Abstract
MicroRNAs are small non coding RNAs that typically inhibit the translation and stability of messenger RNAs, controlling genes involved in cellular processes such as inflammation, cell cycle regulation, stress response, differentiation, apoptosis, and migration. Not surprisingly, microRNAs are also aberrantly expressed in cancer and promote tumorigenesis by disrupting these vital cellular functions. In this review, we first broadly summarize the role of microRNAs in breast cancer and Estrogen Receptor alpha signaling. Then we focus on what is currently known about the role of microRNAs in anti-hormonal therapy or resistance to endocrine agents. Specifically, we will discuss key miRNAs involved in tamoxifen (miR-221/222, 181, 101, 519a, 301, 375, 342, 451, and the let-7 family), fulvestrant (miR-221/222, miR-200 family), and aromatase inhibitor (miR-128 and the let-7 family) resistance.
Introduction
Breast cancer accounts for one in four of all female cancers, making it by far the most common cancer in women in the western world [1]. At least 70% of breast cancers are classified as Estrogen Receptor alpha-positive (ER-positive) breast cancers [2], and interfering with estrogen action has been a mainstay of breast cancer treatment for more than a century [2,3]. Estrogen (E2) is synthesized primarily in the ovaries and regulated by several hormones such as pituitary gonadotrophins, follicle-stimulating hormone and luteinizing hormone [4,5]. The synthesis of estrogens by the ovaries ceases at menopause, Since most cases of breast cancer strike post-menopausal women, the non-ovarian sources of E2, such as skin and adipose tissues, are important in disease progression [6]. There are several ways of blocking the proliferative effects of estrogen in ER-positive breast cancer. The development of the selective estrogen receptor modulators (SERMs) represents the most successful, targeted cancer therapy ever recorded [2,3]. The first anti-estrogen drug developed and used in clinics is tamoxifen, which maximal benefit is achieved with a 5-year treatment leading to a 51% reduction in recurrence and about 28% reduction in deaths during years 0–4 [7,8]. The clinical success of tamoxifen has led to the development and clinical testing of several other effective endocrine therapies that target estrogen synthesis or ER signaling [2]. Anti-estrogen drugs fall into two main classes. The first one is the SERM mentioned earlier, exemplified by tamoxifen, and it acts as an antagonist in the mammary glands by inhibiting E2-dependent activation of ER signaling, which is essential in the breast epithelium; however, it does not modulate the E2-independent activity of ER resulting in a strong agonistic activity in other tissues, such as the uterus. The second class is the selective estrogen receptor down-regulator (SERD), such as fulvestrant, which inhibits ER signaling activation and reduces the half-life of ER, and therefore is anti-estrogenic in all tissues [9]. In the case of the aromatase inhibitors (AIs), they block the peripheral conversion of androgens to estrogens and reduce estrogen levels in both tissue and plasma [10,11]. The primary source of estrogens in postmenopausal women is the conversion of circulating androgens via aromatization at peripheral sites. Consequently the use of AIs in postmenopausal women causes relatively rapid decreases in circulating estrogen levels. Two major classes of AIs have been developed [12]. Type I steroidal drugs, like exemestane, bind competitively and irreversibly to the enzyme and are the ‘‘inactivators’’ [13]. Type II are non-steroidal inhibitors, including anastrozole and letrozole, which bind reversibly to the enzyme. In many cases, however, these therapies fail and women die owing to recurrent, endocrine-resistant tumors [14]. Therefore, two major challenges for the successful treatment of breast cancer are the development of more specific biomarkers that predict therapeutic response to endocrine therapies and the identification of new therapeutic targets for endocrine-resistant disease.
MicroRNAs (miRNAs) are single-stranded RNAs (ssRNAs) of 19–25 nucleotides in length that are generated from endogenous hairpin transcripts [15,16]. They play an important role in the negative regulation of gene expression by base-pairing to partially complementary sites on the target messenger RNAs (mRNAs), usually in the 3’ untranslated region (UTR). Binding of a miRNA to the target mRNA typically leads to translational repression and/or exonucleolytic mRNA decay, although highly complementary targets can be cleaved endonucleolytically [17]. miRNAs have been found to regulate more than 60% of mRNAs and have roles in fundamental processes, such as development, differentiation, cell proliferation, apoptosis, and stress responses [16,18]. Over the past few years, many miRNAs have been implicated in various human cancers and the use of miRNA profiles has shown a large applicability in molecular oncology since their expression can predict tumor initiation, progression and response to therapies [15,16,19–22].
In this review, we summarize and evaluate the recent insights into the roles of miRNAs in breast tumorigenesis as diagnostic and prognostic biomarkers and as modulators of the endocrine therapies. After a general summary of the dysregulation of miRNAs in breast cancer, we will discuss their roles in ER signaling and their involvements in the modulation of the anti-hormonal drug activity and acquisition of endocrine resistance.
microRNA profiling in breast cancer
The first report describing the existence of a miRNA signature characterizing human breast cancer was published in 2005, suggesting the involvement of miRNAs in the pathogenesis of this human neoplasm [23]. The authors described a breast cancer-specific miRNA signature from a genome-wide miRNA expression analysis on a set of 10 normal and 76 tumor breast tissues, resulting in the identification of a list of 29 differentially expressed miRNAs that can classify tumors and normal tissues with an accuracy of 100%. Among the miRNAs differentially expressed, miR-10b, miR-125b and miR-145 were down-modulated, while miR-21 and miR-155 were up-modulated, suggesting that these miRNAs could exert a role as tumor suppressor genes or oncogenes. It is now clear that miRNAs expression can provide important diagnostic and prognostic information similar to mRNA expression profiling.
A study from the Dr. Miska laboratory showed that molecular subtypes of breast cancer (luminal A, luminal B, basal-like, HER2, and normal-like) present different miRNA expression profiles, although a perfect separation of normal and tumor samples was not observed [24]. A set of nine miRNAs was able to classify luminal A from luminal B tumors with the high expression of miR-100 and miR-99a in the luminal A subtype, and miR-103 and miR-107 in the luminal B subtype. The authors also showed high levels of the miR-17-92 cluster and paralogs in the basal tumors and an association of let-7 family members with luminal A tumors, ER positive status and low tumor grade. In another study, a large cohort of breast tumors (1,302 specimens), representing the heterogeneity observed in clinics, as well as 116 adjacent normal breast tissues and a panel of 28 breast cancer cell lines were analyzed to define the global expression of miRNAs in breast cancer [25]. As expected from previous analyses, the first information was the global repression of miRNA expression in tumors compared to adjacent normal tissues, and a gradual decline with increased tumor grade and in breast cancer cell lines. The 853 detectable miRNAs were divided into three main classes: 43% with very low variability, and 22% and 35% with globally high or low expression respectively. In contrast to mRNAs, few miRNAs levels were dependent on copy-number aberrations such as the gain of the miR-17-92 family and loss of the tumor-suppressive miR-31, evaluated in at least 5% of the samples. Although no clinical marker was identified as covariant with miRNA expression, lymphocytic infiltration was shown to be pronouncedly affected by five miRNAs with known functions in immune system regulation: miR-150 [26], miR-155 [27], miR-146a [28], miR-142-3p and miR-142-5p [29]. In contrast to the absence of association between clinical parameters and miRNA expression, several mRNAs representing several key cellular processes in breast cancer biology and survival was identified to be highly interdependent with miRNAs. Tumor-suppressive miRNA families such as miR-143/145 [30], miR-199/214 [31] and miR-127 [32] were identified to be associated with regulatory components of signaling and development, extracellular matrix (ECM), cell adhesion and morphogenesis. The cell cycle gene signature was almost exclusively related to the anti-proliferative miR-139 [33]. Finally, genes related to sex-hormone activity, reflecting the activities of estrogen and progesterone in breast cancer, were related to the ER-dependent miR-342 (see below) and miR-17-92 polycistron [34].
A recent analysis in our lab defined, by analyzing recently published deep-sequencing dataset, a miRNA signature related to the transition from ductal carcinoma in situ (DCIS) to invasive ductal carcinoma (IDC) [35], a key event in breast cancer progression [36]. A nine-miRNA signature was identified that differentiates IDC from DCIS. Specifically, let-7d, miR-210, and miR-221 were down-regulated in DCIS and up-regulated in the invasive transition, thus featuring an expression reversal along the cancer progression path. Indeed, five miRNAs, miR-210, miR-21, miR-106b*, miR-197, and let-7i, were associated with overall survival and time to metastasis and miR-210 was the only one also involved in the invasive transition. In another study, Volinia and Croce identified a survival signature of breast cancer through the analysis of a cohort of 466 patients with primary IDC, the most frequent type of breast cancer, by integrating mRNA, miRNA, and DNA methylation next-generation sequencing data from The Cancer Genome Atlas (TCGA) [37]. Thirty mRNAs and seven miRNAs were associated with the overall survival across different clinical and molecular subclasses of IDC. The prognostic RNAs included PIK3CA, one of the two most frequently mutated genes in invasive breast cancer, and miRNAs such as miR-328, miR-484, and miR-874. The integrated RNA signature was successfully validated on 8 different breast cancer cohorts, comprising a total of 2,399 patients, by showing that a combination of mRNA/miRNA signature has a superior performance for risk stratification with respect to other RNA predictors, including the mRNA signatures used in the commercial MammaPrint and Oncotype DX assays.
It needs to be emphasized that we are beginning to decipher the tumorigenic mechanisms and molecular signaling that miRNAs alter in breast cancer. The biggest challenge ahead to employ miRNAs as biomarkers in clinical decision-making still resides in the validation of these findings in large independent cohorts or by additional preclinical/clinical verification studies.
microRNAs and estrogen receptor signaling
As described above, the first report correlating miRNA expression with ER status in breast tumors was published in 2005 [23]. In the last nine years, a consistent number of publications highlighted two main points related to the interaction of miRNAs with ER signaling: 1) miRNA expression is not only correlated to ER status but is directly controlled by ER itself; 2) in turn, miRNAs control ER expression and estrogen signaling [38,39]. The ability of ER to modulate the expression of miRNAs has been explored in vivo and in vitro. In the first in vivo study, miRNA expression was significantly modulated during E2-induced mammary carcinogenesis in rats [40]. After 6 and 12 weeks of E2 exposure, corresponding to the appearance of atypical hyperplasia of both ducts and alveoli, 15 miRNAs were down-regulated and 19 miRNAs were up-regulated. Of note, the increased expressions of several oncogenic miRNAs, such as miR-21, miR-17/92 cluster, and miR-25/106 cluster were associated with the appearance of lobular hyperplasia. Considering the centrality of ER signaling in the initiation, progression and treatment of breast cancer, the majority of the in vitro studies aimed to identify ER-regulated miRNA expression changes in ER-positive breast cancer MCF7 cells. Different treatment conditions (time of E2 exposure, E2 concentration, hormone deprivation before treatment) and technologies (microarray, RNA sequencing, qRT-PCR array) have been used for the measurement of mature miRNAs, but they have shown low consistency even within the well-studied MCF7 cell line [largely reviewed in 38]. The first global analysis of E2 modulated miRNAs was published by Dr. Nakshatri laboratory in 2009 [41]. After 4 hours of E2 treatment, 21 E2-regulated miRNAs were induced by estrogen, including members of the let-7 family, miR-17/92 cluster, and miR-200b. In a second study, the levels of miRNAs were instead determined by microarray hybridization after early (0–3 h) and delayed (>6 h) E2 stimulation of MCF7 cells [42]. Although the authors did not reveal any miRNAs with expression changes significantly greater than 2-fold comparing 0 hour to 3, 6, and 12 hours of E2 stimulation, they identified the upregulation of the mature miRNAs generated by the processing of 3 paralogous primary miRNAs: pri-mir-17-92, pri-mir-106a-363, and pri-mir-106b-25. The oncogene c-MYC, an ER transcriptional target, was identified to regulate miR-17-92 expression by directly binding to its promoter in response to E2 treatment. The miRNAs (miR-18a, miR-19b, and miR-20b) derived from these pri-miRNAs in turn were again involved in an inhibitory loop and down-regulated ER. However, the miR-17-92 cluster has not yet been associated with endocrine resistant breast cancer. In another study, 172 miRNAs were identified to be up- or down-regulated by ER, of which 52 were in common between the two cell models used, MCF7 and ZR-75-1 [43]. The most consistently deregulated miRNAs after E2 treatment were miR-206, miR-125a/b, miR-17-5p, miR-34a; some members of the let-7 family that act as tumor suppressor genes [44]; and miR-21, miR-155, and miR-10b, which are usually overexpressed in breast cancer and may act as oncogenes [23].
In our recent publication, we profiled MCF7 cells after hormonal starvation and a 6-hour E2 stimulation by using the highly sensitive TaqMan Low Density Arrays, which allows the accurate quantitation of 754 human miRNAs with the sensitivity of the Taqman real-time assay [45]. After 6 days of E2 deprivation, we detected the downregulation of 146 and upregulation of 25 mature miRNAs, organized in 69 different miRNA genes. Of these 69 miRNA genes, 43 genes (85 mature miRNAs) were modulated after 6 hours of E2 stimulation. In our system, we identified a large upregulation of the majority of miRNAs differentially expressed and only few were repressed such as the miR-181 family. Of note, there is a discrepancy between the reported E2-induced upregulation of miR-21 and a previous publication from Dr. Klinge laboratory [46], which showed a reduction of miR-21 after E2 stimulation of MCF7 cells. In our study, we identified a time-dependent reduction of miR-21 during hormone starvation that was reverted after E2 treatment with an 1.78-fold increase of miR-21. This discrepancy can be explained by a biphasic regulation of E2, i.e., induction followed by repression of miR-21. Recently, Hah et al. [47] used GRO-Seq (global nuclear run-on and massively parallel sequencing) to determine estrogen-regulated transcription. They reported changes in expression of 322 miRNA-containing transcripts in MCF7 cells; of these, 119 (37%) were regulated by estrogen.
E2 regulates the expression of miRNAs not only by transcriptional control, but also by affecting the expression and activity of key components of miRNA biogenesis. Several studies suggested that estrogen induced changes in DICER expression may have global effects on miRNA levels by altering their processing rate [48–50]. In another study, decreased AGO1 and AGO2 expressions were detected in ER-positive tumors suggesting that the lower abundance of these argonaute proteins may limit the functional activity of the RNA Induced Silencing Complex (RISC) [51]. However, E2 stimulation enhanced the expression of AGO2 levels in hormone-depleted MCF7 cells and ectopic expression of AGO2 in MCF7 cell lines resulted in increased cell proliferation, reduced cell-cell adhesion and enhanced migratory properties [52]. In addition, activated ER modulated the processing of primary miRNA into pre-miRNA through E2-dependent association with the Drosha complex [53]. It is conceivable that RNA-binding subunits in the Drosha complex dock ER to modulate the efficiency of Drosha-mediated pri-miRNA cleavage and facilitate the production of specific E2-regulated miRNAs.
As stated earlier above, miRNAs control ER expression directly by repressing its translation and sometimes inducing its mRNA degradation, and indirectly by repressing other proteins that cooperate with ER. miR-206 was the first miRNA identified to target ER in breast cancer [54]. Ectopic expression of miR-206 in MCF7 cells has been shown to reduce ER level and also the basal expression levels of ER target genes such as PR, cyclin D1, and pS2, resulting in decreased cell proliferation. Furthermore, miR-206 targets two ER coactivator proteins, SRC-1 and SRC-3, and the transcriptional factor GATA-3 that cooperates with ER [55]. miR-206 continues to suppress estrogenic responses even under the overexpression of ER encoded by mRNA lacking the 3’UTR, confirming that the involvements of additional non-ER targets of miR-206 in the suppression of estrogenic responses.
Another well-studied miRNA family that targets ER is miR-221/222 [56]. We showed that overexpression of miR-221 and 222 in ER-positive cells induces a global change in gene expression that differs from the miR-206 signature and may account for the generation of a more invasive and deadly tumor phenotype [57]. In fact, miR-221 and 222 enforced expression triggers a malignant transformation that confers ability to survive apoptotic pressure under anchorage-independent conditions (decrease in CAV1 and CAV2 expression), induces an increase in cell cycle progression (decrease in PTEN, CDKN1B, and BIM expression) and tumor invasion and metastasis (increase in BMP4, BMP7, and TGFβ3 expression). In another study, the induction of the epithelial-mesenchymal transition in MCF7 cells was associated with increased cancer stem cell-like properties and reduced ER expression, both features that correlate with suppression of miR-200c, miR-203, and miR-205 and overexpression of miR-222 and miR-221, further suggesting that ER is a target of miR-221/222 in breast cancer cells [58].
Introduction of let-7 miRNA in the MCF7 cell line negatively regulated ER activity, inhibited cell proliferation, and induced apoptosis in MCF7 cells [59]. A novel protein lysate microarray-based study identified miR-18a, miR-18b, miR-193b, miR-206, and miR-302c as ER repressors [60]. This is further confirmed by the higher expression levels of miR-18a and miR-18b in ER-negative comparing with ER-positive clinical tumors [60]. Since coregulators modulate ER functions, miRNA that regulate coregulators also influence ER functions in breast cancer cells. miR-17-5p, for instance, represses the translation of AIB1 mRNA, whereby it blocks ER-mediated cell proliferation in MCF7 cells [61].
Overall, the interplay between estrogen signaling and miRNAs provides a better understanding of steroid-related tumorigenesis and the potential to use such information for designing novel anti-hormonal therapeutics as we will discuss below.
microRNAs in endocrine therapy
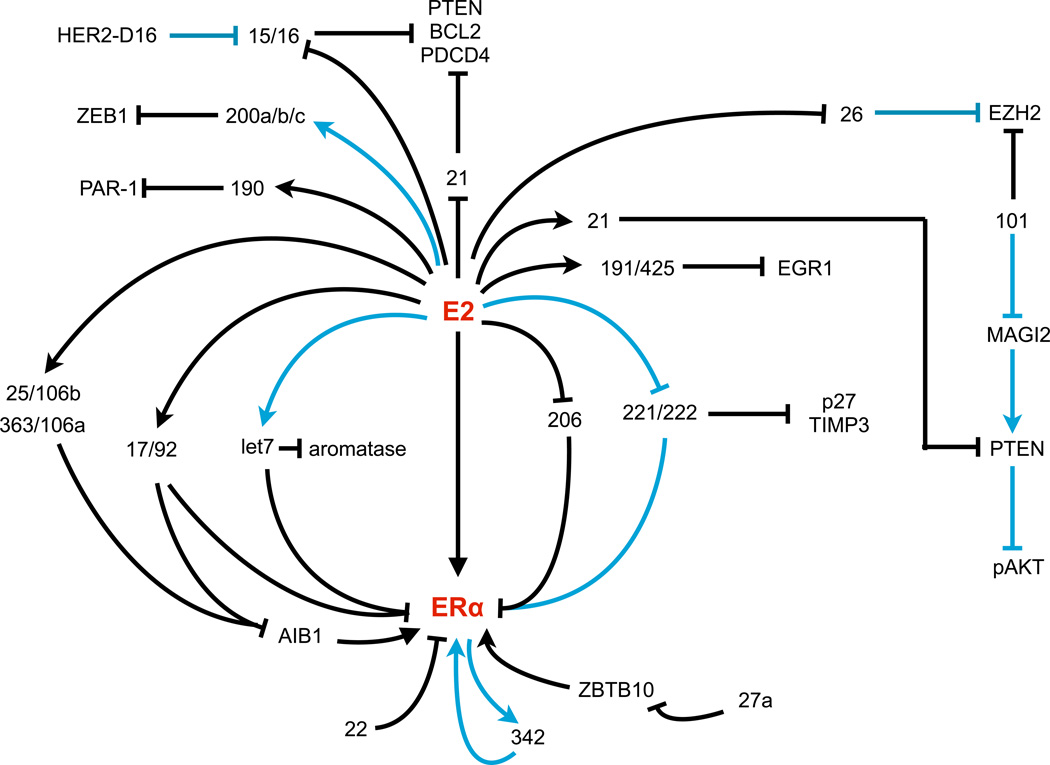
Dysregulation of miRNAs in breast cancer affects not only cellular processes involved in carcinogenesis but can have a direct consequence on the success of therapeutic interventions. In breast cancer, miRNAs have been associated with resistance to almost all forms of treatments, including chemotherapy, endocrine therapy, radiotherapy, and drugs targeting particular signaling molecules. In the following sections we will discuss the involvement of miRNAs in the response and resistance of breast cancer cells to the three main anti-hormonal drugs (Figure 1): tamoxifen, aromatase inhibitors and fulvestrant.
Figure 1. ER-regulated microRNAs network.
Schematic representation of the main E2-regulated microRNAs in breast cancer with an impact on endocrine resistance. The blue lines identified pathways that have been involved in endocrine resistance.
microRNAs and tamoxifen
Tamoxifen is currently the standard adjuvant therapy in women with estrogen receptor-positive breast cancer [62]. Because ~30% of these tumors are resistant to tamoxifen, there is a considerable interest in elucidating the molecular mechanisms of acquiring resistance to this important anticancer drug [63]. The first report assessing the involvement of miRNAs in tamoxifen therapy was published in 2008 by Dr. Majumder and her coworkers and showed that miRNAs were modulated. Specifically, miR-221, 222, 181, 375, 32, 171, 213, and 203 were upregulated, while miR-342, 489, 21, 24, 27, 23, and 200 were downregulated in tamoxifen-resistant cells [64]. Interestingly, they found an increased expression of miR-221 and miR-222 in tamoxifen-resistant cell lines and HER2/neu-positive primary human breast tumors that are resistant to endocrine therapy. Indeed, it was demonstrated that ectopic expression of miR-221/222 in ER-positive cells reduces their sensitivity to tamoxifen attributing this effect to the ability of miR-221/222 to repress tumor-suppressor proteins such as p27 or TIMP3 [64,65]. The same group also evaluated the increased sensitivity (40–45% reduction in tumor volume) of tamoxifen-resistant xenografts to the drug upon combined treatment with the anti-miR inhibitors of miR-221/222 and miR-181, which are markedly up-regulated in tamoxifen-resistant breast cancer cell lines and tumors [65]. In both anti-miR-222- and anti-miR-181-treated tumors, the growth was significantly reduced beginning with the second week of treatment when compared with the control group and that some tumor growth regressed completely after four treatments. One of the major explanations of the miR-221/222 effect on tamoxifen sensitivity came from Dr. Cheng's laboratory who identified, as discussed above, the suppression of the ER by miR-221/222 [56]. Since the lack of ER expression is a major mechanism of tamoxifen resistance in breast cancer, the authors showed that ectopic expression of miR-221 and miR-222 in ER-positive cells, MCF7 and T47D cell lines, reduces ER protein level and renders, as expected, the cells resistant to tamoxifen. These latter studies suggested the importance of miR-221 and miR-222 in the inhibition of tamoxifen-induced cell death and highlighted that the maintenance of low miR-221/222 expression may play a key role in the success of endocrine therapy in breast cancer by increasing the concentration of different tumor-suppressor targets and ER itself.
On the other hand, among the downregulated miRNAs identified in Dr. Majumder’s paper, the role of miR-489 in chemoresistance was most studied. Jiang et al. showned that miR-489 regulates chemoresistance in breast cancer through the epithelial-mesenchymal transition (EMT) pathway [66]. Overexpression of miR-489 inhibits Smad3 expression and its related EMT activities, thus reversing adriamycin resistance in human breast cancer cells. This agrees with previously studies that connect endocrine resistance to EMT. Hiscox et al. showed that tamoxifen-resistant MCF7 cells had an altered morphology similar to cells undergoing EMT [67]. In addition, they showed a loss of association between β-catenin and E-cadherin, increase in the levels of both cytoplasmic and nucleic β-catenin, and increases in the transcriptions of its target genes like c-myc, cyclin D1, CD44, and COX-2. Creighton et al. showed that residual breast cancer cells surviving after letrozole or docetaxel treatment are CD44+/CD24−/low enriched [68]. These cells have an increased mammosphere-forming ability with gene expression patterns that correlate with the “claudin-low” breast cancer, a subtype that shows frequent up-regulation of mesenchymal genes. Therefore, even though there has not been any study showing the direct role of miR-489 in endocrine therapy resistance, miR-489 is likely to be involved in tamoxifen resistance through similar mechanisms.
Contrary to miR-221/222, miR-101 is capable of promoting estrogen-independent growth and conferring tamoxifen resistance without affecting ER expression or activity, implying that dysregulation of miR-101 in tumors could lead to intrinsic resistance to tamoxifen, regardless of ER status [69]. One of the notable molecular alterations caused by miR-101 is the upregulation of pAKT, which can be blocked by anti-miR-101, suggesting the specific effect of miR-101. miR-519a, a member of the miRNA cluster C19MC of around 50 mature miRNAs, was identified to be the most highly upregulated in MCF7 tamoxifen-resistant cells [70]. Overexpression of miR-519a increases viability and S-phase population of the cell cycle but decreases tamoxifen-induced apoptosis. Three tumor suppressor genes involved in PI3K signaling and the cell cycle, PTEN, CDKN1A/p21, and retinoblastoma protein (RB1) were validated to be direct targets of miR-519a. Tamoxifen-treated patients in the poor outcome group (based on random survival forests analysis) had significantly lower expression levels of PTEN, CDKN1A, and RB1, compared to those in the good survival outcome group. Another miRNA potentially involved in tamoxifen resistance is miR-301. It was first associated with tumor recurrence from a global miRNA profiling on 71 archival formalin-fixed, paraffin-embedded blocks of lymph node negative breast cancer patients who participated in a phase III clinical trial comparing tamoxifen versus tamoxifen plus breast radiotherapy [71]. miR-301 attenuation in MCF-7 and T47D cells increase tamoxifen sensitivity, even though only at a specific dose of tamoxifen (300 nmol/L). FOXF2, BBC3, PTEN, and COL2A1 have been verified as direct targets of miR-301, confirming its role in cell proliferation, migration, and tamoxifen resistance.
On the other hand, other miRNAs such as miR-375, miR-342 and miR-451 have been identified to be dramatically suppressed in tamoxifen-resistant breast cancer cells compared to parental cells [72–74]. Enforced expression of these miRNAs in tamoxifen-resistant cells was able to re-sensitize them to the drug. Of note, miR-451 expression was repressed exclusively by tamoxifen whereas E2, SERM raloxifene (Ral), and the SERD fulvestrant had no effect. Loss of miR-451 after tamoxifen treatment induced upregulation of 14-3-3ζ, which is associated to a poor clinical outcome for patients on tamoxifen treatment [75]. Let-7 family members were also identified to be repressed in tamoxifen-resistant cells and inversely correlated with ER-36 [76], the product of a transcript initiated from a promoter located in the first intron of the ER gene [77,78]. Overexpression of let-7b and let-7i miRNAs inhibited the expression of ER-36 and re-senitized tamoxifen-resistant cells to tamoxifen. Overexpression of ER-36 that lacks 3’UTR reverses the phenotype and restores the tamoxifen resistance. In a recent study, an oncogenic isoform of HER2, HER2D16, which promotes constitutive dimerization of the receptor, was identified to be co-expressed in a significant percentage of HER2- and ER-positive breast tumors [79]. HER2D16-expressing xenografts are both tamoxifen resistant and estrogen independent in part due to the upregulation of the antiapoptotic protein, BCL-2. This increased level of BCL-2 in HER2D16 expressing tamoxifen-resistant cells were associated to reduced levels of miR-15a/16 compared with tamoxifen-sensitive cell lines. Reintroduction of miR-15a/16 suppressed BCL-2 expression and sensitized resistant cells to tamoxifen. Conversely, suppression of miR-15a/16 resulted in upregulation of BCL-2 and converted sensitive cells to a tamoxifen-resistant phenotype, confirming that the loss of miR-15a/16 induced BCL-2 upregulation in breast cancer [80,81].
Several studies have demonstrated the association of selected miRNAs with clinical benefit of tamoxifen therapy. In one study, five miRNAs (miR-422a, miR-30a-, miR-187, miR-30c, and miR-182) were differentially expressed between ER-positive, lymph-node negative breast cancer patients who benefited, from those who failed, using tamoxifen monotherapy as the first-line treatment for their relapse [82]. The authors identified an association of high expression levels of miR-30a-3p, miR-30c, and miR-182 with clinical benefit of tamoxifen treatment and longer progression-free survival. However, only miR-30c was independent from traditional predictive markers, suggesting that measuring the expression of this miRNA may have added clinical value. In another study, miR-126 and miR-10a were identified as candidate tumor markers for recurrence in postmenopausal patients with ER-positive early stage breast cancer treated with tamoxifen [83]. Also, miR-26a was associated with better outcome for metastatic breast cancer patients on first-line tamoxifen monotherapy [84]. miR-26a repressed the EZH2 oncogene, which decreased mRNA level was also predictive for favorable outcome in metastatic breast cancer patients treated with tamoxifen.
microRNAs and Aromatase Inhibitors (AI)
Endocrine therapy agents have been developed to target hormone-dependent signaling pathways, either by antagonizing ER through the use of tamoxifen, or by inhibiting estrogen synthesis via AI such as letrozole, anastrozole and exemestane [85,86]. Endocrine therapy agents have shown excellent clinical efficacy, though the main hindrance with these agents is the development of acquired resistance and consequent loss of regulated growth [87]. Currently, few studies have concentrated their attention on the involvement of miRNAs in the response to AI and the acquisition of the resistance phenotype. A large group of miRNAs were identified as differentially expressed in the three AI-resistant cell lines compared to the parental cell line [88]. Among the miRNAs identified, miR-100/99a, miR-125b, miR-205 and miR-181a/b were selectively down-regulated in the AI-resistant cell lines. The authors also showed that miR-128a was strongly expressed in AI-resistant cells and played an important role in regulating TGFβR1 expression, leading to compromised growth inhibitory effects of TGFβ in letrozole-resistant breast cancer cells. Another study analyzed the expression of miRNAs after treatment with letrozole in MCF7 co-cultured with breast cancer stromal cells, which induced a 40% increase in the MCF7 aromatase expression [89]. The authors identified 18 miRNAs modulated after letrozole treatment with a significant increase of the tumor-suppressor let-7f. The increase of let-7f was also identified in two out of three patients who received neoadjuvant letrozole therapy. Although the number of clinical cases evaluated in the study was too small to draw any significant conclusion, these results suggested that the actions of letrozole on the aromatase gene via let-7f restoration may contribute to estrogen depletion and decreased tumor cell proliferation. The authors also demonstrated that let-7f directly represses the aromatase expression in MCF7 cells and let-7f expression was negatively correlated with the level of aromatase mRNA expression in both MCF7 cells and breast cancer tissues [89].
Despite proven clinical efficacy, aromatase inhibitor therapies still have a non-response rate of ~20%. To date, it remains impossible to accurately predict the response to such treatments. Keeping in mind the negative regulation of the aromatase expression by let-7f, the Chiorino’s group investigated the expression of 5 miRNAs predicted to target the aromatase (miR-378, miR-661, miR-1183, miR-1207-5p and miR-98) in a set of 37 breast tumor specimens of which 19 were classified as responder to the anastrozole therapy and 18 as non-responders [90]. Although no statistically significant correlation was observed, with neither aromatase expression nor response to anastrozole neoadjuvant treatment, it should be noted that the number of miRNAs predicted to target the aromatase is much larger (from TargetScan: miRNA families broadly conserved: 25; miRNA families only conserved in mammals: 18) and we can expect more discoveries on the roles of miRNAs in regulating aromatase in the near future.
microRNAs and Fulvestrant
As previously described, Fulvestrant (Faslodex; ICI 182780) is an FDA approved drug for use in postmenopausal patients following failure of first-line endocrine therapies (such as tamoxifen or aromatase inhibitors) [91]. Despite the potent antiestrogen effects of this drug, most breast cancer cases eventually develop resistance to fulvestrant through poorly understood mechanisms [92,93]. The first report on the involvement of miRNAs in fulvestrant resistance compared miRNA expressions in fulvestrant-resistant cells to the parental MCF7 cell line [94]. The authors identified 14 miRNAs downregulated such as miR-191, -181a, and -204, and only two up-modulated miRNAs, miR-222 and miR-221. The authors proposed the use of a combination of these modulated miRNA levels as a diagnostic biomarker to test the efficacy of fulvestrant therapy and evaluate the development of fulvestrant resistant tumors. In a more recent study, the Nephew group in collaboration with our laboratory shed light on the in vitro role played by the up-regulation of miR-222 and miR-221 in the acquisition of fulvestrant resistance [95]. First, we showed that ectopic up-regulation of miR-221 and miR-222 in ER positive cells significantly decreased fulvestrant-induced cell death, and thus it also contributes to hormone-independent growth and fulvestrant resistance. Then, by using genome wide analyses after knocking down miR-221/222 in fulvestrant-resistant cells, we identified multiple oncogenic signaling pathways regulated by miR-221/222 which are essential to the acquisition of fulvestrant resistance. One of the most strikingly regulated is the Wnt pathway, showing upregulation of CTNNB1 (β-catenin) and FZD5 12 hours after miR-221/222 overexpression [95]. Interestingly, even though miR-221/222-activated β-catenin supports estrogen-independent growth, the overexpression of β-catenin alone does not, indicating that miR-221/222 trigger multiple events that are required for the development of fulvestrant resistance. Further studies are needed to define the specific role of miR-221/222 in the development of fulvestrant resistance.
Nephew laboratory has also defined an integrative view of antiestrogen resistance by combining cancer contexts and expression profiles of miRNAs and mRNAs from tamoxifen- and fulvestrant-resistant cell lines in comparison to the parental MCF7 cells [96]. Several important network clusters were identified not only for antiestrogen resistance mechanisms but also for differentiating between resistance to tamoxifen and fulvestrant. In addition to the previously discussed miR-221/222 network, several other miRNA/mRNA clusters were identified to be important in the initiation and maintenance of antiestrogen resistance. An interesting finding is the opposite expression of miR-21/targets cluster in tamoxifen and fulvestrant resistant cell lines [96]. In fact, miR-21 was up-modulated in fulvestrant-resistant, but repressed in tamoxifen-resistant cells, consistent with the cell morphology changes associated with acquired antiestrogen resistance: tamoxifen resistant cells grow as tightly packed colonies with limited cell spreading, while fulvestrant resistant cells, by contrast, show reduced cell-cell contacts and are loosely attached to the culture surface.
In a recent study, Dr. Klinge and her collaborators showed a novel role of the miR-200 family members in increasing the sensitivity of endocrine-resistant LY2 breast cancer cells in response to tamoxifen and fulvestrant [97]. The authors reported a decrease in the expression of miR-200a, miR-200b, and miR-200c in endocrine resistant cells, leading to an increase in ZEB1 expression. The decrease of miR-200 family expression in resistant cells was due to epigenetic changes in the miRNA gene promoter. miR-200b and miR-200c expressions in LY2 were restored from treatment with demethylating agent 5-aza-2’-deoxycytidine and histone deacetylase inhibitor trichostatin A. Interestingly, overexpression of miR-200b and miR-200c enhanced the sensitivity of resistant cells to growth inhibition by antiestrogens 4-OHT and fulvestrant, defining miR-200 as a suppressor of endocrine resistance in breast cancer cells.
Currently, patients with ER-positive breast cancer are treated serially with different endocrine agents resulting in long periods of disease control with no significant toxicity. However, a large number of patients will become refractory to endocrine therapy and progress. The use of miRNA/anti-miRNA-based therapy with existing endocrine therapy may represent an important future step in the treatment of breast cancer to prevent and/or overcome endocrine resistance and benefit these breast cancer patients.
Expert Commentary
Hormonal therapy is the most common and effective therapy for patients with ER-positive breast cancer. However, resistance develops frequently and remains a major clinical problem. miRNAs has been shown in hormonal therapy resistance development and correlate with therapeutic outcome and response. Most previous studies focused on miRNAs in tamoxifen resistance, while studies on the involvement of miRNAs in fulvestrant and aromatase inhibitors, as well as multi-hormonal therapy resistance, are very limited. Further studies should aim at developing combination therapies that involve the use of current hormonal therapy along with the use of miRNAs and their inhibitors to maximize effectiveness and reduce or reverse resistance development.
Five Year View
Better understanding of the inter-regulatory relationship between ER and miRNAs can improve therapy development that helps reverse hormonal therapy resistance due to the loss of ER expression. Measurements of miRNA expressions in tumors and potentially serum may allow us to better predict and monitor response of hormonal therapy.
Conclusion
Breast cancer is not a unique cancer but rather a very heterogenous entity with different molecular profiles, biological behaviors, and clinical outcomes. There is an urgent need of a personalized therapy for the clinical management of early and advanced breast cancer patients. The identification of new prognostic and predictive factors is an essential step for the individualization of breast cancer therapy to guarantee the most active and optimal treatment to patients sparing unwanted side effects. As discussed in this review, miRNAs represent an emerging class of non-coding RNAs that play key roles in virtually all aspects of breast cancer. Abnormalities in their levels can have a widespread and profound effect on cell survival, growth, proliferation and response to adjuvant and endocrine chemotherapies. Yet, most of them have not been characterized for their specific biological activities or potential clinical applications. In this review we have described several works that showed how miRNAs can represent a new set of informative biomarkers for diagnosis and prognosis, and for predicting responses to endocrine therapy in breast cancer in the clinical setting. Unfortunately, the majority of the studies were conducted using in vitro cell lines. They lack interactions with the stroma and may not fully model the activities of patient-derived xenografts or clinical behaviors. Therefore, further studies are needed to confirm the roles of these miRNAs in clinics and whether miRNA profiling provides an advantage for cancer classification over more traditional approaches. As the roles of miRNAs in breast are better understood and novel delivery methods are developed and optimized, both miRNA-based targeted therapies and the manipulation of miRNA levels may emerge as significant novel therapeutic approaches for breast cancer treatment.
Table 1.
Summary of miRNA involved in tamoxifen resistance
| Upregulation | Downregulation | Mechanisms/ gene targets |
Reference |
|---|---|---|---|
| miR-221, 222, 181, 375, 32, 171, 213, 203 |
miR-342, 489, 21, 24, 27, 23, 200 |
miR-221/222: p27, TIMP3, ER; miR- 489: SMAD3 |
[56, 64–66] |
| miR-101 | E2-independent growth |
[69] | |
| miR-519a | PTEN, CDKN1A/p21, RB |
[70] | |
| miR-301 | FOXF2, BBC3, PTEN, COL2A1 |
[71] | |
| miR-375, 342, 451 | 14-3-3ζ | [72–74] | |
| let-7 (let-7b, 7i) | ER-36 | [75] | |
| miR-15a/16 | BCL-2 | [79–81] |
Table 2.
Summary of miRNA involved in AI resistance
Table 3.
Summary of miRNA involved in fulvestrant resistance
Key Issues.
-
-
ER-positive breast cancer accounts for more than 70% of breast cancers.
-
-
Hormonal therapy is a standard therapy for patients with ER-positive breast cancer. Three major types of hormonal therapy are currently available: tamoxifen, fulvestrant, and aromatase inhibitors.
-
-
Hormonal therapy resistance is common, i.e. 30% to tamoxifen and 20% to aromatase inhibitors.
-
-
ER and miRNAs are strongly inter-regulated. ER regulates the expression of numerous miRNAs transcriptionally and the expression of key components in miRNA biogenesis. At the same time, miRNAs regulate ER activity by translation inhibition, mRNA degradation, and targeting other proteins that cooperate with ER.
-
-
miRNAs have been shown to be involved in the resistance to all types of hormonal therapy, and therefore they are attractive therapeutic targets and markers.
Acknowledgments
The authors gratefully acknowledge the entire laboratory of Dr. Croce for helpful discussions and suggestions.
The authors were supported by a grant from National Institutes of Health/National Cancer Institute U01-CA152758 received by CM Croce. DG Cheung was funded by NIH-NIGMS Grant T32-GM086252 and the OSU/HHMI Med into Grad Scholars Program.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Eccles SA, Aboagye EO, Ali S, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15(5):R92. doi: 10.1186/bcr3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 3.Butt AJ, Sutherland RL, Musgrove EA. Live or let die: oestrogen regulation of survival signalling in endocrine response. Breast Cancer Res. 2007;9(5):306. doi: 10.1186/bcr1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou W, Slingerland JM. Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nat. Rev. Cancer. 2014;14(1):26–38. doi: 10.1038/nrc3622. [DOI] [PubMed] [Google Scholar]
- 5.Simpson ER. Sources of estrogen and their importance. J. Steroid. Biochem. Mol. Biol. 2003;86(3–5):225–230. doi: 10.1016/s0960-0760(03)00360-1. [DOI] [PubMed] [Google Scholar]
- 6.Folkerd EJ, Martin LA, Kendall A, Dowsett M. The relationship between factors affecting endogenous oestradiol levels in postmenopausal women and breast cancer. J. Steroid. Biochem. Mol. Biol. 2006;102(1–5):250–255. doi: 10.1016/j.jsbmb.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Visvanathan K, Hurley P, Bantug E, et al. Use of pharmacologic interventions for breast cancer risk reduction: American Society of Clinical Oncology clinical practice guideline. J. Clin. Oncol. 2013;31(23):2942–2962. doi: 10.1200/JCO.2013.49.3122. [DOI] [PubMed] [Google Scholar]
- 8.Jordan VC. Tamoxifen: catalyst for the change to targeted therapy. Eur. J. Cancer. 2008;44(1):30–38. doi: 10.1016/j.ejca.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krell J, Januszewski A, Yan K, Palmieri C. Role of fulvestrant in the management of postmenopausal breast cancer. Expert Rev. Anticancer Ther. 2011;11(11):1641–1652. doi: 10.1586/era.11.138. [DOI] [PubMed] [Google Scholar]
- 10.Chumsri S, Howes T, Bao T, Sabnis G, Brodie A. Aromatase, aromatase inhibitors, and breast cancer. J. Steroid. Biochem. Mol. Biol. 2011;125(1–2):13–22. doi: 10.1016/j.jsbmb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brueggemeier RW, Hackett JC, Diaz-Cruz ES. Aromatase inhibitors in the treatment of breast cancer. Endocr. Rev. 2005;26(3):331–345. doi: 10.1210/er.2004-0015. [DOI] [PubMed] [Google Scholar]
- 12.Ponzone R, Mininanni P, Cassina E, Pastorino F, Sismondi P. Aromatase inhibitors for breast cancer: different structures, same effects? Endocr. Relat. Cancer. 2008;15(1):27–36. doi: 10.1677/ERC-07-0249. [DOI] [PubMed] [Google Scholar]
- 13.Dunn BK, Cazzaniga M, DeCensi A. Exemestane: one part of the chemopreventive spectrum for ER-positive breast cancer. Breast. 2013;22(3):225–237. doi: 10.1016/j.breast.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Geisler J. Differences between the non-steroidal aromatase inhibitors anastrozole and letrozole--of clinical importance? Br. J. Cancer. 2011;104(7):1059–1066. doi: 10.1038/bjc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 18.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 20.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 21.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 24.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvinge H, Git A, Gräf S, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 26.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J. Exp. Med. 2011;208:2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Xu WD, Lu MM, Pan HF, Ye DQ. Association of microRNA-146a with Autoimmune Diseases. Inflammation. 2012;35:1525–1529. doi: 10.1007/s10753-012-9467-0. [DOI] [PubMed] [Google Scholar]
- 29.Andreopoulos B, Anastassiou D. Integrated analysis reveals hsa-miR-142 as a representative of a lymphocyte-specific gene expression and methylation signature. Cancer Inform. 2012;11:61–75. doi: 10.4137/CIN.S9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng X, Guo W, Liu T, et al. Identification of miRs-143 and-145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS ONE. 2011;6:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Han Q, Zhu Y, et al. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J. 2012;279:2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x. [DOI] [PubMed] [Google Scholar]
- 32.Git A, Spiteri I, Blenkiron C, et al. PMC42, a breast progenitor cancer cell line, has normal-like mRNA and microRNA transcriptomes. Breast Cancer Res. 2008;10:R54. doi: 10.1186/bcr2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Hu X, Ge S, Qian G, Zhang J. Regulation of RAP1B by miR-139 suppresses human colorectal carcinoma cell proliferation. Int. J. Biochem. Cell Biol. 2012;44:1465–1472. doi: 10.1016/j.biocel.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Castellano L, Giamas G, Jacob J, et al. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl Acad. Sci. USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volinia S, Galasso M, Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. USA. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. USA. 2013;110(18):7413–7417. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klinge CM. miRNAs and estrogen action. Trends Endocrinol. Metab. 2012;23(5):223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttilla IK, Adams BD, White BA. ERα, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol. Metab. 2012;23(2):73–82. doi: 10.1016/j.tem.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Kovalchuk O, Tryndyak VP, Montgomery B, et al. Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications and aberrant microRNA expression. Cell Cycle. 2007;6(16):2010–2018. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 41.Bhat-Nakshatri P, Wang G, Collins NR, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37(14):4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castellano L, Giamas G, Jacob J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. USA. 2009;106(37):15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cicatiello L, Mutarelli M, Grober OM, et al. Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am. J. Pathol. 2010;176(5):2113–2130. doi: 10.2353/ajpath.2010.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 45.Di Leva G, Piovan C, Gasparini P, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9(3):e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37(8):2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hah N, Danko CG, Core L, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grelier G, Voirin N, Ay AS, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br. J. Cancer. 2009;101(4):673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dedes KJ, Natrajan R, Lambros MB, et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur. J. Cancer. 2011;47(1):138–150. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Hinkal GW, Grelier G, Puisieux A, Moyret-Lalle C. Complexity in the regulation of Dicer expression: Dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br. J. Cancer. 2011;104(2):387–388. doi: 10.1038/sj.bjc.6606022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon SY, Lee JH, Kim B, et al. Complexity in Regulation of microRNA Machinery Components in Invasive Breast Carcinoma. Pathol. Oncol. Res. 2014;20(3):697–705. doi: 10.1007/s12253-014-9750-5. [DOI] [PubMed] [Google Scholar]
- 52.Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150(1):14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamagata K, Fujiyama S, Ito S, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol. Cell. 2009;36(2):340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 54.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007;21(5):1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 55.Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (Eralpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol. Endocrinol. 2009;23(8):1215–1230. doi: 10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 2008;283(45):31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Di Leva G, Gasparini P, Piovan C, et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010;102(10):706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res. Treat. 2012;132(1):75–85. doi: 10.1007/s10549-011-1534-y. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, Deng C, Wang J, et al. Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 2011;127(1):69–80. doi: 10.1007/s10549-010-0972-2. [DOI] [PubMed] [Google Scholar]
- 60.Leivonen SK, Makela R, Ostling P, et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28(24):3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 61.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26(21):8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351(9114):1451–1467. [PubMed] [Google Scholar]
- 63.Osborne CK. Steroid hormone receptors in breast cancer management. Breast Cancer Res. Treat. 1998;51(3):227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- 64.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu Y, Roy S, Nuovo G, et al. Anti-microRNA-222 (anti-miR-222) and-181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J. Biol. Chem. 2011;286(49):42292–42302. doi: 10.1074/jbc.M111.270926. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Jiang L, He D, Yanh D, et al. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett. 2014;588(11):2009–2015. doi: 10.1016/j.febslet.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 67.Hiscox S, Jiang W, Obermeier K, et al. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int. J. Cancer. 2006;118(2):290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- 68.Creighton C, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sachdeva M, Wu H, Ru P, Hwang L, Trieu V, Mo YY. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene. 2011;30:822–831. doi: 10.1038/onc.2010.463. [DOI] [PubMed] [Google Scholar]
- 70.Ward A, Shukla K, Balwierz A, et al. MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J. Pathol. 2014;233(4):368–379. doi: 10.1002/path.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi W, Gerster K, Alajez NM, et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71(8):2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 72.Cittelly DM, Das PM, Spoelstra NS, et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ward A, Balwierz A, Zhang JD, et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32(9):1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 74.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31(1):39–47. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frasor J, Chang EC, Komm B, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66(14):7334–7340. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Y, Deng C, Lu W, et al. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol. Med. 2011;17(11–12):1233–1241. doi: 10.2119/molmed.2010.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi L, Dong B, Li Z, et al. Expression of ER-{alpha}36, a novel variant of estrogen receptor {alpha}, and resistance to tamoxifen treatment in breast cancer. J. Clin. Oncol. 2009;27:3423–3429. doi: 10.1200/JCO.2008.17.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z, Zhang X, Shen P, et al. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen-and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc. Natl. Acad. Sci. USA. 2006;103:9063–9068. doi: 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cittelly DM1, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31(12):2049–2057. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 82.Rodríguez-González FG, Sieuwerts AM, Smid M, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 2011;127(1):43–51. doi: 10.1007/s10549-010-0940-x. [DOI] [PubMed] [Google Scholar]
- 83.Hoppe R, Achinger-Kawecka J, Winter S, et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur. J. Cancer. 2013;49(17):3598–3608. doi: 10.1016/j.ejca.2013.07.145. [DOI] [PubMed] [Google Scholar]
- 84.Jansen MP, Reijm EA, Sieuwerts AM, et al. High miR-26a and low CDC2 levels associate with decreased EZH2 expression and with favorable outcome on tamoxifen in metastatic breast cancer. Breast Cancer Res. Treat. 2012;133(3):937–947. doi: 10.1007/s10549-011-1877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Santen RJ, Brodie H, Simpson ER, Siiteri PK, Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr. Rev. 2009;30(4):343–375. doi: 10.1210/er.2008-0016. [DOI] [PubMed] [Google Scholar]
- 86.Lønning PE, Eikesdal HP. Aromatase inhibition 2013: clinical state of the art and questions that remain to be solved. Endocr. Relat. Cancer. 2013;20(4):R183–R201. doi: 10.1530/ERC-13-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beelen K, Zwart W, Linn SC. Can predictive biomarkers in breast cancer guide adjuvant endocrine therapy? Nat. Rev. Clin. Oncol. 2012;9(9):529–541. doi: 10.1038/nrclinonc.2012.121. [DOI] [PubMed] [Google Scholar]
- 88.Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res. Treat. 2010;124(1):89–99. doi: 10.1007/s10549-009-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shibahara Y, Miki Y, Onodera Y, et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J. Pathol. 2012;227(3):357–366. doi: 10.1002/path.4019. [DOI] [PubMed] [Google Scholar]
- 90.Ghimenti C, Mello-Grand M, Grosso E, et al. Regulation of aromatase expression in breast cancer treated with anastrozole neoadjuvant therapy. Exp. Ther. Med. 2013;5(3):902–906. doi: 10.3892/etm.2012.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ciruelos E, Pascual T, Arroyo Vozmediano ML, et al. The therapeutic role of fulvestrant in the management of patients with hormone receptor-positive breast cancer. Breast. 2014;23(3):201–208. doi: 10.1016/j.breast.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 92.Howell A. Preliminary experience with pure antiestrogens. Clin. Cancer Res. 2001;7(12 Suppl):4369s–4375s. [PubMed] [Google Scholar]
- 93.Howell A, Abram P. Clinical development of fulvestrant ("Faslodex") Cancer Treat. Rev. 2005;31(Suppl 2):S3–S9. doi: 10.1016/j.ctrv.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 94.Xin F, Li M, Balch C, et al. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25(4):430–434. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rao X, Di Leva G, Li M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30(9):1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nam S, Long X, Kwon C, Kim S, Nephew KP. An integrative analysis of cellular contexts, miRNAs and mRNAs reveals network clusters associated with antiestrogen-resistant breast cancer cells. BMC Genomics. 2012;13:732. doi: 10.1186/1471-2164-13-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manavalan TT, Teng Y, Litchfield LM, Muluhngwi P, Al-Rayyan N, Klinge CM. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLoS One. 2013;8(4):e62334. doi: 10.1371/journal.pone.0062334. [DOI] [PMC free article] [PubMed] [Google Scholar]