Summary
The QseBC two‐component system (TCS) is associated with quorum sensing and functions as a global regulator of virulence. Based on sequence similarity within the sensor domain and conservation of an acidic motif essential for signal recognition, QseBC is primarily distributed in the Enterobacteriaceae and Pasteurellaceae. In Escherichia coli, QseC responds to autoinducer‐3 and/or epinephrine/norepinephrine. Binding of epinephrine/norepinephrine is inhibited by adrenergic antagonists; hence QseC functions as a bacterial adrenergic receptor. Aggregatibacter actinomycetemcomitans QseC is activated by a combination of epinephrine/norepinephrine and iron, whereas only iron activates the Haemophilus influenzae sensor. QseC phosphorylates QseB but there is growing evidence that QseB is activated by non‐cognate sensors and regulated by dephosphorylation via QseC. Interestingly, the QseBC signaling cascades and regulons differ significantly. In enterohemorrhagic E. coli, QseC induces expression of a second adrenergic TCS and phosphorylates two non‐cognate response regulators, each of which induces specific sets of virulence genes. This signaling pathway integrates with other regulatory mechanisms mediated by transcriptional regulators QseA and QseD and a fucose‐sensing TCS and likely controls the level and timing of virulence gene expression. In contrast, A. actinomycetemcomitans QseC signals through QseB to regulate genes involved in anaerobic metabolism and energy production, which may prime cellular metabolism for growth in an anaerobic host niche. QseC represents a novel target for therapeutic intervention and small molecule inhibitors already show promise as broad‐spectrum antimicrobials. Further characterization of QseBC signaling may identify additional differences in QseBC function and inform further development of new therapeutics to control microbial infections.
Keywords: catecholamine, iron, QseBC, two‐component system, virulence
Introduction
The ability to quickly respond and adapt to environmental flux is central for microbial colonization and persistence in a given niche or host organism. Bacteria sense and respond to changes in their environment using a variety of mechanisms such as two‐component sensing and quorum sensing. In two‐component systems (TCS), an environmental ‘signal' is detected by a sensor histidine kinase, which initiates a signal transduction cascade that in many cases results in transcriptional regulation of target genes. Although TCS are broadly distributed in bacteria, for many TCS the specific signal that activates the sensor kinase and the target genes that are regulated upon its activation have not been identified. The QseBC TCS is conserved in a broad spectrum of bacterial species (Clarke et al., 2006) and is closely associated with quorum‐sensing mechanisms (Sperandio et al., 2002; Novak et al., 2010) and generally functions as a global regulator of virulence. As shown in Fig. 1, the QseC histidine kinase is comprised of a periplasmic sensor domain, two membrane‐spanning domains, and a cytoplasmic kinase domain. The QseB response regulator is comprised of a receiver domain and a helix‐turn‐helix (HTH) DNA binding domain. QseBC is one of the few TCS where the signal that activates the sensor has been identified for several organisms and the regulon that is controlled by the system has been characterized. This review focuses on the distribution of the QseBC TCS, the mechanisms of activation of the sensor kinase and response regulator, the signal transduction pathways that are initiated upon activation of the system, and the regulons and functional outcomes of QseBC. It will also highlight the similarities and differences of this system in pathogens that colonize mucosal surfaces in the human oral cavity and in the gastrointestinal and urogenital tracts.
Figure 1.
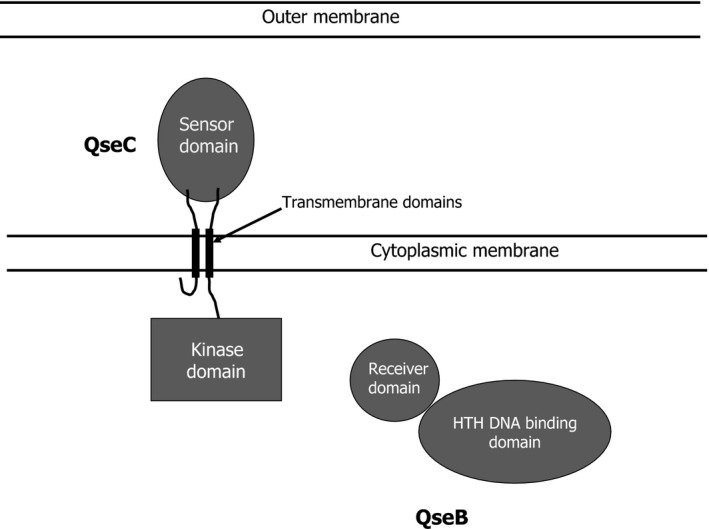
Schematic illustrating the structural domains of the QseC histidine kinase and the QseB response regulator.
Distribution and structure of the QseBC TCS
The National Center for Biotechnology Information (NCBI) Protein Database contains over 12,000 proteins that exhibit similarity to the Conserved Protein Domain Family PRK10337, which is designated as sensor protein QseC. However, PRK10337 encompasses the cytoplasmic kinase and ATPase domains that are highly conserved across a wide range of sensor kinase polypeptides. Hence, it is likely that many of these 12,000 proteins are not paralogs of QseC but instead represent sensor kinases of other TCS. In contrast, a search of the NCBI Gene Database for genes that are annotated as qseC yielded a more limited set of only 164 genes. Of these, 159 are encoded by α‐, β‐ and γ‐proteobacteria and the remaining five are contained in eukaryotic genomes. Consistent with this, a protein BLAST search carried out by Clarke et al. (2006) using only the periplasmic sensor domain of Escherichia coli QseC identified related sequences in the genera Shigella, Salmonella, Erwinia, Haemophilus, Pasteurella, Actinobacillus, Chromobacterium, Rubrivivax, Thiobacillus, Ralstonia, Psychrobacter and Aspergillus. Similar results were obtained using the periplasmic sensor domain of Aggregatibacter actinomycetemcomitans QseC (residues 37–169) as a probe. This search identified QseC‐like sequences primarily in the α‐, β‐ and γ‐proteobacteria (see Table 1). As shown in Fig. 1, a further comparison of the periplasmic sensor domains of QseC‐like sequences encoded by organisms in the Enterobacteriaceae and Pasteurellaceae families showed that the EYRDD motif (boxed in Fig. 2) which was previously shown to be essential for QseC signal recognition in A. actinomycetemcomitans (Weigel et al., 2015) and Haemophilus influenzae (Steele et al., 2012) was conserved in all sequences. A similar acidic motif is also present in multiple copies in the PmrB sensor of Salmonella enterica, which is closely related to QseC. Further examination of the QseC‐like sequences in Fig. 2 identified two additional acidic motifs that are highly conserved (shown in red).
Table 1.
Comparison of QseC‐like proteins with the periplasmic sensor domain of Aggregatibacter actinomycetemcomitans QseC
| % identity | % coveragea | Acidic motifsb | |||
|---|---|---|---|---|---|
| EDD | EDDDE | EYRDD | |||
| γ‐Proteobacteria | |||||
| Enterobacteriaceae | 45–52 | 97–100 | +c | + | + |
| Pasteurellaceae | 45–100 | 96–100 | + | + | + |
| Pseudomonadales | 24–38 | 19–96 | ± | – | ± |
| Alteromonadales | 22–38 | 25–99 | – | – | ± |
| β‐Proteobacteria | |||||
| Burkholderiaceae | 25–32 | 42–100 | – | – | – |
| Alcaligenaceae d | 27–54 | 54–100 | ± | ± | ± |
| Comamonadaceae | 26–48 | 15–79 | – | – | – |
| Neisseriaceae | 27–48 | 23–97 | ± | ± | ± |
| α‐Proteobacteria | |||||
| Rhizobiales | 26–47 | 36–99 | – | – | – |
| Rhodobacterales | 25–45 | 32– | – | – | – |
| Eukaryotes | |||||
| Animals | 43e | 61 | + | + | + |
| Fungi | 48–56 | 13–16 | – | – | – |
| Green plants | 44 | 19 | – | – | – |
Coverage represents the portion (in per cent) of the probe sequence that exhibits similarity to the target sequence.
The acidic motifs present in the periplasmic signal domain of A. actinomycetemcomitans QseC are highlighted in red in Fig. 1.
'+' indicates that the motif was conserved in all of the sequences that were examined; ‘±' indicates that the motif was conserved in some of the sequences; ‘–' indicates that the motif was not conserved in the sequences that were examined.
Only one organism in this family, Basilea psittacipulmonis, exhibited 54% sequence identity with the periplasmic signal domain of A. actinomycetemcomitans QseC and contained all three acidic motifs.
The Mediterranean fruit fly, Ceratitis capitata, exhibited 43% sequence identity across 80 residues in the periplasmic domain and contains the three acidic motifs present in QseC of Enterobacteria and Pasteurellales.
Figure 2.

Alignment of sequences derived from the QseC periplasmic sensor domains of the following organisms: Aa, Aggregatibacter actinomycetemcomitans; Ec, Escherichia coli; Se, Salmonella enterica; Sf, Shigella flexneri; Pa, Pectobacterium atrosepticum; Ecl, Enterobacter cloacae; Pan, Pantoea ananatis; Sm, Serratia marsescens; Kp, Klebsiella pneumonia; Ap, Actinobacillus pleuropneumoniae; Hi, Haemophilus influenzae; Pm, Pasteurella multocida; Bp, Basilea psittacipulmonis; Cc, Ceratitis capitatta; and Lh, Laribacter hongkongensis. Three conserved acidic motifs are shown in red text and the motif that is essential for signal binding is boxed.
Only two QseC‐like sequences outside the Enterobacteriaceae and Pasteurellaceae families showed similar high levels of sequence identity of the periplasmic sensor domain and conservation of the acidic motifs described above; QseC of Basilea psittacipulmonis in the family Alcaligenaceae and, surprisingly, QseC encoded by the Mediterranean Fruit Fly, Ceratitis capitata. Both of these proteins also possess the cytoplasmic kinase, dimer interface and ATPase domains that are present in the other QseC polypeptides. As shown in Table 1, the QseC‐like proteins encoded by the remaining families and orders of organisms either exhibited lower overall sequence identity (typically < 40%), exhibited similarity to only a portion of the A. actinomycetemcomitans sensor domain or both. These QseC‐like proteins also lacked the conserved acidic motif(s) that is essential for QseC signal recognition. As a representative example, the QseC‐like sequence from Laribacter hongkongensis (family Neisseriaceae) is included in Fig. 2. This sequence exhibits only 30% identity to the sensor domain of A. actinomycetemcomitans QseC and lacks the EYRDD motif involved in signal recognition.
Together, the comparisons described above suggest that QseC is structurally and functionally conserved mainly in the Enterobacteriaceae and Pasteurellaceae and in a limited number of organisms outside of these groups. Furthermore, the observation that the EYRDD motif involved in QseC signal recognition is not conserved in the QseC‐like sequences in the other organisms suggests that these sensors may respond to different stimuli. However, it is also possible that sensor proteins exist that are functionally related to QseC but without significant sequence similarity in the sensory domain. For example, the QseC‐like sensor of Francisella tularensis is reported to be functionally interchangeable with QseC of E. coli (Rasko et al., 2008) but the periplasmic domain of this protein exhibits no homology with the sensor domain of the E. coli or A. actinomycetemcomitans QseC proteins. This review will focus primarily on the properties and activities of QseC in the Enterobacteriaceae and Pasteurellaceae.
In many organisms, the qseBC locus is associated with another gene, designated ygiW, that encodes a putative periplasmic protein in the OB fold family (Ginalski et al., 2004), but transcription and genetic organization of ygiW relative to the qseBC operon varies. As shown in Fig. 3A, ygiW resides upstream from and is co‐transcribed with qseBC in many Pasteurellaceae. Steele et al. (2012) and Juarez‐Rodriguez et al. (2013) have shown that an attenuator stem loop exists in the intergenic region between ygiW and qseBC in H. influenzae and A. actinomycetemcomitans, respectively. Hence, primary transcripts encoding ygiW alone and ygiW‐qseBC are produced by these organisms and the overall expression of qseBC is reduced relative to ygiW (Steele et al., 2012). A similar attenuation of qseBC transcription probably occurs in Actinobacillus pleuropneumoniae and B. psittacipulmonis as both of these operons also contain an inverted repeat in the ygiW‐qseB intergenic region that may form a stem loop resembling a rho‐independent terminator. ygiW also resides upstream of qseBC in many of the available Enterobacteriaceae genome sequences, but in these organisms it is transcribed from the opposite strand (see Fig. 3B). A similar gene configuration is present in the fruit fly, C. capitata. Finally, in organisms such as Serratia marsescens, Pasteurella multocida and L. hongkongensis, ygiW is not adjacent to the qseBC locus (see Fig. 3C) but in some cases is present elsewhere in the genome.
Figure 3.
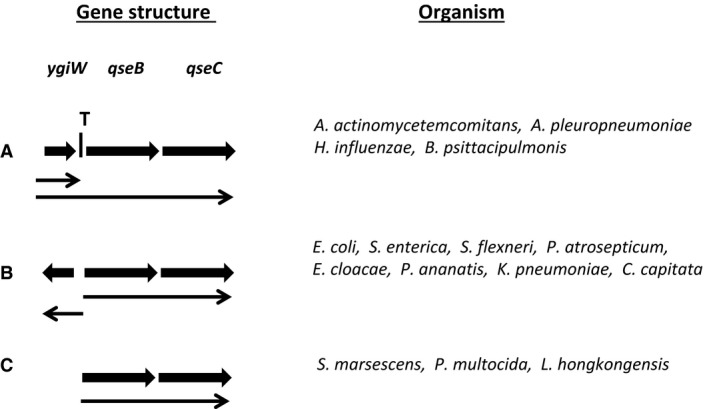
Comparison of the organization of the qseBC locus. In many organisms in the Enterbacteriaceae and Pasteurellaceae families, ygiW is adjacent to qseBC and is either co‐transcribed with qseBC (A) or transcribed from the opposite strand as qseBC (B). For other organisms, ygiW is not adjacent to qseBC (C) and is either located elsewhere in the genome or is not present at all. The transcriptional attenuator located between ygiW and qseBC is indicated by the letter ‘T'. Arrows indicate the direction of transcription.
Transcription of ygiW has been shown to be regulated by the QseB response regulator S. enterica (Merighi et al., 2009), A. actinomycetemcomitans (Juarez‐Rodriguez et al., 2013) and H. influenzae (Steele et al., 2012), and presumably in the other organisms shown in Fig. 3A,B. In S. enterica, ygiW has been associated with virulence through several mechanisms. Moreira et al. (2013) showed that visP (ygiW) encodes a protein that binds to the sugar moiety of peptidoglycan and inhibits Fe2+/α‐ketoglutarate‐dependent dioxygenase (LpxO). This results in decreased LpxO‐dependent modification of lipopolysaccharide and increased resistance to stressors within the vacuole during intramacrophage replication. However, VisP was also shown to function independently of LpxO in a murine colitis model and conferred resistance to cationic antimicrobial peptides (Moreira et al., 2013). Similarly in E. coli, YgiW has been suggested to be part of a stress response circuit that confers resistance to hydrogen peroxide, cadmium and acid stress (Lee et al., 2010). The function of YgiW in other organisms has not been determined.
Activation of the QseC sensor
Sperandio et al. (1999) showed that expression of the Locus of Enterocyte Effacement (LEE) operon encoding the type III secretion system (TTSS) of enterohaemorrhagic E. coli (EHEC) serotype O157:H7 was induced by autoinducer‐2 (AI‐2)‐dependent quorum sensing and to identify other potential quorum‐sensing‐regulated virulence genes and regulatory components, a gene array was hybridized with cDNA derived from E. coli O157:H7 or an isogenic luxS mutant. The quorum‐sensing E. coli regulator B and C (QseBC) was identified in these studies as a TCS that was regulated by AI‐2‐dependent quorum sensing (Sperandio et al., 2002). This study also showed that the motility of a luxS, but not qseC, mutant could be restored by exogenous AI‐2, suggesting that QseBC is necessary to respond to AI‐2. Consistent with these early observations, Gonzalez Barrios et al. (2006) subsequently showed that AI‐2 increased biofilm growth of wild‐type E. coli MG1655 but not an isogenic qseBC mutant. In addition, Novak et al. (2010) showed that qseBC expression in A. actinomycetemcomitans was induced by exogenous AI‐2 and that induction required the putative AI‐2 receptors, LsrB and RbsB. Athough each of these studies suggested that QseBC may represent a part of the AI‐2 signaling circuit, they did not directly demonstrate that AI‐2 functions as a signal that activates the QseC sensor. Indeed, AI‐2 that was partially purified by chromatography on a C‐18 Sep Pack column was shown to induce bioluminescence of a Vibrio harveyi reporter strain, but this preparation failed to induce expression of the LEE1 operon, restore type III secretion, or induce qseBC expression in EHEC. In contrast, a fraction that was eluted with methanol increased transcription of LEE1 and qseBC (Sperandio et al., 2003) but did not induce V. harveyi bioluminescence. The structure of this compound, designated autoinducer‐3 (AI‐3) has not yet been determined but electrospray mass spectroscopy demonstrated a major peak of 213.1 Da and several minor peaks between 109.1 Da and 222.9 Da. Gonzalez Barrios et al. (2006) also suggested that AI‐2 does not directly activate the QseC sensor and showed that stimulation of E. coli biofilm growth by AI‐2 requires the protein encoded by open reading frame b3022, which in turn regulates qseBC. B3022 was designated MqsR (motility quorum‐sensing regulator) and is now known to be a CGU‐specific mRNA interferase (Yamaguchi & Inouye, 2009). This signaling mechanism may also function in A. actinomycetemcomitans, which encodes a homolog of MqsR (Novak et al., 2010), but not in E. coli O157:H7 which lacks this gene. Hence, AI‐2 is not the signal that activates QseC and it is possible that the link between AI‐2 quorum sensing and QseBC function may exist only in organisms that also express MqsR. In contrast, AI‐3 represents an autoinducer that directly signals through QseC but its initial identification was confounded by the fact that AI‐3 production is reduced as a result of the metabolic deficiency that is caused by the inactivation of the AI‐2 synthase luxS (Walters et al., 2006). Walters et al. (2006) also showed that AI‐3 is produced by other enterobacteria such as Shigella sp., Salmonella sp., Klebsiella pneumonia and Enterobacter cloacae. However, it is not known if AI‐3 is more widely produced by other groups of organisms, as is AI‐2. For example, we have so far been unable to identify AI‐3 in A. actinomycetemcomitans (Weigel et al., 2015).
Sperandio et al. (2003) also showed that the ability of the EHEC luxS mutant (deficient in AI‐3 production) to generate attaching and effacing (A/E) lesions on HeLa cells was indistinguishable from the parent strain. This was surprising because in the absence of AI‐3, the expression of the LEE operon encoding the TTSS required for the development of A/E lesions should be significantly reduced. This result suggested that HeLa cells may produce a factor that complements AI‐3 deficiency. Consistent with this, type III secretion of the luxS mutant was restored when bacteria were incubated with conditioned medium from HeLa cells that were size fractionated for compounds > 1 kDa. It was subsequently shown that purified epinephrine (Ep) or norepinephrine (Ne) at physiologic concentrations (i.e. 50 μm) increased expression of LEE1 and activated type III secretion in the luxS mutant. Furthermore, complementation of AI‐3 deficiency by these hormones was blocked by α‐ and β‐adrenergic antagonists (Sperandio et al., 2003). In addition, a qseC mutant was not complemented by exogenous AI‐3 or Ep whereas each compound complemented the EHEC luxS mutant, suggesting that both AI‐3 and Ep may signal via the QseC sensor. This was subsequently demonstrated directly by Clarke et al. (2006), who showed that Ne and AI‐3 bound to the periplasmic domain of QseC and induced autophosphorylation of the sensor. Binding and autophosphorylation of QseC were significantly reduced in the presence of an α‐adrenergic receptor antagonist. Hence, the E. coli QseC sensor mediates both interspecies and interkingdom signaling and is activated by signals that are produced by bacteria (AI‐3) and the eukaryotic host (Ep, Ne). In addition, Moreira & Sperandio (2012) suggest that the QseC paralog in S. enterica (PreB) functions as an adrenergic receptor whereas Merighi et al. (2009) report that the PreB does not respond to catecholamine hormones.
Although biochemical identification of the signals that activate many of the other QseC paralogs shown in Fig. 2 has not been reported, some surprising differences exist for those that have been characterized. Weigel et al. (2015) showed that A. actinomycetemcomitans QseC is activated by a combination of catecholamine hormones and iron, but not by either compound individually. This suggests that a catecholamine–iron complex may be the signal that is recognized by the A. actinomycetemcomitans sensor, or alternatively that catecholamines and iron interact individually with the sensor and activation of QseC occurs only when both are bound. In addition, as production of AI‐3 by A. actinomycetemcomitans has not yet been demonstrated (Weigel et al., 2015), catecholamine hormones and iron may be the only signals that activate QseC in this organism. Interestingly, qseBC expression in E. coli is also induced by elevated levels of Fe3+ but QseC is not directly activated by iron. Instead, iron activates the PmrB sensor, which in turn phosphorylates response regulator PmrA. Activated PmrA then binds to the qseBC promoter and induces expression of the operon. Furthermore, Guckes et al. (2013) showed that the PmrB sensor phosphorylates the non‐cognate QseB response regulator, which autoregulates the qseBC operon (see below). The pmrAB genes are not present in the A. actinomycetemcomitans genome but the QseB binding site in the ygiW‐qseBC promoter is identical to the consensus PmrA binding sequence in the pmrAB operon of E. coli (Juarez‐Rodriguez et al., 2014). Hence, it is possible that in the absence of PmrAB, the A. actinomycetemcomitans QseC sensor may have evolved to integrate the iron and catecholamine sensory functions of the PmrAB and QseBC TCSs of E. coli.
In contrast with QseC from E. coli and A. actinomycetemcomitans, the QseC paralog of H. influenzae is activated only by ferrous iron or zinc, and does not appear to respond to Ep or Ne. Hence this TCS was designated as a ferrous‐iron‐responsive system (FirS) (Steele et al., 2012). Iron activation of the FirS also differed from iron activation of A. actinomycetemcomitans QseC in that both ferrous and ferric iron activates A. actinomycetemcomitans QseC in the presence of catecholamines (Weigel et al., 2015) whereas only ferrous iron activates H. influenzae FirS (Steele et al., 2012). This is interesting because a DYRED motif in the periplasmic domain of FirS is conserved in A. actinomycetemcomitans QseC (EYRDD, see Fig. 2) and both sequences have been shown to be essential for activation of the respective sensors (Steele et al., 2012; Weigel et al., 2015). Hence, as summarized in Table 2, a variety of signals including AI‐3, catecholamine hormones, iron and zinc, have been shown to activate QseC and the responsiveness of QseC to each of these signals can differ among organisms. Furthermore, there is increasing evidence that other signaling mechanisms that are independent of QseC may mediate the response of some organisms to catecholamine hormones (Karavolos et al., 2013) and it is possible that H. influenzae responds to catecholamines via one of these pathways rather than QseC (FirS). Finally, qseBC expression in Actinobacillus pleuropneumoniae is also induced by Ep and Ne (Li et al., 2012), suggesting that catecholamine hormones may function to activate the QseC sensor in this organism, but this has not yet been directly demonstrated.
Table 2.
Signals that activate the QseC sensor
| Organism | Signal | Reference |
|---|---|---|
| Escherichia coli | Ep, Ne; AI‐3 | Clarke et al. (2006) |
| Aggregatibacter actinomycetemcomitans | Ep,Ne/Fe2+ or Fe3+ a | Weigel et al. (2015) |
| Haemophilus influenzae | Fe2+, Zn | Steele et al. (2012) |
| Salmonella enterica | Ep, Ne | Moreira & Sperandio (2012), Merighi et al. (2009)b |
Activation of QseC occurs only in the presence of both the catecholamine and iron.
Merighi et al. reported that S. enterica PreAB (QseBC) did not respond to catecholamines.
Activation of QseB and transcriptional regulation of qseBC
Similar to most other TCS, the activation of QseC leads to phosphorylation of the QseB response regulator and several studies have confirmed this either by directly demonstrating phosphate transfer (Clarke et al., 2006) or by showing that site‐specific mutation of the conserved Asp51 inhibits QseB function (Kostakioti et al., 2009; Juarez‐Rodriguez et al., 2014). In addition, using a mobility shift assay, Clarke & Sperandio (2005a,b) demonstrated that phosphorylation of QseB is required for it to bind to QseB‐regulated promoters in vitro. One of the initial outcomes resulting from activation of QseB is the auto‐induction of the qseBC operon and ygiW and the induction of ygiW expression occurs regardless of the architecture of the ygiW‐qseBC locus (see Fig. 3). In A. actinomycetemcomitans, the promoter that drives ygiW‐qseBC expression resides completely within a fragment of 138 bp upstream from the ygiW start codon (Juarez‐Rodriguez et al., 2013) and two transcriptional start sites at nucleotides ‐15 and ‐53 have been mapped in this region (Juarez‐Rodriguez et al., 2014). The distal initiation site at nucleotide ‐53 is the main transcriptional start site and although both sites are associated with putative ‐10 and ‐35 elements, only the upstream promoter is regulated by QseBC. Within this promoter, QseB binds to the direct repeat sequence CTTAA‐N6‐CTTAA where the CTTAA repeats flank the ‐35 element. Similar to A. actinomycetemcomitans, the qseBC promoter of EHEC also contains two transcriptional initiation sites at nucleotides ‐27 and ‐77 relative to the qseB start codon; each is associated with putative ‐10 and ‐35 elements and the distal promoter is regulated by QseBC. However, using DNAse footprinting, two QseB binding sites were mapped in this promoter, a high‐affinity site that appears to overlap the distal transcriptional start site and a low‐affinity site located upstream between nucleotides ‐409 and ‐423 (Clarke & Sperandio, 2005b). Comparison of these sites with the promoter sequence of the QseB‐regulated flhDC operon generated a QseB binding consensus sequence, CAATTACGAATTA, where the underlined residues are most highly conserved (Clarke & Sperandio, 2005a). However, a direct repeat CTTAA‐N6‐CTTAA identical to the QseB binding site in A. actinomycetemcomitans overlaps the ‐10 element of the QseB‐regulated distal promoter and this site was suggested by Guckes et al. (2013) to be the site in the qseBC promoter that is bound by the PmrA response regulator. Given the proximity of this repeat sequence to the protected region identified by DNAse footprinting (Clarke & Sperandio, 2005b), it is possible that this direct repeat also represents the QseB binding site in EHEC.
There is also growing evidence that activation of QseB may be modulated by several additional mechanisms. Juarez‐Rodriguez et al. (2014) showed that lacZ expression from the ygiW‐qseBC promoter was significantly reduced in a ΔqseBC strain of A. actinomycetemcomitans and that expression was complemented to wild‐type levels by a single‐copy chromosomal insertion of qseBC. Interestingly, partial complementation of lacZ expression was obtained by a single copy insertion of qseB but not by qseB‐D51A. This suggests that QseB can be phosphorylated and partially activated in the absence of QseC. A similar reaction occurs in E. coli (Kostakioti et al., 2009). In E. coli, the absence of QseC results in constitutively high qseB transcription that arises from bidirectional cross‐regulation between the structurally related QseBC and PmrAB TCS (Guckes et al., 2013). Without QseC, the non‐cognate PmrB sensor phosphorylates QseB and this reaction exhibits kinetics similar to phosphorylation of QseB by its cognate sensor (Guckes et al., 2013). As a result, transcription of qseBC can be activated by PmrA resulting in constitutive expression of qseB. In contrast, transcription of ygiW‐qseBC is not constitutively high in the absence of QseC in A. actinomycetemcomitans. One explanation for this is that A. actinomycetemcomitans genome does not encode the PmrAB TCS. Partial activation of QseB in the absence of QseC in A. actinomycetemcomitans may instead result from inefficient phosphorylation by other non‐cognate sensors, or alternatively by the transfer of a phosphate from the small phosphate donor acetyl‐phosphate (Wolfe, 2005). Consistent with the latter possibility, the genes that encode enzymes required for the production of acetyl phosphate are present in A. actinomycetemcomitans genome.
QseB activation can also be modulated by dephosphorylation mediated by either QseC or PmrB (Kostakioti et al., 2009; Guckes et al., 2013). Incubation of phospho‐QseB with vesicles containing QseC resulted in the rapid loss of phosphate from the response regulator and a concomitant increase in phospho‐QseC, indicating that QseC can reverse phosphate flow via dephosphorylation of QseB. A similar reaction occurs with PmrB, but although the kinetics of PmrB‐mediated activation of QseB are similar to QseC, the kinetics of PmrB‐mediated dephosphorylation are significantly slower than QseC. Together, these results suggest that QseB activation can occur through interaction with either cognate or non‐cognate sensors but is modulated primarily by the phosphatase activity of the cognate QseC. In addition, Hughes et al. (2009) suggested that unphosphorylated QseB may play an active role in regulating E. coli motility by repressing flhDC expression. Overexpression of QseB in a ΔqseC strain resulted in a decrease in cell motility and a five‐fold reduction in lacZ activity from a reporter construct containing the flhDC promoter, presumably arising from an overabundance of unphosphorylated QseB. Unphosphorylated QseB was also shown by motility shift experiments to bind to the flhDC promoter at a site between the high‐ and low‐affinity QseB binding sites that are bound by phospho‐QseB (Hughes et al., 2009). Together, this suggests that QseC controls transcription of flhDC via QseB and that QseB plays a dual role in its phosphorylated and unphosphorylated forms to fine tune this process. These results also suggest the possibility that QseB may play a broader regulatory role in gene expression by controlling the transcription of other genes in its unphosphorylated form.
Reading et al. (2009) also suggest that the QseC sensor is capable of activating several non‐cognate response regulators. By screening a panel of 31 purified E. coli response regulators, QseC was shown to phosphorylate two additional non‐cognate proteins, QseF and KdpE. QseF is a response regulator that is also activated by its cognate sensor, the QseE adrenergic receptor (Reading et al., 2009), indicating that cross talk occurs between the two catecholamine responsive TCS of E. coli. KdpE is also activated by its cognate sensor KdpD, which responds to changes in osmolarity and various metabolites (Hughes et al., 2009). Of the 344 genes induced by QseC that have been identified (see below), 336 are present in the regulons of these three response regulators that are activated by phosphorylation via QseC.
Finally, two recent studies suggest that the transcription of the qseBC operon is also regulated by nucleoid‐associated proteins. Sharma & Casey (2014) demonstrated that deletion of hha resulted in a decrease in motility of EHEC and compared the contribution and hierarchy of Hha and QseBC in controlling EHEC motility. Using single and double gene deletion mutants, they showed that transcription of qseC was significantly reduced in a hha‐deficient background relative to wild‐type or a complemented strain and that hha was hierarchically superior to qseBC in regulating motility. This suggests that Hha functions as a net positive regulator of qseBC expression. In addition, Juarez‐Rodriguez et al. (2014) identified three integration host factor (IHF) binding sites in the ygiW‐qseBC promoter region of A. actinomycetemcomitans. One of these sites was located just upstream from the QseB binding direct repeat sequence and deletion of this region reduced ygiW‐qseBC expression by approximately 2.5‐fold, suggesting that binding of IHF to this site positively regulates transcription of ygiW‐qseBC. The other IHF binding sites mapped to the region between the ‐10 and ‐35 elements of the proximal promoter and a sequence near the 5′‐end of the ygiW open reading frame. Presumably, one or both of these sites function as negative regulators of expression since deleting ihfA or ihfB in A. actinomycetemcomitans results in a net two‐fold increase in ygiW‐qseBC expression.
Functional outcomes of QseBC activation: biofilms and virulence
In many organisms for which QseBC has been characterized, this TCS functions as a global regulator that controls complex phenotypes such as biofilm formation and virulence. For example, Novak et al. (2010) showed that inactivation of qseC in A. actinomycetemcomitans reduced total biofilm biomass and average biofilm depth by greater than 90% and complementation of the mutant with a plasmid‐borne copy of qseC restored biomass and biofilm depth to levels even greater than the wild‐type, presumably due to the presence of qseC in multicopy. Juarez‐Rodriguez et al. (2014) also obtained a similar biofilm phenotype using a strain of A. actinomycetemcomitans that expressed QseC with an in‐frame deletion of the periplasmic sensor domain. This suggests that the interaction of catecholamines and iron with the sensor domain of QseC is required for stimulation of biofilm growth and consistent with this, an increase in biofilm biomass occurs when A. actinomycetemcomitans is cultured in the presence of Ep or Ne and iron (W.A. Weigel and D.R. Demuth, unpublished). Similarly, Yang et al. (2014) showed that Ep and Ne increased E. coli K‐12 biofilm formation by approximately 50% over control cultures that were grown without the hormone. Furthermore, an isogenic ΔqseC mutant exhibited a reduction in biofilm growth of approximately 80% in control medium and exhibited little increase in biofilm biomass when cultured in the presence of catecholamines. Unal et al. (2012) also showed that a qseC‐deficient strain of H. influenzae exhibited a significant decrease in biofilm formation relative to wild‐type when cultured under static, semi‐static or open flow conditions. Hence, for these organisms, the activation of QseBC clearly promotes sessile growth. Catecholamine hormones have also been shown to stimulate the planktonic growth of other organisms such as Bordetella, E. coli, and S. enterica (Freestone et al., 2000, 2008), suggesting that activation of QseBC by catecholamines may promote sessile growth of these organisms as well. However, it is also important to note that induction of Actinobacillus pleuropneumoniae growth by Ep and Ne has recently been demonstrated to occur independently of QseBC (Li et al., 2015), suggesting that alternative mechanisms may exist to promote growth by catecholamines. Indeed, Karavolos et al. (2013) suggest that other sensor kinases such as QseE, BasS and CpxA may also play a role in catecholamine interkingdom communication.
Comparing wild‐type and qseBC mutant strains using a variety of in vitro and in vivo model systems has also clearly demonstrated the important role that this TCS plays in regulating virulence. QseBC has been shown to regulate the expression of genes associated with production of flagella and motility of EHEC (Sperandio et al., 2002) and a qseC mutant exhibited attenuated virulence after intragastric inoculation in a rabbit model (Clarke et al., 2006). The LEE pathogenicity island encoding a TTSS and various effector proteins are also under QseC‐dependent regulation but this does not occur through direct activation by QseB (Hughes et al., 2009). Instead, QseC activates a non‐cognate response regulator, KdpE, which in turn induces the expression of Ler, the main activator of the LEE loci. Finally, QseC also regulates the expression of a second TCS, QseEF, which is essential for pedestal formation and the formation of attaching and effacing (A/E) lesions (Reading et al., 2007). Indeed, as discussed in greater detail below, QseBC initiates a complex signaling cascade that integrates both positive and negative controls of EHEC virulence factors (Hughes et al., 2009; Pifer & Sperandio, 2014). In uropathogenic E. coli, deletion of qseC resulted in reduced bladder titers and decreased formation of intracellular bacterial communities in a mouse acute infection model (Kostakioti et al., 2009), and a decreased ability to establish chronic cystitis (Kostakioti et al., 2012). This was partially explained by a reduction in the production of the type 1 pilus, which led to decreased adherence and invasion of bladder epithelial cells. Restoration of type 1 pilus production in the qseC mutant increased adherence, invasion, and the ability to establish a chronic infection. However, the complemented strain still exhibited a fitness disadvantage relative to wild‐type (Kostakioti et al., 2012), suggesting that a functional QseC sensor is necessary for maintaining chronic uropathogenic E. coli infections.
The role of QseBC in virulence of Salmonella is less clear. Several studies have shown that catecholamine hormones enhance Salmonella virulence (Williams et al., 2006; Methner et al., 2008) but there is some controversy over whether QseBC mediates this process. In S. enterica serovar Typhimurium, qseBC was shown to be required for invasion of HeLa cells and for intracellular replication in J774 macrophages (Moreira et al., 2010; Moreira & Sperandio, 2012). In addition, a qseC mutant exhibited attenuated virulence in a mouse systemic infection model (Rasko et al., 2008; Moreira et al., 2010) and decreased colonization of the swine gastrointestinal tract (Bearson & Bearson, 2008). Similarly, a qseC mutant of S. enterica serovar Dublin was reported to be attenuated after oral infection of cattle (Pullinger et al., 2010b). In contrast, virulence of a qseC mutant of S. enterica serovar Typhimurium did not differ from wild‐type in a bovine ligated ileal loop model (Pullinger et al., 2010a). This inconsistency has not yet been fully explained but may arise from differences in the strains, media, assay systems and/or constructs used in these studies.
There is also differing evidence to correlate the function of qseBC and virulence in oral and respiratory pathogens. Using a mouse model of periodontitis, Novak et al. (2010) showed that after oral infection, wild‐type A. actinomycetemcomitans significantly induced the resorption of alveolar bone, one of the main clinical symptoms of periodontal disease in humans. In contrast, a qseC mutant was avirulent and bone loss in the group of mice infected with the mutant strain was indistinguishable from that in the sham‐infected controls. Complementation of the qseC mutant increased alveolar bone resorption back to wild‐type levels. Hence, these results clearly indicate that qseBC regulates A. actinomycetemcomitans virulence. Similar to A. actinomycetemcomitans, expression of the qseBC operon in Actinobacillus pleuropneumoniae is induced by both Ep and Ne (Li et al., 2012) and catecholamines induce several other virulence factors as well, including apxlA (encoding a toxin in the RTX family), pgaB (production of extracellular matrix carbohydrate) and APL_0443 (autotransporter adhesion). Inactivation of the qseBC operon also resulted in reduced expression of 17 Actinobacillus pleuropneumoniae genes including hugZ, encoding a putative heme/iron utilization protein, and pilM encoding a Tfp pilus assembly protein (Liu et al., 2015). PilM mediates adherence of Actinobacillus pleuropneumoniae to porcine lung cells and QseB was shown to directly bind to the pilM promoter (Liu et al., 2015). However, following intratracheal challenge, virulence of the ΔqseBC mutant did not significantly differ from wild‐type in a porcine pneumonia model (Liu et al., 2015). Interestingly, although pilM expression is reduced in the ΔqseBC strain, deletion of pilM resulted in a significant attenuation of virulence. Hence, qseBC regulates an essential virulence factor of Actinobacillus pleuropneumoniae but deletion of qseBC does not generate a detectable phenotype. One explanation for this may be that Actinobacillus pleuropneumoniae possesses redundant mechanisms that respond to catecholamine hormones and the loss of qseBC is complemented by other pathways. Alternatively, pilM expression may be reduced but not completely eliminated in the ΔqseBC mutant and it is possible that the basal level of pilM expression is sufficient to still allow Tfp pilus assembly to occur.
The QseBC TCS of the fish pathogen Eduardsiella tarda also responds to catecholamine hormones and is phylogenetically related to QseBC of EHEC and S. enterica. This TCS exhibits functional similarity to QseBC as well (Wang et al., 2011) and Eduardsiella tarda causes intramuscular infections exhibiting hemorrhagic necrotic lesions and suppurative abscesses, suggesting that the organism is capable of cell invasion and systemic spreading. Similar to EHEC, deletion of qseC or qseB significantly reduced the expression of flagellar genes fldH, fliA and motA and the mutated strain exhibited impaired motility. The qseC mutant also showed reduced intracellular survival in J774 macrophages. Replication of Eduardsiella tarda in macrophages is dependent on a TTSS and consistent with this, expression of the TTSS genes esaC and eseB were significantly downregulated in the mutated strain. Furthermore, the qseB and qseC mutants were attenuated in virulence by 8‐ to 16‐fold in a zebra fish infection model and were out‐competed by the wild‐type strain (Wang et al., 2011). Together, these results clearly indicate that the QseBC two TCS of Eduardsiella tarda also functions as a global regulator of virulence. Finally, the association of QseBC and virulence is not limited to organisms that infect animal hosts, but plant hosts as well. Pectobacterium carotovorum causes soft rot in a variety of crop plants including cabbage, onion, radish, and potato and a screen of a transposon library for mutations exhibiting reduced virulence in Chinese cabbage identified 14 different loci, including qseC (Lee et al., 2013). Subsequent characterization of the mutant showed that in addition to reduced virulence, the loss of QseC function resulted in an 80% reduction in biofilm growth. In addition, although motility is important for Pectobacterium carotovorum virulence (Cui et al., 2008), qseC did not regulate the expression of flagellar genes as in EHEC and the qseC mutant exhibited motility similar to the wild‐type strain.
QseBC regulons and signal cascades
Given the range of organisms that express qseBC, the broad scope of host organisms that these bacteria infect, and the different virulence strategies tha t are used by these pathogens, it is likely that the QseBC regulon and signaling cascade may be species‐specific. At present, the QseBC regulons for many of the organisms listed in Fig. 2 have not been thoroughly characterized. However, QseC‐regulated genes and its signaling cascade have been characterized in EHEC (see Fig. 4) and in the oral pathogen A. actinomycetemcomitans (see Fig. 5). This section will focus on comparing these systems as a model to highlight how the QseBC regulons and signaling cascades may differ.
Figure 4.
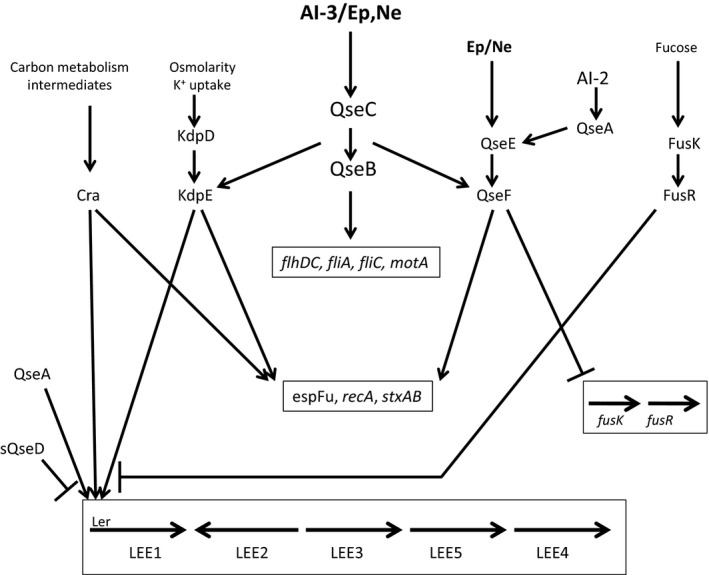
The QseC signaling cascade of enterohemorrhagic Escherichia coli. The QseC sensor is activated by autoinducer‐3 (AI‐3) and/or catecholamine hormones [epinephrine (Ep), norepinephrine (Ne)] and phosphorylates its cognate response regulator QseB and two non‐cognate response regulators, QseF and KdpE. Each of the response regulators induces the expression of specific sets of genes shown in boxes. The non‐cognate regulators QseF and KdpE can also be activated by their cognate sensors, QseE and KdpD, respectively. Like QseC, QseE is activated by Ep,Ne but it does not interact with AI‐3, and KdpD senses osmolarity and potassium. In addition, two quorum sensing‐regulated LysR‐type transcriptional regulatory proteins, QseA and sQseD, contribute to the regulation of the LEE locus and QseA also induces the expression of QseEF. FusKR senses fucose and functions to downregulate LEE expression and the expression of the fusKR operon itself can be downregulated by the QseF response regulator.
Figure 5.
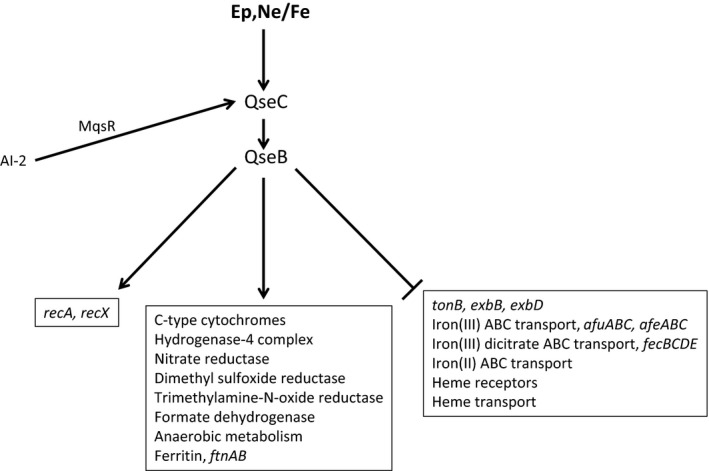
The QseC signaling cascade of Aggregatibacter actinomycetemcomitans. The QseC sensor is activated by both epinephrine (Ep) and norepinephrine (Ne), and iron and primarily signals through its cognate response regulator QseB to induce genes associated with anaerobic metabolism and energy production and downregulate genes encoding high‐affinity iron acquisition proteins (shown in boxes). Expression of qseBC is induced by autoinducer‐2 (AI‐2) and may involve the MqsR regulator, similar to Escherichia coli K‐12 MG1655.
Using a microarray that represented all of the genes in E. coli K‐12 MG1655, EHEC strains EDL933 and Sakai, uropathogenic E. coli strain CFT073, and 700 probes representing intergenic regions, Hughes et al. (2009) showed that when EHEC was grown in Luria–Bertani broth, 708 genes were upregulated and 126 genes were downregulated in a qseC mutant relative to wild‐type. Most of the differentially expressed genes represent genes that are in the core E. coli genome (Rasko et al., 2008) and are present in all pathovars. Many of these genes code for proteins with metabolic functions. However, 260 of the differentially expressed genes were specific for EHEC, indicating that the QseBC regulon differs among the E. coli pathovars. These results show that in Luria–Bertani medium, the majority of differentially expressed genes are repressed upon activation of QseC. In contrast, when EHEC was cultured under conditions that are conducive to virulence gene expression, e.g. in Dulbecco's modified Eagle's medium containing AI‐3, Ep, or a combination of both, the overall pattern of differentially expressed genes changed dramatically. Under these growth conditions in the presence of AI‐3, 106 genes were upregulated and 273 were downregulated, whereas in medium containing both AI‐3 and Ep, 70 genes were upregulated and 311 were downregulated. Hence, in Dulbecco's modified Eagle's medium, the expression of the majority of QseC‐regulated genes was induced. Included in the genes that were induced in a QseC‐dependent manner are the qseBC operon itself, flagellar genes (flhDC, fliA, fliC and motA) and the LEE1 operon which encodes ler, the transcriptional activator of the other operons in the LEE pathogenicity island. In addition, many genes encoding effectors that are translocated by the TTSS were induced by QseC as was stxAB, encoding Shiga toxin. The stxAB genes are late genes encoded by the λ‐bacteriophage and expressed during the lytic phase only after induction of the bacterial SOS response (Neely & Friedman, 1998). Consistent with this, QseC also induced recA expression. Initially, Hughes et al. (2009) presumed that the induction of the QseC‐regulated genes was mediated by the QseB response regulator and indeed, QseB was known to bind to the promoters of qseBC and flhDC, the master regulator of the flagella regulon (Clarke & Sperandio, 2005a,b). However, although QseB interacts with the flhDC promoter, deletion of qseB had no effect on motility and the ΔqseB strain expressed flagella at the same level as wild‐type. Hence, the phenotype exhibited by the qseC mutant differs from the strain in which its cognate response regulator was deleted. It has subsequently been demonstrated that in EHEC, QseB has dual regulatory functions that are dependent upon its state of phosphorylation. Non‐phosphorylated QseB functions to repress flhDC transcription by interacting with a site in the flhDC promoter that resides between nucleotides ‐300 and ‐650 whereas phospho‐QseB induces transcription by binding to sites located between nucleotides −650 to −950 and −300 and +50 (Hughes et al., 2009). In the absence of QseB, basal levels of flhDC expression (i.e. QseC independent) allow for production of flagella and motility whereas in the absence of QseC, QseB remains unphosphorylated and represses flhDC transcription.
Deletion of qseB also had no effect on the expression of ler or stxAB, suggesting that QseC‐dependent regulation of these genes may be indirect. Although it is generally believed that cross talk between a sensor kinase and a non‐cognate response regulator is rare, Yamamoto et al. (2005) showed that trans‐phosphorylation of non‐cognate response regulators can occur in vitro. Using a similar approach, Hughes et al. (2009) showed that QseC was capable of activating two non‐cognate response regulators, KdpE and QseF, and a functional role for these response regulators was demonstrated using gene deletion mutants. Deletion of kdpE reduced the expression of ler but had no effect on stxAB expression or motility and deletion of QseF reduced stxAB expression but had no effect on ler expression or motility. This suggests that QseC‐dependent regulation of ler and stxAB occurs via QseC‐mediated activation of the KdpE and QseF response regulators, respectively.
As shown in Fig. 4, activation of QseC by AI‐3 and/or Ep results in the phosphorylation of the QseB, KdpE and QseF response regulators, each of which induces the expression of distinct sets of virulence genes. QseF is also activated by its cognate sensor QseE. Expression of qseEF is stimulated by Ep in a QseC‐dependent manner but the QseE sensor itself is also activated by catecholamine hormones. However, QseE is not activated by AI‐3 (Reading et al., 2009). This central signaling cascade is also integrated with several additional signaling mechanisms. The expression of ler is repressed in a glycolytic environment and induced under gluconeogenic conditions and this regulation is mediated by the catabolite repressor/activator protein, Cra. KdpE and Cra each bind to distinct sites in the ler promoter and their binding is diminished under glycolytic conditions (Njoroge & Sperandio, 2012;). In addition, the interaction of KdpE and Cra facilitates binding to their respective sites in the ler promoter. Cra and KdpE also function coordinately to facilitate pedestal formation by EHEC and regulate the expression of espFu, an effector required for the formation of AE lesions (Njoroge et al., 2013). Finally, Cra may also play a role in the post‐transcriptional regulation of the LEE4 operon. Together, these results indicate that the sensing of carbon metabolites via Cra integrates with QseC signaling to control the expression of EHEC virulence genes. The functions of two additional LysR‐type transcriptional regulators are also integrated with QseBC signaling. As shown in Fig. 4, QseA is induced by AI‐2 and in turn it induces the expression of qseEF and also binds to the ler promoter to promote ler expression (Kendall et al., 2010). In addition, a second LysR‐type transcriptional regulator designated QseD is present in a truncated form in EHEC and functions to repress ler expression as deletion of this gene results in increased expression of all of the LEE operons (Habdas et al., 2010). QseD expression is repressed in a QseBC‐dependent manner. Finally, a fucose sensing system encoded by fusKR is also integrated with the QseBC cascade. Fucose is generated in the gut by cleavage from mucin by commensal organisms such as Bacteroides thetaiotaomicron. Activation of FusKR represses the expression of ler and other fucose utilization genes, which may allow EHEC to conserve energy by limiting virulence gene expression as it transits the mucus layer and also avoid competition with commensals for using fucose as a carbon source (Pacheco et al., 2012; Pacheco & Sperandio, 2015). Conversely, under conditions where virulence gene expression is required and the QseBC signaling cascade is activated, QseF functions to downregulate the expression of fusKR. Together, the QseBC cascade and the other signaling mechanisms that integrate with this pathway may function coordinately to allow EHECC to fine tune and optimize virulence gene expression.
As shown in Fig. 5, the QseBC regulon and signaling cascade of A. actinomycetemcomitans is significantly different from that of EHEC, which may reflect the different niches and virulence strategies that are used by these organisms. The A. actinomycetemcomitans genome does not code for a TTSS and the organism is non‐motile and lacks the flagellar apparatus. Hence, many of the QseC‐regulated virulence genes encoded by the LEE and flagellar loci of EHEC are not present in A. actinomycetemcomitans. Overall, 235 genes (> 11% of the genome) are differentially expressed when A. actinomycetemcomitans was grown in a chemically defined medium containing Ne and iron relative to cultures in the absence of signal (Weigel et al., 2015). Of these, 99 genes are induced and 136 are downregulated in a QseC‐dependent manner. In contrast to EHEC, none of the genes encoding the well‐characterized virulence factors of A. actinomycetemcomitans, e.g. the RTX leukotoxin, cytolethal distending toxin, tad fimbriae, autotransporter epithelial cell adhesins, EmaA, or the pga matrix biogenesis components were upregulated by QseBC. Instead, the majority of the induced genes encode proteins associated with anaerobic metabolism or respiration. This group includes electron transport components such as a hydrogenase complex and proteins involved in the reduction of nitrate, DMSO, trimethylamine‐N‐oxide, fumarate and formate. In addition, enzymes associated with the metabolism of aspartate, fumarate, malate, oxaloacetate, pyruvate, and formate were significantly induced upon activation of QseBC in A. actinomycetemcomitans. Novak et al. (2010) showed that qseBC was required for A. actinomycetemcomitans virulence but the results of Weigel et al. (2015) suggest that the shift in cellular metabolism and energy production that occurs upon activation of QseBC, rather than the direct regulation of virulence factors, may be the primary link between this TCS and A. actinomycetemcomitans virulence. Hence, the function of QseBC in A. actinomycetemcomitans may be to prime the organism to persist in an anaerobic host environment. Consistent with this, it is striking that many of the genes induced by the activation of QseBC were also identified by Jorth et al. (2013) as being induced during subcutaneous growth in vivo in a mouse abscess model relative to A. actinomycetemcomitans biofilm growth in vitro. Finally, the recA and recX genes are induced by QseBC, suggesting that the TCS may influence the SOS response in A. actinomycetemcomitans as well as EHEC.
The genes of the QseBC regulon that are downregulated encode a variety of metabolic functions but a significant number of these genes are associated with iron uptake (Weigel et al., 2015). In contrast, the expression of ftnAB encoding ferritin is strongly induced, indicating that activation of QseBC increases iron storage capacity and decreases high‐affinity acquisition of iron. Recent studies suggest that activated neutrophils, polymorphonuclear cells and macrophages release catecholamines and lactoferrin in response to inflammatory stimuli (Brown et al., 2003; Flierl et al., 2007, 2008, 2009) and catecholamines can function as pseudosiderophores capable of extracting iron from transferrin and lactoferrin (Freestone et al., 2000; Anderson & Armstrong, 2008; Bearson & Bearson, 2008; Sandrini et al., 2010). Hence, the inflamed subgingival pocket may be an iron‐replete environment and QseBC may play a dual role of priming cellular metabolism as well as allowing A. actinomycetemcomitans to detect and exploit the production of catecholamines by host cells to facilitate the acquisition of iron from lactoferrin or other host iron‐binding proteins during infection.
The QseBC signal cascade of A. actinomycetemcomitans shown in Fig. 5 is significantly less complex than what occurs in EHEC. There is no current evidence to suggest that A. actinomycetemcomitans produces AI‐3 (Weigel et al., 2015), so QseC may be activated only by a combination of catecholamines and iron. In A. actinomycetemcomitans, the expression of qseBC is also induced by AI‐2, which may occur via the MqsR regulator as described for E. coli K‐12 MG1655 by Gonzalez Barrios et al. (2006). In addition, A. actinomycetemcomitans does not possess genes encoding the qseEF or kdpDE TCS, so in contrast with EHEC, QseB is the only response regulator that is currently known to be phosphorylated by QseC. Similarly, A. actinomycetemcomitans also lacks genes encoding QseA, QseD and FusKR, indicating that the additional signaling pathways that integrate with the QseBC cascade in EHEC do not exist in this organism. It is possible that some genes in the A. actinomycetemcomitans QseBC regulon are acted upon indirectly by transcriptional regulators other than QseB, but these additional putative regulatory proteins have not yet been identified.
QseBC as a potential vaccine or therapeutic target
The initial identification of catecholamines as the activating signal for QseC rapidly led to reports that existing α‐ and/or β‐adrenergic antagonists functioned to inhibit QseC‐mediated signaling and suggested that QseC may represent a novel therapeutic target. To identify more potent inhibitors, Rasko et al. (2008) screened a library of 150,000 small organic molecules for candidates that were capable of reducing lacZ expression of a LEE1::lacZ reporter. Ultimately, 75 compounds were identified and one, designated LED209 (N‐phenyl‐4‐(3‐phenylthioureido)benzenesulfonamide) was chosen for further study. LED209 at a concentration of 5 pm was shown to inhibit binding of Ne by QseC and to abolish A/E lesion formation on cultured epithelial cells. However, the compound did not influence ligand binding or the function of QseE (Curtis et al., 2014), indicating that it is specific for QseC. In addition, LED209 did not affect the growth of EHEC, which is a desirable trait because without survival pressure, it is less likely that EHEC will develop resistance to the compound. Although it failed to reduce EHEC intestinal colonization of infant rabbits, oral administration in mice before and subsequent to intraperitoneal injection of a lethal dose of S. typhimurium resulted in increased survival and reduced recovery of viable bacteria from the spleens and livers of treated animals. Similarly, LED209 was shown to reduce F. tularensis virulence in a variety of in vitro and in vivo model systems (Rasko et al., 2008). Interestingly, LED209 was subsequently shown by Curtis et al. (2014) to function as a prodrug and is cleaved within the bacterial cell to generate the active component, which labels cytoplasmic lysine residues 256 and 427 of QseC. Substituting Arg for Lys256 or Lys427 rendered QseC inactive, confirming their functional importance. Presumably, the modification of these cytoplasmic residues alters the conformation of the periplasmic domain to prevent binding of Ep or Ne, but this has not yet been demonstrated experimentally. In addition, Lys256 and Lys427 are not conserved in the QseC proteins expressed by organisms in the family Pasteurellaceae, or in Serratia marsescens, Pantoea ananatis or B. psittacipulmonis. Indeed, in many of these organisms, the residue at position 256 is Arg, suggesting that these QseC proteins may be functionally distinct from EHEC and that LED209 may not be active against these bacteria. LED209 has also recently been conjugated with poly(amidoamine) dendrimers (PAMAM) to successfully improve their selectivity against Gram‐negative bacteria and reduce the cytotoxic activity of unconjugated PAMAM against mammalian cells (Xue et al., 2015).
Dean & van Hoek (2015) also conducted a small molecule screen to identify potential new therapeutics targeting QseC of Francisella novicida. A screen of 420 FDA‐approved drugs was conducted and three drugs, toremifene, chlorpromazine and maprotiline, were identified that inhibited QseC‐dependent formation of biofilms. Of these, toremifine and chlorpromazine had additional undesirable activities and were subsequently excluded and maprotiline was chosen for further study. Maprotiline significantly reduced the expression of virulence factor lglC encoded on the Francisella pathogenicity island and rescued wax worm larvae infected with F. novicida. Additional in vivo studies showed that treatment with maprotiline prolonged the time of disease onset and increased the overall survival of F. novicida‐infected mice. These results suggest that the FDA‐approved polycyclic antidepressant maprotiline may have additional use against Francisella infections by targeting the QseC sensor.
Plant secondary metabolites encompass a broad range of chemical scaffolds and are known to contribute to plant defense systems against bacterial, fungal and insect infections (Langenheim, 1994). Vikram et al. (2012) screened various limonoids derived from citrus species for anti‐biofilm and TTSS‐inhibitory activity of EHEC and identified five compounds that reduced EHEC biofilm formation, the most potent of which was isolimonic acid. This compound also reduced EHEC adhesion to Caco‐2 cells by approximately three‐fold without influencing cell viability. As the adherence of EHEC to epithelial cells requires a variety of QseC‐regulated factors, Vikram et al. (2012) subsequently examined the effect of isolimonic acid on the expression of flhC, ler and several other genes encoded by the LEE1 and LEE2 operons and showed that treatment of bacteria resulted in a 5‐fold to 12‐fold reduction of all virulence genes tested. Inhibition of gene expression and biofilm formation was subsequently shown to be dependent on QseBC signaling and to also require QseA; however, its mechanism of action remains to be determined. Nonetheless, isolimonic acid likely represents a lead compound for the development of additional, more potent agents that reduce EHEC virulence by targeting the QseBC signaling cascade. Finally, Chaudhari & Kariyawasam (2014) showed that treatment of avian macrophage‐like cells with purified recombinant QseC stimulated the expression of interferon‐γ, Toll‐like receptor‐4 and Toll‐like receptor‐15 and that conditioned medium from these cells reduced the expression of virulence genes of avian pathogenic E. coli O78. This suggests that QseC may induce host innate immune factors that downregulate the expression of important E. coli virulence factors. Hence, in addition to representing a viable therapeutic target, it is possible that QseC may also have utility as a potential subunit vaccine candidate. Additional functional characterization of QseBC signaling in E. coli, A. actinomycetemcomitans and other organisms may highlight additional similarities and differences in this TCS and should be informative to further develop new drug candidates and therapeutic approaches to circumvent the increase in antibiotic resistance and control microbial infections.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgements
The preparation of this manuscript was supported by National Institutes of Health grants DE014605 and DE023206.
References
- Anderson, M.T. and Armstrong, S.K. (2008) Norepinephrine mediates acquisition of transferrin‐iron in Bordetella bronchiseptica . J Bacteriol 190: 3940–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson, B.L. and Bearson, S.M. (2008) Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine‐enhanced growth. Microbes Infect 10: 807–816. [DOI] [PubMed] [Google Scholar]
- Brown, S.W. , Meyers, R.T. , Brennan, K.M. et al (2003) Catecholamines in a macrophage cell line. J Neuroimmunol 135: 47–55. [DOI] [PubMed] [Google Scholar]
- Chaudhari, A.A. and Kariyawasam, S. (2014) Innate immunity to recombinant QseC, a bacterial adrenergic receptor, may regulate expression of virulence genes of avian pathogenic Escherichia coli . Vet Microbiol 171: 236–241. [DOI] [PubMed] [Google Scholar]
- Clarke, M.B. and Sperandio, V. (2005a) Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli . Mol Microbiol 57: 1734–1749. [DOI] [PubMed] [Google Scholar]
- Clarke, M.B. and Sperandio, V. (2005b) Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol Microbiol 58: 441–455. [DOI] [PubMed] [Google Scholar]
- Clarke, M.B. , Hughes, D.T. , Zhu, C. , Boedeker, E.C. and Sperandio, V. (2006) The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103: 10420–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Yang, H. and Chatterjee, A.K . (2008) Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J Bacteriol 190: 4610–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.M. , Russel, R. , Moreira, C.G. et al (2014) QseC inhibitors as an antivirulence approach for Gram‐negative pathogens. MBio 5: e02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, S.N. and van Hoek, M.L. (2015) Screen of FDA‐approved drug library identifies maprotiline, an antibiofilm and antivirulence compound with QseC sensor‐kinase dependent activity in Francisella novicida . Virulence 6: 487–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl, M.A. , Rittirsch, D. , Nadeau, B.A. et al (2007) Phagocyte‐derived catecholamines enhance acute inflammatory injury. Nature 449: 721–725. [DOI] [PubMed] [Google Scholar]
- Flierl, M.A. , Rittirsch, D. , Huber‐Lang, M. , Sarma, J.V. and Ward, P.A. (2008) Catecholamines‐crafty weapons in the inflammatory arsenal of immune/inflammatory cells or opening pandora's box? Mol Med 14: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl, M.A. , Rittirsch, D. , Nadeau, B.A. et al (2009) Upregulation of phagocyte‐derived catecholamines augments the acute inflammatory response. PLoS ONE 4: e4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone, P.P. , Lyte, M. , Neal, C.P. , Maggs, A.F. , Haigh, R.D. and Williams, P.H. (2000) The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J Bacteriol 182: 6091–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone, P.P. , Sandrini, S.M. , Haigh, R.D. and Lyte, M. (2008) Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol 16: 55–64. [DOI] [PubMed] [Google Scholar]
- Ginalski, K. , Kinch, L. , Rychlewski, L. and Grishin, R.L. (2004) BOF: a novel family of bacterial OB‐fold proteins. FEBS Lett 567: 297–301. [DOI] [PubMed] [Google Scholar]
- Gonzalez Barrios, A.F. , Zuo, R. , Hashimoto, Y. , Yang, L. , Bentley, W.E. and Wood, T.K. (2006) Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum‐sensing regulator (MqsR, B3022). J Bacteriol 188: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckes, K.R. , Kostakioti, M. , Breland, E.J. et al (2013) Strong cross‐system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci U S A 110: 16592–16597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habdas, B.J. , Smart, J. , Kaper, J.B. and Sperandio, V. (2010) The LysR‐type transcriptional regulator QseD alters type three secretion in enterohemorrhagic Escherichia coli and motility in K‐12 Escherichia coli . J Bacteriol 192: 3699–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, D.T. , Clarke, M.B. , Yamamoto, K. , Rasko, D.A. and Sperandio, V. (2009) The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog 5: e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth, P. , Trivedi, U. , Rumbaugh, K. and Whiteley, M. (2013) Probing bacterial metabolism during infection using high‐resolution transcriptomics. J Bacteriol 195: 4991–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez‐Rodriguez, M.D. , Torres‐Escobar, A. and Demuth, D.R. (2013) ygiW and qseBC are co‐expressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology 159: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez‐Rodriguez, M.D. , Torres‐Escobar, A. and Demuth, D.R. (2014) Transcriptional regulation of the Aggregatibacter actinomycetemcomitans ygiW‐qseBC operon by QseB and integration host factor proteins. Microbiology 160: 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos, M.H. , Winzer, K. , Williams, P. and Khan, C.M. (2013) Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol Microbiol 87: 455–465. [DOI] [PubMed] [Google Scholar]
- Kendall, M.M. , Rasko, D.A. and Sperandio, V. (2010) The LysR‐type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 76: 1306–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti, M. , Hadjifrangiskou, M. , Pinkner, J.S. and Hultgren, S.J. (2009) QseC‐mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli . Mol Microbiol 73: 1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti, M. , Hadjifrangiskou, M. , Cusumano, C.K. , Hannan, T.J. , Janetka, J.W. and Hultgren, S.J. (2012) Distinguishing the contribution of type 1 pili from that of other QseB‐misregulated factors when QseC is absent during urinary tract infection. Infect Immun 80: 2826–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheim, J.H. (1994) Higher plant terpenoids: a phytocentric overview of their ecological roles. J Chem Ecol 20: 1223–1280. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Hiibel, S.R. , Reardon, K.F. and Wood, T.K. (2010) Identification of stress‐related proteins in Escherichia coli using the pollutant cis‐dichloroethylene. J Appl Microbiol 108: 2088–2102. [DOI] [PubMed] [Google Scholar]
- Lee, D.H. , Lim, J.A. , Lee, J. et al (2013) Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology 159: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Xu, Z. , Zhou, Y. et al (2012) Global effects of catecholamines on Actinobacillus pleuropneumoniae gene expression. PLoS One 7: e31121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Chen, Z. , Bei, W. et al (2015) Catecholamines promote Actinobacillus pleuropneumoniae growth by regulating iron metabolism. PLoS One 10: e0121887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Hu, L. , Xu, Z. et al (2015) Actinobacillus pleuropneumoniae two‐component system QseB/QseC regulates the transcription of PilM, an important determinant of bacterial adherence and virulence. Vet Microbiol 177: 184–192. [DOI] [PubMed] [Google Scholar]
- Merighi, M. , Septer, A.N. , Carroll‐Portillo, A. et al (2009) Genome‐wide analysis of the PreA/PreB (QseB/QseC) regulon of S. enterica serovar Typhimurium. BMC Microbiol. 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methner, U. , Rabsch, W. , Reissbrodt, R. and Williams, P.H. (2008) Effect of norepinephrine on colonisation and systemic spread of Salmonella enterica in infected animals: role of catecholate siderophore precursors and degradation products. Int J Med Microbiol 298: 429–439. [DOI] [PubMed] [Google Scholar]
- Moreira, C.G. and Sperandio, V. (2012) Interplay between the QseC and QseE bacterial adrenergic sensor kinases in Salmonella enterica serovar Typhimurium pathogenesis. Infect Immun 80: 4344–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, C.G. , Weinshenker, D. and Sperandio, V. (2010) QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun 78: 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, C.G. , Herrera, C.M. , Needham, B.D. et al (2013) Virulence and stress‐related periplasmic protein (VisP) in bacterial/host associations. Proc Natl Acad Sci U S A 110: 1470–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely, M.N. and Friedman, D.I. (1998) Functional and genetic analysis of regulatory regions of coliphage H‐19B: location of shiga‐like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol 28: 1255–1267. [DOI] [PubMed] [Google Scholar]
- Njoroge, J. and Sperandio, V. (2012) Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun 80: 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge, J.W. , Gruber, C. and Sperandio, V. (2013) The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli . J Bacteriol 195: 2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, E.A. , Shao, H. , Daep, C.A. and Demuth, D.R. (2010) Autoinducer‐2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans . Infect Immun 78: 2919–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco, A.R. and Sperandio, V. (2015) Enteric pathogens exploit the microbiota‐generated nutritional environment of the gut. Microbiol Spectr 3(3):MBP‐0001‐2014. doi: 10.1128/microbiolspec.MBP‐0001‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco, A.R. , Curtis, M.M. , Ritchie, J.M. et al (2012) Fucose sensing regulates bacterial intestinal colonization. Nature 492: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifer, R. and Sperandio, V . (2014) The interplay between the microbiota and enterohemorrhagic escherichia coli . Microbiol Spectr. 2(5):EHEC‐0015‐2013. [DOI] [PubMed] [Google Scholar]
- Pullinger, G.D. , Carnell, S.C. , Sharaff, F.F. et al (2010a) Norepinephrine augments Salmonella enterica‐induced enteritis in a manner associated with increased net replication but independent of the putative adrenergic sensor kinases QseC and QseE. Infect Immun 78: 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullinger, G.D. , van Diemen, P.M. , Dziva, F. and Stevens, M.P. (2010b) Role of two‐component sensory systems of Salmonella enterica serovar Dublin in the pathogenesis of systemic salmonellosis in cattle. Microbiology 156: 3108–3122. [DOI] [PubMed] [Google Scholar]
- Rasko, D.A. , Rosovitz, M.J. , Myers, G.S. et al (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190: 6881–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading, N.C. , Torres, A.G. , Kendall, M.M. , Hughes, D.T. , Yamamoto, K. and Sperandio, V. (2007) A novel two‐component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol 189: 2468–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading, N.C. , Rasko, D.A. , Torres, A.G. and Sperandio, V. (2009) The two‐component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci U S A 106: 5889–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrini, S.M. , Shergill, R. , Woodward, J. et al (2010) Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J Bacteriol 192: 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V.K. and T.A. Casey. T.A. (2014) Determining the relative contribution and hierarchy of hha and qseBC in the regulation of flagellar motility of Escherichia coli O157:H7. PLoS One 9: e85866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio, V. , Mellies, J.L. , Nguyen, W. , Shin, S. and Kaper, J.B. (1999) Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic. Proc Natl Acad Sci U S A 96: 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio, V. , Torres, A.G. and Kaper, J.B. (2002) Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two‐component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli . Mol Microbiol 43: 809–821. [DOI] [PubMed] [Google Scholar]
- Sperandio, V. , Torres, A.G. , Jarvis, B. , Nataro, J.P. and Kaper, J.B. (2003) Bacteria‐host communication: the language of hormones. Proc Natl Acad Sci U S A 100: 8951–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, K.H. , O'Connor, L.H. , Burpo, N. , Kohler, K. and Johnston, J.W. (2012) Characterization of a ferrous iron‐responsive two‐component system in nontypeable Haemophilus influenzae . J Bacteriol 194: 6162–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal, C.M. , Singh, B. , Fleury, C. et al (2012) QseC controls biofilm formation of non‐typeable Haemophilus influenzae in addition to an AI‐2‐dependent mechanism. Int J Med Microbiol 302: 261–269. [DOI] [PubMed] [Google Scholar]
- Vikram, A. , Jesudhasan, P.R. , Pillai, S.D. and Patil, B.S. (2012) Isolimonic acid interferes with Escherichia coli O157:H7 biofilm and TTSS in QseBC and QseA dependent fashion. BMC Microbiol 12: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, M. , Sircili, M.P. and Sperandio, V. (2006) AI‐3 synthesis is not dependent on luxS in Escherichia coli . J Bacteriol 188: 5668–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wang, Q. , Yang, M. et al (2011) QseBC controls flagellar motility, fimbrial hemagglutination and intracellul ar virulence in fish pathogen Edwardsiella tarda . Fish Shellfish Immunol 30: 944–953. [DOI] [PubMed] [Google Scholar]
- Weigel, W.A. , Demuth, D.R. , Torres‐Escobar, A. and Juarez‐Rodrigues, M.D. (2015) Aggegatibacter actinomycetemcomitans QseBC is activated by catecholamines and iron and regulates genes encoding proteins associated with anaerobic respiration and metabolism. Mol Oral Microbiol. 30: 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.H. , Rabsch, W. , Methner, U. , Voigt, W. , Tschape, H. and Reissbrodt, R. (2006) Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 24: 3840–3844. [DOI] [PubMed] [Google Scholar]
- Wolfe, A.J. (2005) The acetate switch. Microbiol Mol Biol Rev 69: 12–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X.Y. , Mao, X.G. , Li, Z. et al (2015) A potent and selective antimicrobial poly(amidoamine) dendrimer conjugate with LED209 targeting QseC receptor to inhibit the virulence genes of Gram negative bacteria. Nanomedicine 11: 329–339. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. and Inouye, M. (2009) mRNA interferases, sequence‐specific endoribonucleases from the toxin‐antitoxin systems. Prog Mol Biol Transl Sci 85: 467–500. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K. , Hirao, K. , Oshima, T. , Aiba, H. , Utsumi, R. and Ishihama, A. (2005) Functional characterization in vitro of all two‐component signal transduction systems from Escherichia coli . J Biol Chem 280: 1448–1456. [DOI] [PubMed] [Google Scholar]
- Yang, K. , Meng, J. , Huang, Y.C. et al (2014) The role of the QseC quorum‐sensing sensor kinase in epinephrine‐enhanced motility and biofilm formation by Escherichia coli . Cell Biochem Biophys 70: 391–398. [DOI] [PubMed] [Google Scholar]


