Abstract
The dynamic nature of actin polymers is modulated to facilitate a diverse range of cellular processes. These dynamic properties are determined by different isoforms of tropomyosin which are recruited to distinct subpopulations of actin polymers to differentially regulate their functional properties. This makes tropomyosin an attractive target for labelling discrete actin populations. We have assessed the effect of different fluorescent labelling strategies for this protein. Although tropomyosin–fluorescent fusions decorate actin in vivo, they are either nonfunctional or perturb regulation of actin nucleation and cell cycle timings. Thus, conclusions and physiological relevance should be carefully evaluated when using tropomyosin fusions.
Keywords: acetylation, actin cytoskeleton, Cdc8, fission yeast, Schizosaccharomyces pombe
Abbreviations
CAR, contractile actomyosin ring
FP, fluorescent proteins
Tpm, tropomyosins
Many dynamic processes within the eukaryotes are driven by the actin cytoskeleton. Actin polymers can generate force to remodel membrane architecture as well as act as tracks for molecular motors. It is the dynamic nature of the actin polymers that is critical for their cellular function. These key properties are regulated by diverse actin‐binding proteins, which drive nucleation, stabilisation, cross‐linking or even depolymerisation of actin complexes 1.
One such class of actin‐binding proteins are the tropomyosins (Tpm), a highly conserved class of dimeric α‐helical coiled‐coil proteins which associate with and stabilise actin 2. Tpm dimers interact in an end‐to‐end fashion to generate filaments that associate with and coil around the F‐actin polymer. These Tpm cofilaments play a central role in stabilising actin filament dynamics in muscle cells and therefore regulate muscle contraction 3. In nonmuscle cells, Tpm plays a critical role in stabilising and also defining the biophysical properties of a specific actin polymer as well as modulating its ability to interact with other proteins and thereby regulate its function 4.
Yeast and animal cells express multiple isoforms of Tpm, each of which associate with specific actin structures at a specific place and time, in an exquisitely orchestrated manner, to fine tune the biophysical nature of each actin polymer in a specific way 2. This makes the different Tpms attractive targets as markers during the analysis of specific actin structures and functions. There is sequence diversity along the length of the different Tpm isoforms, but it is the ends of the protein that seem to be key in defining their function. The termini of the Tpm dimer not only define the structural and charge landscape at the end of the actin‐Tpm copolymer but also determine the nature of the end to end contacts between adjacent dimers. This overlap region can vary significantly between different Tpm isoforms and is essential for defining the stability of the Tpm polymer and its ability to bind cooperatively to F‐actin. Indeed, seemingly innocuous modifications to the charge on the polypeptide in these overlapping regions can have a profound effect upon the stability of the actin‐Tpm complex. For example, in many cell types the addition of acetyl group to the amino‐terminal residue of the Tpm stabilises the alpha‐helical structure of each coiled‐coil structure to facilitate its interaction with carboxyl region of adjacent Tpm molecules 5, 6. Thus, small changes in the charge and shape of the Tpm protein have a dramatic impact upon protein interactions and function.
The molecular‐genetic plasticity of yeast model systems provides an excellent setting for studying the impact protein modifications have upon its cellular structure. Yeasts express a smaller number of Tpm isoforms in comparison to metazoan cells, which makes them an attractive organism in which to characterise specific Tpm functions. The fission yeast Schizosaccharomyces pombe has a single tropomyosin, TpmCdc8, which exists in amino‐terminally acetylated and nonacetylated forms within the cell 7, 8. Depending upon its acetylation status, TpmCdc8 associates with the actomyosin ring during mitosis to facilitate myosin II motor activity and cytokinesis, or with cytoplasmic actin filaments upon which myosin V motors move 9, 10, 11. It thus provides a simple system for studying the regulation of function of this essential cytoskeletal protein.
Fluorescent Proteins (FP) provide an attractive method for following protein dynamics within a live cell 12. FPs have been developed with a variety of spectral properties to facilitate diverse functional analysis. However, FP fusions can impact protein function by inhibiting normal protein folding or affecting interactions with other proteins. Not surprisingly both microtubule and actin cytoskeletons are acutely sensitive to fluorescent labelling of protein components 13. Diverse FP‐labelled markers exist to follow the dynamics of the actin cytoskeleton in a live cell context, however, many have subsequently been shown to alter the behaviour and organisation of the polymers within cells 14, 15, 16. Tpms are attractive candidates for markers of specific actin structures and have been used to follow filament dynamics in diverse cell types, in a live cell context 17, 18, 19, 20, 21. However, it is unclear how fusing a FP to the termini of the Tpm protein impacts its normal function.
Here, we describe in vitro and in vivo analyses of amino‐ and carboxyl‐terminal fusions between the fission yeast tropomyosin, TpmCdc8, and a monomeric fluorescent protein. We establish that while TpmCdc8‐carboxyl terminal fusions disrupted the ability of the protein to polymerise or associate with actin in vitro, the amino‐terminal fusion formed filaments and associated with actin in a manner similar to amino‐terminally acetylated endogenous TpmCdc8. However, while this protein facilitated the formation of a functional contractile actomyosin ring (CAR) and cell growth in cells lacking functional endogenous TpmCdc8, it disrupted the normal timing of cell division and normal myosin V movements. Thus, each Tpm‐FP fusion has a significant impact upon the function of this critical cytoskeletal protein.
Materials and methods
Molecular biology
pJC20cdc8 + and pREP41cdc8 + were described previously 8. cdc8‐Gly 3 ‐Cerulean3‐His 6 and His 6 ‐Cerulean3‐Gly 3 ‐cdc8 terminal fusions were synthesised as Nde1‐BamH1 fragments (Thermo Fisher Scientific, Waltham, MA, USA) and cloned into pJC20 22 and pREP41 23 bacterial and fission yeast expression vectors.
Cell culture
The yeast strains used in the study were h− leu1.32; h− cdc8.110 myo2.mCherry leu1.32 and h− cdc8.110 myo52.mNeongreen leu1.32. Cell culture and maintenance were carried out according to 24 using Edinburgh minimal medium with Glutamic acid nitrogen source (EMMG). Growth rates were determined from growth curves generated using a BMG Spectrostar Nano plate reader while shaking cells at 36 °C. All cells were maintained as early to midlog phase cultures for 48 h before being used for all analyses.
Microscopy
Imaging was undertaken as described previously 9. Timelapse images of > 10 cells undergoing cytokinesis were used to calculate each CAR constriction rate.
Protein purification
Unlabelled TpmCdc8 protein was purified as described previously 8. TpmCdc8 Cerulean3 fusions were expressed from pJC20 Tpm cdc8 ‐Cerulean3‐His 6 and pJC20His 6 ‐Cerulean3‐Tpm cdc8 in either BL21 DE3 or BL21 DE3 pNatB 25 cells. Midlog cultures were grown for 3 h with 100 mg·L−1 IPTG. Cells were harvested, resuspended in 30 mL lysis buffer (20 mm Tris pH 7.5, 100 mm NaCl, 2 mm EGTA and 5 mm MgCl2), lysed by sonication. Debris and insoluble components were removed by centrifugation and the resulting supernatant was incubated with 10 mg·L−1 DNase and 10 mg·L−1 RNase at 4 °C for 1 h before isolating His6‐labelled fusions on nickel‐agarose columns and eluting with imidazole. After buffer exchange into FPLC loading buffer (5 mm Tris pH 7.0, 100 mm NaCl) the FP‐labelled TpmCdc8 was further purified with FPLC using 2 × 5 mL Pharmacia HiTrap‐Q columns in tandem, by elution with a 100–900 mm NaCl gradient. Fusions were isolated from appropriate fractions, and concentrated in 5 mm Tris pH 7.0. The purity and mass of the proteins were determined by mass spectroscopy, while parallel Bradford, gel densitometry and spectroscopic analyses were used in parallel to confirm protein concentrations. Rabbit actin was purified as described previously 26.
Circular dichroism
Measurements were made in 1‐mm quartz cuvettes using a Jasco 715 spectropolarimeter. Cdc8 proteins were diluted in CD buffer (10 mm Potassium phosphate, 500 mm NaCl, 5 mm MgCl2 pH 7.0) to a concentration of 0.4 mg·mL−1. Thermal unfolding data were obtained by monitoring the CD signal at 222 nm with a heating rate of 1 °C·min−1. At completion of the melting‐curve the sample was cooled at a rate of 20 °C·min−1. CD data are presented as differential absorption (∆A).
Viscomentry
A Cannon‐Manning semimicroviscometer was used to determine the viscosity of 20 μm Cdc8 samples at 20 ± 1 °C in 1 mL of viscometry buffer (20 mm MOPS, 5 mm MgCl2 pH 7.0). NaCl concentration was increased from 0 to 250 mm. Kinematic viscosity was calculated using the manufacturer's predetermined microviscometer kinematic viscosity constant (0.03235) and the average efflux time (typically 30–40 s), calculated from five observations per sample at each NaCl concentration.
Actin‐binding assay
Cosedimentation assays were performed at 25 °C by mixing 10 μm actin with increasing concentrations of Tpm as described previously 27.
Results
In order to determine the impact fusion of a fluorescent protein to the termini had upon the ability of tropomyosin to self‐polymerise, associate with actin and function within a cell, bacteria and fission yeast expression constructs were generated which allowed encoding of amino and carboxyl terminal fusions between the fission yeast tropomyosin, TpmCdc8, and the monomeric fluorescent protein, Cerulean3 28, each juxtaposed a poly‐glycine linker (Fig. 1A). The Cerulean3 fluorophore is a relatively bright, photo‐ and thermostable monomeric fluorescent protein, and due to its low emission wavelength, allows excellent spatial resolution in live cell imaging applications.
Figure 1.
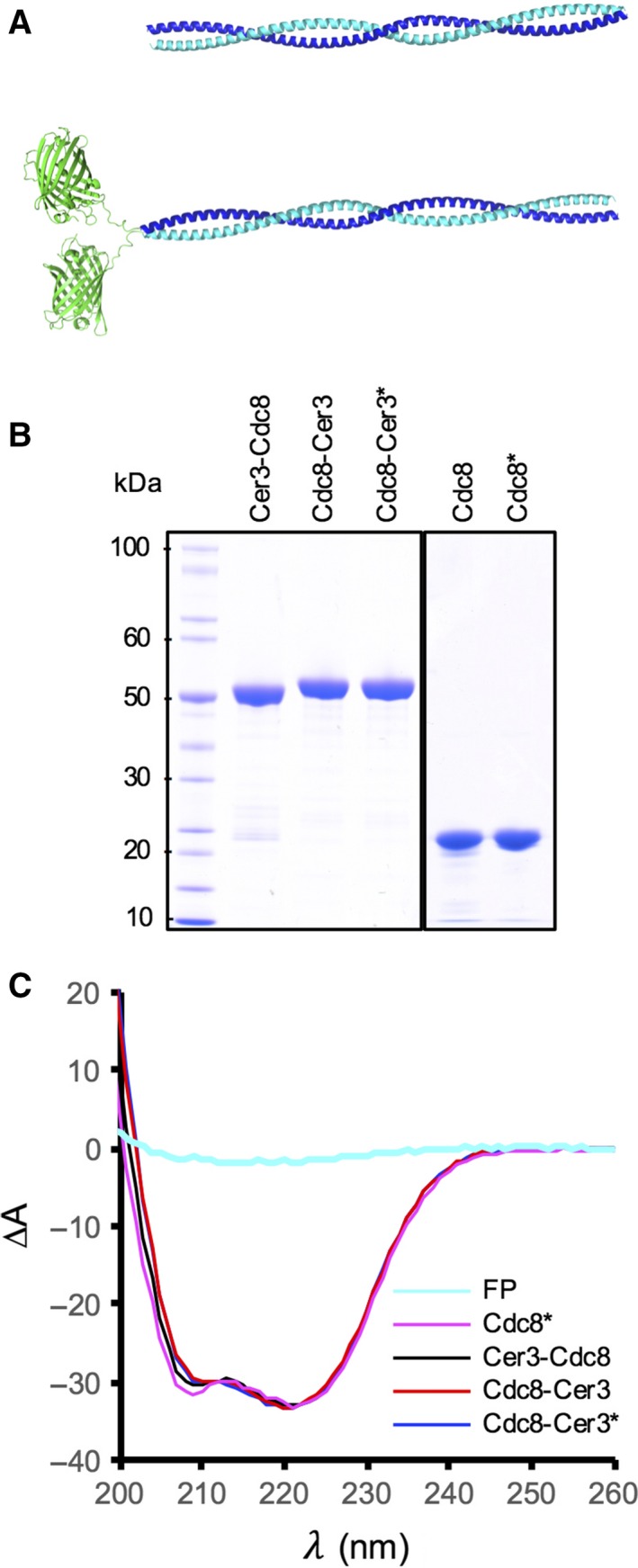
Tropomyosin proteins used in this study. (A) Predictive models of TpmCdc8 (upper panel) and Cerulean3‐TpmCdc8 (lower panel) dimers. (B) Coomassie blue‐stained SDS/PAGE analysis of purified wild‐type and Cerulean3‐tagged TpmCdc8 proteins. (C) CD Spectra of purified acetylated wt (magenta), amino‐terminally Cerulean3‐tagged (black), and both acetylated (blue) and unacetylated (red) carboxyl‐terminally Cerulean3‐tagged TpmCdc8 proteins. A CD spectrum of a fluorescent protein (cyan) is included for comparison. *Denotes amino‐terminally acetylated protein.
To facilitate biochemical analysis of the Tpm proteins, each Cerulean3 fusion (Cerulean3‐TpmCdc8 and TpmCdc8‐Cerulean3) together with the wild‐type protein was expressed and purified from E. coli cells (Fig. 1B). In addition, wild‐type and the TpmCdc8‐Cerulean3 carboxyl fusion which had been acetylated upon the amino‐methionine resides were isolated using a recombinant NatB acetylation system 25 (Fig. 1B – acetylated proteins denoted by *). The mass and purity of each protein was confirmed using mass spectroscopy analysis (Table 1).
Table 1.
Summary of data presented
| Protein | Expected Mass (kDa) | Determined mass (kDa) | T m (°C) | Viscosity (cSt) | K 50% (μM) | Growth ratea (∆OD600 h−1) | CAR ∅ constriction rate (μm·min−1)a | Cell length μm ( ± SD)a , b | Myo52 movementsa |
|---|---|---|---|---|---|---|---|---|---|
| TpmCdc8 | 18 964 | 18 963.9 | 33.4 | 1.08 | 2.7 | – | – | – | – |
| ace‐TpmCdc8 | 19 006 | 19 006.4 | 35.4 | 1.22 | 0.6 | 0.08 | 0.088 ± 0.014 | 11.8 ± 3.0 | +++ |
| Cerulean3‐TpmCdc8 | 47 002 | 47 001.6 | 35.5 | 1.17 | 0.69 | 0.063 | 0.09 ± 0.007 | 13.9 ± 4.3 | + |
| TpmCdc8‐Cerulean3 | 46 871 | 46 870.4 | 34.3 | 0.99 | n.a. | – | 0 | 24.2 ± 9.7 | – |
| ace‐TpmCdc8‐Cerulean3 | 46 913 | 46 912.4 | 35.7 | 1.02 | n.a. | 0 | 0 | – | – |
Determined in cdc8.110 cells at 36 °C.
GFP alone: 18.9 ± 4.3 μm.
As a first step, Circular dichroism (CD) analyses were performed upon each protein to assess the impact the addition of Cerulean3 had on the thermal stability of the dimer. As reported previously 27, broad negative CD spectra peaks at 208 and 222 nm, consistent for α‐helical structures were observed for the wild‐type and modified forms of the TpmCdc8 protein (Fig. 1C). The presence of the Cerulean3 fusion had no significant impact upon the overall CD spectra, as the relative CD signal of an equivalent amount of FP alone is 5% that of that observed for the dimeric coiled‐coil TpmCdc8 proteins (Fig. 1C). Normalised melting curves for absorbance at 222 nm of each protein are shown in Fig. 2A. Once each melting curve was acquired, the protein was cooled and subjected to two further runs. CD spectra were acquired at the start of each run, and were identical for each protein (not shown), indicating each TpmCdc8 was able to rapidly refold to its original state. Figure 2B illustrates the first derivative plots for these data, from which mid‐point melting temperatures (T m) were calculated (Table 1). The T m of unmodified and amino‐terminally acetylated TpmCdc8 were equivalent to values established previously 27. The addition of Cerulean3 to the carboxyl end of TpmCdc8 had a minimal effect on the thermostabiltiy of TpmCdc8, increasing the T m from 33.4 °C to 34.3 °C. However, fusing Cerulean3 to the amino‐terminal of the TpmCdc8 increased the thermal stability of the protein to equal that of acetylated wild‐type TpmCdc8, 35.5 °C. Similarly, acetylating the amino‐terminal methionine of TpmCdc8‐Cerulean3 increased the T m to an equivalent value (T m = 35.7 °C). These data confirm that the addition of the Cerulean3 polypeptide to either end of TpmCdc8 had no significant detrimental effect upon stability of the TpmCdc8 coiled‐coil protein.
Figure 2.
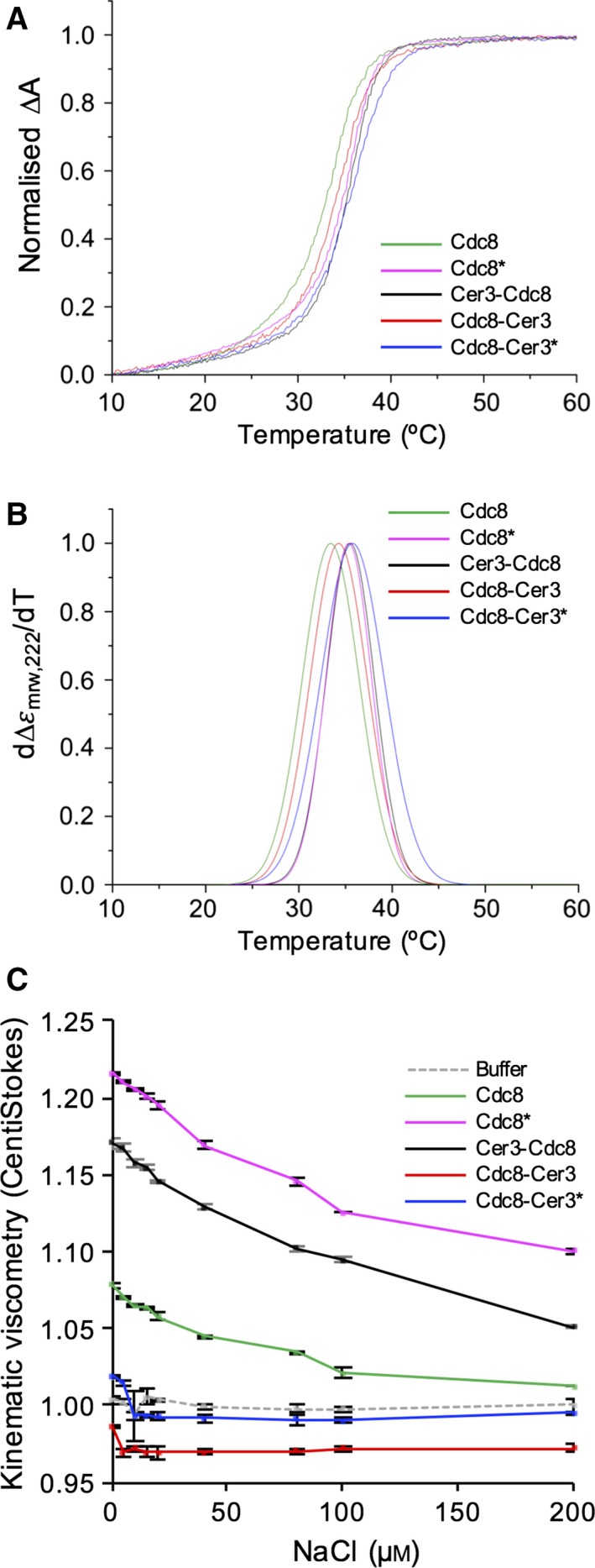
FP Fusions affect Tpm filament formation not themostability. (A) Normalised differential CD absorbance data for unacetylated TpmCdc8 (green), acetylated TpmCdc8 (magenta), Cerulean3–TpmCdc8 (black), aceteylated TpmCdc8–Cerulean3 (blue) and unaceylated TpmCdc8–Cerulean3 (red) at 222 nm. (B) First derivative plots at 222 nm of data described in (A). (C) Viscosity of 20 μm TpmCdc8 proteins and buffer (grey dashed) at increasing NaCl concentrations (0–200 mm) at 23 °C.
The additional mass from Cerulean3 at the termini of TpmCdc8 has the potential to impact the ability to form end‐to‐end interactions between the amino and carboxyl termini of TpmCdc8 dimers and therefore allow the protein to polymerise. Viscometry was used to investigate the effect the addition of the fluorescent protein had upon the ability of TpmCdc8 to form filaments. In this assay, a higher viscosity correlates with a greater capacity to form filaments. Upon the addition of NaCl the strength of ionic interactions between the ends of TpmCdc8 proteins was reduced, which is reflected in a reduction in viscosity. Figure 2C shows viscosity data for each TpmCdc8 and TpmCdc8 fluorescent protein fusion. The viscosity of each protein in the absence of NaCl is shown in Table 1. Acetylation increases the propensity of TpmCdc8 to form filaments, which was reflected in an increase in the viscosity from 1.08 centiStokes (cSt) for the unmodified protein, to 1.22 cSt for the amino‐terminally modified form. The viscosity data for tagged TpmCdc8 indicates the end‐to‐end interactions are affected by the addition of the Cerulean3 fluorophore to either terminus of the protein. In the absence of salt, amino‐terminally tagged TpmCdc8 formed end‐to‐end interactions and gave a viscosity reading of 1.17 cSt, which is lower than the wild‐type acetylated protein. The carboxyl terminal fusion had a more significant impact upon TpmCdc8 filament formation, and abolished the ability of the protein to form end‐to‐end interactions, as the viscosity values for the unacetylated and acetylated TpmCdc8‐Cerulean3 proteins of 0.99 and 1.02 cSt, respectively, are equivalent to buffer only control samples. These data indicate that fusion of the Cerulean3 fluorescent protein to either end of TpmCdc8 impacts the end‐to‐end interactions between adjacent tropomyosin dimers.
In addition to effects on Cdc8 stability and end‐to‐end contacts, we performed cosedimentation experiments to determine whether the addition of the Cerulean3 fusion had an impact upon the ability of TpmCdc8 interactions with actin. Examples of Coomassie‐stained SDS/PAGE gels used to determine the binding affinity of Cerulean3‐TpmCdc8 and both unmodified and amino‐terminally acetylated TpmCdc8‐Cerulean3 are shown in Fig. 3. In each experiment shown the lower faster migrating bands correspond to actin, with the density remaining approximately constant between each sample. The upper bands are TpmCdc8 Cerulean3 fusions and the density increases as dimer concentrations increase from left to right. Figure 3A shows a typical example of a binding experiment for the amino‐terminal Cerulean3‐TpmCdc8 fusion. This protein can be seen to bind strongly to actin as a faint band can be observed in the pellet with as little as 0.5 μm Cerulean3‐TpmCdc8, with a much denser band visible at 12 μm Tpm. A typical binding curve from a Cerulean3‐TpmCdc8‐binding experiment is shown in Fig. 3D. The K 50% for this fusion was calculated to be 0.69 μm, which is similar to the previously reported K 50% value for acetylated TpmCdc8 (0.46 μm) 8, 27. In contrast, binding data for both unacetylated (Fig. 3B) and acetylated (Fig. 3C) TpmCdc8‐Cerulean3 illustrate both forms of the carboxyl terminal fusion bind weakly to actin. Even at 20 μm Tpm, only a small proportion of the protein is in the pellet fraction, while a large amount of Tpm remains in the supernatant. Both acetylated and unacetylated forms of TpmCdc8‐Cerulean3 bound extremely weakly to actin and binding curves could not be generated, as the binding coefficients were estimated to be greater than 20 μm.
Figure 3.
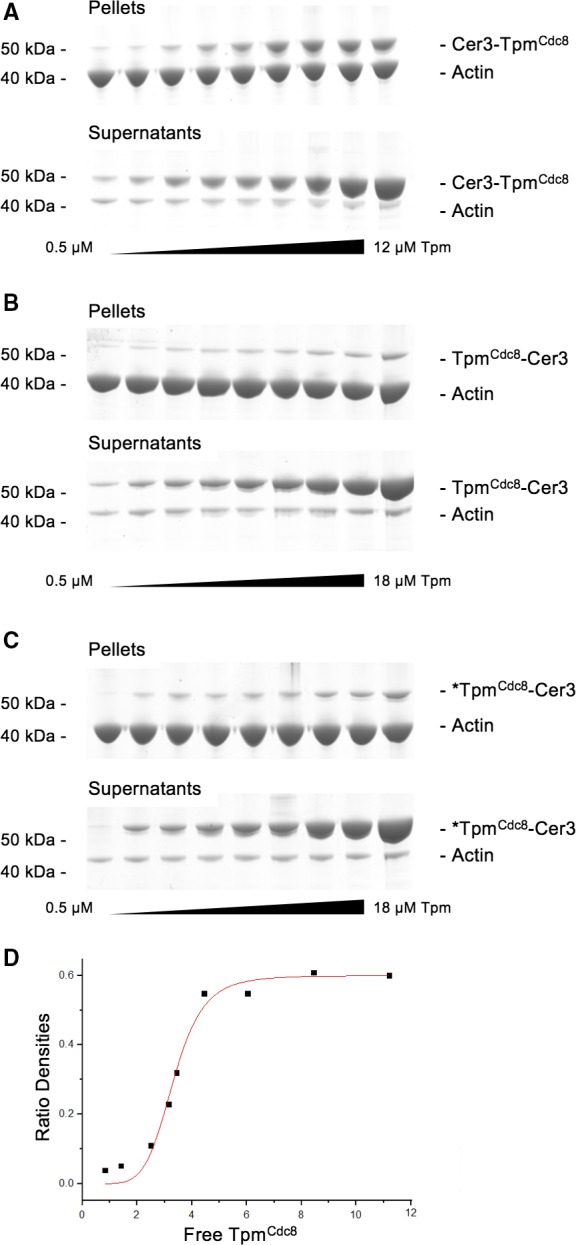
Actin‐binding assays of FP Tpm fusions. SDS/PAGE gels of pellet and supernatant fractions from cosedimentation assays of (A) Cerulean3‐TpmCdc8, (B) TpmCdc8‐Cerulean3 and (C) Nt‐acetylated TpmCdc8‐Cerulean3. (D) Binding curve of the free TpmCdc8 concentration against the ratio of density of actin, for Cerulean3‐TpmCdc8, measured by densitometry of cosedimentation SDS/PAGE gels. Curves represent Hill equation lines of best fit.
In conclusion, in vitro analyses show that the addition of a fluorescent protein fusion to either end of the TpmCdc8 did not impact the α‐helical content or thermal stability of the protein. However, carboxyl terminal fusions inhibited both the ability of Tpm to form end‐to‐end contacts, and ability to associate with actin. In contrast, the amino‐terminal Cerulean3‐TpmCdc8 fusion protein has properties that closely mimic the acetylated endogenous protein.
We next explored the ability of each fusion to localise and complement the function of the endogenous TpmCdc8 protein within the fission yeast cell. Having previously established that expression levels of GFP‐TpmCdc8 from the nmt41 promoter reflect TpmCdc8 levels from the endogenous locus 8, we chose to express the TpmCdc8 fusions using the same repressible promoter 23. We first examined the localisation of the Cerulean3‐TpmCdc8 and TpmCdc8‐Cerulean3 fusion proteins in wild‐type cells. Each protein was seen to associate with the actin polymers within the contractile CAR, when endogenous TpmCdc8 was present in the cell (Fig. 4A,B). In contrast, only the Cerulean3‐TpmCdc8 was seen to localise to and support the formation of the actin ring in cells possessing the temperature‐sensitive cdc8‐110 allele when held at the restrictive temperature (Fig. 4C, D) Consistent with these findings comparison of growth kinetics between cdc8‐110 cells containing plasmid born copies of wild‐type Tpm cdc8, Tpm cdc8 ‐Cerulean3 or Cerulean3‐Tpm cdc8 showed that only the amino‐terminal fusion permitted growth, at a rate approaching that of wild‐type at the restrictive temperature of 36 °C (Fig. 4E, Table 1). However, while Cerulean3‐TpmCdc8 facilitated the formation of an actin ring, cells were significantly longer than wild‐type (Fig. 4C, Table 1) demonstrating the normal timing of cell division was disrupted. These data suggest that the fusions may be able to form heterodimers with endogenous unlabelled tropomyosin, as only the amino‐terminal Cerulean3‐TpmCdc8 fusion could form dimers with the capacity to form filaments and associate with actin in the absence of the wild‐type protein. This is consistent with the in vitro analysis (Figs 2 and 3).
Figure 4.
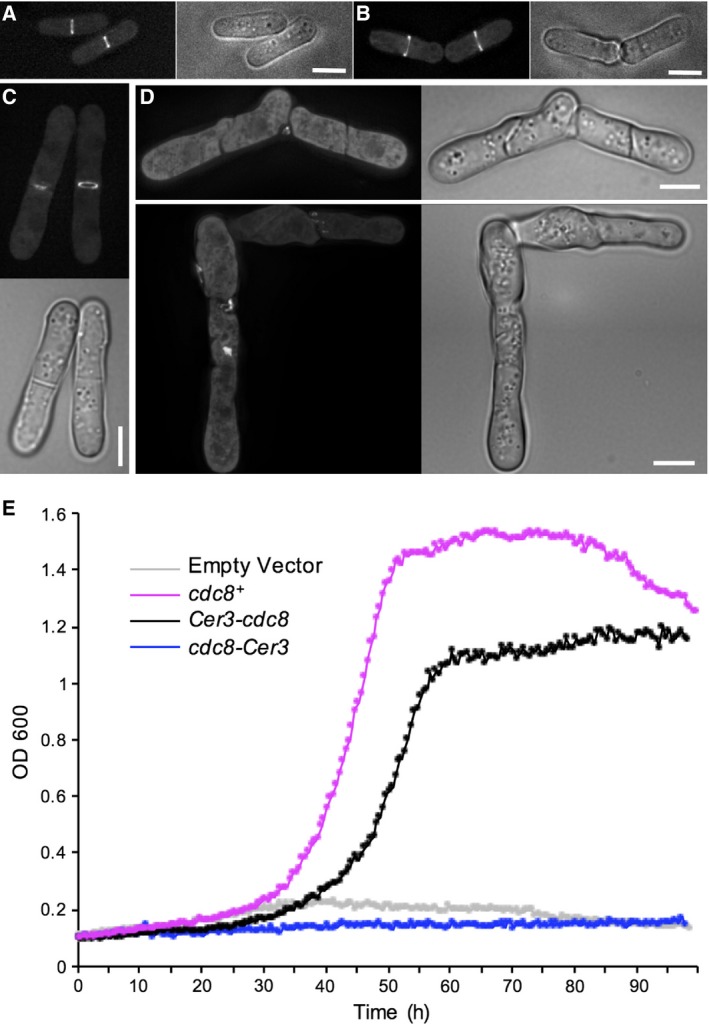
Ability of FP Tpm fusions to associate with and support CAR function. Localisation of Cerulean‐TpmCdc8 (A and C) and TpmCdc8‐Cerulean3 (B and D) in wild‐type (A and B) and cdc8.110 (C and D) cells at at 36 °C. (E) Growth curves of cdc8.110 cells containing plasmids encoding for wild‐type Tpm cdc8+ (magenta), Cerulean3‐Tpm cdc8 (black), Tpm cdc8 ‐Cerulean3 (blue) or an empty vector (grey) cultured at 36 °C. Scale bars: 5 μm.
As TpmCdc8 plays a key role in regulating the motor activity of both class II and V myosins 10, 11, 27, we next determined how the fusions affected the ability of the coiled‐coil protein to regulate myosin motor activity in vivo. Myo2 is the only essential myosin within the S. pombe cell, and is required for the assembly of the CAR from actomyosin containing medial nodes 29, 30, as well as providing the contractile force required for this organelle's constriction 31, 32, 33. As a readout on the ability of each fusion protein to regulate the contractile activity of myosin II 8, 15, 34, we measured the rate on CAR constriction in cdc8.110 myo2.mCherry cells at 36 °C. Consistent with our previous observation that the myo2.mCherry allele is functional at this temperature 9, these cells formed normal contractile rings which constricted at an average rate of 0.088 ± 0.014 μm·min−1 (mean ∆CAR‐diameter rate ± SD), when wild‐type TpmCdc8 was expressed from an episomal plasmid (Fig. 5A). In comparable cells expressing Cerulean3‐TpmCdc8, the CAR constricted at a rate of 0.09 ± 0.007 μm·min−1. However, in more than 30% of these mitotic cdc8.110 myo2.mCherry Cerulean3‐Tpm cdc8 cells, we observed MyoII incorporated into multiple contractile actomyosin rings (Fig. 5B). Thus, expression of Cerulean3‐TpmCdc8 disrupted the regulation of node‐dependent formation of the CAR, but once formed the actin‐Cerulean3‐TpmCdc8 copolymers were capable of regulating normal myosin II‐dependent contraction. In contrast, cdc8.110 myo2.mCherry cells expressing TpmCdc8‐Cerulean3 failed to form a stable CAR when cultured at 36 °C. Instead Myo2 foci concentrated to the cell equator and incorporated into randomly organised contractile filaments or aggregates (Fig. 5C). Thus, unable to form a functional CAR these cells failed to complete cytokinesis, leading to a loss of viability (Fig. 5C).
Figure 5.
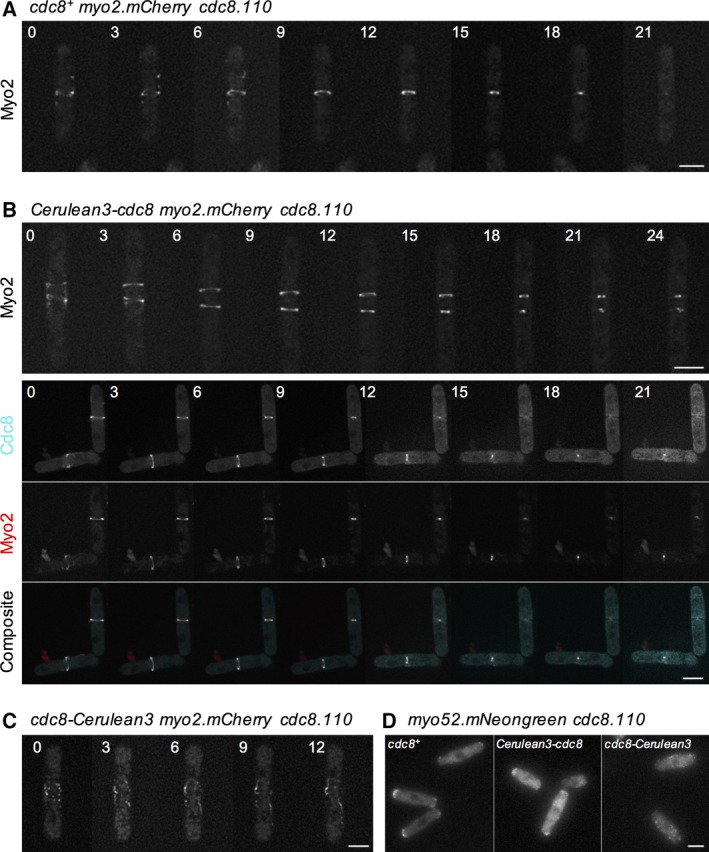
Myosin regulatory function of FP Tpm fusions. Montages of timelapse frames of (A) cdc8.110 myo2.mCherry pREP41Tpm cdc8+, (B) cdc8.110 myo2.mCherry pREP41Cerulean3‐Tpm cdc8, and (C) cdc8.110 myo2.mCherry pREP41Tpm cdc8‐Cerulean3 cells grown at 36 °C. Images show mCherry (A–C) and Cerulean (B‐lower panels) signals Timings are shown in minutes. (D) Micrographs of mNeongreen signal from cdc8.110 myo52.mNeongreen cells expressing different Tpm cdc8 constructs. Scale bars: 5 μm.
Finally, we explored the impact tagging TpmCdc8 had upon the ability of the protein to regulate myosin V function. The fission yeast myosin V, Myo52, associates with interphase actin‐TpmCdc8 copolymers, and moves towards the barbed end of these filaments to deliver cellular cargoes to the growing tips of the cell 35, 36, 37. In yeasts Tpm plays a critical role in modulating the duty‐ratio of this dimeric motor protein to promote processive movement upon actin 11, 38. Using a cdc8.110 myo52.mNeogreen strain, we tested the ability of each TpmCdc8 fusion to facilitate the actin‐dependent movement of the myosin V protein. Myo52 fails to accumulate at the growing ends of fission yeast cells lacking functional TpmCdc8 39. Consistent with an inability to associate with formin‐nucleated tropomyosin associated actin polymers, Myo52 movements were not observed in cdc8.110 myo52.mNeogreen cells expressing TpmCdc8‐Cerulean3 and the myosin V was not seen to accumulate at ends of cells (Fig. 5D), but instead localise to randomly dispersed foci throughout the cell. In contrast, Myo52 was seen at the ends of cells expressing wild‐type TpmCdc8 or the amino‐terminal Cerulean3‐TpmCdc8 fusion. However, in comparison to TpmCdc8‐containing cells, fewer Myo52 movements were observed within the cytosol in the presence of Cerulean3‐TpmCdc8 (not shown). Thus, while this fusion is capable of forming interphase actin polymers, it does not promote normal processive movement of myosin V within the cell.
Discussion
Over the last decade tropomyosin–fluorescent protein fusions have been used to study the organisation of actin polymers within diverse cell types 17, 18, 19, 20, 21. In each of these studies either the wild‐type protein is present within the cell or the Tpm being labelled is nonessential, thus the ability of the fusion‐protein to complement its function has not been tested. Given the terminal regions of a Tpm are critical for modulating its ability to (a) form end‐to‐end contacts; (b) regulate cooperative movements on the actin filament; (c) interact with regulatory proteins; and (d) interact with specific actin polymers at precise cellular locations, we decided to investigate the impact FP‐Tpm fusions had upon the biophysical properties and function of this essential cytoskeletal protein. Localisation studies of globular proteins have used tagging strategies where an FP is inserted at a surface loop within the protein sequence, resulting in fusion proteins with varying degrees of functionality 40, 41, 42. However, this strategy is not suitable for use with tropomyosins as they are single alpha‐helices, and insertion of an FP with associated flexible linkers would not only disrupt its packing on the actin surface but would also change the flexibility and shape of the tropomyosin filament, and thereby disrupt its ability to regulate actin function 43.
Having a single Tpm protein, TpmCdc8, makes the fission yeast an attractive model for studying the regulation and function of this conserved dimeric protein. We show that fusing Cerulean3 to the carboxyl terminus of this Tpm disrupts the ability of this protein to form end‐to‐end contacts and abolishes its ability to interact with actin and function within a cell, even when amino‐terminally acetylated. In contrast, the addition of the monomeric FP to the amino‐terminal of TpmCdc8 confers the resultant protein with physical properties equivalent to the acetylated wild‐type protein. This is consistent with previous studies on vertebrate tropomyosins which indicate amino‐terminal fusions do not interfere with head‐to‐tail self‐association and actin‐binding properties of tropomyosin 19, 44, 45. However, modifications to the amino‐terminus of the Tpm protein not only affect interactions with other actin‐binding proteins 9, 45 but also change the structure at the end of actin‐Tpm copolymers.
We have previously shown the cellular distribution of TpmCdc8 and its interactions with actin and myosin are modulated by amino‐terminal acetylation 9, 27, a modification common to all Tpms studied to date 4. This is consistent with the findings presented here. While Cerulean3‐TpmCdc8 binds to actin and regulates myosin II activity during CAR constriction to the same extent as wild‐type protein, its expression often disrupts initial polymerisation events and results in the disruption of normal actin ring formation and a subsequent delay in cell division. This supports models where the Tpm amino‐terminus is crucial during actin‐Tpm cofilament nucleation and growth. Thus, the addition of FP tags to the end of the Tpm changes the landscape at the end of the actin polymer, disrupting normal amino‐terminal signalling. Consistent with this idea, a single amino‐acid substitution within the amino‐terminus of TpmCdc8 (E6K) has been shown to biophysically mimic the effect of amino‐terminal acetylation, however, unlike an amino‐terminal fusion, can fully complement endogenous cdc8 function 8, 27. The terminus of the TpmCdc8–E6K is almost identical to the wild‐type protein, and is acetylated in vivo, again supporting the hypothesis that the biophysical landscape at the amino‐terminus of the TpmCdc8 protein is crucial for its proper regulation and function.
Both TpmCdc8 fusion are capable of localising to the CAR in cells possessing the wild‐type cdc8 + allele. However, only Cerulean3‐TpmCdc8 was able to localise and support growth in the absence of functional endogenous protein. This indicates the carboxyl‐terminal fusion, which is unable to form end‐to‐end polymers or associate with actin alone, is capable of forming heterodimers with endogenous Tpm protein. Similarly, GFP fusions with budding yeast Tpm1 localises to the S. pombe CAR, but do not complement TpmCdc8 function. Thus, the ability of a FP‐Tpm fusion to decorate actin polymers in a cell does not provide any indication of its functionality at any level.
In conclusion, while the impact of fusing a fluorescent protein to the terminus of a Tpm is likely to vary between different Tpm isoforms and cell types, it is clear from data presented here that their functionality and biophysical properties should be determined and shown to reflect that of the wild‐type protein. Unless this is the case, care should be taken in drawing conclusions from data generated from using them.
Author contributions
HRB performed the experiments; HRB, MAG and DPM conceived and designed the experiments; HRB, MAG and DPM analyzed the data; DPM wrote the manuscript.
Acknowledgements
We thank Drs Gerald Offer for predictive structures of TpmCdc8, Ben Goult for generating Fig. 1A and Dr Natali Fili for comments on the manuscript. This work was supported by the University of Kent, funding from the Biotechnology and Biological Sciences Research Council (BB/K012045/1) and Royal Society Industry Fellowship to DPM.
Edited by Dietmar Manstein
The copyright line for this article was changed on 15 September 2016 after original online publication.
References
- 1. Pollard TD, Blanchoin L and Mullins RD (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29, 545–576. [DOI] [PubMed] [Google Scholar]
- 2. Gunning PW, Schevzov G, Kee AJ and Hardeman EC (2005) Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 15, 333–341. [DOI] [PubMed] [Google Scholar]
- 3. Ebashi S and Ebashi F (1964) A new protein component participating in the superprecipitation of myosin B. J Biochem 55, 604–613. [DOI] [PubMed] [Google Scholar]
- 4. Gunning PW, Hardeman EC, Lappalainen P and Mulvihill DP (2015) Tropomyosin – master regulator of actin filament function in the cytoskeleton. J Cell Sci 128, 2965–2974. [DOI] [PubMed] [Google Scholar]
- 5. Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock‐DeGregori SE and Cohen C (2001) Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci USA 98, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greenfield NJ, Huang YJ, Swapna GVT, Bhattacharya A, Rapp B, Singh A, Montelione GT and Hitchcock‐DeGregori SE (2006) Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol 364, 80–96. [DOI] [PubMed] [Google Scholar]
- 7. Balasubramanian MK, Helfman DM and Hemmingsen SM (1992) A new tropomyosin essential for cytokinesis in the fission yeast S. pombe . Nature 360, 84–87. [DOI] [PubMed] [Google Scholar]
- 8. Skoumpla K, Coulton AT, Lehman W, Geeves MA and Mulvihill DP (2007) Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe . J Cell Sci 120, 1635–1645. [DOI] [PubMed] [Google Scholar]
- 9. Johnson M, East DA and Mulvihill DP (2014) Formins determine the functional properties of actin filaments in yeast. Curr Biol 24, 1525–1530. [DOI] [PubMed] [Google Scholar]
- 10. Clayton JE, Sammons MR, Stark BC, Hodges AR and Lord M (2010) Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol 20, 1423–1431. [DOI] [PubMed] [Google Scholar]
- 11. Clayton JE, Pollard LW, Sckolnick M, Bookwalter CS, Hodges AR, Trybus KM and Lord M (2013) Fission yeast tropomyosin specifies directed transport of myosin‐V along actin cables. Mol Biol Cell 25, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaner NC, Steinbach PA and Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2, 905–909. [DOI] [PubMed] [Google Scholar]
- 13. Snaith HA, Anders A, Samejima I and Sawin KE (2010) New and old reagents for fluorescent protein tagging of microtubules in fission yeast; experimental and critical evaluation. Methods Cell Biol 97, 147–172. [DOI] [PubMed] [Google Scholar]
- 14. Courtemanche N, Pollard TD and Chen Q (2016) Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat Cell Biol 18, 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coulton AT, East DA, Galinska‐Rakoczy A, Lehman W and Mulvihill DP (2010) The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci 123, 3235–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du J, Fan Y‐L, Chen T‐L and Feng X‐Q (2015) Lifeact and Utr230 induce distinct actin assemblies in cell nuclei. Cytoskeleton (Hoboken) 72, 570–575. [DOI] [PubMed] [Google Scholar]
- 17. Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y and Pellman D (2006) Polo‐like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313, 108–111. [DOI] [PubMed] [Google Scholar]
- 18. Goins LM and Mullins RD (2015) A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila . Mol Biol Cell 26, 2491–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin C, Schevzov G and Gunning P (2010) Alternatively spliced N‐terminal exons in tropomyosin isoforms do not act as autonomous targeting signals. J Struct Biol 170, 286–293. [DOI] [PubMed] [Google Scholar]
- 20. McElhinny AS, Schwach C, Valichnac M, Mount‐Patrick S and Gregorio CC (2005) Nebulin regulates the assembly and lengths of the thin filaments in striated muscle. J Cell Biol 170, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davidson AE, Siddiqui FM, Lopez MA, Lunt P, Carlson HA, Moore BE, Love S, Born DE, Roper H, Majumdar A et al (2013) Novel deletion of lysine 7 expands the clinical, histopathological and genetic spectrum of TPM2‐related myopathies. Brain 136, 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clos J and Brandau S (1994) pJC20 and pJC40–two high‐copy‐number vectors for T7 RNA polymerase‐dependent expression of recombinant genes in Escherichia coli . Protein Expr Purif 5, 133–137. [DOI] [PubMed] [Google Scholar]
- 23. Maundrell K (1993) Thiamine‐repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127–130. [DOI] [PubMed] [Google Scholar]
- 24. Moreno S, Klar A and Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 25. Johnson M, Coulton AT, Geeves MA and Mulvihill DP (2010) Targeted amino‐terminal acetylation of recombinant proteins in E. coli . PLoS One 5, e15801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spudich JA and Watt S (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin‐troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246, 4866–4871. [PubMed] [Google Scholar]
- 27. East DA, Sousa D, Martin SR, Edwards TA, Lehman W and Mulvihill DP (2011) Altering the stability of the Cdc8 overlap region modulates the ability of this tropomyosin to bind co‐operatively to actin and regulate myosin. Biochem J 438, 265–273. [DOI] [PubMed] [Google Scholar]
- 28. Watkins JL, Kim H, Markwardt ML, Chen L, Fromme R, Rizzo MA and Wachter RM (2013) The 1.6 A resolution structure of a FRET‐optimized Cerulean fluorescent protein. Acta Crystallogr D Biol Crystallogr 69, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu J‐Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR and Pollard TD (2006) Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol 174, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B and Pollard TD (2008) Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319, 97–100. [DOI] [PubMed] [Google Scholar]
- 31. Kitayama C, Sugimoto A and Yamamoto M (1997) Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol 137, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lord M, Laves E and Pollard TD (2005) Cytokinesis depends on the motor domains of myosin‐II in fission yeast but not in budding yeast. Mol Biol Cell 16, 5346–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naqvi NI, Eng K, Gould KL and Balasubramanian MK (1999) Evidence for F‐actin‐dependent and ‐independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J 18, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stark BC, Sladewski TE, Pollard LW and Lord M (2010) Tropomyosin and myosin‐II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell 21, 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Win TZ, Gachet Y, Mulvihill DP, May KM and Hyams JS (2001) Two type V myosins with non‐overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J Cell Sci 114, 69–79. [DOI] [PubMed] [Google Scholar]
- 36. Motegi F, Arai R and Mabuchi I (2001) Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol Biol Cell 12, 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. East DA and Mulvihill DP (2011) Regulation and function of the fission yeast myosins. J Cell Sci 124, 1383–1390. [DOI] [PubMed] [Google Scholar]
- 38. Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE and Trybus KM (2012) Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr Biol 22, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grallert A, Martin‐Garcia R, Bagley S and Mulvihill DP (2007) In vivo movement of the type V myosin Myo52 requires dimerisation but is independent of the neck domain. J Cell Sci 120, 4093–4098. [DOI] [PubMed] [Google Scholar]
- 40. Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K et al (2009) AAA+ ring and linker swing mechanism in the dynein motor. Cell 136, 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu C‐F, Chiou J‐G, Minakova M, Woods B, Tsygankov D, Zyla TR, Savage NS, Elston TC, Lew DJ and Balasubramanian M (2015) Role of competition between polarity sites in establishing a unique front. eLife 4, e11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bendezú FO, Vincenzetti V, Vavylonis D, Wyss R, Vogel H and Martin SG (2015) Spontaneous Cdc42 polarization independent of GDI‐mediated extraction and actin‐based trafficking. PLoS Biol 13, e1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holmes KC and Lehman W (2008) Gestalt‐binding of tropomyosin to actin filaments. J Muscle Res Cell Motil 29, 213–219. [DOI] [PubMed] [Google Scholar]
- 44. Urbancikova M and Hitchcock‐DeGregori SE (1994) Requirement of amino‐terminal modification for striated muscle alpha‐tropomyosin function. J Biol Chem 269, 24310–24315. [PubMed] [Google Scholar]
- 45. Hitchcock‐DeGregori SE and Heald RW (1987) Altered actin and troponin binding of amino‐terminal variants of chicken striated muscle alpha‐tropomyosin expressed in Escherichia coli . J Biol Chem 262, 9730–9735. [PubMed] [Google Scholar]


