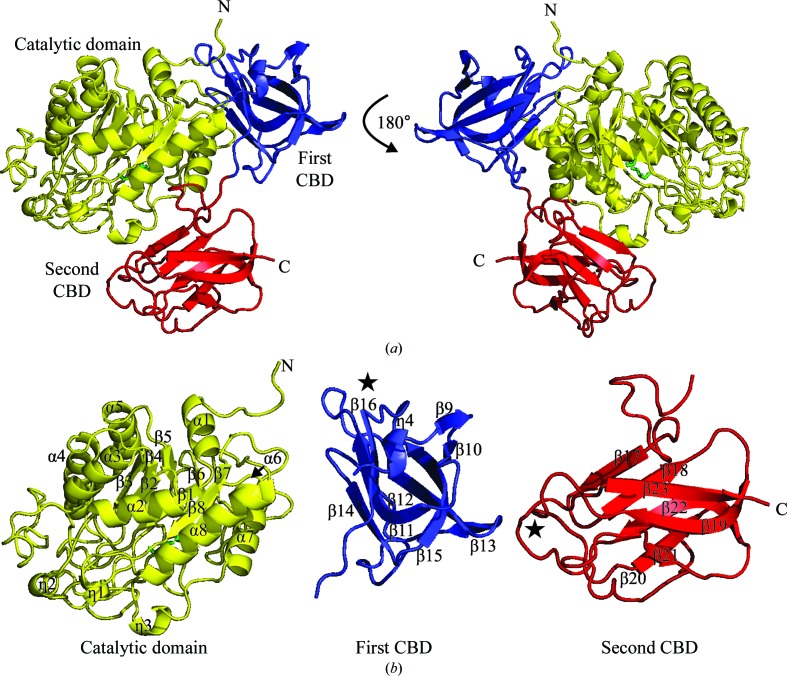
The structure of GH29_0940 is presented in Fig. 2 ▸. GH29_0940 is a monomer and has approximate dimensions of 74 × 62 × 42 Å. There is no evidence from the crystal packing of an oligomer. This is in contrast to the related PDB structures (PDB entries 3eyp, 3ues, 3gza, 2wvs and 1hl8), where there is significant buried surface area that is predicted to be involved in the dimerization and assembly of these macromolecules.
Figure 2.
The structure of GH29_0940. (a) Cartoon representation of the GH29 monomer. The catalytic domain is coloured yellow, the first carbohydrate-binding domain (CBD) blue and the second CBD red. Two glycerol molecules (shown as green sticks) are present in the active site of the catalytic domain. (b) Three-dimensional structure of each domain of GH29_0940 individually. The α-helical segments and β-strands are labelled. The 310-helices are indicated by η. The filled stars indicate the region containing the predicted substrate-binding residues. All figures were produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger).
The structure of GH29_0940 comprises three domains [catalytic domain, residues 1–328; first carbohydrate-binding domain (CBD), residues 329–440; second CBD, residues 441–562; Figs. 2 ▸ a and 2 ▸ b], in comparison with the majority of the related structures that only have two domains. All loops in the GH29_0940 structure are well defined by their electron density. The structure of GH29_0940 is similar to PDB entry 4zrx, which also has three domains and is also a monomer. In comparison to dimeric GH29 structures, it is likely that the second C-terminal CBD blocks oligomerization.
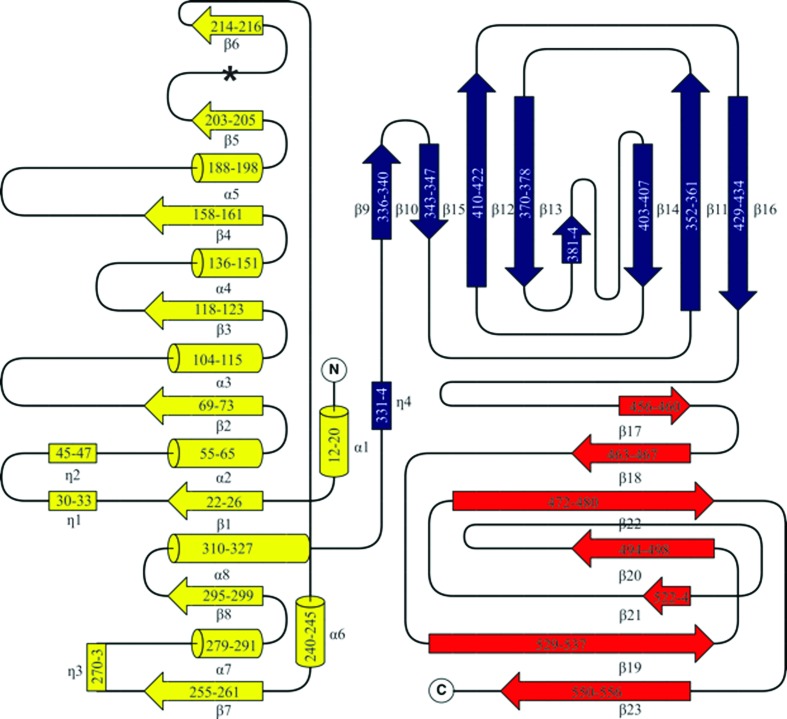
The topology of GH29_0940 is shown in Fig. 3 ▸. GH29_0940 consists of a total of eight α-helices, 23 β-strands and four 310-helices, comprising an N-terminal TIM-barrel-like fold (catalytic domain), a β-sandwich domain (first CBD) and an additional C-terminal β-sandwich domain (second CBD). The domains follow one another in series with no crossover regions. The catalytic domain forms a discontinuous β-barrel with eight parallel β-strands (around a central axis), surrounded by seven α-helices, with an N-terminal α-helix ‘cap’ (a total of eight α-helices). There are three 310-helices positioned at the C-terminal end of the barrel and a depression at the C-terminal ends of the β-strands hosts the putative active site.
Figure 3.
Topology of GH29_0940. β-Strands are represented by arrows, 310-helices by rectangles and α-helices are shown as cylinders. The topology is coloured according to the domains: the catalytic domain is coloured yellow, the first CBD is in blue and the second CBD is in red. The numbering reflects the residues involved in each secondary structure. The asterisk represents the location of the missing α-helix usually positioned as the fifth helix surrounding the β-barrel (which typically has a total of nine α-helices including the N-terminal α-helix ‘cap’). This figure was drawn with TopDraw (within CCP4) with manual modifications (Bond, 2003 ▸; Winn et al., 2011 ▸).
The topology of the active site is created by the length and the orientation of the loops that follow after the β-strands (Wierenga, 2001 ▸). These loops are important for the function of the enzyme and it is noted that the length, orientation and inclusion of additional short α-helices varies between the closely related GH29-family members (Fig. 6a).
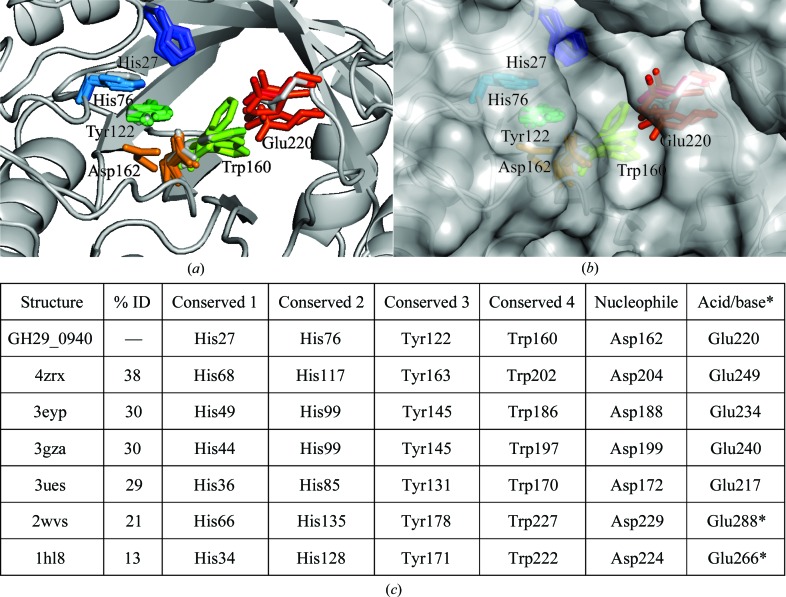
Five out of 21 strictly conserved residues (His27, His76, Tyr122, Trp160 and Asp162; Fig. 1 ▸) are found in the active site (along with a sixth highly conserved residue, Glu220). A superposition of GH29_0940 and all related GH29 structures illustrates how similarly positioned these residues are in the active site (Figs. 4 ▸ a, 4 ▸ b and 4 ▸ c), in particular, the two residues that function as a proton donor (Glu220) and a nucleophile (Asp162) (Davies & Henrissat, 1995 ▸).
Figure 4.
A catalytic domain comparison of residues found to be highly conserved (as determined by a structural alignment; Fig. 1 ▸) in the active pocket of GH29_0940 and its related structures. Six (out of 21) conserved residues are located in the active site. (a, b) A cartoon representation of a superposition of related GH29 structures (PDB entries 4zrx, 3eyp, 3ues, 3gza, 2wvs and 1hl8), showing the conserved residues as coloured sticks (GH29_0940 in grey) and a grey surface representation. Residues are numbered according to GH29_0940 numbering. (c) A table detailing the position of each conserved residue in each structure and the percentage sequence identity (residues marked with an asterisk are not found in the same structural position in the sequence alignment and hence are conservative but not identical). The structures were superimposed using Coot (Emsley et al., 2010 ▸) and all figures were produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger).
GH29 protein family active sites exhibit a ‘pocket’ topology that is found in enzymes such as exopolysaccharidases that are adapted to substrates with a large number of available chain ends (Davies & Henrissat, 1995 ▸). While there is strict conservation of the residues in the ‘pocket’ active site of GH29 enzymes, there is great diversity in the overall shape owing to a variation in the surrounding active-site loops. There is evidence that these loops are dynamic and undergo large conformational changes upon substrate binding, playing a critical role in the catalytic process (Sakurama et al., 2012 ▸). The flexibility of specific residues and loops in and surrounding the active site during catalysis has been demonstrated structurally (Lammerts van Bueren et al., 2010 ▸).
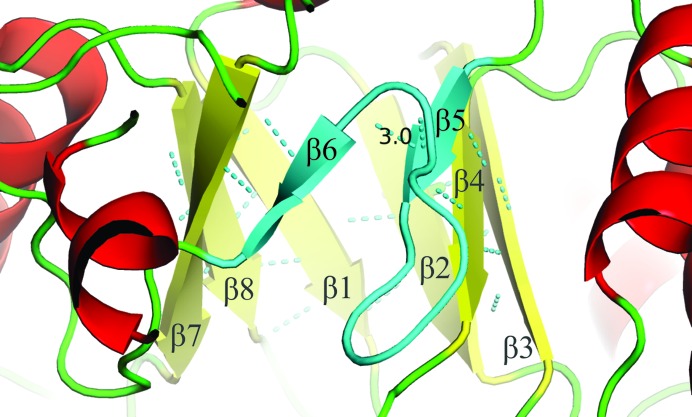
The catalytic N-terminal domain exhibits the familiar TIM-barrel fold reported for other GH29 enzymes (Guillotin et al., 2014 ▸; Lammerts van Bueren et al., 2010 ▸). However, β-strands 5 and 6 are offset, with only one intra-main-chain hydrogen bond between them. This makes the barrel discontinuous at this point (Fig. 5 ▸). Five of the six related GH29 structures also have the corresponding β-strands offset, with the exception of PDB entry 1hl8, which has β-strand 7 offset. There is also a lack of the distinctive repetitive eightfold strand/helix layout typified by the TIM-barrel core. There is a missing α-helix which would typically be situated between β-strands 5 and 6 (and positioned as the fifth α-helix in the core, not including the α-helix ‘cap’), which is instead replaced by a loop (Fig. 5 ▸). This may be a feature related to the discontinuity of the barrel at this point.
Figure 5.
The GH29_0940 enzyme features an apparent ‘open’ β-barrel appearance. β-Strands 5 and 6 (coloured cyan) are offset, with only one intra-main-chain hydrogen bond between them. The α-helix that should be present between β-strands 5 and 6 is instead replaced by a loop (coloured cyan) which alters the typical TIM-barrel form. Helices are coloured red, β-strands yellow and loops green. The TIM-barrel β-strands are numbered. This figure was produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger).
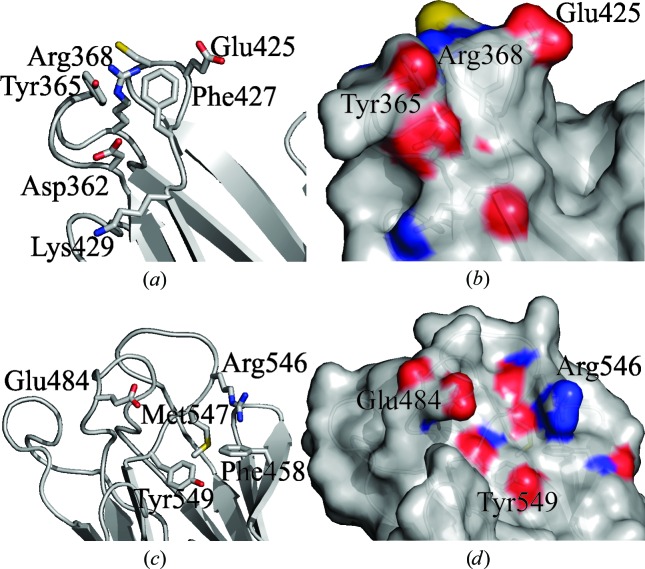
The central first CBD of GH29_0940 is constructed of one 310-helix and eight antiparallel β-strands packed into two β-sheets of five and three strands, respectively, forming a two-layer β-sandwich containing a jelly-roll motif. The first CBD is the equivalent of the C-terminal carbohydrate-binding domain of most other related GH29 structures. This domain is poorly characterized in terms of GH29 family functionality and the residues implicated in the binding of substrates have not been identified in the GH29 family. However, this carbohydrate-binding domain (also known as CBM32 or f5/f8-type C domain) has been well studied in glycoside hydrolase (GH) family enzymes from C. perfringens. The multidomain enzyme GH89 from C. perfringens comprises a catalytic GH89 domain, six CBM32 domains and six other modules. Functional assays and X-ray crystallography of the individual CBM32 domains reveals that four out of six of these domains can bind carbohydrates (Ficko-Blean et al., 2012 ▸). Key amino-acid residues identified in galactose binding include the aromatic residues tyrosine and phenylalanine and the polar residues histidine, arginine and asparagine. These five residues make up the classical galactose-binding motif in the CBM32 family (Abbott et al., 2008 ▸; Ficko-Blean & Boraston, 2006 ▸). Examination of the first CBD (CBM32-1) of GH29_0940 reveals a number of candidate residues (Asp362, Tyr365, Arg368, Glu425, Phe427 and Lys429) for carbohydrate binding along the edge of the β-sandwich (Figs. 6 ▸ a and 6 ▸ b). The structure of GH29_0940 CBM32-1 suggests that it has a type C ‘small-sugar-binding’ function, which means it could bind monosaccharides, disaccharides or trisaccharides using hydrogen bonding between the protein and the ligand, as it lacks the extended binding-site groove that type B ‘glycan-chain-binding’ CBMs exhibit, for example (Boraston et al., 2004 ▸).
Figure 6.
Cartoon representations of the two carbohydrate-binding (CBM32) domains of GH29_0940 show residues that may be involved in substrate binding. (a) CBM32-1 (first CBD) potential substrate-binding residues shown as sticks and (b) the surface of the region; (c) CBM32-2 (second CBD) potential substrate-binding residues shown as sticks and (d) the surface of the region. The sticks and surface are coloured by element. Residues are labelled. All figures were produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger).
The C-terminal second CBD consists of seven antiparallel β-strands packed into two β-sheets of four and three strands, respectively, forming a two-layer β-sandwich containing a jelly-roll motif. The second CBD is also a CBM32 domain that has a similar architecture to the first CBD (CBM32-1); however, its sequence was found to align closely with the structures of CBM32 domains from B. ovatus (32% identity; PDB entry 4zrx), C. perfringens (26% identity; PDB entry 2j1a) and Y. enterocolitica (22% identity; PDB entry 2jd9) (Fig. 7 ▸ b). Structural comparison of the second CBD module (CBM32-2) of GH29_0940 with PDB entries 2j1a (a galactose-binding domain) and 2jd9 (a galacturonic acid-binding domain) does not immediately identify the same functional residues on the same edge of the β-sandwich. Instead, the aromatic amino acids (Phe458 and Tyr549) and polar amino acids (Glu484, Arg546 and Met547) that are consistent with the properties of carbohydrate-binding sites (Ficko-Blean et al., 2012 ▸) are found on an adjacent loop along the same edge of the β-sandwich (Figs. 6 ▸ c and 6 ▸ d). It is common that the binding-site topology in CBMs varies in the location of aromatic residues and loop structures so as to alter the shape to mirror the conformation of the substrate. The structure of GH29_0940 CBM32-2 suggests that it has type C ‘small-sugar-binding’ function. A mixture of charged states of binding pockets can be found in examples from the CBM32 family (Abbott et al., 2008 ▸). This variation may reflect the diversity in substrate binding.
Figure 7.
(a) Cartoon representation of all related GH29 structures individually superimposed by secondary structure with GH29_0940 (in yellow). PDB entry 3eyp was used for molecular replacement. (b) A cartoon representation of superimposed carbohydrate-binding module (CBM) structures with significant sequence identity to the duplicate CBM of GH29_0940. Structures are identified by their PDB code. Amino-acid sequence identity and query coverage are detailed in the tables. The structures were superimposed using Coot (Emsley et al., 2010 ▸) or PyMOL and all figures were produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger). The sequence identity was calculated with Geneious (v.6.0.3; Biomatters Ltd) and percentage query coverage with PSI-BLAST against the PDB.
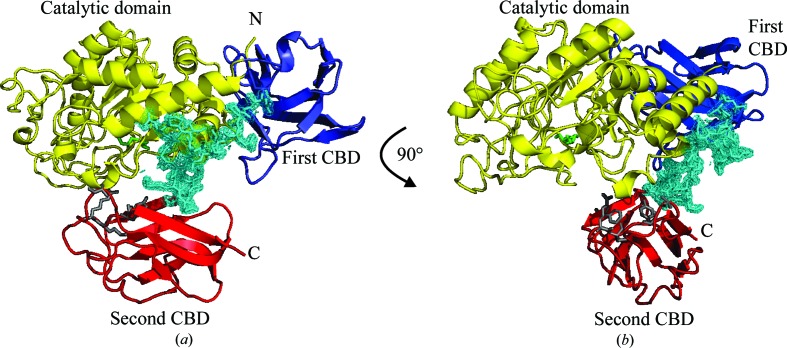
The presence of multiple CBM32-family domains in an enzyme is common in other glycoside hydrolases, such as GH20, GH31, GH84 and GH89 from C. perfringens (Ficko-Blean & Boraston, 2006 ▸). For example, the hydrolase GH89 from C. perfringens has six CBM32 modules (Ficko-Blean et al., 2012 ▸). At the time of solving the structure of GH29_0940, it revealed a variation in the GH29 family, members which normally comprise a catalytic domain followed by a single carbohydrate-binding domain. The CBM32-2 domain is positioned in close proximity to the catalytic domain in the crystal structure. CBM32-2 is linked to the preceding CBM32-1 domain via a long loop comprising 20 predominantly hydrophilic residues (green box; Fig. 1 ▸). Electron density for this loop was clearly visible in the structure, which indicated it was immobile within the confines of the crystal lattice (Fig. 8 ▸). The predicted substrate-binding residues of CBM32-2 are on a loop at the face of the β-sandwich which is positioned alongside the catalytic pocket of the catalytic domain. The proximity of the two domains in the crystal structure indicates that there may be a role for the CBM32-2 domain in delivering the substrate to the active site. A recently deposited GH29 structure (PDB entry 4zrx from B. ovatus) also has a duplicate CBM32 domain. Like our structure, the second CBM32 domain of PDB entry 4zrx links to the previous domain via a long 15-amino-acid hydrophilic loop. However, the duplicate domain is positioned further away from the catalytic domain and has the loop regions (with potential binding residues) orientated away from the catalytic site (Fig. 7 ▸ a), providing evidence of the potential dynamic functional role of a second CBM32 domain.
Figure 8.
A 20-residue hydrophilic loop (shown in cyan) between the first and second carbohydrate-binding domains (CBDs; coloured blue and red, respectively) positions the predicted binding face of the second CBD alongside the catalytic pocket (coloured yellow). (a) A cartoon representation of the GH29_0940 structure and the electron density surrounding the hydrophilic loop which was clearly visible in the structure (indicating that it was immobile within the confines of the crystal lattice). (b) A 90° rotation of GH29_0940 shows how the position of the second CBD and its predicted binding residues (shown as grey sticks) places it in the immediate vicinity of the active pocket of the catalytic domain. Two glycerol molecules in the active pocket are shown as green sticks. This figure was produced using PyMOL v.1.3 (http://www.pymol.org; Schrödinger).
GH29_0940 is predicted to be an α-l-fucosidase based on its amino-acid similarity to other proteins in the database; however, no α-l-fucosidase activity has been experimentally determined to date. 13 relevant pNP-conjugated substrates were tested (at 37°C) and no detectable activity was observed (by measuring liberated pNP at 405 nm; results not shown). A reducing-ends assay (30 min at 37°C; Talbot & Sygusch, 1990 ▸) using seven candidate substrates showed no detectable activity (results not shown).
GH29 enzymes can be classified into two subgroups according to specificity and sequence homology (Ashida et al., 2009 ▸). Subgroup A (GH29-A) comprises enzymes that act on a chromogenic substrate (e.g. pNP-α-l-fucopyranoside) such as the enzymes from T. maritima and B. thetaiotaomicron (PDB entries 1hl8 and 2wvs; Sulzenbacher et al., 2004 ▸; Lammert van Bueren et al., 2010 ▸). In contrast, members of subgroup B (GH29-B) are poor catalysts of pNP-α-l-fucopyranoside, such as the enzymes from B. thetaiotaomicron and B. longum subsp. infantis (PDB entries 3eyp and 3ues; Guillotin et al., 2014 ▸; Sakurama et al., 2012 ▸). As GH29_0940 does not exhibit any activity towards any of the pNP substrates tested, this suggests that it may belong to the GH29-B subgroup.
4. Conclusions
The crystal structure of a putative α-l-fucosidase from the glycoside hydrolase (GH) 29 family, cloned from metagenomic DNA isolated from the rumen of a cow, was determined to a resolution of 2.04 Å. The enzyme has three distinct domains: a catalytic GH family 29 domain (catalytic domain), a family 32 carbohydrate-binding module (CBM32-1; first CBD) and a second family 32 carbohydrate-binding module (CBM32-2; second CBD). Currently, only six other GH29 enzyme structures exist, only one of which contains the second CBD. Our structure has similar features to the recently solved three-domain GH29 enzyme but shares only 38% sequence identity. The catalytic domain exhibits a TIM-barrel-like fold, which normally consists of an eightfold repeat of a β/α unit, such that eight parallel β-strands are folded into a closed barrel in the centre, surrounded by eight α-helices. In this structure, there are eight β-strands and only seven α-helices surrounding the TIM-barrel-like core (with an additional α-helix provided by the α-helix ‘cap’). β-Strands 5 and 6 are offset, with only one intra-main-chain hydrogen bond between them, making the barrel discontinuous and giving it an ‘open’ conformation. The α-helix that should be observable between β-strands 5 and 6 is missing and is replaced by a loop, which alters the typical TIM-barrel form, providing a large meandering loop instead of the usual combination of β-strands and α-helices. This divergence from the canonical TIM-barrel structure suggests that this enzyme may have inherent flexibility in this region. This structural departure also appears to be a feature of all other GH29-family enzymes. Structural comparison with other GH29 enzymes reveals six characteristic (structurally conserved) active-site residues including the catalytic nucleophile (Asp162) and proton donor (Glu220) in the active pocket.
The first and second CBDs are carbohydrate-binding modules (CBM32), each forming a two-layer β-sandwich containing a jelly-roll motif. The residues implicated in the binding of substrates have not been identified in the GH29 family; however, they can be elucidated from studies of the CBM32 domains of other GH-family enzymes. Structural similarities place the two CBDs of GH29_0940 in the type C ‘small sugar binding’ functional group and identify potential ligand-binding sites at the edge of the β-sandwich. A number of potential carbohydrate-binding residues are present in the loop regions of the β-sandwich, which gives the appearance of a pocket-like area. This pocket shape supports the classification of these domains as ‘small sugar binding’ as opposed to, for example, ‘glycan chain binding’ which requires a deep binding groove. The first CBD is similar in structure to other GH29-enzyme CBM32 domains, whereas the second CBD shows structural similarities to the third domain of the recently deposited GH29 structure with PDB code 4zrx (from B. ovatus) as well as domains from other bacterial species such as C. perfringens and Y. enterolitica. Intriguingly, the binding pocket of the second CBD is in close proximity to the catalytic pocket (in the crystal structure) and this domain is ‘suspended’ by a long hydrophilic loop, indicating a potential degree of mobility. This suggests a possible mechanism for shuttling a substrate between the binding domain and the catalytic domain or perhaps a mechanism to allow the enzyme to adhere to a carbohydrate-rich surface, leaving the catalytic domain free to act upon a substrate. The other existing three-domain GH29 structure also reveals this ‘pendulum-like’ configuration with the duplicate CBM positioned not only further away from the catalytic domain but also in a flipped orientation with its potential binding sites distal to the catalytic site. The arrangement of these two structures supports our hypothesis of a large range of movement of the third domain to assist substrate shuttling between domains.
Supplementary Material
PDB reference: GH29_0940, 5k9h
Acknowledgments
The research was funded by the New Zealand Ministry of Business, Innovation and Employment’s New Economy Research Fund (contract C10X0803 to Dr Graeme Attwood). We thank Bill Kelly (AgResearch) for examining gene-sequence data to identify novel fosmid clone candidates.
References
- Abbott, D. W., Eirín-López, J. M. & Boraston, A. B. (2008). Mol. Biol. Evol. 25, 155–167. [DOI] [PubMed]
- Abbott, D. W., Hrynuik, S. & Boraston, A. B. (2007). J. Mol. Biol. 367, 1023–1033. [DOI] [PubMed]
- Adams, P. D. et al. (2011). Methods, 55, 94–106.
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Ashida, H., Miyake, A., Kiyohara, M., Wada, J., Yoshida, E., Kumagai, H., Katayama, T. & Yamamoto, K. (2009). Glycobiology, 19, 1010–1017. [DOI] [PubMed]
- Bond, C. S. (2003). Bioinformatics, 19, 311–312. [DOI] [PubMed]
- Boraston, A. B., Bolam, D. N., Gilbert, H. J. & Davies, G. J. (2004). Biochem. J. 382, 769–781. [DOI] [PMC free article] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Davies, G. & Henrissat, B. (1995). Structure, 3, 853–859. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Ficko-Blean, E. & Boraston, A. B. (2006). J. Biol. Chem. 281, 37748–37757. [DOI] [PubMed]
- Ficko-Blean, E., Stuart, C. P., Suits, M. D., Cid, M., Tessier, M., Woods, R. J. & Boraston, A. B. (2012). PLoS One, 7, e33524. [DOI] [PMC free article] [PubMed]
- Ficko-Blean, E., Stubbs, K. A., Nemirovsky, O., Vocadlo, D. J. & Boraston, A. B. (2008). Proc. Natl Acad. Sci. USA, 105, 6560–6565. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp 571–607. Totowa: Humana Press.
- Guillotin, L., Lafite, P. & Daniellou, R. (2014). Biochemistry, 53, 1447–1455. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. L. (2001). J. Mol. Biol. 305, 567–580. [DOI] [PubMed]
- Lammerts van Bueren, A., Ardevol, A., Fayers-Kerr, J., Luo, B., Zhang, Y., Sollogoub, M., Bleriot, Y., Rovira, C. & Davies, G. J. (2010). J. Am. Chem. Soc. 132, 1804–1806. [DOI] [PubMed]
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. (2014). Nucleic Acids Res. 42, D490–D495. [DOI] [PMC free article] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Notredame, C., Higgins, D. G. & Heringa, J. (2000). J. Mol. Biol. 302, 205–217. [DOI] [PubMed]
- Plewczynski, D., Slabinski, L., Tkacz, A., Kajan, L., Holm, L., Ginalski, K. & Rychlewski, L. (2007). Polymer, 48, 5493–5496.
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rye, C. S. & Withers, S. G. (2000). Curr. Opin. Chem. Biol. 4, 573–580. [DOI] [PubMed]
- Sakurama, H., Fushinobu, S., Hidaka, M., Yoshida, E., Honda, Y., Ashida, H., Kitaoka, M., Kumagai, H., Yamamoto, K. & Katayama, T. (2012). J. Biol. Chem. 287, 16709–16719. [DOI] [PMC free article] [PubMed]
- Slabinski, L., Jaroszewski, L., Rychlewski, L., Wilson, I. A., Lesley, S. A. & Godzik, A. (2007). Bioinformatics, 23, 3403–3405. [DOI] [PubMed]
- Sulzenbacher, G., Bignon, C., Nishimura, T., Tarling, C. A., Withers, S. G., Henrissat, B. & Bourne, Y. (2004). J. Biol. Chem. 279, 13119–13128. [DOI] [PubMed]
- Talbot, G. & Sygusch, J. (1990). Appl. Environ. Microbiol. 56, 3505–3510. [DOI] [PMC free article] [PubMed]
- Wierenga, R. K. (2001). FEBS Lett. 492, 193–198. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F. & Xu, Y. (2012). Nucleic Acids Res. 40, W445–W451. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: GH29_0940, 5k9h