Abstract
Neglected tropical diseases caused by parasitic nematodes inflict an immense health and socioeconomic burden throughout much of the developing world. Current estimates indicate that more than two billion people are infected with nematodes, resulting in the loss of 14 million disability‐adjusted life years per annum. Although these parasites cause significant mortality, they primarily cause chronic morbidity through a wide range of severe clinical ailments. Treatment options for nematode infections are restricted to a small number of anthelmintic drugs, and the rapid expansion of anthelmintic mass drug administration raises concerns of drug resistance. Preservation of existing drugs is necessary, as well as the development of new treatment options and methods of control. We focus this review on how the democratization of CRISPR/Cas9 genome editing technology can be enlisted to improve our understanding of the biology of nematode parasites and our ability to treat the infections they cause. We will first explore how this robust method of genome manipulation can be used to newly exploit the powerful model nematode Caenorhabditis elegans for parasitology research. We will then discuss potential avenues to develop CRISPR/Cas9 editing protocols in filarial nematodes. Lastly, we will propose potential ways in which CRISPR/Cas9 can be used to engineer gene drives that target the transmission of mosquito‐borne filarial nematodes.
Keywords: CRISPR, filariasis, gene drives, mosquito, neglected tropical diseases
Abbreviations
- Cas9
CRISPR associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeats
- GPCRs
G protein‐coupled receptors
- HDR
homology directed repair
- LF
lymphatic filariasis
- MCR
mutagenic chain reaction
- mf
microfilariae
- NHEJ
non‐homologous end joining
- NTD
neglected tropical disease
- PAM
protospacer adjacent motif
Introduction
Neglected tropical diseases (NTDs) are a collection of communicable diseases that challenge global human health and perpetuate poverty in economically deprived areas of the world 1. Parasitic nematodes (roundworms) account for the greatest share of the overall neglected disease burden. Over two billion humans are currently infected with nematodes 2, causing upwards of 14 million disability‐adjusted life years (DALYs) lost per annum 3. Medically important nematode parasites that have been prioritized for elimination include vector‐borne filarial parasites and soil‐transmitted helminths (STHs) 4. Although these parasites cause significant mortality, their primary damage is exacted through chronic morbidity and a wide range of severe clinical manifestations that include physical disfigurement, blindness, anemia, persistent inflammation and pain, fatigue, malnutrition, tissue damage, and childhood stunting 5, 6, 7.
Efforts to control these devastating pathogens rely mostly on the distribution of anti‐parasitic drugs (anthelmintics) in endemic areas 8. The number of anthelmintics currently available for clinical use is disconcertingly small, and attempts to develop vaccines that provide immunity against parasitic nematodes have been largely unsuccessful. The likely emergence of anthelmintic resistance seriously threatens future nematode parasite disease control in human populations 9, 10, 11, 12, 13, 14, 15, 16. This threat is underscored by the rapid development and widespread presence of drug‐resistant nematode populations in veterinary medicine following mass drug administration (MDA) 17, 18, 19. Therefore, we have significant motivation to protect the efficacy of existing treatments and to develop new treatments that target human parasitic nematodes 20, 21, 22. We focus this review on how the democratization of CRISPR/Cas9 genome editing technology can be enlisted to improve our understanding of the biology of nematode parasites and our ability to treat the infections they cause. We will first explore how this robust method of genome manipulation can be used to newly exploit the powerful model nematode Caenorhabditis elegans for parasitology research. We will then discuss potential avenues to develop CRISPR/Cas9 editing protocols in filarial nematodes. Lastly, we will propose potential ways in which CRISPR/Cas9 can be used to engineer gene drives that target the transmission of mosquito‐borne filarial nematodes.
Caenorhabditis elegans as a model for parasitic nematodes
The hermaphroditic free‐living nematode C. elegans has long been established as a highly tractable model system for the study of metazoan biology 23. Many fundamentally important biological discoveries were enabled by the experimental tractability of this model organism. Caenorhabditis elegans is easy to maintain in the laboratory and has a short life cycle (∼ 3.5 days) where it progresses from embryo through four larval stages (L1–L4) before reaching adulthood. Transitions out of each larval stage occur via turnover or ‘molting’ of the cuticular hypodermis. Adverse environmental conditions can elicit developmental arrest at the second molt, referred to as the dauer stage. The adult animal contains 959 somatic nuclei with completely defined cell lineages 24, and the 100 Mb C. elegans genome 25 is fully sequenced and exceptionally well annotated in comparison with other multicellular species.
By contrast, parasitic nematodes have complex life cycles and a severely limited genetic and molecular toolkit for direct experimentation. The phylogenetic proximity of C. elegans to human, animal, and plant parasitic nematodes has encouraged the use of this model system for a range of medically and agriculturally important parasites 26, 27, 28, 29. Despite the considerable diversity that exists between and among free‐living and parasitic nematodes, we know of appreciable conservation of life‐cycle structure, gene content, and physiology between C. elegans and parasitic nematodes. In fact, it is hypothesized that the dauer stage of free‐living nematodes is a pre‐adaptation to the third infective larval stage (L3) in parasitic nematodes 30. Historically, C. elegans has been indispensable to the task of understanding modes of action (MOA) for the major classes of anthelmintics. Molecular targets for the macrocyclic lactones, nicotinic agonists, and benzimidazoles were first characterized using genetic approaches in C. elegans 31, 32, 33. These mechanisms are directly relevant to drug activity in parasitic nematodes. Caenorhabditis elegans has been used to validate specific genetic variants that can give rise to anthelmintic resistance 34, 35 and to express and functionally characterize parasite transgenes 34, 36. However, C. elegans has thus far been ineffective as a screening tool for the discovery of new anthelmintics using whole‐organism phenotypic screens 37. We posit that new developments in C. elegans research and advances in parasite genomics can help overcome many of these limitations and further the utility of the C. elegans model system where it has already been proved to be useful.
Helminth genomic and transcriptomic resources have quickly scaled in both quantity and quality since the publication of the first parasitic nematode genome over a decade ago 38. New parasite draft genomes are being assembled and annotated at an accelerated pace 39, and these resources allow for more accurate appraisals of genome synteny and sequence conservation among nematodes. Knowledge of variation among different species can help to identify gene families and pathways that are best conserved between C. elegans and different parasitic nematodes, as well as unique features of parasite genomes that are more likely associated with parasitic adaptations to host environments. Additionally, knowledge of variation 40 within the C. elegans species can be used to investigate the genetic basis for complex nematode traits. The natural genotypic and phenotypic diversity that exists among wild C. elegans strains remains a new and largely untapped resource in parasitology. This natural variation can help uncover genetic determinants of drug action 41, place gene structure–function analyses in an evolutionary context, and improve upon C. elegans drug screening pipelines that depend on phenotypes elicited in a single laboratory genetic background. These nematode genome diversity data and the timely development of high‐throughput phenotyping assays 42, 43, 44 and CRISPR/Cas9 genome editing 45 combine to greatly strengthen the future translational potential of C. elegans for parasitology.
CRISPR/Cas genome editing in C. elegans as a platform to study parasite biology
The CRISPR/Cas9 genome editing platform arose from the dissection of adaptive immunity systems in bacteria and archae 46, 47, 48, 49. In type II systems, clustered regularly interspaced short palindromic repeat (CRISPR) arrays are transcribed and processed into CRISPR‐RNAs (crRNAs) that interact with trans‐activating CRISPR RNA (tracrRNA) and a CRISPR‐associated protein (Cas) endonuclease to target foreign nucleic acids 50, 51. The minimal components of this RNA‐guided DNA endonuclease system form the basis of a powerful molecular biology toolkit for eukaryotic genome engineering. Site‐specific genome editing can be achieved with the simple delivery of a ‘guide’ RNA (gRNA) and Streptococcus pyogenes Cas9 protein 52, 53, 54. gRNAs contain 17–20 nucleotides of a target genome sequence and are synthetically fused to an RNA scaffold that assumes the role of tracrRNA. The targeted genome region must occur immediately upstream of a proto‐spacer adjacent motif (PAM) (5′‐NGG‐3′) 47. gRNAs direct Cas9 nuclease activity to specific genomic loci via Watson–Crick base pairing, inducing double‐stranded breaks (DSBs) and activation of DNA repair mechanisms. In eukaryotic cells, a DSB will lead to heterogenous insertion/deletion (indel) polymorphisms through the non‐homologous end joining (NHEJ) pathway. Alternatively, DNA repair templates with homology flanking the DSB location can be used to introduce specific mutations through the homology‐directed repair (HDR) pathway.
The establishment of type II CRISPR/Cas9 DNA cleavage in vitro 52, 55 and in mammalian cells 53, 54, 56, 57 has led to the rapid development of this genome editing mechanism in other heterologous systems. The ability to simply and precisely edit genomes is rapidly transforming the landscape of basic and translational biomedical science research. In model organism systems, easily adaptable genome editing protocols have opened up exciting new research possibilities. A number of CRISPR/Cas9 genome editing protocols have been established in C. elegans 45, including co‐CRISPR 58, co‐conversion 59, bacterial feeding 60, SapTrap 61, and self‐excising cassettes 62. These protocols can be used to efficiently introduce genome modifications through both the NHEJ and HDR pathways. Importantly, CRISPR/Cas9 editing in C. elegans opens new doors to studying the biology of closely related nematode parasites. Genome editing in this model nematode system can build upon past approaches and lead to a better elucidation of parasite gene function, understanding of parasite cellular and physiological pathways, and a grasp of anthelmintic MOA and resistance.
CRISPR/Cas9 can be used to introduce nematode parasite genes into the experimentally tractable and anatomically and physiologically relevant C. elegans model system. Studies have shown that C. elegans knock‐out strains can be functionally rescued with transformation of parasite transgenes 34, 36, 63. The precise insertion of parasite genetic information into the C. elegans genome using HDR templates provides significant advantages over transformation approaches 64 resulting in large extra‐chromosomal transgene arrays that require selection to maintain or randomly integrated transgenes that can result in deleterious gene disruption. CRISPR/Cas9 theoretically avoids these complications and allows for site‐directed conservation of genome synteny and context, as well as greater control of the spatiotemporal aspects of gene expression. In cases of one‐to‐one orthology between parasitic nematode and C. elegans genes, we have a higher likelihood of functional parasite gene expression in C. elegans. Inferred evolutionary relationships among helminths and free‐living nematodes can help guide identification of orthologous and paralogous gene groupings. Comparative transcript localization can also help filter for parasite genes suitable for heterologous expression. Entire parasite genes, gene domains, or regulatory elements can be swapped into orthologous sites in the C. elegans genome (Fig. 1). Successful expression of parasite genes or chimeric parasite‐C. elegans genes can be coupled to assays of protein function with convenient reporters (e.g., changes in fluorescence or electrophysiological outputs) and phenotypic detection schemes. CRISPR/Cas9 HDR templates can be used to introduce specific de novo variants and to carry out scanning mutagenesis to probe parasite protein structure–function relationships in phylogenetically related cell types. Optimized endpoint assays of parasite protein activity in C. elegans cells and tissues provide a direct pathway to drug and vaccine target validation, as well as new target‐based compound screening approaches for anthelmintic discovery. Mutations in potentially druggable parasite transgenes that lead to stark and deleterious nematode phenotypes can be used to prioritize targets for in vivo screening. Screens can be engineered to select for pharmacological agents that phenocopy both loss‐of‐function or gain‐of‐function mutant alleles. Given that most anthelmintics are ion channel agonists, it would be advantageous to pair expression schemes with assays capable of detecting both agonist 65 or antagonist activity.
Figure 1.
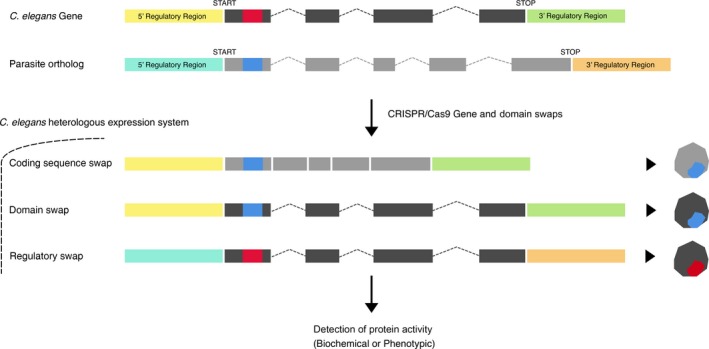
CRISPR‐mediated structure–function studies of parasite genes in C. elegans. CRISPR/Cas9 HDR templates can be used to precisely place parasite genetic information into the genomic and physiological context of C. elegans. Entire parasite genes, coding sequences, functional domains, or regulatory elements can be swapped into orthologous sites in the C. elegans genome. Heterologous expression can be interfaced with established biochemical or phenotypic endpoints of protein activity.
It is likely that some parasite genes that lack C. elegans orthologs have unique functions that cannot be reconstituted in the surrogate C. elegans expression framework. Parasite proteins may not express or behave correctly in the absence of required complements of chaperones and other accessory proteins, signaling proteins, post‐translational modifiers, or other parasite‐specific molecules. Further, it will be unclear as to where to express parasite genes with no obvious C. elegans homolog. In these cases, expression in nematode cells could still provide certain advantages over using surrogate cell culture systems derived from other phyla. It is reasonable to expect that parasite‐specific proteins will better approximate native activity in nematode cell types that categorically resemble their original environment (e.g., neuronal versus muscle cells). However, measuring this activity might necessitate the co‐expression of parasite accessory and signaling proteins, parasite substrates upon which the protein enzymatically acts or chimeric proteins that can divert signaling or activity to existing C. elegans pathways 66. The development of robust assays that can directly measure parasite protein activity is a potential bottleneck and will require different solutions for different proteins. Where functional expression of a parasite gene proves difficult in the canonical N2 laboratory strain, it will be worthwhile to consider alternative strains within the C. elegans wild isolate collection.
Another arena where this toolkit will be immediately useful is the study of anthelmintic MOA and mechanisms of resistance. Conserved anthelmintic targets in C. elegans can be replaced by wild‐type and mutant parasite orthologs. Measurements of anthelmintic responses in ‘parasitized’ strains can be used to uncover new determinants of drug action and to elucidate the number and locations of potential variants that can confer resistance to a given anthelmintic. In cases where the canonical N2 laboratory‐adapted strain does not elicit a noticeable drug‐exposure phenotype, wild C. elegans isolates that are sensitive to a given drug can be used for expression. The tractability of the C. elegans platform for parasite gene or protein expression will likely reveal both conserved and species‐specific mechanisms of anthelmintic resistance and MOA.
Extending genome‐editing technology to filarial parasites
The genetic toolkit available for use in parasitic nematodes is notably underdeveloped. RNA interference (RNAi) is stage‐restricted and unreliable in nearly all helminth species where it has been reported 67, 68. Although progress has been made in establishing parasite transgenesis, only a single example of stable germline transgenesis has been published 69. The ability to directly edit the nuclear genomes of parasites in an efficient and heritable manner would be a giant leap forward in helminthology. We have a strong basis for attempting this feat in the soil‐transmitted and filarial nematode species for which experimental techniques and culture systems are the furthest developed. A number of avenues for the translation of CRISPR/Cas9 technology from C. elegans to Strongyloides spp., Ascaris suum, Brugia malayi, and Haemonchus contortus have been recently outlined 70, 71. We build upon these ideas and focus on establishing heritable genome editing in the filarial nematode B. malayi, a causative agent of human lymphatic filariasis (LF).
Lymphatic filariasis is a mosquito‐borne NTD caused by thread‐like parasitic nematodes. Over 120 million humans are infected with LF in 81 endemic countries, and an estimated 1.3 billion people live at active risk of infection 72. Of those infected with LF, 40 million are clinically symptomatic and suffer from severe disfigurement and physical incapacitation. The World Health Organization (WHO) Global Program to Eliminate Lymphatic Filariasis (GPELF) aims to eradicate the debilitating socioeconomic burden of this disease through expanded MDA of anthelmintics 73. Although this approach is a necessary and important course of action, increased drug selection pressures will undoubtedly also increase the odds that anthelmintic resistance will develop and spread in parasitic nematode populations. Therefore, it is important to develop new strategies and tools to complement and diversify the current approach to LF control. CRISPR/Cas9 technology offers one powerful approach to experimentally dissect filarial nematode biology better.
Filarial parasites have a complex life cycle that involves an intermediate mosquito vector and a definitive mammalian host (Fig. 2). Laboratory‐adapted strains of B. malayi can be maintained using the mosquito species Aedes aegypti and jird or cat models of infection 74. Delivery of the CRISPR/Cas9 machinery to the parasite germline is a requisite for establishing heritable genome editing in B. malayi. Delivery and expression of promoter‐driven DNA reporter constructs has been achieved through biolistic transformation of isolated intrauterine embryos and infective larval stage (L3) parasites and through the biolistic transformation and microinjection of adult female parasites in culture 75, 76. However, these mechanical delivery methods result in damaged embryos that cannot be propagated. Intraperitoneal chemical delivery of DNA to L3 parasites within a naïve jird host can lead to the recovery of developmentally competent parasites but with low transfection efficiency 77. Although transgenes can be maintained extrachromosomally, stable germline integration of transgenes has not yet been achieved in B. malayi. Brugia RNAi can be carried out via ex vivo soaking of L3 78 or adult parasites 79, 80, electroporation of adult females 80, and intra‐mosquito delivery to larval stages 81. Further optimization of these methods of ex vivo and in vivo transfection could lead to the successful delivery of CRISRP/Cas9 nucleic acid constructs or ribonucleoprotein (RNP) complexes to Brugia without loss of parasite viability. Alternative chemical transfection reagents 82, 83, 84, nanoparticle or viral delivery approaches 85, 86, and potentially less disruptive forms of membrane deformation 87 can be considered for different parasite life stages. The transfection of active CRISPR constructs into isolated embryos, adult‐contained embryos, or microfilariae (mf) would potentially improve germline access and provide selection checkpoints that occur on reasonable experimental timescales both before and after development within the mosquito host.
Figure 2.
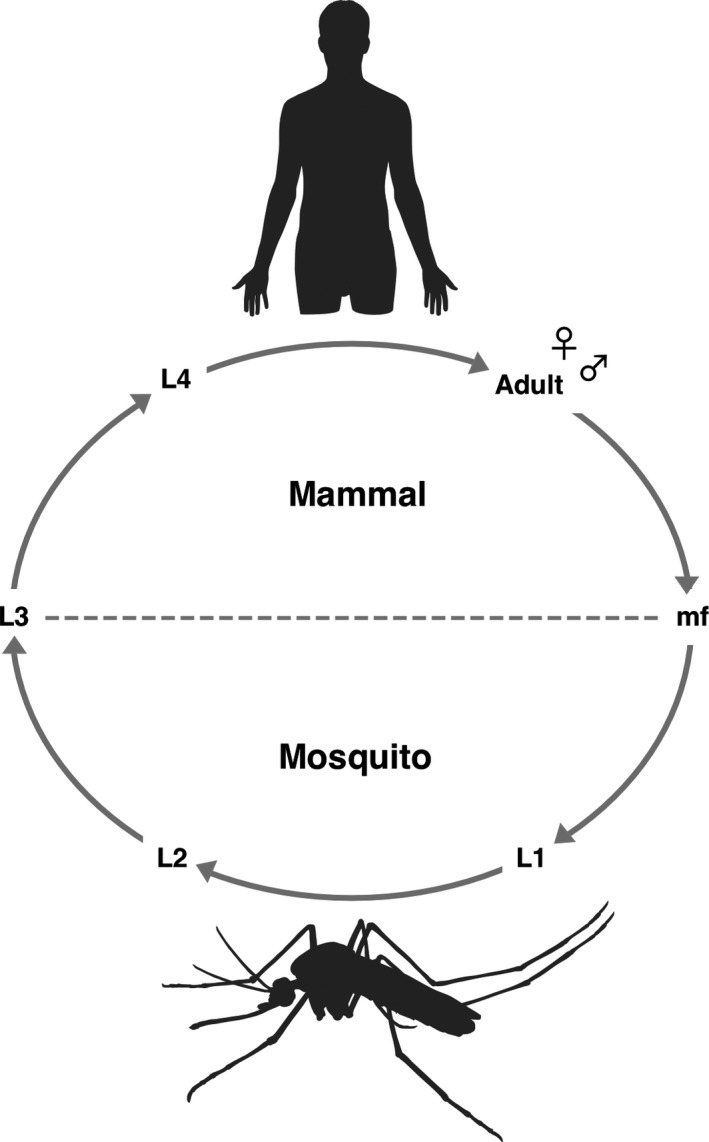
Life cycle of the causative parasites of human LF. Sheathed mf are taken up by mosquitoes in the form of a blood meal from an infected human host. Ingested mf undergo molting and development within the mosquito and are deposited as infective larvae onto human skin during a blood meal. The larvae penetrate skin through the mosquito bite wound and migrate to the lymphatics, where they develop into male and female adult parasites. Female parasites release large numbers of mf into host circulation to complete the life cycle. The entire life cycle of B. malayi can be maintained in the laboratory using the Aedes aegypti black‐eyed Liverpool strain as an intermediate vector and Mongolian jirds (Meriones unguiculatus) (or cats) as definitive hosts. Mosquitoes can be fed microfilaria‐infected blood and infective stage larvae can be dissected from mosquitoes and used to establish mammalian infection through either intraperitoneal or subcutaneous routes.
CRISPR editing via nucleic acid constructs in Brugia will require identification of suitable RNA polymerase II promoters for Cas9 expression and U6 promoters for gRNA expression. Candidate promoters can likely be identified using bioinformatics analyses of existing sequence data. It is possible that non‐native RNA polymerase III promoters will also be sufficiently active 88. Delivery of Cas9 protein and in vitro transcribed gRNAs does not require knowledge of promoter activity. Even with efficient delivery of CRISPR constructs, it is likely that editing events will be rare, often heterozygous, and occur in small mosaic subsets of the soma and germline of individual parasites. Therefore, enrichment for intended genome editing events will require development of transformation markers and selection strategies. Work done in C. elegans illuminates potential routes of achieving this aim. HDR templates could be used to knock‐in fluorescent reporter proteins that can be interfaced with automated methods of optical selection. The validation of mutant alleles that confer dominant phenotypes and that do not interrupt parasite developmental competence could provide a more laborious method of phenotypic selection. Resolving stage‐specific parasite responses to various antibiotics and anthelmintics could lead to the use of drug resistance genes or alleles that can be used for ex vivo and in vivo transformant selection. Care will have to be taken to understand the confounding effects of antibiotics and anthelmintic metabolites on the Wolbachia endosymbiont of B. malayi 89, 90. Selected transformants can be followed by bulk mating within a jird host for detection of progeny with germline homozygous edits.
An entirely different approach to conversion of low frequency heterozygous editing events is through a limited RNA‐guided parasite gene drive. CRISPR/Cas9 gene drives are discussed for mosquito vectors in the following section, but the same principles can potentially be applied to drive edited loci through a population of laboratory parasites. Once established, CRISPR/Cas9 in filarial and other parasitic nematode species can be used to validate drug and vaccine targets, directly test alleles that confer anthelmintic resistance, and even potentially to engineer non‐pathogenic parasites for protection against subsequent infection or for use in helminthic therapies 91.
Engineered CRISPR/Cas9 gene drives for control of lymphatic filariasis
Gene drive systems are selfish genetic elements that spread themselves throughout populations. These genetic elements depend on a variety of fascinating mechanisms to bias inheritance in their favor thereby ensuring their transmission to progeny at frequencies that exceed expectations of Mendelian independent assortment. Gene drive systems found in nature include homing endonuclease genes (HEGs) 92, autosomal and sex‐linked meiotic drives 93, and heritable bacteria 94. HEGs copy themselves to the homologous chromosome using site‐specific endonuclease activity and HDR. Meiotic drive elements disable or destroy homologous chromosomes at loci heterozygous for the drive element. Strains of the maternally inherited bacteria Wolbachia promote their transmission through cytoplasmic incompatibility. The study of these and other systems led to a recognition that natural gene drive mechanisms could be harnessed to artificially manipulate wild populations through the engineered spread of genetic traits 95, 96, 97, 98, 99, 100. Synthetic gene drives are currently the subject of intense research activity and significant public interest as a potential means to combat major ecological and medical challenges.
Importantly, synthetic gene drives represent a potential pathway to control or eradicate vector‐borne protozoal, viral, and helminth diseases that significantly challenge global human health. Synthetic gene drives that have been proposed for vector‐based disease control distort genetic inheritance patterns in wild vector populations to favor transmission of alleles that lead to two specific goals: population suppression or population modification 96, 100. In population suppression drives, the goal is to promote eventual population crash of the disease‐carrying vectors. Eradication of the mosquito vector is an example of a population suppression gene drive for the control of filarial nematodes. In population modification drives, the goal is to rapidly propagate alleles that confer pathogen resistance or prevent pathogen transmission from vector to definitive host. Any gene drive that makes mosquito vectors more recalcitrant to filarial nematode infection or reduces the rate of parasite transmission to humans would constitute a population modification gene drive. Although natural gene drive mechanisms have inspired the current conceptual framework for synthetic gene drives 97, robust adaptation of these systems 98, 99 has proved slow and difficult. CRISPR/Cas9 presents an immediate pathway to overcoming many of the technical challenges that have hampered previous efforts to design and to implement synthetic gene drives. Theoretical implementations of CRISPR/Cas9 gene drives, as well as concerns relating to gene drive efficacy, safety, and ecological impact have been reviewed elsewhere 101, 102, 103.
Here, we will focus our attention on potential routes and challenges to enlisting CRISPR/Cas9 gene drives to combat mosquito‐borne filarial nematode infections. The prospects of targeting human pathogenic nematodes through mosquito genetic control are promising given the recent demonstration of the CRISPR/Cas9 mutagenic chain reaction (MCR) in Drosophilamelanogaster 104 and the extension of this homing‐like method to achieve proof‐of‐concept gene drives in malaria mosquito vectors. MCR allows for autocatalytic CRISPR/Cas9‐mediated conversion of heterozygous loci into homozygous loci. MCR was used to spread antimalarial effector genes in Anopheles stephensi, compromising the ability of this vector to transmit malaria 105. Additionally, MCR was employed to spread recessive female sterility genes in the principal Plasmodium falciparum vector Anopheles gambiae, compromising the reproductive potential of this important vector 106. These efficient RNA‐guided homing drives provide elegant examples of both population modification and population suppression approaches for targeting Plasmodium transmission, suggesting powerful new avenues for filarial nematode control. Malaria parasites and filarial nematode parasites have sometimes overlapping geographic distributions and are transmitted by common mosquito species in the genus Anopheles. The utility of CRISPR/Cas9 gene drives for mosquito‐based filariasis control is worthy of more detailed exploration of the parasite biology with specific recognition of the challenges unique to these multicellular parasites. Additionally, these techniques offer a novel strategy to eradicate LF in wild mosquito populations that are responsible for filarial nematode transmission.
The etiological agents of LF in humans are the filarial nematodes Wuchereria bancrofti, Brugia malayi, and Brugia timori. These parasites can be transmitted by a diverse range of mosquito species in the genera Aedes, Anopheles, Culex, and Mansonia. In competent mosquitoes, human filarial parasites are ingested as mf in a blood meal and travel through the midgut and hemocoel to arrive at the thoracic muscles, the primary site of development. In this phase of development, first larval‐stage parasites complete two molts and emerge as third larval stage parasites. The larvae migrate to the mosquito head and proboscis, where they are positioned to infect their vertebrate hosts. Significant variation in susceptibility to filarial infection exists among mosquito species, resulting from differences in mosquito physiology, anatomy, and innate immunity 107, 108. Physiological and physical factors that prevent parasite development define mosquito refractoriness. By contrast, canonical immune responses that act to suppress parasite infection define mosquito resistance. Natural variation in these determinants underlies observed differences in mosquito transmission competence for filarial nematodes. Throughout this review, we use the term resistance to broadly encompass all genetic factors that decrease mosquito transmission potential where the mechanism is unknown or hypothetical. It is unlikely that a single universal gene drive approach could either suppress all relevant mosquito species or reduce the capacity for mosquito vectors to transmit disease for all naturally occurring pairings of mosquito and nematode parasite species. Therefore, gene drives that target LF parasites will have to be developed individually for different mosquito species and mosquito–nematode pairings.
The objective of population suppression drives for LF control is to spread genes that impair reproductive capacity and precipitate population crashes in the mosquito populations that transmit Wuchereria and Brugia. Many competent vectors of LF are capable of transmitting other pathogens. For example, suppression drives targeting the LF vector Ae. aegypti can help prevent transmission of Zika virus, and suppression drives targeting LF vectors in the genus Anopheles can aid malaria control. Therefore, population suppression schemes devised to eradicate LF can lead to global health synergies. CRISPR/Cas9 suppression drives include two primary methods: (a) disruption of genes that control viability or fertility, whereby an accumulation of carrier heterozygotes leads to a critical threshold of sterile homozygosity, or (b) sex‐biasing drives (e.g., X‐shredder) that deplete wild populations of females. To expand these and similar strategies to the range of mosquito species that contribute to LF, we must understand the conserved and/or species‐specific genes that cause a recessive sterility phenotype when targeted, chromosomal regions that can be targeted to significantly bias transmission of sex chromosomes or molecular factors that govern sex determination 109. The development of RNA‐guided suppression technologies will be primarily influenced by an improved species‐specific understanding of mosquito biology and population dynamics with less consideration of the biology of the filarial nematode species. Given the potential and unknown ecological concerns associated with targeted mosquito extinction, it is important to also explore CRISPR/Cas9 population modification drives that target LF parasites but do not necessitate mosquito eradication.
The objective of population modification drives for LF control is to spread genes throughout susceptible mosquito populations to interfere with filarial nematode development and to interrupt transmission to vertebrate hosts. The first task in building CRISPR/Cas9 population modification systems for LF control will be to identify or engineer genes that are deleterious to the development or survival of filarial parasites within their mosquito hosts. These antifilarial genetic elements can be packaged within RNA‐guided drive cassettes and introduced into wild mosquito populations to spread mosquito resistance to nematode infection and transmission. In contrast to suppression drives, these approaches are specific to the interactions of mosquitoes and nematodes and, therefore, are not generically applicable to other pathogens. Two broad strategies present themselves for antifilarial allele development: (a) exploitation of natural variation in mosquito genetic factors and pathogen response mechanisms that confer parasite resistance or refractoriness and (b) creation of new alleles and mechanisms that inhibit parasite migration, development, and transmission.
The genetic factors that contribute to natural variation in LF susceptibility within and among mosquito species can provide the basis for population modification strategies. Existing alleles that reduce the ability of particular mosquito species or strains to transmit LF can be spread throughout a population of competent vectors using gene drives. The genetic basis for mosquito susceptibility to both Wuchereria and Brugia has been investigated in laboratory infection models using Ae. aegypti. Early studies revealed a sex‐linked gene (f m) associated with Ae. aegypti susceptibility to B. malayi, Brugia pahangi, and W. bancrofti 110, 111. Later work revealed two quantitative trait loci (QTL) that underlie Ae. aegypti susceptibility to B. malayi: one corresponding to f m (fsb1) and an additional additive and smaller effect QTL (fsb2) 112. Investigators looking at mf ingestion and midgut penetration in the same model system identified a single QTL linked to fsb2 113. Experiments using improved genomic sequence data 114 and genetic linkage maps for Ae. aegypti 115 and an exome‐wide association mapping using wild Ae. aegypti populations 116 recapitulated the primary known sex‐linked locus controlling resistance to B. malayi. Although narrowing of this QTL to causative gene(s) has proved difficult, transcriptome analysis correlates genetic variation in this locus to a reduced expression of immune response genes 116.
These studies are immensely valuable, but the road to the identification of natural susceptibility alleles that can be effectively used in gene drives is littered with caveats. The extent to which QTL discovered for a particular interaction of a mosquito species and filarial nematode species will be relevant to other naturally occurring combinations of mosquitoes and filarial nematodes is uncertain. Susceptibility can vary among parasite species and even within different strains of the same parasite species 117. Additionally, the susceptibility trait does not have a single genetic cause 118. These observations suggest a pronounced and unexplored role for genetic interactions between parasite and host, and indicate that mosquito–filarial susceptibility is controlled by multiple genes. QTL mapped in model infection systems or with narrowly defined laboratory traits will have to be carefully investigated to establish the relevance to field transmission and other mosquito–nematode pairings, along with validating that effect sizes are meaningful and robust to environmental variation. Importantly, mapping to smaller genomic intervals will require the development of higher throughput or automated mosquito phenotyping pipelines, improved genetic linkage and physical maps, and the scaled application of marker‐based or whole‐genome sequencing technologies. QTL can then be more systematically narrowed using CRISPR/Cas9 to precisely identify causal coding and non‐coding genetic variants. Specific gene variants or QTL intervals that do not exceed the template length limitations of efficient HDR can be engineered into mosquito genomes using CRISPR/Cas9, and the individual or combinatorial spread of this genetic cargo in RNA‐guided drives can be used to confer mosquito population resistance to one or more parasite species that cause LF.
A potentially more tractable approach to antifilarial gene drive is to use our understanding of nematode biology to identify biomolecules that target developing parasites within the mosquito and disrupt their transmission. This approach will ideally lead to the discovery of broad‐spectrum anthelmintic effector molecules that act on substrates conserved among filarial nematode species and strains. We will examine the prospects of using both RNA and protein molecules as putative antifilarial effectors to be deployed in RNA‐guided mosquito population modification gene drives.
The discovery of RNA effectors that successfully inhibit parasite transmission will be a valuable step toward engineering population modification drives that target LF (Fig. 3A). RNA effectors acting through the canonical mosquito RNAi pathway can confer mosquito resistance to viral pathogens 119, and it is therefore conceivable that similar transgene constructs can be used to knock‐down the expression of genes critical to the transmission of filarial parasites. Brugia malayi is amenable to RNAi, and the B. malayi genome contains the canonical RNAi pathway along with a limited complement of proteins involved in the amplification and spread of RNAi triggers 120. Antifilarial RNA effectors, expressed as short hairpin (sh) or inverted repeat (IR) RNAs from mosquito transgenes, can be designed to target filarial nematode genes with high specificity. The success of this approach will depend on the careful selection of parasite target genes and the effective delivery of active RNAi triggers to parasite cells.
Figure 3.
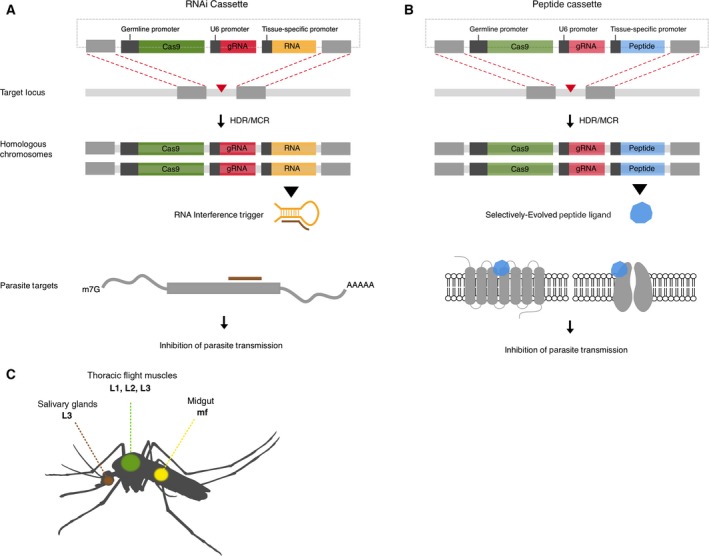
CRISPR/Cas9 gene drives targeting the transmission of filarial parasites. CRISPR cassettes are introduced using the HDR pathway and spread to homologous chromosomes using the MCR. (A) CRISPR gene drive cassette carrying an RNA effector that suppresses the expression of a specific parasite gene through the canonical RNAi pathway, leading to the inhibition of parasite transmission. (B) CRISPR gene drive cassette carrying a peptide effector selectively evolved to modulate parasite protein activity to inhibit parasite transmission. (C) Migratory course of LF‐causing parasites in the intermediate mosquito host. Effector molecules can be delivered to larval stage parasites using tissue‐specific mosquito promoters that are active in the midgut and thoracic flight muscles. The salivary glands may provide an additional but brief exposure window.
The promise of this approach is strengthened by an in vivo RNAi protocol demonstrating robust knock‐down of B. malayi transcripts within the intermediate mosquito host Ae. aegypti 81. This protocol leverages the mosquito as a conduit for the manual delivery of RNA and allows for the protracted exposure of healthy larval parasites to gene‐specific RNAi triggers. Intrathoracic injection of small interfering RNA (siRNA) into Ae. aegypti leads to systemic dispersion, and injection of both siRNA and double‐stranded RNA (dsRNA) triggers into mosquitoes infected with B. malayi elicit equivalently strong levels of parasite target transcript knock‐down. Using this assay, suppression of a cathepsin L‐like cysteine protease (Bm‐cpl‐1), known to be involved in molting and highly expressed in L3 larvae 121, led to stark motility phenotypes and a complete inhibition of parasite migration to the mosquito head 81. It is reasonable to expect that silencing of parasite target genes can be achieved if RNAi‐based effectors can be transcribed by mosquito promoters and made locally available at abundances that approach working RNAi injection concentrations. RNA effectors transcribed within mosquito cells will potentially undergo cell‐to‐cell transport in either or both mosquito or nematode cells. Transport can theoretically occur for both pre‐processed dsRNAs and mature siRNAs in either cell type. The determinants of cell‐to‐cell RNA dispersion and amplification within mosquito tissues are not well characterized 122, and the primary mechanism of RNA uptake by parasitic nematodes remains unknown. However, the presence of RNA‐dependent RNA polymerases in B. malayi suggests that the active silencing triggers are likely to be amplified within parasite cells.
The selection of putative parasite gene targets for RNA effector development can be guided by a number of important factors. First, loss of parasite gene function should be significantly deleterious to intra‐mosquito migration and development. The RNAi method described above is currently the best known approach to associate parasite gene loss‐of‐function with specific in vitro and intra‐mosquito phenotypes. Candidate nematode genes can be screened readily as targets for RNA effector design using this in vivo RNAi paradigm. Further, the target must be robustly expressed in one or more intra‐mosquito life stages. Growing transcriptomic resources 123 can be used to explore gene expression profiles to identify potential targets. Human‐infective Brugia and Wuchereria spend the greatest fraction of their developmental time within the thoracic muscles, arguing for the prioritization of effector targets upregulated or constitutively active across the L1–L3 stages. Flight muscle‐specific promoters have been identified and may be suitable for driving RNA‐effector expression 81, 124. This crucial developmental window coincides with the completion of two molts and active mosquito tissue consumption. Cuticle turnover and host cell lysis along with uptake through the parasite gut should improve the efficiency of transgenic RNA delivery.
In addition to Bm‐cpl‐1, for which a potent intra‐mosquito transmission interruption phenotype is already established, important targets include genes that encode other enzymes in the molting pathway 125, known anthelmintic targets 126, abundant larval proteins 127, surface proteins 128, secreted proteins 129, 130, exosomal proteins 131, chemoreceptors, and members of classically druggable protein families with fundamental roles in parasite physiology and neuromuscular control. The latter includes neuropeptidergic and aminergic G protein‐coupled receptors (GPCRs) 132, 133, as well as parasite‐specific ion channels 134. Caenorhabditis elegans RNAi screens can also help identify pan‐nematode targets and phenotypes relevant to larval parasite fitness but will bias against parasite‐specific genes critical to mosquito–filarial nematode interactions. It is preferable that chosen targets be (a) highly diverged from the mosquito gene complement to minimize off‐target silencing, (b) non‐redundant to minimize parasite compensatory mechanisms, and (c) conserved across different LF parasites to exhibit broad‐spectrum activity. Although any given gene can ultimately prove recalcitrant to RNAi‐mediated suppression, many potential new targets exist. To help mitigate potential nematode resistance to RNA effectors delivered through a CRISPR/Cas9 gene drive, an array of effectors can be used to target multiple parasite genes as well as different regions of the same gene(s).
Peptide effectors with antifilarial activity represent an alternative to RNA‐based effectors for CRISPR/Cas9‐driven population modification (Fig. 3B). Peptide effectors can exert biochemical and pharmacological actions on parasite protein substrates and interrupt normal parasite development. One of the best understood examples of endogenous peptide effectors are neuropeptides that modulate neuromuscular signaling and that can produce deleterious locomotory and pharyngeal pumping phenotypes 135, 136, 137, 138. Bioactive peptides can be expressed in mosquitoes to prevent movement and intra‐mosquito migration of nematodes. We should also consider selecting for additional peptide effectors to target the parasites that cause LF. Importantly, phage display technology has been a powerful tool used to produce peptide effectors that disrupt malaria transmission 139, 140, 141, 142. Even though phage display has been used to screen third larval stage cDNA to identify vaccine antigens 143, iterative protein evolution techniques have yet to be exploited for antifilarial effector discovery. A number of protocols are available for this mode of protein engineering 144, whereby randomized peptide libraries are filtered and enriched for substrate affinity over successive rounds of peptide diversification and selection. mRNA display is the most prominent in vitro method devised for this task and has been used successfully to generate peptide ligands for a range of cytosolic and membrane proteins 145, 146, 147. A variant of mRNA display, cDNA display 148, provides the added benefit of greater stability in the information‐carrying molecules and can be extended to a wider bandwidth of chemical conditions. Because library transformation is not required, in vitro display methods like mRNA and cDNA display can be used with larger library sizes than phage or yeast display. The choice of suitable parasite proteins as substrates for these platforms is non‐trivial, but prioritization can be guided by reasonable parameters.
Many of the same considerations in RNA effector target selection carry over to peptide target selection. The targets of peptide effectors can also be prioritized using temporal expression data, but the dynamics of protein expression with respect to mRNA expression must be considered. Directed peptide evolution in the absence of functional selection will more likely produce inhibitors and antagonists of native parasite protein function, and therefore, loss‐of‐function of candidate targets should drastically impair parasite development. Parasite RNAi screens within the mosquito can serve as a validation tool, but a potential disconnect exists between the timing and nature of defects elicited by transcript suppression and defects resulting from biochemical or pharmacological inhibition. Proteins secreted at the host–parasite interface, involved in parasite survival, critical to chemosensation and migratory behaviors, or central to neuromuscular control, are promising targets for peptide effector development. For cytosolic or extracellular protein targets, full‐length proteins or protein domains can be immobilized and used in both mRNA and cDNA display. Although mRNA display has been used previously to identify ligands for immobilized receptor ectodomains 149, the extension of cDNA display to full‐length transmembrane proteins in a cell culture environment 150, 151 makes it more useful to identify class A GPCR and ion channel effectors. Parasitic nematode metabotropic and ionotropic receptors have been heterologously expressed and pharmacologically characterized in cell systems amenable to cDNA display, including mammalian 152 and yeast cell lines 153. As with antifilarial RNA effector development, the low hanging fruit for display‐based peptide effector discovery is Bm‐CPL‐1. This target is localized to the glandular esophagus and released during molting 121 and is therefore theoretically accessible to an exogenous peptide inhibitor.
It is critical that parasite targets are accessible to exogenous peptides delivered within mosquito tissues, and preferably, these targets should be highly diverged from host mosquito proteins, non‐redundant, and conserved across different LF parasites. Putative peptide effectors can be tested in vitro for anthelmintic phenotypes, including paralytic effects on parasite musculature and impaired protein secretion. Effectors can also be injected into mosquito tissues relevant to the migratory status of target‐expressing parasites to test for inhibition of transmission potential. Peptides eliciting strong anthelmintic activity can then be introduced into transgenic mosquitoes to screen for gene drive utility. The mosquito midgut and thoracic muscles are attractive sites for peptide delivery along the migratory pathway of filarial nematodes (Fig. 3C). Candidate tissue‐specific promoters are available for driving effector expression at these locations 154, 155, as well as potential signal peptides 156, 157 that can be fused to direct peptide secretion. As with RNA effector delivery, the thoracic flight muscles present the longest peptide effector exposure time, possibly aided by the molting process and protein uptake through the parasite gut. To help mitigate potential nematode resistance to peptide effectors delivered through a CRISPR/Cas9 gene drive, an array of effectors can be used to target multiple parasite proteins. Additionally, single effectors can hypothetically be evolved to simultaneously target more than one parasite protein. Regardless of the target, these population modification gene drives have the potential to be powerful tools in our arsenal to fight lymphatic filariases borne by mosquito vectors.
Concluding remarks
The advent of the CRISPR/Cas system has transformed genetic techniques in a variety of organisms by providing methods for transgenesis, loss‐ and gain‐of‐function genetics, and tagging of endogenous genes. As outlined, this technology can be applied to the control of helminth‐borne NTDs, specifically LF. The approach to limit the spread of these nematodes could be through a better understanding of parasite biology directly or through the powerful toolkit of the model nematode C. elegans. Gene replacement to ‘parasitize’ C. elegans will likely be a fruitful approach. Lastly, the power of CRISPR/Cas‐mediated gene drives could be applied to the control of the nematodes that cause LF. We suggest some addressable approaches to limit the ability of nematodes to spread through mosquito hosts. Most importantly, control of mosquito populations can lead to global health synergies by limiting LF and the pathogenic agents of malaria. We believe that focused efforts over the next decade can use CRISPR/Cas to effect lasting changes on the control of NTDs.
Acknowledgements
We thank members of the Andersen Laboratory for helpful comments and critical reading of this review. The authors thank Lyric Bartholomay for insightful comments on the manuscript. MZ was funded by a Bill & Melinda Gates Foundation Grand Challenges Explorations Award [OPP1098456]. ECA is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. Funding for ECA is also provided by a Basil O'Connor Starter Research Award from the March of Dimes Foundation.
Author contributions
MZ and ECA conceived of the ideas, wrote the manuscript, and made manuscript revisions.
References
- 1. Hotez PJ (2013) Ntds v. 2.0: blue marble health neglected tropical disease control and elimination in a shifting health policy landscape. PLoS Negl Trop Dis 7, e2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pullan RL, Smith JL, Jasrasaria R & Brooker SJ (2014) Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hotez PJ, Alvarado M, Basáñez M, Bolliger I, Bourne R, Boussinesq M, Brooker SJ, Brown AS, Buckle G, Budke CM et al (2014) The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Negl Trop Dis 8, e2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization (2015) Investing to Overcome the Global Impact of Neglected Tropical Diseases: Third WHO Report on Neglected Diseases 2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5. Shang Y, Tang LH, Zhou SS, Chen YD, Yang YC & Lin SX (2010) Stunting and soil‐transmitted‐helminth infections among school‐age pupils in rural areas of southern China. Parasit Vectors 3, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D & Hotez PJ (2006) Soil‐transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367, 1521–1532. [DOI] [PubMed] [Google Scholar]
- 7. Taylor MJ, Hoerauf A & Bockarie M (2010) Lymphatic filariasis and onchocerciasis. Lancet 376, 1175–1185. [DOI] [PubMed] [Google Scholar]
- 8. Keenan JD, Hotez PJ, Amza A, Stoller NE, Gaynor BD, Porco TC & Lietman TM (2013) Elimination and eradication of neglected tropical diseases with mass drug administrations: a survey of experts. PLoS Negl Trop Dis 7, e2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geerts S & Gryseels B (2000) Drug resistance in human helminths: current situation and lessons from livestock. Clin Microbiol Rev 13, 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennett A & Guyatt H (2000) Reducing intestinal nematode infection: efficacy of albendazole and mebendazole. Parasitol Today 16, 71–77. [DOI] [PubMed] [Google Scholar]
- 11. Geerts S & Gryseels B (2001) Anthelmintic resistance in human helminths: a review. Trop Med Int Health 6, 915–921. [DOI] [PubMed] [Google Scholar]
- 12. Albonico M, Engels D & Savioli L (2004) Monitoring drug efficacy and early detection of drug resistance in human soil‐transmitted nematodes: a pressing public health agenda for helminth control. Int J Parasitol 34, 1205–1210. [DOI] [PubMed] [Google Scholar]
- 13. Flohr C, Tuyen LN, Lewis S, Minh TT, Campbell J, Britton J, Williams H, Hien TT, Farrar J & Quinnell RJ (2007) Low efficacy of mebendazole against hookworm in Vietnam: two randomized controlled trials. Am J Trop Med Hyg 76, 732–736. [PubMed] [Google Scholar]
- 14. Churcher TS & Basáñez M‐G (2009) Sampling strategies to detect anthelmintic resistance: the perspective of human onchocerciasis. Trends Parasitol 25, 11–17. [DOI] [PubMed] [Google Scholar]
- 15. Churcher TS, Pion SDS, Osei‐Atweneboana MY, Prichard RK, Awadzi K, Boussinesq M, Collins RC, Whitworth JA & Basáñez M‐G (2009) Identifying sub‐optimal responses to ivermectin in the treatment of River Blindness. Proc Natl Acad Sci USA 106, 16716–16721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osei‐Atweneboana MY, Awadzi K, Attah SK, Boakye DA, Gyapong JO & Prichard RK (2011) Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus . PLoS Negl Trop Dis 5, e998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolstenholme AJ, Fairweather I, Prichard R, von Samson‐Himmelstjerna G & Sangster NC (2004) Drug resistance in veterinary helminths. Trends Parasitol 20, 469–476. [DOI] [PubMed] [Google Scholar]
- 18. Kaplan RM (2004) Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol 20, 477–481. [DOI] [PubMed] [Google Scholar]
- 19. Kaplan RM & Vidyashankar AN (2012) An inconvenient truth: global worming and anthelmintic resistance. Vet Parasitol 186, 70–78. [DOI] [PubMed] [Google Scholar]
- 20. Prichard R, von Samson‐Himmelstjerna G, Blackhall W & Geary T (2007) Foreword: towards markers for anthelmintic resistance in helminths of importance in animal and human health. Parasitology 134, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 21. Prichard RK, Basáñez MG, Boatin BA, McCarthy JS, García HH, Yang GJ, Sripa B & Lustigman S (2012) A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS Negl Trop Dis 6, e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCarthy JS, Lustigman S, Yang G‐J, Barakat RM, García HH, Sripa B, Willingham AL, Prichard RK & Basáñez M‐G (2012) A research agenda for helminth diseases of humans: diagnostics for control and elimination programmes. PLoS Negl Trop Dis 6, e1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blaxter M (1998) Caenorhabditis elegans is a nematode. Science 282, 2041–2046. [DOI] [PubMed] [Google Scholar]
- 24. Sulston JE, Schierenberg E, White JG & Thomson J (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev Biol 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 25. C. elegans Sequencing Consortium. (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 26. Bürglin TR, Lobos E & Blaxter ML (1998) Caenorhabditis elegans as a model for parasitic nematodes. Int J Parasitol 28, 395–411. [DOI] [PubMed] [Google Scholar]
- 27. Geary TG & Thompson DP (2001) Caenorhabditis elegans: how good a model for veterinary parasites? Vet Parasitol 101, 371–386. [DOI] [PubMed] [Google Scholar]
- 28. Gilleard JS (2005) The use of Caenorhabditis elegans in parasitic nematode research. Parasitology 128, S49–S23. [DOI] [PubMed] [Google Scholar]
- 29. Jones LM, De Giorgi C & Urwin PE (2011) C. elegans as a resource for studies on plant parasitic nematodes In Genomics and Molecular Genetics of Plant‐Nematode Interactions (Jones JT, Gheysen G. & Fenoll C, eds), pp. 175–220. Springer Netherlands, Dordrecht, the Netherlands. [Google Scholar]
- 30. Crook M (2014) The dauer hypothesis and the evolution of parasitism: 20 years on and still going strong. Int J Parasitol 44, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Driscoll M, Dean E, Reilly E, Bergholz E & Chalfie M (1989) Genetic and molecular analysis of a Caenorhabditis elegans beta‐tubulin that conveys benzimidazole sensitivity. J Cell Biol 109, 2993–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cully DF, Vassilatis DK, Liu KK, Paress PS, Van der Ploeg LH, Schaeffer JM & Arena JP (1994) Cloning of an avermectin‐sensitive glutamate‐gated chloride channel from Caenorhabditis elegans . Nature 371, 707–711. [DOI] [PubMed] [Google Scholar]
- 33. Boulin T, Gielen M, Richmond JE, Williams DC, Paoletti P & Bessereau J‐L (2008) Eight genes are required for functional reconstitution of the Caenorhabditis elegans levamisole‐sensitive acetylcholine receptor. Proc Natl Acad Sci USA 105, 18590–18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwa MS, Veenstra JG, Van Dijk M & Roos MH (1995) β‐tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans . J Mol Biol 246, 500–510. [DOI] [PubMed] [Google Scholar]
- 35. Dent JA, Smith MM, Vassilatis DK & Avery L (2000) The genetics of ivermectin resistance in Caenorhabditis elegans . Proc Natl Acad Sci USA 97, 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glendinning SK, Buckingham SD, Sattelle DB, Wonnacott S & Wolstenholme AJ (2011) Glutamate‐gated chloride channels of Haemonchus contortus restore drug sensitivity to ivermectin resistant Caenorhabditis elegans . PLoS One 6, e22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geary TG (2012) Mechanism‐based screening strategies for anthelmintic discovery In Parasitic Helminths: Targets, Screens, Drugs and Vaccines (Caffrey CR, eds), pp. 121–134. Wiley‐VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 38. Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda‐Saavedra D et al (2007) Draft genome of the filarial nematode parasite Brugia malayi . Science 317, 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, Done J, Down T, Gao S & Grove C et al (2016) WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res 44, D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix M‐A & Kruglyak L (2012) Chromosome‐scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet 44, 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghosh R, Andersen EC, Shapiro JA, Gerke JP & Kruglyak L (2012) Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans . Science 335, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wählby C, Kamentsky L, Liu ZH, Riklin‐Raviv T, Conery AL, O'Rourke EJ, Sokolnicki KL, Visvikis O, Ljosa V, Irazoqui JE et al (2012) An image analysis toolbox for high‐throughput C. elegans assays. Nat Methods 9, 714–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andersen EC, Shimko TC, Crissman JR, Ghosh R, Bloom JS, Seidel HS, Gerke JP & Kruglyak L (2015) A powerful new quantitative genetics platform, combining Caenorhabditis elegans high‐throughput fitness assays with a large collection of recombinant strains. G3 (Bethesda) 5, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burns AR, Luciani GM, Musso G, Bagg R, Yeo M, Zhang Y, Rajendran L, Glavin J, Hunter R, Redman E et al (2015) Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat Commun 6, 7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM & Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR‐Cas9 system. Nat Methods 10, 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mojica FJM & Garrett RA (2013) Discovery and seminal developments in the CRISPR field. In CRISPR‐Cas Systems (Barrangou R, van der Oost J, eds), pp. 1–31 Springer, Berlin, Germany. [Google Scholar]
- 47. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA & Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- 48. Sorek R, Kunin V & Hugenholtz P (2008) CRISPRA widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6, 181–186. [DOI] [PubMed] [Google Scholar]
- 49. Marraffini LA & Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sander JD & Joung JK (2014) CRISPR‐Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH et al (2015) An updated evolutionary classification of CRISPR‐Cas systems. Nat Rev Microbiol 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA & Charpentier E (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE & Church GM (2013) RNA‐guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gasiunas G, Barrangou R, Horvath P & Siksnys V (2012) Cas9‐crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jinek M, East A, Cheng A, Lin S, Ma E & Doudna J (2013) RNA‐programmed genome editing in human cells. eLife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cho SW, Kim S, Kim JM & Kim J‐S (2013) Targeted genome engineering in human cells with the Cas9 RNA‐guided endonuclease. Nat Biotechnol 31, 230–232. [DOI] [PubMed] [Google Scholar]
- 58. Kim H, Ishidate T, Ghanta KS, Seth M, Conte D, Shirayama M & Mello CC (2014) A co‐CRISPR strategy for efficient genome editing in Caenorhabditis elegans . Genetics 197, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS & Fire AZ (2014) Efficient marker‐free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans . Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu P, Long L, Xiong K, Yu B, Chang N, Xiong J‐W, Zhu Z & Liu D (2014) Heritable/conditional genome editing in C. elegans using a CRISPR‐Cas9 feeding system. Cell Res 24, 886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwartz ML & Jorgensen EM (2016) SapTrap, a toolkit for high‐throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans . Genetics 202, 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dickinson DJ, Pani AM, Heppert JK, Higgins CD & Goldstein B (2015) Streamlined genome engineering with a self‐excising drug selection cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sloan MA, Reaves BJ, Maclean MJ, Storey BE & Wolstenholme AJ (2015) Expression of nicotinic acetylcholine receptor subunits from parasitic nematodes in Caenorhabditis elegans . Mol Biochem Parasitol 204, 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mello C & Fire A (1995) DNA transformation. Methods Cell Biol 48, 451–482. [PubMed] [Google Scholar]
- 65. Law W, Wuescher LM, Ortega A, Hapiak VM, Komuniecki PR & Komuniecki R (2015) Heterologous expression in remodeled C. elegans: a platform for monoaminergic agonist identification and anthelmintic screening. PLoS Pathog 11, e1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coward P, Chan SD, Wada HG, Humphries GM & Conklin BR (1999) Chimeric G proteins allow a high‐throughput signaling assay of Gi‐coupled receptors. Anal Biochem 270, 242–248. [DOI] [PubMed] [Google Scholar]
- 67. Geldhof P, Visser A, Clark D, Saunders G, Britton C & Gilleard J, Berriman M & Knox D (2007) RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitology 134, 609–619. [DOI] [PubMed] [Google Scholar]
- 68. Knox DP, Geldhof P, Visser A & Britton C (2007) RNA interference in parasitic nematodes of animals: a reality check? Trends Parasitol 23, 105–107. [DOI] [PubMed] [Google Scholar]
- 69. Shao H, Li X, Nolan Thomas J, Massey HC Jr, Pearce EJ & Lok JB (2012) Transposon‐mediated chromosomal integration of transgenes in the parasitic nematode Strongyloides ratti and establishment of stable transgenic lines. PLoS Pathog 8, e1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ward JD (2015) Rendering the intractable more tractable: tools from Caenorhabditis elegans ripe for import into parasitic nematodes. Genetics 201, 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Britton C, Roberts B & Marks N (2016) Functional Genomics Tools for Haemonchus contortus and Lessons From Other Helminths. Adv Parasitol 93, 599–623. [DOI] [PubMed] [Google Scholar]
- 72. Rebollo MP & Bockarie MJ (2013) Toward the elimination of lymphatic filariasis by 2020: treatment update and impact assessment for the endgame. Exp Rev Anti Infect Ther 11, 723–731. [DOI] [PubMed] [Google Scholar]
- 73. Ichimori K, King JD, Engels D, Yajima A, Mikhailov A, Lammie P & Ottesen EA (2014) Global programme to eliminate lymphatic filariasis: the processes underlying programme success. PLoS Negl Trop Dis 8, e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Michalski ML, Griffiths KG, Williams SA, Kaplan RM & Moorhead AR (2011) The NIH‐NIAID filariasis research reagent resource center. PLoS Negl Trop Dis 5, e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Higazi TB, Merriweather A, Shu L, Davis R & Unnasch TR (2002) Brugia malayi: transient transfection by microinjection and particle bombardment. Exp Parasitol 100, 95–102. [DOI] [PubMed] [Google Scholar]
- 76. Shu L, Katholi CR, Higazi T & Unnasch TR (2003) Analysis of the Brugia malayi HSP70 promoter using a homologous transient transfection system. Mol Biochem Parasitol 128, 67–75. [DOI] [PubMed] [Google Scholar]
- 77. Xu S, Liu C, Tzertzinis G, Ghedin E, Evans CC, Kaplan R & Unnasch TR (2011) In vivo transfection of developmentally competent Brugia malayi infective larvae. Int J Parasitol 41, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kushwaha S, Singh PK, Shahab M, Pathak M & Bhattacharya SM (2012) In vitro silencing of Brugia malayi trehalose‐6‐phosphate phosphatase impairs embryogenesis and in vivo development of infective larvae in jirds. PLoS Negl Trop Dis 6, e1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Landmann F, Foster JM, Slatko BE & Sullivan W (2012) Efficient in vitro RNA interference and immunofluorescence‐based phenotype analysis in a human parasitic nematode, Brugia malayi . Parasit Vectors 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Singh M, Singh PK & Misra‐Bhattacharya S (2012) RNAi mediated silencing of atpase RNA helicase gene in adult filarial parasite Brugia malayi impairs in vitro microfilaria release and adult parasite viability. J Biotechnol 157, 351–358. [DOI] [PubMed] [Google Scholar]
- 81. Song C, Gallup JM, Day TA, Bartholomay LC & Kimber MJ (2010) Development of an in vivo RNAi protocol to investigate gene function in the filarial nematode, Brugia malayi . PLoS Pathog 6, e1001239–e1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liang S, Knight M & Jolly ER (2013) Polyethyleneimine mediated DNA transfection in schistosome parasites and regulation of the wnt signaling pathway by a dominant‐negative smmef2. PLoS Negl Trop Dis 7, e2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li L, He Z‐Y, Wei X‐W, Gao G‐P & Wei Y‐Q (2015) Challenges in CRISPR/cas9 delivery: potential roles of nonviral vectors. Hum Gene Ther 26, 452–462. [DOI] [PubMed] [Google Scholar]
- 84. Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen Z‐Y & Liu DR (2015) Cationic lipid‐mediated delivery of proteins enables efficient protein‐based genome editing in vitro and in vivo. Nat Biotechnol 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Maggio I, Holkers M, Liu J, Janssen JM, Chen X & Gonçalves MA (2014) Adenoviral vector delivery of RNA‐guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep 4, 5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yin H, Song C‐Q, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A et al (2016) Therapeutic genome editing by combined viral and non‐viral delivery of CRISPR system components in vivo. Nat Biotechnol 34, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z, Li N, Zu Y & Qin L (2015) CRISPR‐Cas9 delivery to hard‐to‐transfect cells via membrane deformation. Science Adv 1, e1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Duvoisin R, Ayuk MA, Rinaldi G, Suttiprapa S, Mann VH & Lee CM, Harris N & Brindley PJ (2012) Human u6 promoter drives stronger shRNA activity than its schistosome orthologue in Schistosoma mansoni and human fibrosarcoma cells. Transgenic Res 21, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rao R & Weil GJ (2002) In vitro effects of antibiotics on Brugia malayi worm survival and reproduction. J Parasitol 88, 605–611. [DOI] [PubMed] [Google Scholar]
- 90. Serbus LR, Landmann F, Bray WM, White PM, Ruybal J, Lokey RS, Debec A & Sullivan W (2012) A cell‐based screen reveals that the albendazole metabolite, albendazole sulfone, targets Wolbachia . PLoS Pathog 8, e1002922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Khan AR & Fallon PG (2013) Helminth therapies: translating the unknown unknowns to known knowns. Int J Parasitol 43, 293–299. [DOI] [PubMed] [Google Scholar]
- 92. Edgell DR (2009) Selfish DNA: homing endonucleases find a home. Curr Biol 19, R115–R117. [DOI] [PubMed] [Google Scholar]
- 93. Zimmering S, Sandler L & Nicoletti B (1970) Mechanisms of meiotic drive. Annu Rev Genet 4, 409–436. [DOI] [PubMed] [Google Scholar]
- 94. Stouthamer R, Breeuwer JA & Hurst GD (1999) Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53, 71–102. [DOI] [PubMed] [Google Scholar]
- 95. Hamilton WD (1967) Extraordinary sex ratios. A sex‐ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 156, 477–488. [DOI] [PubMed] [Google Scholar]
- 96. Burt A (2003) Site‐specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci 270, 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sinkins SP & Gould F (2006) Gene drive systems for insect disease vectors. Nat Rev Genet 7, 427–435. [DOI] [PubMed] [Google Scholar]
- 98. Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ, Burt A et al (2011) A synthetic homing endonuclease‐based gene drive system in the human malaria mosquito. Nature 473, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, Burt A, Stoddard BL, Windbichler N & Crisanti A (2014) A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun 5, 3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Burt A (2014) Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci 369, 20130432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Esvelt KM, Smidler AL, Catteruccia F & Church GM (2014) Concerning RNA‐guided gene drives for the alteration of wild populations. eLife 3, e03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Oye KA, Esvelt K, Appleton E, Catteruccia F, Church G, Kuiken T, Lightfoot SB‐Y, McNamara J, Smidler A & Collins JP (2014) Regulating gene drives. Science 345, 626–628. [DOI] [PubMed] [Google Scholar]
- 103. Champer J, Buchman A & Akbari OS (2016) Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet 17, 146–159. [DOI] [PubMed] [Google Scholar]
- 104. Gantz VM & Bier E (2015) Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E & James AA (2015) Highly efficient Cas9‐mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi . Proc Natl Acad Sci USA 112, E6736–E6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S et al (2016) A CRISPR‐Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae . Nat Biotechnol 34, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Beerntsen BT, James AA & Christensen BM (2000) Genetics of mosquito vector competence. Microbiol Mol Biol Rev, 64, 115–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bartholomay LC (2014) Infection barriers and responses in mosquito‐filarial worm interactions. Curr Opin Insect Sci 3, 37–42. [DOI] [PubMed] [Google Scholar]
- 109. Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MAE, Chen X‐G et al (2015) Sex determination. A male‐determining factor in the mosquito Aedes aegypti . Science 348, 1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Macdonald WW (1962) The genetic basis of susceptibility to infection with semi‐periodic Brugia malayi in Aedes aegypti . Ann Trop Med Parasit 56, 373–382. [Google Scholar]
- 111. Macdonald WW & Ramachandran CP (1965) The influence of the gene fm (filarial susceptibility, Brugia malayi) on the susceptibility of Aedes aegypti to seven strains of Brugia, Wuchereria and Dirofilaria . Ann Trop Med Parasit 59, 64–73. [DOI] [PubMed] [Google Scholar]
- 112. Severson DW, Mori A, Zhang Y & Christensen BM (1994) Chromosomal mapping of two loci affecting filarial worm susceptibility in Aedes aegypti . Insect Mol Biol 3, 67–72. [DOI] [PubMed] [Google Scholar]
- 113. Beerntsen BT, Severson DW, Klinkhammer JA, Kassner VA & Christensen BM (1995) Aedes aegypti: a quantitative trait locus (QTL) influencing filarial worm intensity is linked to QTL for susceptibility to other mosquito‐borne pathogens. Exp Parasitol 81, 355–362. [DOI] [PubMed] [Google Scholar]
- 114. Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M et al (2007) Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316, 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Juneja P, Osei‐Poku J, Ho YS, Ariani CV, Palmer WJ, Pain A & Jiggins FM (2014) Assembly of the genome of the disease vector Aedes aegypti onto a genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl Trop Dis 8, e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Juneja P, Ariani CV, Ho YS, Akorli J, Palmer WJ, Pain A & Jiggins FM (2015) Exome and transcriptome sequencing of Aedes aegyptiidentifies a locus that confers resistance to Brugia malayi and alters the immune response. PLoS Pathog 11, e1004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bartholomay LC & Christensen BM (2002) Vector‐parasite interactions in mosquito‐borne filariasis In The Filaria (Klei T. & Rajan TV, eds), pp. 9–19. Springer US, Boston, MA. [Google Scholar]
- 118. Farid HA, Hammad RE, Kamal SA & Christensen BM (2004) Selection of a strain of Culex pipiens highly susceptible to Wuchereria bancrofti . Egypt J Biol 2, 125–131. [Google Scholar]
- 119. Franz AWE, Sanchez‐Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA & Olson KE (2006) Engineering RNA interference‐based resistance to dengue virus type 2 in genetically modified Aedes aegypti . Proc Natl Acad Sci USA 103, 4198–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dalzell JJ1, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, Fleming CC, Day TA, Mousley A & Marks NJ et al (2011) Rnai effector diversity in nematodes. PLoS Negl Trop Dis 5, e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Guiliano DB, Hong X, McKerrow JH, Blaxter ML, Oksov Y, Liu J, Ghedin E & Lustigman S (2004) A gene family of cathepsin L‐like proteases of filarial nematodes are associated with larval molting and cuticle and eggshell remodeling. Mol Biochem Parasitol 136, 227–242. [DOI] [PubMed] [Google Scholar]
- 122. Barnard A‐C, Nijhof AM, Fick W, Stutzer C & Maritz‐Olivier C (2012) RNAi in arthropods: insight into the machinery and applications for understanding the pathogen‐vector interface. Genes 3, 702–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Choi Y‐J, Aliota MT, Mayhew GF, Erickson SM & Christensen BM (2014) Dual RNA‐seq of parasite and host reveals gene expression dynamics during filarial worm‐mosquito interactions. PLoS Negl Trop Dis 8, e2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Allen ML & Christensen BM (2004) Flight muscle‐specific expression of act88f: GFP in transgenic Culex quinquefasciatus Say (Diptera: Culicidae). Parasitol Int 53, 307–314. [DOI] [PubMed] [Google Scholar]
- 125. Page AP, Stepek G, Winter AD & Pertab D (2014) Enzymology of the nematode cuticle: a potential drug target? Int J Parasitol Drugs Drug Resist 4, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Moreno Y, Nabhan JF, Solomon J, Mackenzie CD & Geary TG (2010) Ivermectin disrupts the function of the excretory‐secretory apparatus in microfilariae of Brugia malayi . Proc Natl Acad Sci USA 107, 20120–20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gregory WF, Atmadja AK, Allen JE & Maizels RM (2000) The abundant larval transcript‐1 and‐2 genes of Brugia malayi encode stage‐specific candidate vaccine antigens for filariasis. Infect Immun 68, 4174–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hirzmann J, Hintz M, Kasper M, Shresta TR, Taubert A, Conraths FJ, Geyer R, Stirm S, Zahner H & Hobom G (2002) Cloning and expression analysis of two mucin‐like genes encoding microfilarial sheath surface proteins of the parasitic nematodes Brugia and Litomosoides . J Biol Chem 277, 47603–47612. [DOI] [PubMed] [Google Scholar]
- 129. Moreno Y & Geary TG (2008) Stage‐and gender‐specific proteomic analysis of Brugia malayi excretory‐secretory products. PLoS Negl Trop Dis 2, e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bennuru S, Semnani R, Meng Z, Ribeiro JMC, Veenstra TD & Nutman TB (2009) Brugia malayi excreted/secreted proteins at the host/parasite interface: stage‐and gender‐specific proteomic profiling. PLoS Negl Trop Dis 3, e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, Bartholomay LC & Kimber MJ (2015) Release of small RNA‐containing exosome‐like vesicles from the human filarial parasite Brugia malayi . PLoS Negl Trop Dis 9, e0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, Marks NJ, Maule AG & Mousley A (2014) New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom 3, 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Anderson RC, Newton CL, Millar RP & Katz AA (2014) The Brugia malayi neuropeptide receptor‐4 is activated by FMRFamide‐like peptides and signals via Gα i. Mol Biochem Parasitol 195, 54–58. [DOI] [PubMed] [Google Scholar]
- 134. Williamson SM, Walsh TK & Wolstenholme AJ (2007) The cys‐loop ligand‐gated ion channel gene family of Brugia malayi and Trichinella spiralis: a comparison with Caenorhabditis elegans . Invert Neurosci 7, 219–226. [DOI] [PubMed] [Google Scholar]
- 135. Brownlee D & Walker R (1999) Actions of nematode FMRFamide‐related peptides on the pharyngeal muscle of the parasitic nematode, Ascaris suum . Ann N Y Acad Sci, 897, 228–238. [DOI] [PubMed] [Google Scholar]
- 136. Reinitz CA, Herfel HG, Messinger LA & Stretton AO (2000) Changes in locomotory behavior and camp produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans . Mol Biochem Parasitol 111, 185–197. [DOI] [PubMed] [Google Scholar]
- 137. McVeigh P, Leech S, Marks NJ, Geary TG & Maule AG (2006) Gene expression and pharmacology of nematode NLP‐12 neuropeptides. Int J Parasitol 36, 633–640. [DOI] [PubMed] [Google Scholar]
- 138. Mousley A, Novozhilova E, Kimber MJ, Day TA & Maule AG (2010) Neuropeptide physiology in helminths In Neuropeptide Systems as Targets for Parasite and Pest Control (Geary TG. & Maule A, eds), pp. 78–97. Springer US, New York, NY. [Google Scholar]
- 139. Ghosh AK, Ribolla PE & Jacobs‐Lorena M (2001) Targeting Plasmodium ligands on mosquito salivary glands and midgut with a phage display peptide library. Proc Natl Acad Sci USA 98, 13278–13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lanzillotti R & Coetzer TL (2008) Phage display: a useful tool for malaria research? Trends Parasitol 24, 18–23. [DOI] [PubMed] [Google Scholar]
- 141. Isaacs AT, Li F, Jasinskiene N, Chen X, Nirmala X, Marinotti O, Vinetz JM & James AA (2011) Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi . PLoS Pathog 7, e1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Isaacs AT, Li F, Jasinskiene N, Chen X, Nirmala X, Marinotti O, Vinetz JM & James AA (2012) Transgenic Anopheles stephensi coexpressing single‐chain antibodies resist Plasmodium falciparum development. Proc Natl Acad Sci USA 109, E1922–E1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gnanasekar M, Rao KVN, He Y‐X, Mishra PK, Nutman TB, Kaliraj P & Ramaswamy K (2004) Novel phage display‐based subtractive screening to identify vaccine candidates of Brugia malayi . Infect Immun 72, 4707–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bradbury AR, Sidhu S, Dübel S & McCafferty J (2011) Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 29, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gold L (2001) mRNA display: diversity matters during in vitro selection. Proc Natl Acad Sci USA 98, 4825–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ja WW & Roberts RW (2004) In vitro selection of state‐specific peptide modulators of G protein signaling using mRNA display. Biochemistry 43, 9265–9275. [DOI] [PubMed] [Google Scholar]
- 147. Seelig B (2011) mRNA display for the selection and evolution of enzymes from in vitro‐translated protein libraries. Nat Protoc 6, 540–552. [DOI] [PubMed] [Google Scholar]
- 148. Ueno S & Nemoto N (2012) cDNA display: rapid stabilization of mRNA display. Methods Mol Biol 805, 113–135. [DOI] [PubMed] [Google Scholar]
- 149. William WJ, West AP, Delker SL, Bjorkman PJ, Benzer S & Roberts RW (2007) Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor. Nat Chem Biol 3, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Doi N, Yamakawa N, Matsumoto H, Yamamoto Y, Nagano T, Matsumura N, Horisawa K & Yanagawa H (2012) DNA display selection of peptide ligands for a full‐length human G protein‐coupled receptor on cho‐k1 cells. PloS One 7, e30084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ueno S, Yoshida S, Mondal A, Nishina K, Koyama M, Sakata I, Miura K, Hayashi Y, Nemoto N, Nishigaki K et al (2012) In vitro selection of a peptide antagonist of growth hormone secretagogue receptor using cDNA display. Proc Natl Acad Sci USA 109, 11121–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Forrester SG, Prichard RK & Beech RN (2002) A glutamate‐gated chloride channel subunit from Haemonchus contortus: expression in a mammalian cell line, ligand binding, and modulation of anthelmintic binding by glutamate. Biochem Pharmacol 63, 1061–1068. [DOI] [PubMed] [Google Scholar]
- 153. Kimber MJ, Sayegh L, El‐Shehabi F, Song C, Zamanian M, Woods DJ, Day TA & Ribeiro P (2009) Identification of an Ascaris G protein‐coupled acetylcholine receptor with atypical muscarinic pharmacology. Int J Parasitol 39, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Moreira LA, Edwards MJ, Adhami F, Jasinskiene N, James AA & Jacobs‐Lorena M (2000) Robust gut‐specific gene expression in transgenic Aedes aegypti mosquitoes. Proc Natl Acad Sci USA 97, 10895–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pham D‐D & Chavez C (2005) The ferritin light‐chain homologue promoter in Aedes aegypti . Insect Mol Biol 14, 263–270. [DOI] [PubMed] [Google Scholar]
- 156. Stracker TH, Thompson S, Grossman GL, Riehle MA & Brown MR (2002) Characterization of the AeaHP gene and its expression in the mosquito Aedes aegypti (Diptera: Culicidae). J Med Entomol 39, 331–342. [DOI] [PubMed] [Google Scholar]
- 157. Riehle MA, Fan Y, Cao C & Brown MR (2006) Molecular characterization of insulin‐like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides 27, 2547–2560. [DOI] [PubMed] [Google Scholar]


