Abstract
Drug development targeting fibroblast growth factor receptors (FGFRs) represents an emerging theme in the field of medicinal chemistry. Considering the fact that most of the currently identified FGFR agonists are long chain peptides with limited stability, the discovery of novel non-peptide FGFR ligands is still highly demanded. A linear one-bead-one-compound peptoid (oligomers of N-substituted glycine units) library with a theoretical diversity of 106 was designed and synthesized. Microarray-based screening led to the identification of four hit sequences 1-4 as FGFR1α ligands, which were further confirmed using both solution-phase and solid-phase binding assays. Western blot results indicated that peptoids 2-4 activated FGFR signaling pathways, resulting in increased levels of p-Akt and p-ERK in different cell lines. Our work discovered novel peptoid ligands as FGFR agonists, shedding new light on FGFR-based drug discovery.
Introduction
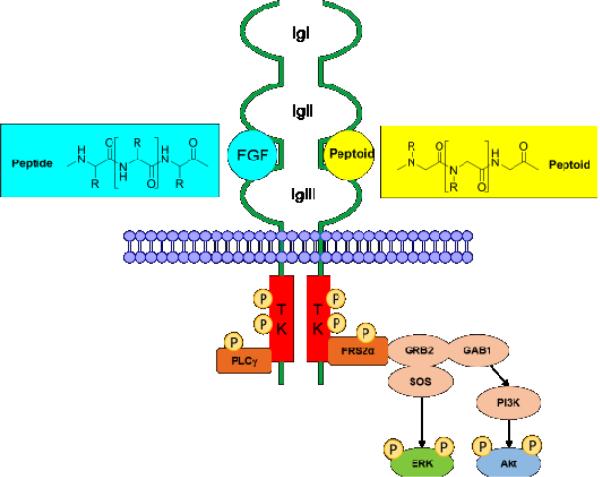
Fibroblast growth factor receptors (FGFRs) consist of an extracellular ligand-binding region composed of three immunoglobulin (Ig)-like modules (IgI-III), a single transmembrane (TM) domain, and a split tyrosine-kinase (TK) domain (Fig. 1). There are four FGFRs in mammals, and alternative splicing of FGFR 1, 2, and 3 in the third Ig-like loop domain further gives rise to IIIb and IIIc isoforms.1
Fig. 1. FGF-FGFR structure and downstream signaling pathway.
The extracellular domain of FGFRs consists of three ligand binding Ig domains (IgI-III). Binding of FGFs to the FGFRs results in receptor dimerization and autophosphorylation of the tyrosine kinase (TK) domains. Upon phosphorylation, the major FGFR effector FRS2 acts as a docking site for GRB2, which then activates extracellular signal-regulated kinase (ERK) and Akt via SOS and GAB1.
Binding of FGFs to the FGFRs results in receptor dimerization, followed by kinase activation, and autophosphorylation of tyrosine residues in the intracellular domain of FGFR. The phosphorylation of tyrosine residues activates FGFR substrate 2α (FRS2α), which further phosphorylates the mitogen-activated protein kinase (MAPK) and phosphoinositide-3 kinase (PI3K) pathways, leading to increased phosphorylation of ERK and Akt (Fig. 1). Other effector proteins including phospholipase-Cγ (PLCγ), Src homologous and collagen A (ShcA), and STAT transcription factors are also activated by FGFRs.2-4
In humans, 22 members of the FGF family have been identified, all of which are structurally and functionally related polypeptide growth factors with the molecular mass ranges from 17 to 34 kDa. FGFs are closely involved in angiogenesis, wound healing, embryonic development and various endocrine signaling pathways.5 In addition to the cognate FGFs, FGFRs also interact with a number of cell adhesion molecules, such as the neural cell adhesion molecule (NCAM), which plays multiple roles in nervous system development and maintenance, and modulates neuronal plasticity.6, 7
Due to the essential roles of FGF/FGFR signaling in angiogenesis,8-10 wound healing, cell migration, neural outgrowth and embryonic development, drug development targeting FGFRs becomes an emerging theme in the field of medicinal chemistry. During the past few years, several peptide agonists of FGFRs have been identified, such as canofins, hexafins, and dekafins, all of which are based on the crystal structures of FGFs-FGFR complexes.11-13 Upon binding to and activating FGFRs, these peptide agonists lead to induction of neurite outgrowth and protection against apoptosis in vitro.14 Recently, FGF-P, a small peptide derived from FGF-2, was found to be a potential mitigator of acute gastrointestinal syndrome (AGS) due to ionizing radiation (IR).15, 16 In addition, peptides agonists of FGFR derived from NCAM have also been reported, such as Encamin, FGL peptide, BCL peptide.17-19 Considering the complexity and high cost of the synthesis of long-chain peptide ligands, as well as the instability of peptides toward proteolysis, the identification of novel non-peptide FGFR ligands in a straightforward and low-cost manner will certainly shed new light on FGFR-targeted drug discovery. However, the development of non-peptide ligands of FGFR remains largely unexplored.
Peptoids are oligomers of N-substituted glycine (NSG) units (Fig. 1), which represent a class of polymers as peptide mimics. Compared with peptides, the side chains in peptoids are attached to the nitrogen instead of the α-carbon.20 As a result, peptoids are incapable of forming hydrogen bonds characteristic of the secondary structures found in peptides. Distinctive advantages of peptoids over peptides include 1) enhanced stability toward proteolysis; 2) resistance to denaturation induced by solvent, temperature, or chemicals; 3) better cell penetration; and 4) low immunogenicity. Peptoids can be easily synthesized based on the two-step submonomer solid-phase synthetic method including acylation and amination; this method uses diverse primary amines as building blocks with high availability at low cost.21 More importantly, large one-bead-one-compound (OBOC) peptoid libraries can be efficiently synthesized using the “split-and-pool” method,22 and the high compatibility with existing screening methods for OBOC libraries makes peptoid libraries an ideal tool for identifying ligands toward various biological targets.23 Previously, two peptoids as orexin receptor (OxR1) antagonists discovered from a 3 million library provided a platform to identify G protein-coupled receptor (GPCR) ligands using quantum dot labeled and cell-based screening protocol.24 To expand our research scope to other biological targets of great importance, herein, we reported the discovery of novel FGFR agonists from peptoid library using microarray-based screening.
Chemical microarrays composed of peptides, carbohydrates, small molecules and other biological molecules enable investigators to rapidly analyze molecular interactions between immobilized molecules and complex biological mixtures in parallel.25-27 Combinatorial chemistry and chemical microarray techniques have already proven to be effective tools for biomedical research, including protein binding studies, enzyme-substrate and inhibitor studies, and diagnostic studies.28-33
Experimental
Peptoid library design and synthesis
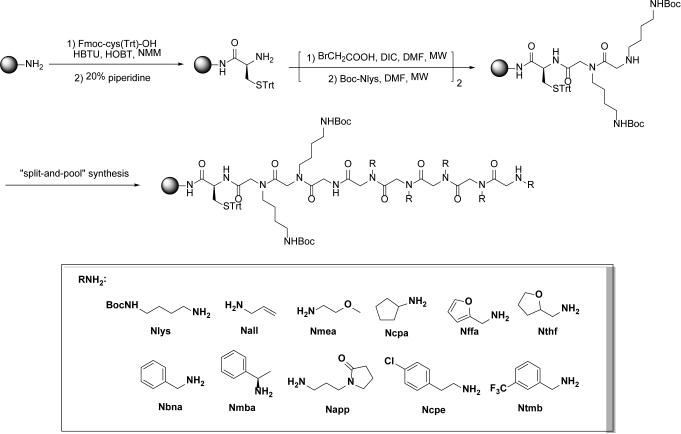
A one-bead-one-compound peptoid library was constructed by split-and-pool synthesis on Rink-Amide beads. An invariable sequence (Cys-Nlys-Nlys), which precedes the variable region, was linked to the bead. The thiol residue in Cysteine was utilized to spot the peptoids onto maleimide-modified glass slides; the Nlys-Nlys linker provides a positive charge to aid in ionization in the mass spectrometer. Ten amines were used to construct the variable region. The theoretical diversity of the library was 106.
The peptoid library was synthesized starting from Rink-Amide aminomethyl polystyrene resins (H50056023, particle size: 500 μm, capacity: 0.53 mmol/g, Rapp Polymere, Germany). Rink-Amide beads were swelled in dimethylformamide (DMF) for 1 h, and were treated twice with 20% piperidine in DMF for 1 h to remove the Fmoc group. Fmoc-Cys (Trt)-OH was coupled with 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole (HOBT), N-methylmorpholine (NMM) in DMF, and the Fmoc group was removed with 20% piperidine in DMF. The two Nlys sequences were attached using traditional peptoid synthetic method assisted by microwave. The beads were treated with 2 M bromoacetic acid and 2 M diisopropylcarbodiimide (DIC) in DMF, and the reaction was performed in a microwave oven set to 10% power for a total of 30 s. After thoroughly washed with DMF, the beads were then treated with Nlys solution (2 M in anhydrous DMF) in microwave oven as described above. The variable peptoid region was constructed using split-and-pool protocol and the same acylation and amination procedures as Nlys were used and repeated until the desired length was achieved.
To test the quality of the library, 30 beads were randomly selected. Each bead was treated with 20 μL of cleavage cocktail containing 95% trifluoroacetic acid (TFA), 2.5% triisopropylsilane, and 2.5% water for 1 h to cleave peptoids from the resins as well as to remove all the side chain protecting groups. The cleavage mixture was evaporated, and the resulting residue was dissolved in acetonitrile/water (80:20). The structure of each peptoid was decoded using the 3200 QTRAP® System (AB Sciex, Redwood City, CA, USA). The parent peak was identified using Q1 mode, and the sequence of each peptoid was determined using Enhanced Product Ion (MS/MS) mode.
Immobilization of peptoid library to glass slides
The above prepared peptoid library was thoroughly washed with dichloromethane to remove any DMF, and each bead was isolated into a single well of 96-well plates. Each well was then treated with 95% TFA cleavage cocktail as described above. The cleavage mixture was dried, and the resulting residue in each well was dissolved in 20 μL of DMSO. After that, 10 μL of the DMSO solution in each well of the 96-well plates was transferred into 384-well master plates using multichannel pipettes. Peptoid arrays were then prepared by immobilizing each peptoid in the 384-well plates onto maleimide-modified glass slide using Arrayit SpotBot® Extreme Microarray Spotters (Arrayit Corporation, Sunnyvale, CA, USA). Each peptoid was spotted onto the slide in triplicates using 946MP4 pins, and the glass slide was allowed to dry overnight after printing.
Screening for FGFR1α ligands
The glass slide was washed with 1X TBST buffer and blocked with 5% bovine serum albumin (BSA) in TBST for 1 h at room temperature. The slide was then incubated with 1 μg/mL of recombinant human FGFR1α (IIIc) Fc chimera (658-FR-050, R&D, Minneapolis, MN, USA) in TBST at 4 °C for 1 h with gentle shake. Unbound FGFR was thoroughly washed off with TBST (3 × 15 min). The slide was further incubated with 3 μg/mL of FITC-labeled anti-Fc (F9512, Sigma) at 4 °C for 1 h. After that, the slide was washed with TBST (3 × 15 min), dried, and visualized using GenePix 4000B Microarray Scanner (Molecular Devices, Sunnyvale, CA, USA). Hit peptoids were selected according to the intensity of the spots’ fluorescence. The structures of selected peptoids were elucidated by MS/MS spectrometry using their corresponding stock solutions.
On-bead validation of binding affinity
Hit peptoids and two randomly-selected control peptoids were resynthesized on TentaGel beads (MB160002, particle size: 150 μm, capacity: 0.3 mmol/g, Rapp Polymere, Germany) using the same method as described above. The beads were washed with DMF/H2O with a successive ratio of 90:10, 50:50, 0:100 and then with TBST. The beads were blocked with 5% BSA in TBST at 4 °C overnight with gentle shake. After blockade, the beads were incubated with 1 μg/mL of recombinant human FGFR1α (IIIc) Fc chimera (658-FR-050, R&D, Minneapolis, MN, USA) in TBST at 4 °C for 1 h, washed with TBST, and treated with 2 μg/mL of Texas Red-labeled anti-Fc (SAB3701287, Sigma) in TBST at 4 °C for 1 h. After washing with TBST, beads were visualized using the Olympus IX73 inverted microscope system with a Texas Red filter.
Resynthesized of soluble peptoids
Resynthesis of hit peptoids and two randomly-selected control peptoids was conducted on Rink-Amide AM resin (855004, 200-400 mesh, Novabiochem, Germany). The general peptoid synthesis protocol as described in the library synthesis section was used to build the peptoid portion. After the synthesis, peptoids were cleaved off the resin using a 95% TFA, 2.5% triisopropylsilane, and 2.5% water mixture for 2 h at room temperature, and the crude product was purified by reverse-phase preparative HPLC using acetonitrile-water (0.1% TFA) as the eluents.
Cell culture
MCF-7, MDA-231, and NIH3T3 cell lines were purchased from ATCC (Manassas, VA, USA). Dulbecco's Modified Eagle Medium (DMEM), and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NE, USA). All the three cell lines were cultured in DMEM medium supplied with 10% FBS and 1% antibiotic in a humidified atmosphere at 37 °C with 5% CO2.
Cell lysates preparation
Cells were plated in 100-mm dishes with 10 mL DMEM containing 10% FBS and 1% antibiotic. After growing to a confluence of around 80%, cells were subjected to serum deprivation with 9 mL DMEM containing 1% antibiotic overnight. Cells were then treated with 1 mL of DMEM with or without peptoids for indicated time. Cells were washed PBS and lysed in 400 μL lysis buffer containing 50 mM Tris-HCl (pH 8.0), 137 mM NaCl, 1 mM EDTA, 50 mM NaF, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1% Triton X-100, and 10% glycerl. Lysates were collected and centrifuged for 15 min at 25,000 × g at 4 °C. The protein concentrations were measured using Bradford assay. All the cell lysates were stored at −80 °C.
Western blot
Cell lysates containing 20-40 μg total protein were mixed with protein loading buffer, heated for 5 min at 95 °C, and subjected to SDS-polyacrylamide (12%) gel electrophoresis. Proteins were transferred onto nitrocellulose mini membrane using Trans-Blot® Turbo™ Transfer System (Bio-Rad, CA, USA). Membranes were blocked with 5% BSA in TBST for 1 h at room temperature, incubated with specific primary antibody (concentrations according to suppliers) at 4 °C overnight, washed with TBST (3 × 15 min), incubated with horseradish peroxidase (HRP)-linked secondary antibody for 1 h at room temperature, and washed with TBST (3 × 15 min). Enhanced chemiluminescence detection was carried out using Supersignal® West Pico Chemiluminescent Substrate Kits (1856135, 1856136, Thermo Scientific), and the blots were visualized using ChemiDoc™ MP Imaging System (Bio-Rad, CA, USA). The following primary antibodies were used: Phospho-p44/42 MAPK XP® Rabbit mAb (#4370, Cell Signaling, Danvers, MA, USA), phosphor-Akt XP® Rabbit mAb (#4060, Cell Signaling, Danvers, MA, USA), and β-Actin Mouse mAb (#3700, Cell Signaling, Danvers, MA, USA).
RESULTS AND DICUSSION
Preparation of OBOC peptoid library
To explore the potential of discovering novel FGFR agonists from peptoids, a linear OBOC peptoid library was constructed on 500 μm Rink Amide beads (Fig. 2). Briefly, Rink Amide beads were first attached with a triphenylmethyl (Trt)-protected cysteine residue, which was utilized to spot the peptoids onto maleimide-modified glass slides in the following procedure.34 Next, an Nlys-Nlys linker was introduced as the invariant region, which provides a positive charge to aid in ionization in the mass spectrometer. Following the invariant linker, split-and-pool synthesis using the standard sub-monomer method for peptoids was employed to construct the variable region of the OBOC library.21,22,35 The split-and-pool protocol ensures that each bead expresses only one compound and each compound exhibits equal distribution in the library. The variable region was diversified at six positions using random substitutes (R groups) derived from 10 different primary amines (Fig. 2), affording a library with a theoretical diversity of one million (106) compounds. The quality of the constructed library was tested by cleaving 30 beads randomly selected from the library and sequencing the cleaved products using MS/MS. In each case, a single and strong molecular ion was observed, and the molecules could be sequenced unambiguously by MS/MS. The spectrum of 10 peptoids were presented in the Supporting Information as examples.
Fig. 2. Preparation of the OBOC peptoid library.
The diversity of the library: 106 = 1,000,000 compounds. Beads: Rink-Amide aminomethyl polystyrene resins, particle size: 500 μm, capacity: 0.53 mmol/g.
Microarray-based screening targeting FGFR
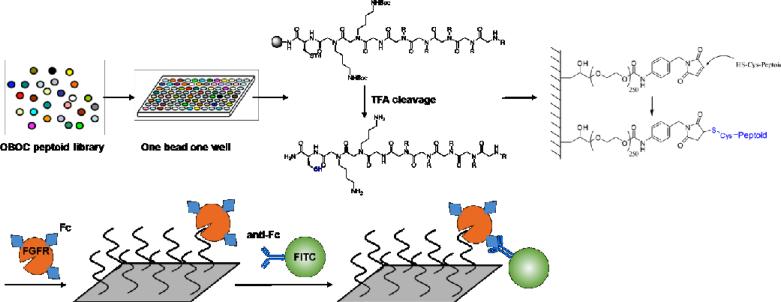
The procedure of microarray-based screening of the OBOC library targeting FGFR was illustrated in Fig. 3. Each bead in the library was isolated into a single well of a 96-well plate and treated with cleavage cocktail to release the peptoid from each bead. Both the Trt and Boc protecting groups were also removed simultaneously under this condition. The cleavage mixture was evaporated and redissolved in dimethyl sulfoxide (DMSO). Half of the solution in each well was transferred into 384-well master plates, and the remaining solution in 96-well plates was kept for structure decoding after screening.
Fig. 3.
Schematic representation of identifying FGFR ligands from peptoid OBOC library based on microarray technology.
The most commonly used solid support for microarray printing is a standard microscope glass slide, and automatic arrays have been developed to print chemical microarrays through the surface contact between the solid support and the tip of the needle or pin. In this study, using the Arrayit SpotBot® Extreme Microarray Spotters, peptoids in 384-well master plates were immobilized onto a maleimide-modified glass slide through Michael reaction between the thiol group on the C-terminal cysteine of the peptoid and the maleimide residue on the slide.36 Each peptoid was printed onto the slide in triplicates.
FGFR1α, which plays important roles in angiogenesis, embryonic development, cell proliferation, cell differentiation, and wound healing,37 was employed as the target for subsequent screening as a proof of concept. After washing with TBST buffer and blocked with 5% bovine serum albumin (BSA), the glass slide immobilized with peptoid array was incubated with Fc-modified FGFR1α (1 μg/mL in TBST) at 4 °C for 1 h with gentle shake. The slide was thoroughly rinsed with TBST to wash off any unbound FGFR1α. Then the slide was further incubated with FITC-labeled anti-Fc (3 μg/mL in TBST) at 4 °C for 1 h. Thus, positive peptoids showing FGFR1α binding affinities were labeled with FITC through the Fc/anti-Fc interaction (Fig. 3).
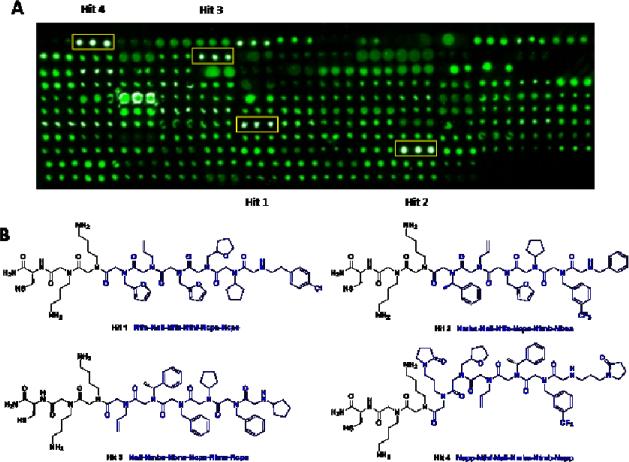
After the slide was prepared, high-throughput screening was conducted using GenePix Microarray Scanner according to the intensity of the spots’ fluorescence on the slide. As shown in Fig. 4A, four peptoids (1-4) displayed significantly more prominent fluorescence compared with others, and the fluorescence intensity of each of these peptoids showed good consistency among the triplicate spots, indicating the molecules attached at these spots as hit peptoids toward FGFR1α. Thus, the stock solutions of these four hit peptoids were subjected to MS/MS spectrometry to elucidate their chemical structures. The sequence of all the four peptoids were unambiguously interpreted (see supporting information), and their structures were shown in Fig. 4B.
Fig. 4.
(A) Micro array Scanner image of the peptoid-immobilized slide after incubation with Fc-modified FGFR1α and then FITC-labeled anti-Fc. Peptoids were printed onto the slide in triplicates. Four hit peptoids which showed stronger fluorescence compared with surrounding spots are framed in yellow rectangles. (B) Chemical structures of the four hit peptoids (1-4) as confirmed by MS/MS, the structure and sequence of the variable region of each peptoid was shown in blue.
Validation of the binding ability of hit peptoids
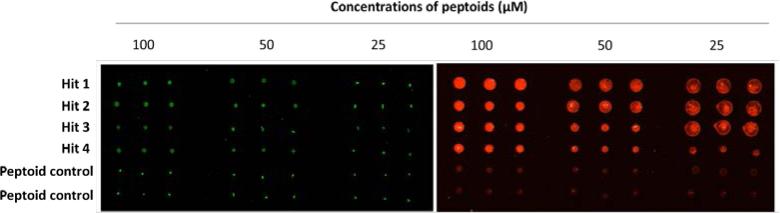
In order to confirm the binding affinity of the four hit peptoids toward FGFR1α, peptoids 1-4 as well as two randomly selected negative peptoids (see supporting information) were reprinted onto maleimide-modified glass slide at three concentrations (100, 50, and 25 μM) in triplicates. The slide was visualized under GenePix Microarray Scanner, and the image was shown in Fig. 5 (left). The green color indicated the intrinsic florescence from peptoids, and equal intensity was observed for both hit peptoids and peptoid controls at all the three concentrations. The same incubation procedure as shown in Fig. 3 was then applied to the slide, except that Texas Red-labeled anti-Fc was used instead of FITC-labeled anti-Fc. The slide was re-visualized and the image was shown on the right panel of Fig. 5. Apparently, the spots representing the four hit peptoids exhibited much stronger florescence than the two peptoid controls at all the three concentrations, demonstrating the high affinity of peptoids 1-4 toward FGFR1α.
Fig. 5. Confirmation of the binding affinity of peptoids 1-4 toward FGFR1 α.
Hit peptoids 1-4 and two randomly selected negative peptoids were printed onto maleimide-modified slide in triplicates at three concentrations (100, 50, and 25 μM). The slide was visualized under GenePix Microarray Scanner before (left) and after (right) incubation with Fc-modified FGFR1α and Texas Red-labeled anti-Fc.
To further validate the binding ability of the hit peptoids toward FGFR1α, on-bead binding assay was conducted after peptoids 1-4 were resynthesized on TentalGel beads (150 μM). Beads bearing one of the four hit peptoids were washed with TBST, blocked with 5% BSA, and incubated with Fc-tagged FGFR1α (1 μg/mL in TBST) and Texas Red-labeled anti-Fc (2 μg/mL in TBST) sequentially. Meanwhile beads bearing a randomly selected peptoid were subjected to the same treatment as above as a control. Beads were then observed directly under fluorescence microscope using Texas Red filter, and the image was shown in Fig. 6 using 2 and 4 as an example. It is evident that beads bearing peptoid 2 or 4 are much brighter than the control under the same condition, further confirming the binding affinity of the hit peptoids toward FGFR1α.
Fig. 6. On-bead validation of the binding affinities of peptoids 2 and 4 toward FGFR1α.
Peptoids were synthesized on TentalGel beads. Beads were washed with TBST, blocked with 5% BSA, and incubated with Fc-tagged FGFR1α and Texas Red-labeled anti-Fc sequentially. Another pool of beads bearing randomly selected peptoid was subjected to the same treatment as a negative control. Beads were observed under microscope (4X) using Texas Red filter.
Effects of hit peptoids on FGFR signaling pathways
After the binding affinity of the four hit peptoids was confirmed by both solution-phase (Fig. 5) and solid-phase (Fig. 6) assays, we next aimed to investigate whether the binding of the peptoids to FGFR were able to activate the receptor. As is introduced above, FGFR agonists lead to the activation of various intracellular signaling pathways, resulting in the phosphorylation of several downstream protein targets, such as Akt and ERK (Fig. 1). Therefore, we examined the expression levels of p-Akt and p-ERK upon peptoids treatment using Western blot.
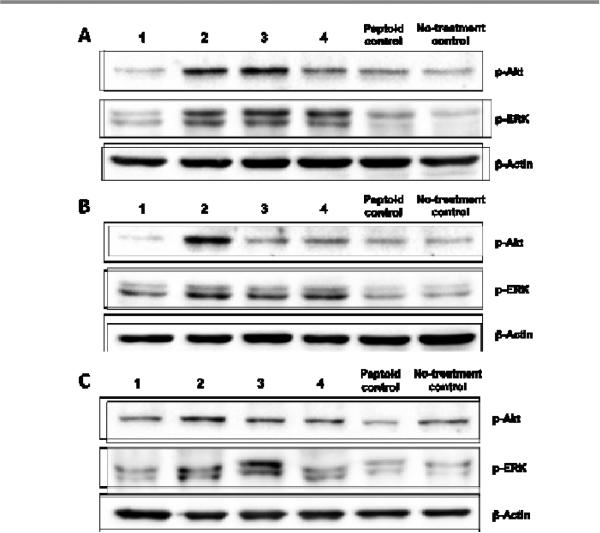
Three cell lines were selected including human breast cancer cell lines MCF-7 and MDA-231 and mouse embryonic fibroblast cell NIH-3T3. It was previously reported that MCF-7 cells have greater levels of FGFR1 and that MDA-231 cells have greater relative levels of FGFR2.38 In addition, NIH-3T3 cells are known to express predominantly FGFR1,39,40 and are commonly used in studying FGFR signaling pathways.41,42 After serum starvation, all the three cell lines were treated with peptoids 1-4 and a randomly selected peptoid (peptoid control 1, supporting information) at a concentration of 15 μM for 10 min. Cell lysates were then prepared, and the expression levels of p-Akt and p-ERK were detected using Western blot. Generally speaking, the effects of 1-4 on p-Akt and p-ERK were consistent among the three cell lines. As shown in Fig. 7, peptoid 1 exhibited negligible effects on the expression levels of p-Akt and p-ERK in all of the three cell lines, suggesting that peptoid 1, although acts as a FGFR1α ligand, cannot activate the FGFR1α downstream signaling pathways. However, treatment with peptoids 2-4 resulted in apparently increased phosphorylation of Akt and ERK to different extents in different cells. The most prominent effects of 2-4 on the levels of p-Akt and p-ERK were observed in MCF-7 cells (Fig. 7A), which predominantly express FGFR1, indicating that 2-4 might have higher specificity for FGFR1. In addition, peptoid 2 led to a significantly increased phosphorylation of Akt in MDA-231 cells compared to other peptoid ligands (Fig. 7B). Taken together, the Western blot results demonstrated that peptoids 2-4 identified using microarray-based screening showed FGFR downstream signaling activating effects.
Fig. 7.
Effects of hit peptoids 1-4 on the expression levels of p-Akt and p-ERK in (A) MCF-7, (B) MDA-231, and (C) NIH-3T3 cells. After serum starvation , all of the three cell lines were treated with peptoids 1-4 and a randomly selected peptoid control at a concentration of 15 μM for 10 min. The expression levels of p-Akt and p-ERK were detected by Western blot using specific antibodies.
Conclusion
Mounting evidence nowadays suggests the potential role of FGF/FGFR signaling pathway as an attractive target in drug discovery. The close involvement of FGF/FGFR in angiogenesis, wound healing, embryonic development, and various endocrine signaling pathways highlights the necessity of discovering novel FGFR ligands with diverse chemical structures, although numerous FGFR peptide ligands have been documented. Against this backdrop, and as a continuing exploration in our lab to identify peptoid ligands toward biologically significant targets, a linear OBOC peptoid library was designed and synthesized with high efficiency using combinatorial chemistry method (Fig. 2). Microarray-assisted high-throughput screening (Fig. 3) led to the rapid identification of four peptoids that were able to bind to FGFR1α (Fig. 4). The binding affinity of the hit peptoids were further validated in both solution-phase (Fig. 5) and solid-phase (Fig. 6) assays. The effects of the hit peptoids on FGFR signaling pathways were investigated in different cell lines using Western blot. It was found that peptoids 2-4 led to increased phosphorylation of Akt and ERK (Fig. 7). Particularly, peptoid 2 proved to be the most potent p-Akt and p-ERK activator, which elevate the expression of p-Akt and p-ERK in both a dose- and time- dependent manner and compete with recombinant human FGF-2 in competitive ELISA assay, suggesting that 2 is a potent FGFR agonist (unpublished data). Taken together, our results demonstrated the feasibility of identifying non-peptide FGFR ligands using both chemical and biological methods. Future studies are aimed to further validate the agonist effects of hit peptoids on FGFR and evaluate the biological activities of these novel peptoid FGFR agonists. These work are now actively pursued in our lab to explore their potential application in clinical use.
Supplementary Material
Acknowledgements
This work was supported by grants from American Cancer Society Chris DiMarco Institutional Research Grant to XQ, UF Interdisciplinary Center for Biotechnology Research (ICBR) Agilent Microarray Program Award to XQ, the Seed Funds from the Emerging Pathogens Institute to XQ and in part by the NIH/NCATS Clinical and Translational Science Award to the University of Florida UL1 TR00064.
Footnotes
The authors declare no competing interests.
Electronic Supplementary Information (E SI) available: and experimen tal details. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Mohammadi M, Olsen SK, Ibrahimi OA. Cytokine Growth F. Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Eswarakumar VP, Lax I, Schlessinger J. Cytokine Growth F. Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Cytokine Growth F. Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Beenken A, Mohammadi M. Nat. Rev. Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh N, Ornitz DM. J. Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinsby AM, Berezin V, Bock E. Front. Biosci. 2004;9:2227–2244. doi: 10.2741/1393. [DOI] [PubMed] [Google Scholar]
- 7.Cambon K, Hansen SM, Venero C, Herrero AI, Skibo G, Berezin V, Bock E, Sandi C. J. Neurosci. 2004;24:4197–4204. doi: 10.1523/JNEUROSCI.0436-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andre F, Cortes J. Breast Cancer Res. Treat. 2015;150:1–8. doi: 10.1007/s10549-015-3301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong L, Han Y, Liu J, Brain L. Expert Rev. Gastroent. 2013;7:759–765. doi: 10.1586/17474124.2013.837804. [DOI] [PubMed] [Google Scholar]
- 10.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, Helfrich BA, Bunn PA, Jr., Heasley LE. Mol. Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manfe V, Kochoyan A, Bock E, Berezin V. J. Neurochem. 2010;114:74–86. doi: 10.1111/j.1471-4159.2010.06718.x. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Christensen C, Kohler LB, Kiselyov VV, Berezin V, Bock E. Dev. Neurobiol. 2009;69:837–854. doi: 10.1002/dneu.20740. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Christensen C, Kiselyov VV, Kohler LB, Bock E, Berezin V. J. Neurochem. 2008;104:667–682. doi: 10.1111/j.1471-4159.2007.05070.x. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Bock E, Berezin V. Int. J. Mol. Sci. 2010;11:2291–2305. doi: 10.3390/ijms11062291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Sun W, Wang J, Zhang M, Yang S, Tian Y, Vidyasagar S, Pena LA, Zhang K, Cao Y, Yin L, Wang W, Zhang L, Schaefer KL, Saubermann LJ, Swarts SG, Fenton BM, Keng PC, Okunieff P. Int. J. Radiat. Oncol. Biol. Phys. 2010;77:261–268. doi: 10.1016/j.ijrobp.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Tian Y, Yin L, Zhang M, Beck LA, Zhang B, Okunieff P, Zhang L, Vidyasagar S. Int. J. Radiat. Oncol. Biol. Phys. 2011;81:248–254. doi: 10.1016/j.ijrobp.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen SM, Kohler LB, Li S, Kiselyov V, Christensen C, Owczarek S, Bock E, Berezin V. J. Neurochem. 2008;106:2030–2041. doi: 10.1111/j.1471-4159.2008.05544.x. [DOI] [PubMed] [Google Scholar]
- 18.Jacobsen J, Kiselyov V, Bock E, Berezin V. Neurochem. Res. 2008;33:2532–2539. doi: 10.1007/s11064-008-9680-2. [DOI] [PubMed] [Google Scholar]
- 19.Leong QM, Lai HK, Lo RG, Teo TK, Goh A, Chow PK. J. Vasc. Interv. Radiol. 2009;20:833–836. doi: 10.1016/j.jvir.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Zuckermann RN. Biopolymers. 2011;96:545–555. doi: 10.1002/bip.21573. [DOI] [PubMed] [Google Scholar]
- 21.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Am. Chem. Soc. 1992;114:10646–10647. [Google Scholar]
- 22.Figliozzi GM, Goldsmith R, Ng S, Banville SC, Zuckermaru RN. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 23.Fu J, Lee T, Qi X. Future Med. Chem. 2014;6:809–823. doi: 10.4155/fmc.14.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi X, Astle J, Kodadek T. Mol. Biosyst. 2010;6:102–107. doi: 10.1039/b915611a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumaresan PR, Lam KS. Mol. Biosyst. 2006;2:259–270. doi: 10.1039/b602004f. [DOI] [PubMed] [Google Scholar]
- 26.Lam KS, Renil M. Curr. Opin. Chem. Biol. 2002;6:353–358. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 27.Ma H, Horiuchi KY. Drug Discov. Today. 2006;11:661–668. doi: 10.1016/j.drudis.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Uttamehandani M, Sun H, Yao SQ. QSAR Comb. Sci. 2006;25:1009–1019. [Google Scholar]
- 29.Barry CE, Wilson M, Lee R, Schoolnik GK. Int. J. Tuberc. Lung Dis. 2000;4:S189–S193. [PubMed] [Google Scholar]
- 30.Lam KS, Liu RW, Miyamoto S, Lehman AL, Tuscano JM. Acc. Chem. Res. 2003;36:370–377. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]
- 31.Puckett JW, Muzikar KA, Tietjen J, Warren CL, Ansari AZ, Dervan PB. J. Am. Chem. Soc. 2007;129:12310–12319. doi: 10.1021/ja0744899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho D, Dose C, Albrecht CH, Severin P, Falter K, Dervan PB, Gaub HE. Biophys. J. 2009;96:4661–4671. doi: 10.1016/j.bpj.2009.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu XB, Talukder P, Bhattacharya C, Fahmi NE, Lines JA, Dedkova LM, LaBaer J, Hecht SM, Chen SX. Bioorg. Med. Chem. Lett. 2014;24:5699–5703. doi: 10.1016/j.bmcl.2014.10.058. [DOI] [PubMed] [Google Scholar]
- 34.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Chem. Biol. 2010;17:38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. J. Am. Chem. Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 36.MacBeath G, Koehler AN, Schreiber SL. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 37.Groth C, Lardelli M. Int. J. Dev. Biol. 2002;46:393–400. [PubMed] [Google Scholar]
- 38.Nurcombe V, Smart CE, Chipperfield H, Cool SM, Boilly B, Hondermarck H. J. Biol. Chem. 2000;275:30009–30018. doi: 10.1074/jbc.M003038200. [DOI] [PubMed] [Google Scholar]
- 39.Maher PA. J. Cell Biol. 1996;134:529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhen Y, Sorensen V, Jin Y, Suo Z, Wiedlocha A. Oncogene. 2007;26:6372–6385. doi: 10.1038/sj.onc.1210473. [DOI] [PubMed] [Google Scholar]
- 41.Ren M, Qin H, Ren R, Tidwell J, Cowell JK. Cancer Res. 2011;71:7312–7322. doi: 10.1158/0008-5472.CAN-11-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Hutter B, Jager N. Nat. Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.