The number of human protein kinases is greater than 500, and they can be classified as tyrosine kinases, serine or threonine kinases, and protein kinases with dual specificity for both tyrosine and serine/threonine phosphorylation. Our work and others’ have expanded the protein kinome by adding a new branch of protein kinases classified as metabolic enzymes. Pyruvate kinase M2 (PKM2), which is a glycolytic enzyme that catalyzes conversion of phosphoenolpyruvate and ADP to pyruvate and ATP, respectively, is the first glycolytic enzyme identified as a protein kinase.1 PKM2 phosphorylates substrate proteins at tyrosines, serines, or threonines in a signaling context- and subcellular compartment-dependent manner. It is known that PKM2 phosphorylates many critical cellular proteins, including histone H3 at T11, at STAT3 at Y705, Bub3 at Y207, myosin light chain 2 (MLC2) at Y118, and mTORC1 inhibitor AKT1 substrate 1 (AKT1S1) at S202/203, thereby regulating the Warburg effect, gene transcription, G1-S phase transition, mitosis, cytokinesis, and mTORC1 activation.2 Recently, we demonstrated for the first time that phosphoglycerate kinase 1 (PGK1), the first ATP-generating glycolytic enzyme known to catalyze the conversion of 1,3-diphosphoglycerate and ADP to 3-phosphoglycerate and ATP, respectively, acts as a protein kinase, phosphorylating pyruvate dehydrogenase kinase-1 (PDHK1) at T338 in mitochondria. This phosphorylation activates PDHK1, inhibits mitochondrial pyruvate metabolism, and promotes glycolysis.3 Thus, we established that PKM2 and PGK1, the only 2 ATP-generating glycolytic enzymes, function as protein kinases and play instrumental roles in tumor development.
In addition to glycolysis, fructose metabolism is a critical component of cell metabolism, and aberrant fructose metabolism leads to liver diseases.4 In the fructose metabolic pathway, fructokinase, which is also known as ketohexokinase (KHK) and is a rate-limiting metabolic enzyme, catalyzes the conversion of fructose and ATP into fructose 1-phosphate and ADP. Fructose 1-phosphate is metabolized into dihydroxyacetone phosphate and glyceraldehyde by aldolase and subsequently converge in the glycolysis pathway.5 Mutually exclusive splicing of adjacent exons 3A and 3C in KHK precursor RNA has resulted in expression of KHK isoform A (KHK-A or fructokinase A) or C (KHK-C or fructokinase C).6 Whereas KHK-A is ubiquitously expressed at low levels, KHK-C is predominantly highly expressed in liver, kidney, and pancreatic cells. KHK-C, which has high binding affinity to fructose compared to that of KHK-A, exhibits much greater activity toward fructose phosphorylation than does KHK-A.4 In a recent study, we demonstrated that KHK-A is the dominant KHK isoform expressed in hepatocellular carcinoma (HCC) cells, resulting in much lower fructose catabolism rates in HCC cells than in normal hepatocytes.
To identify the mechanism of switching of expression from KHK-C to KHK-A in HCC cells, we used biotinylated RNA containing the sequence spanning the 3' end of exon 3A or 3C and its adjacent intron as bait to identify the splicing regulators associated with these regions. We found that 2 splicing-regulatory proteins, hnRNPH1 and hnRNPH2, bind to a motif located in the intron adjacent to the 3' end of exon 3C. Depletion of hnRNPH1 and hnRNPH2 by expression of their short hairpin RNA inhibited the inclusion of exon 3A and reversed the expression from that of KHK-A to that of KHK-C in HCC cells. Examination of the promoters of hnRNPH1 and hnRNPH2 revealed a noncanonical E-box that can be bound by the oncogenic transcription factor c-Myc. Knockdown of c-Myc expression resulted in the same effect induced by depletion of hnRNPH1 and hnRNPH2 on KHK isoform expression, indicating that c-Myc upregulates hnRNPH1/2 expression and subsequently induces KHK-A expression in HCC cells.
To characterize the functions of KHK-A in HCC cells, we performed mass spectrometric analysis of KHK-associated proteins and found that KHK-A but not KHK-C interacted with phosphoribosyl pyrophosphate synthetase 1 (PRPS1), a rate-limiting enzyme in the de novo nucleic acid synthesis pathway. Importantly, an in vitro phosphorylation assay demonstrated that KHK-A functions as a protein kinase and directly phosphorylates PRPS1 at T225. This phosphorylation blocked the binding of the allosteric inhibitor ADP to PRPS1, thereby abrogating the feedback inhibition by ADP and leading to elevated de novo nucleic acid synthesis by constitutively activating PRPS1 in HCC cells (Fig. 1). Suppression of a splicing switch from KHK-C to KHK-A via depletion of hnRNPH1 and hnRNPH2 or expression of the phosphorylation-resistant PRPS1 T225A mutant dramatically inhibited HCC proliferation and tumor growth in mouse livers. Immunohistochemical staining of human HCC tissue demonstrated that KHK-A and PRPS1 pT225 were positively correlated with each other and that the levels of KHK-A and PRPS1 pT225 staining were inversely correlated with survival duration in HCC patients, supporting a pivotal role for KHK-A–dependent PRPS1 phosphorylation in HCC progression.7
Figure 1.
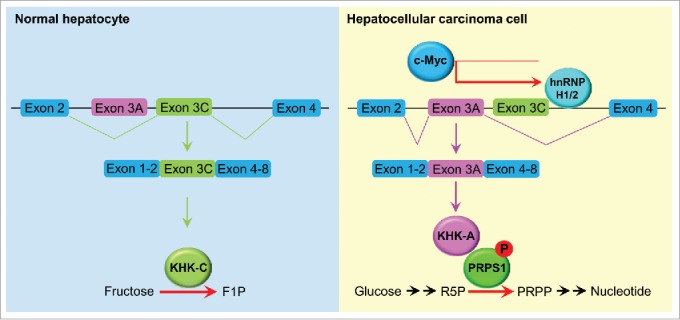
A splicing switch from KHK-C to KHK-A promotes HCC formation. KHK-C, the isoform of KHK with high activity toward fructose phosphorylation, was predominantly expressed in normal hepatocytes. In HCC cells, high expression of c-Myc enhanced the expression of hnRNPH1 and hnRNPH2, which bound to the exon 3C-3'/intron region of KHK, leading to a splicing switch from KHK-C to KHK-A and reduced fructose metabolism. KHK-A phosphorylated and activated PRPS1, resulting in increased de novo nucleic acid synthesis for hepatocellular tumorigenesis. F1P, fructose 1-phosphate; R5P, ribose-5-phosphate; PRPP, phosphoribosyl pyrophosphate.
In summary, we provided important insight into integrated regulation of cancer cell metabolism and other critical cell activities by identifying a new class of protein kinases with the dual roles of metabolic enzymes and protein kinases and by characterizing their functions in regulation of tumor cell proliferation. Disrupting the activity of these newly identified protein kinases, including PKM2, PGK1, and KHK-A, may be an effective approach to cancer treatment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Lu Z. PKM2 functions as a histone kinase. Cell Cycle 2012; 11:4101-2; PMID:23070542; http://dx.doi.org/ 10.4161/cc.22325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li X, Zheng Y, Lu Z. PGK1 is a new member of the protein kinome. Cell Cycle 2016; 15(14):1803-4; PMID:27105392; http://dx.doi.org/ 10.1080/15384101.2016.1179037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Zia Y, Aldape K, He J, Hunter T, Wang L, Lu Z.. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell 2016; 61:705-19; PMID:26942675; http://dx.doi.org/ 10.1016/j.molcel.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ishimoto T, Lanaspa MA, Le MT, Garcia GE, Diggle CP, MacLean PS, Jackman MR, Asipu A, Roncal-Jimenez CA, Kosugi T, et al.. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proc Natl Acad Sci U S A 2012; 109:4320-5; PMID:22371574; http://dx.doi.org/ 10.1073/pnas.1119908109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lyssiotis CA, Cantley LC. Metabolic syndrome: F stands for fructose and fat. Nature 2013; 502:181-2; PMID:24108049; http://dx.doi.org/ 10.1038/502181a [DOI] [PubMed] [Google Scholar]
- [6].Bonthron DT, Brady N, Donaldson IA, Steinmann B. Molecular basis of essential fructosuria: molecular cloning and mutational analysis of human ketohexokinase (fructokinase). Hum Mol Genet 1994; 3:1627-31; PMID:7833921; http://dx.doi.org/ 10.1093/hmg/3.9.1627 [DOI] [PubMed] [Google Scholar]
- [7].Li X, Qian X, Peng L-X, Jiang Y, Hawke DH, Zheng Y, Xia Y, Lee J-H, Cote G, Wang H, et al.. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol 2016; 18:561-71; PMID:27088854; http://dx.doi.org/ 10.1038/ncb3338 [DOI] [PMC free article] [PubMed] [Google Scholar]


