ABSTRACT
PICH is an SNF2-family DNA translocase that appears to play a role specifically in mitosis. Characterization of PICH in human cells led to the initial discovery of “ultra-fine DNA bridges” (UFBs) that connect the 2 segregating DNA masses in the anaphase of mitosis. These bridge structures, which arise from specific regions of the genome, are a normal feature of anaphase but had escaped detection previously because they do not stain with commonly used DNA dyes. Nevertheless, UFBs are important for genome maintenance because defects in UFB resolution can lead to cytokinesis failure. We reported recently that PICH stimulates the unlinking (decatenation) of entangled DNA by Topoisomerase IIα (Topo IIα), and is important for the resolution of UFBs. We also demonstrated that PICH and Topo IIα co-localize at the rDNA (rDNA). In this Extra View article, we discuss the mitotic roles of PICH and explore further the role of PICH in the timely segregation of the rDNA locus.
KEYWORDS: Chromosome condensation, DNA translocases, mitosis, topoisomerase II, ultra-fine bridges
Introduction
A number of daunting challenges must be overcome every time a cell commits to entering a new cell division cycle. For example, the more than 3 billion base pairs of DNA in a human cell must be copied exactly once, and the resulting sister chromatids must be faithfully segregated to each daughter cell. Mitosis presents a particular challenge for several reasons. First, if any unresolved DNA catenation arising during DNA replication persists between the sister chromatids when the cell enters mitosis, disjunction of sisters can be compromised (reviewed in1). Second, the DNA must be condensed evenly in early mitosis in order to ensure that the chromosomes acquire sufficient structural rigidity to withstand the forces of the mitotic spindle (reviewed in2). Third, the mitotic spindle has to attach correctly to each pair of kinetochores to allow the sister chromatids to be accurately and evenly disjoined (reviewed in3). Finally, the cohesin protein that embraces the sisters after S-phase must be dissociated to permit successful disjunction.4 If any of these processes encounters problems or delays, then the cell needs to be able to respond through the rapid mobilization of genome maintenance factors.
Even though cell division is a remarkably efficient process, problems often arise in mitosis, and chromosome segregation can be compromised. It was recognized decades ago that defective chromosome segregation was quite common. Examples of aberrant DNA structures that are detectable in the anaphase of mitosis include chromatin bridges and lagging chromosomes. While these can also occur in unperturbed cells, they are strongly induced by genome stressors, or in cells defective in genome maintenance factors (reviewed in5). One such factor, PICH, is the focus of this article.
PICH is a member of the SWI/SNF2 family of proteins, which play diverse roles in all aspects of chromosome structure and function.6 PICH belongs to a sub-class of SNF2 proteins related to the RAD54 protein, which is involved in homologous recombination.7 In addition to a canonical SNF2-like ATPase domain, PICH contains a so-called PICH family domain of unknown function.8 What makes PICH unusual among SNF2 family proteins is the fact that it is excluded from the nucleus during interphase, and only gains access to chromatin after nuclear envelope breakdown in early prometaphase.8 Thereafter, PICH apparently localizes to various ‘problematic’ regions of chromosomes in prometaphase and metaphase, and then to unprocessed DNA structures in anaphase. Indeed, PICH is a well-established marker of ultra-fine DNA bridges (UFBs), which are thread-like structures that connect the 2 separating sister masses in anaphase.8-11 UFBs are composed of DNA but, unlike the more widely studied chromatin bridges, cannot be detected using conventional DNA dyes and do not contain histones. Despite being discovered relatively recently, UFBs are now known to be far more prevalent than chromatin bridges in cultured cells. UFBs arise from a number of specific chromosomal loci, including centromeres8, telomeres11 and common fragile sites.10 These loci have in common a tendency to show atypical patterns of DNA replication, largely due to their ‘late’ timing of DNA replication, or their intrinsic propensity to stall replication forks. Irrespective of their origin, most, if not all, UFBs are bound by PICH. A curious feature of this association is that PICH often decorates each UFB along its entire length, irrespective of the stage of anaphase.8,9 One explanation for this unusual behavior is that PICH has an inherent capacity to bind to DNA that is stretched – such as might be expected for UFBs subjected to the tension on DNA created by mitotic spindle forces.12 Recent evidence suggests that UFB-like structures might also exist that lack PICH, which are defined by the localization of the DNA repair and replication protein, TOPBP1.13,14 Hence, it will be intriguing to define in the future the nature of these apparently novel DNA structures.
A number of studies have characterized the phenotype of cells with compromised PICH function. Using RNAi-based protein depletion approaches, a consensus has emerged that PICH is required for chromosome maintenance, but some phenotypic variation has been reported following PICH depletion.8,15-20 In an effort to clarify any discrepancies, our laboratory, in collaboration with the Nigg and Wang laboratories, generated chicken and human cell lines completely lacking PICH via targeted gene inactivation.21,22 Consistent with a role for PICH in faithful genome maintenance, the PICH-deficient chicken cells had increased levels of chromatin bridging, micronuclei, and polyploidy.21 These PICH−/− cells were also hypersensitive to the Topo II inhibitor, ICRF-193. Through the analysis of PICH−/− chicken cells expressing either wild-type or ATPase-dead human PICH (K128A) fused to mCherry, we characterized a number of mitotic roles for PICH.21 First, we demonstrated that the ATPase activity of PICH is required for efficient UFB resolution. Importantly, we were also able to observe the fate of cells with persistent/unprocessed UFBs that are decorated with PICH-K128A, but cannot be resolved effectively. These cells became binucleated and, in the subsequent S-phase, polyploid. Hence, it would appear that unprocessed UFBs lead to cytokinesis failure. Second, we revealed that PICH localizes to the rDNA locus in the nucleolar organizer region (NOR). This novel mitotic localization pattern of PICH had previously been overlooked in human cell lines, and was only revealed through analysis of chicken DT40 cells. This was because the rDNA is tightly clustered in the chicken genome, unlike in human cells where it is dispersed over the short arms of the acrocentric chromosomes. This allowed us to observe large PICH-positive ‘bodies’ in the chicken cells, which we subsequently identified as the rDNA cluster. We then confirmed that PICH also localized with rDNA loci in human cells. Finally, we demonstrated that Topo IIα co-localizes with PICH at the rDNA, and also on UFBs in anaphase, suggesting that the ICRF-193 sensitivity of the PICH−/− cells is due to incomplete resolution of these structures. To further explore this relationship, we purified human PICH and Topo IIα and demonstrated that PICH strongly stimulates the decatenation activity of Topo IIα in vitro. Nevertheless, this stimulation was independent of the ATPase activity of PICH, and therefore the functional role of the ATP-driven translocation activity of PICH remains unclear. Based on these observations, we proposed that PICH and Topo IIα functionally cooperate to resolve UFBs and catenated rDNA in mitosis.
Our previous study21 left several open questions regarding the role of PICH at the rDNA locus. For instance, why do PICH and Topo IIα co-localize at the rDNA locus, and how does PICH deficiency affect the integrity of the rDNA cluster in chicken cells? Here, we elaborate on our previous findings, and provide additional evidence for a role of PICH in the maintenance of rDNA stability.
Results
PICH and UBF occupy different regions of the rDNA
Previously,21 we demonstrated that PICH and the rDNA-specific marker, upstream binding factor (UBF), co-localize in large foci (PICH bodies) in the NORs in prometaphase and metaphase chicken DT40 cells. In anaphase, the PICH bodies segregate into the 2 daughter cells.21 In cells expressing PICH, UBF foci disappear during late anaphase, whereas cells expressing ATPase-dead PICH-K128A exhibit persistent foci, which often remain throughout mitosis until the next G1 phase. Two DAPI-negative PICH bodies/UBF foci can usually be observed in prometaphase and metaphase, with one focus being consistently larger than the other (which we refer to as the major and the minor locus).
To investigate more precisely how PICH interacts with NORs, we employed super resolution imaging using structured illumination microscopy and 3D rendering to provide a more detailed image of the composition of PICH bodies. Interestingly, we observed that PICH and UBF exhibited a mutually exclusive localization pattern at both the major and the minor rDNA locus (Fig. 1). Nevertheless, PICH and UBF appeared to form an intimately associated structure, with UBF concentrated in the center (Fig. 1). In prometaphase, metaphase and anaphase cells, the staining patterns observed for PICH and PICH-K128A were generally very similar. In late anaphase and telophase cells, however, PICH staining at the UBF foci generally became much weaker, while staining for PICH-K128A persisted.21
Figure 1.
PICH and UBF occupy different regions of the rDNA. PICH−/− cells ectopically expressing either hPICH (left) or hPICH-K128A (right) were arrested in prometaphase using nocodazole, released from this arrest, and then imaged at different stages of mitosis using structured illumination microscopy. Images were 3D rendered. PICH (red) and UBF (green) were detected using specific antibodies, and DNA (grayscale) was stained with DAPI. The small white boxes denote zoomed images of the minor NOR, and the large white boxes denote the major NORs. Scale bars, 5 μm.
PICH does not regulate rDNA transcription
UBF localizes only at rDNA sites that are transcriptionally active, irrespective of whether the cells are in interphase or mitosis.23,24,25 This is because UBF is believed to assist in the loosening of compacted, transcriptionally silent, rDNA units.23 The silencing of rDNA transcription that occurs in mitosis is induced by phosphorylation and deacetylation of the rDNA transcription factor complex SL1 governed by CDK1/cyclin B26,27 and SIRT1,28 respectively. These modifications prevent SL1 from interacting with UBF and the rDNA promoter.26,28,29 Thus, the occupancy of SL1 and RNA Pol I at the rDNA promoter decreases during mitosis.28 However, UBF is still maintained23-25,30 at rDNA units that were transcribed in the preceding interphase.
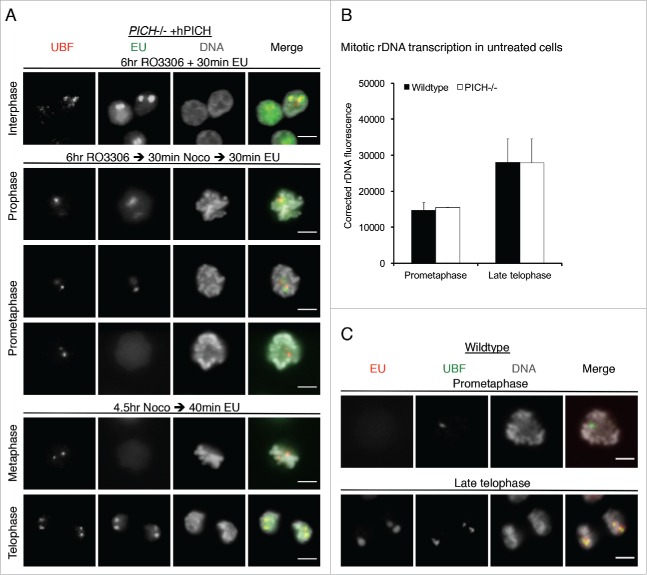
One possible explanation for the mutually exclusive pattern of UBF and PICH localization at the rDNA locus is that PICH is required for modulation of rDNA transcription. To test this hypothesis, we measured incorporation of 5-ethynyl-uridine (EU) at rDNA loci, which labels sites of ongoing transcription, in PICH deficient cells expressing hPICH or hPICH-K128A. To investigate transcription occurring specifically in mitosis, cells were synchronised in late G2 with the CDK1 inhibitor, RO3306, and then released synchronously into prophase. In the RO3306-arrested G2 cells, we observed that all rDNA sites marked by UBF were EU positive (Fig. 2A; top panel). In cells incubated with EU only after they were released from a G2 arrest, rDNA transcription began to decline during prophase and prometaphase, and was undetectable by late prometaphase (Fig. 2A; middle). Therefore, rDNA transcription is progressively repressed as cells traverse prophase and prometaphase under our experimental conditions. In cells arrested in prometaphase with nocodazole, and then released into medium containing EU, rDNA transcription was not detectable from late prometaphase onwards, and only resumed again when cells entered late telophase (Fig. 2A; bottom). The EU foci in these telophase cells co-localized with UBF foci, confirming that they denote NORs and the active rDNA loci. The silencing of transcription in mid-mitosis was still detectable in PICH-deficient cells (not shown), which indicates that PICH is not required for this effect.
Figure 2.
PICH does not appear to regulate rDNA transcription. (A) PICH−/− +hPICH cells were incubated with EU and imaged by immunofluorescence microscopy. EU (green) was detected using Alexa Fluor 488 azide, UBF (to denote the rDNA; red) was detected using a specific antibody, and the DNA (grayscale) was stained with DAPI. Scale bars, 5 μm. In the top panel, cells were arrested in G2 by RO3306 treatment for 6 hours before labeling with EU. In the middle panels, cells were released from the RO3306 arrest into medium containing nocodazole for 30 mins before labeling with EU. Cells in either prophase or prometaphase are shown. In the bottom panels, cells were arrested in prometaphase for 4.5 hours using nocodazole, and then released for 30 mins before labeling with EU. Cells in either metaphase or telophase are shown. (B/C) Asynchronously growing wild-type and PICH−/− cells were incubated with EU for 15 mins, and then imaged by immunofluorescence microscopy. rDNA transcription around the major DT40 rDNA locus was quantified in prometaphase and in late telophase cells (panel B) as described in the Materials and Methods. More than 100 prometaphase or late telophase cells were quantified in 3 independent experiments. Data points are averages ± s.d. (C) Representative examples of wild-type cells in prometaphase and late telophase stained for EU (red) and UBF (green). DNA (grayscale) was stained with DAPI. Scale bars, 5 μm.
To confirm that PICH is not involved in regulating rDNA transcription, we quantified EU incorporation in asynchronous wild-type DT40 and PICH−/− cells using UBF as a marker of the rDNA. In this analysis, the average fluorescence intensity of EU at the major rDNA locus was very similar between wild-type and PICH−/− cells in prometaphase or late telophase (Fig. 2B). Representative examples of images of wild-type cells are shown in Figure 2C. We conclude that, although PICH co-localizes with Topo IIα at the rDNA in prometaphase, it is very unlikely to modulate rDNA transcription. This contrasts with a proposed role for Topo IIα in this process.31
The rDNA marks a novel class of UFBs
In chicken cells in anaphase, we observed that PICH bodies were often elongated and lagging behind the bulk chromosomal DNA, suggestive of DNA bridging of the locus. Using super resolution microscopy, we could readily observe DAPI-negative DNA bridges at the rDNA loci in early anaphase (Fig. 3A). Thus the rDNA defines a fourth class of UFBs, along with centromeric, telomeric and fragile-site-associated UFBs. On these rDNA bridge structures, the mutually exclusive localization pattern of PICH and UBF was still evident, with each protein occupying independent domains along the extended locus (Fig. 3A). UBF localization did not require the ATPase activity of PICH (Fig. 3A).
Figure 3.
The rDNA marks a novel class of UFBs. (A,C-E) PICH−/− cells ectopically expressing either hPICH or hPICH-K128A were arrested in prometaphase using nocodazole then released into drug-free medium or medium containing 0.25 μM ICRF-193 for 40 min, as indicated. (A-C) PICH (red) and UBF (green) were detected using specific antibodies and the DNA (grayscale) was stained with DAPI. Scale bars, 5 μm. In panel A, cells were imaged using structured illumination microscopy and the images were 3D rendered. White boxes denote zoomed images. (B) An example of a DAPI-negative rDNA-UFB in an untreated PICH−/− cell. (C) Representative examples of rDNA-UFBs arising from either the major or the minor rDNA locus in PICH deficient cells expressing either wild-type hPICH or hPICH-K128A (as indicated). (D/E) Quantification of total UFBs per anaphase (left), of rDNA-UFBs/anaphase (middle), and of rDNA-UFBs/UFB (right) in PICH deficient cells expressing either wild-type hPICH or hPICH-K128A (as indicated). Each data point is an average of at least 3 independent experiments. Significance levels were calculated using a Students t-test for parametric observations and are indicated as *P < 0.05, **P < 0.01 and ***P < 0.001.
Next, we investigated whether UBF might reveal the presence of rDNA-UFBs in cells lacking PICH. Indeed, rDNA-UFBs marked by UBF were detectable in PICH−/− cells (Fig. 3B) suggesting that rDNA-UFBs arise independently of PICH protein. We also observed that the rDNA could still be connected by UFBs in late anaphase (Fig. 3C), and that these UFBs could originate from either the minor or the major rDNA locus (Fig. 3C). Quantification of UFBs revealed that more than 10% of the spontaneous UFBs in chicken cells were derived from the rDNA. Interestingly, the hPICH-K128A cells exhibited twice as many rDNA-UFBs as compared to the hPICH cells, suggesting that the ATPase activity of PICH is important for resolution of rDNA-UFBs (Fig. 3D). When cells were released from nocodazole arrest into medium containing ICRF-193, the overall number of UFBs increased markedly both in cells expressing hPICH and those expressing hPICH-K128A (Figs. 3D and 3E). This increase in the frequency of ICRF193-induced UFBs has been attributed previously8,10 to an increase in centromeric UFBs. However, we observed that the frequency of rDNA-UFBs was also increased by ICRF-193 (Figs. 3D/E), suggesting that Topo IIα modulates the formation or resolution of both centromeric and rDNA-UFBs.
Discussion
Our recent findings have revealed a novel role for PICH in the maintenance of rDNA stability. Chicken cells were shown previously to harbour one cluster of 400 rDNA repeats on micro-chromosome 16.32 However, it appears that 2 rDNA clusters are detectable in the DT40 cell clone that we utilized in our studies (one ‘major’ and one ‘minor’). Interestingly, PICH is recruited to both of these rDNA clusters in mitosis, but the functional significance of this association was not clear previously. To address this, we used super resolution microscopy to further analyze the composition and behavior of PICH bodies at the DT40 rDNA cluster in mitosis.
In lower eukaryotes, the rDNA is frequently the most problematic genomic locus to segregate in anaphase. For example, the rDNA array on ChrXII in budding yeast is often the last genomic region to segregate and can be stranded in the bud neck while the remaining chromosomes have visibly segregated between the mother and daughter cells.33 One potential explanation for this phenomenon is that rDNA transcription continues until the last possible moment in mitosis in order to maintain a continuous supply of rRNA and hence ribosomes.34 Thus, the rDNA initiates DNA condensation later than the bulk of the genome, and hence has less time to prepare for anaphase.33,35 However, ChrXII is the longest chromosome in yeast and it was shown recently that the length of the chromosome alone determines the timing of segregation of a chromosome lacking rDNA repeats.36 These data, therefore, challenge the notion that segregation of ChrXII is delayed because of the presence of the rDNA array.
In human cells, global gene expression is repressed from early prophase. Transcription of the rDNA ceases by late prophase,25 and resumes again in telophase.28,37 The silencing of rDNA transcription is believed to take place prior to nuclear envelope breakdown, and is accompanied by disassembly of the nucleolus.37,38 In contrast, in chicken DT40 cells, we could still detect ongoing transcription around UBF foci after nucleolar disassembly in early prometaphase cells. Whether this reflects a DT40-specific phenomenon, or is due to our synchronisation procedure (i.e. CDK1 inhibition) remains to be determined. In agreement with previous reports, we confirmed that rDNA transcription re-starts in late telophase. Based on the mutually exclusive localization pattern of PICH and UBF in PICH bodies, we hypothesized that PICH might be required for transcriptional silencing at the rDNA. However, we observed that rDNA transcriptional silencing in mid-mitosis could still occur normally in PICH-deficient cells. Likewise, the re-initiation of rDNA transcription in late telophase was unaffected by PICH-deficiency.
Another important event that follows transcriptional silencing is DNA condensation. Due to the late transcriptional shut-off at the rDNA loci, vertebrate cells have only a relatively short period between early prometaphase and anaphase to effect DNA condensation and prepare for sister chromatid disjunction. Consistent with active condensation occurring in PICH bodies, we demonstrated previously that Topo IIα is also present at the rDNA cluster in mitosis. Topo IIα is known to stimulate DNA condensation,39 and condensation-induced DNA supercoiling has been proposed to stimulate decatenation by Topo IIα.2,40 Incomplete condensation during late mitosis might explain why rDNA-UFBs are common in cells. In support of this proposal, depletion of the condensin subunit, SMC2, drastically increases the incidence of UFBs.41 Furthermore, even though active NORs are under-condensed in early mitosis, condensin I is still concentrated at the rDNA, presumably to ensure successful condensation during the mid-stages of mitosis.42
In human cells, the rDNA is ∼43 kb,43 but not all of the rDNA repeat units are actively transcribed. Silenced rDNA repeats can be distinguished from active ones by their distinct histone modifications. Because UBF binds only to active rDNA repeats (reviewed in44), one explanation for the mutually exclusive localization patterns of PICH and UBF in mitosis might have been that PICH associates only with silenced rDNA repeats. However, the results presented here argue against this being the case. Moreover, silent rDNA clusters are DAPI-positive,44 whereas PICH bodies and rDNA-UFBs are not.
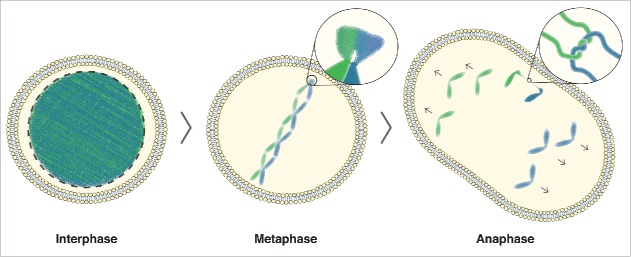
In Figure 4, we summarize our findings. We propose that PICH bodies assemble at the rDNA in early mitosis in preparation for the ensuing steps of rDNA segregation. As this region remains under condensed until transcriptional silencing in mid-mitosis, these protein ‘factories’ might be pre-assembled to facilitate the rapid condensation and segregation of the rDNA during the narrow window after transcription is shut-down. PICH may then perform a number of critical functions to facilitate rDNA segregation. In addition to directly stimulating Topo II activity and therefore facilitating condensation, PICH may also buffer the forces of the mitotic spindle as it attempts to stretch out the uncondensed rDNA.12 In doing so, PICH may provide the necessary rigidity to hold the DNA entanglements in a configuration that is optimal for decatenation. Like centromeres and telomeres, rDNA clusters are highly repetitive.45 The repetitive nature of the rDNA loci makes them prone to aberrant recombination, and this could also promote sister chromatid entanglement, and hence UFBs. Future studies should be aimed at identifying the mechanisms and signaling processes that lead to the recruitment of PICH and its partners to these types of ‘troublesome’ regions of the genome in early mitosis.
Figure 4.
rDNA-UFB model. In interphase cells, DNA is uncondensed. As cells enter mitosis, the DNA begins to condense, with the exception of the rDNA. In metaphase cells, the condensed chromosomes are aligned at the equator of the cell. The zoomed image shows the rDNA where delayed condensation causes a delay in decatenation of any DNA entanglement that arose due to DNA replication or homologous recombination. In anaphase, this can lead to DNA bridging, as shown in the zoomed image of a catenated DNA bridge. Ongoing condensation, in combination with the tension building up on unresolved catenanes by the spindle pulling forces, allows efficient decatenation of any rDNA-UFBs during anaphase.
Materials and methods
Cell culture and drug treatments
Chicken DT40 Cre1 B cell lymphoma cells were grown at 39.5°C in RPMI-1640 GlutaMAX medium supplemented with 10% fetal bovine serum, 2% chicken serum, penicillin/streptomycin (Gibco/Life sciences) and 50 mM 2-mercaptoethanol (Sigma). Synchrony in G2 was achieved by treating cells with 15 μM RO3306 (Sigma) for 6 hrs. Synchrony in prometaphase was achieved by treating cells with 0.05 μg ml−1 Nocodazole (Sigma) for 4.5 hrs. ICRF-193 was from Sigma.
Indirect immunofluorescence microscopy
DT40 cells were fixed (4% paraformaldehyde in PBS), permeabilized (0.25% Triton-X 100, 1% BSA in PBS), washed and incubated with antibodies on Polysine-coated slides (Thermo Fisher). Slides were mounted in Vectashield with DAPI (Vector laboratories). Primary antibodies were visualized with Alexa Fluor-conjugated secondary antibodies (Life Technologies). For regular fluorescence microscopy, slides were imaged on an Olympus BX63 fluorescence microscope, and the images were analyzed with ImageJ.46 For super-resolution microscopy, slides were imaged on an Elyra PS.1 super resolution microscope (Zeiss) using structured illumination microscopy, and the images were analyzed and 3D rendered with ZEN 2010 (Zeiss). Z-stack images for quantification of rDNA transcription (Fig. 2B–C) were taken using a DeltaVision imaging system (Applied Precision, GE Healthcare).
Antibodies, transcription imaging and quantification
Primary antibodies for immunofluorescence were as follows: Mouse anti-UBF (sc-13125, Santa Cruz 1:250), Mouse anti-PICH (04–1540, Millipore, 1:100), guinea pig anti-PICH (in-house,21 1:400). Transcription was visualized using the Click-iT® RNA Alexa Fluor® imaging kit (Thermo Fisher) according to the manufacturer's protocol. rDNA transcription was quantified using ImageJ.46 The background was subtracted as described previously47 using 3 background readings from areas adjacent to the cells. It should be noted that we chose to only measure the major rDNA locus, marked by UBF foci, since not all DT40 cells seemed to have a readily discernible minor rDNA locus.21 Corrected rDNA fluorescence (CRF) was calculated according to the formula: CRF = Integrated Density of rDNA locus – (Area of rDNA locus × Mean fluorescence of background readings).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Anna Bizard and Hocine Mankouri for helpful comments on the manuscript. We acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen. We acknowledge Rikke Winther-Sorensen for designing the graphical layout of Figure 4.
Funding
Work in the authors' laboratory is supported by The Danish National Research Foundation (DNRF115). CFN is supported by a fellowship from the Carlsberg Foundation.
References
- [1].Mankouri HW, Huttner D, Hickson ID. How unfinished business from S-phase affects mitosis and beyond. EMBO J 2013; 32:2661–71; PMID:24065128; http://dx.doi.org/ 10.1038/emboj.2013.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baxter J, Aragon L. A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet 2012; 28:110–7; PMID:22236810; http://dx.doi.org/ 10.1016/j.tig.2011.11.004 [DOI] [PubMed] [Google Scholar]
- [3].Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 2013; 14:25–37; PMID:23258294; http://dx.doi.org/ 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 2002; 9:515–25; PMID:11931760; http://dx.doi.org/ 10.1016/S1097-2765(02)00473-2 [DOI] [PubMed] [Google Scholar]
- [5].Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 2011; 26:125–32; PMID:21164193; http://dx.doi.org/ 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- [6].Meier K, Brehm A. Chromatin regulation: how complex does it get? Epigenetics 2014; 9:1485–95; PMID:25482055; http://dx.doi.org/ 10.4161/15592294.2014.971580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ceballos SJ, Heyer WD. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochimica et Biophysica Acta 2011; 1809:509–23; PMID:21704205; http://dx.doi.org/ 10.1016/j.bbagrm.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 2007; 128:101–14; PMID:17218258; http://dx.doi.org/ 10.1016/j.cell.2006.11.041 [DOI] [PubMed] [Google Scholar]
- [9].Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 2007; 26:3397–409; PMID:17599064; http://dx.doi.org/ 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 2009; 11:753–60; PMID:19465922; http://dx.doi.org/ 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- [11].Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res 2012; 40:7358–67; PMID:22576367; http://dx.doi.org/ 10.1093/nar/gks407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Biebricher A, Hirano S, Enzlin JH, Wiechens N, Streicher WW, Huttner D, Wang LH, Nigg EA, Owen-Hughes T, Liu Y, et al.. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol Cell 2013; 51:691–701; PMID:23973328; http://dx.doi.org/ 10.1016/j.molcel.2013.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Germann SM, Schramke V, Pedersen RT, Gallina I, Eckert-Boulet N, Oestergaard VH, Lisby M. TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J Cell Biol 2014; 204:45–59; PMID:24379413; http://dx.doi.org/ 10.1083/jcb.201305157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Broderick R, Nieminuszczy J, Blackford AN, Winczura A, Niedzwiedz W. TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat Commun 2015; 6:6572; PMID:25762097; http://dx.doi.org/ 10.1038/ncomms7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Leng M, Bessuso D, Jung SY, Wang Y, Qin J. Targeting Plk1 to chromosome arms and regulating chromosome compaction by the PICH ATPase. Cell Cycle 2008; 7:1480–9; PMID:18418076; http://dx.doi.org/ 10.4161/cc.7.10.5951 [DOI] [PubMed] [Google Scholar]
- [16].Hubner NC, Wang LH, Kaulich M, Descombes P, Poser I, Nigg EA. Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 2010; 119:149–65; PMID:19904549; http://dx.doi.org/ 10.1007/s00412-009-0244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kurasawa Y, Yu-Lee LY. PICH and cotargeted Plk1 coordinately maintain prometaphase chromosome arm architecture. Mol Biol Cell 2010; 21:1188–99; PMID:20130082; http://dx.doi.org/ 10.1091/mbc.E09-11-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ke Y, Huh JW, Warrington R, Li B, Wu N, Leng M, Zhang J, Ball HL, Yu H. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J 2011; 30:3309–21; PMID:21743438; http://dx.doi.org/ 10.1038/emboj.2011.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kaulich M, Cubizolles F, Nigg EA. On the regulation, function, and localization of the DNA-dependent ATPase PICH. Chromosoma 2012; 121:395–408; PMID:22527115; http://dx.doi.org/ 10.1007/s00412-012-0370-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rouzeau S, Cordelieres FP, Buhagiar-Labarchede G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Gueret M. Bloom's syndrome and PICH helicases cooperate with topoisomerase IIalpha in centromere disjunction before anaphase. PLoS One 2012; 7:e33905; PMID:22563370; http://dx.doi.org/ 10.1371/journal.pone.0033905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, Bjerregaard VA, Liu Y, Nigg EA, Wang LH, et al.. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun 2015; 6:8962; PMID:26643143; http://dx.doi.org/ 10.1038/ncomms9962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al.. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819–23; PMID:23287718; http://dx.doi.org/ 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kermekchiev M, Workman JL, Pikaard CS. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol Cell Biol 1997; 17:5833–42; PMID:9315641; http://dx.doi.org/ 10.1128/MCB.17.10.5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sobol M, Yildirim S, Philimonenko VV, Marasek P, Castano E, Hozak P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013; 4:478–86; PMID:24513678; http://dx.doi.org/ 10.4161/nucl.27154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gebrane-Younes J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci 1997; 110(Pt 19):2429–40; PMID:9410881 [DOI] [PubMed] [Google Scholar]
- [26].Kuhn A, Vente A, Doree M, Grummt I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J Mol Biol 1998; 284:1–5; PMID:9811537; http://dx.doi.org/ 10.1006/jmbi.1998.2164 [DOI] [PubMed] [Google Scholar]
- [27].Sirri V, Roussel P, Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J Cell Sci 1999; 112(Pt 19):3259–68; PMID:10504331 [DOI] [PubMed] [Google Scholar]
- [28].Voit R, Seiler J, Grummt I. Cooperative action of Cdk1/cyclin B and SIRT1 is required for mitotic repression of rRNA synthesis. PLoS Genet 2015; 11:e1005246; PMID:26023773; http://dx.doi.org/ 10.1371/journal.pgen.1005246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J 1998; 17:7373–81; PMID:9857193; http://dx.doi.org/ 10.1093/emboj/17.24.7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol 1996; 133:235–46; PMID:8609158; http://dx.doi.org/ 10.1083/jcb.133.2.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ray S, Panova T, Miller G, Volkov A, Porter AC, Russell J, Panov KI, Zomerdijk JC. Topoisomerase IIalpha promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat Commun 2013; 4:1598; PMID:23511463; http://dx.doi.org/ 10.1038/ncomms2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bloom SE, Bacon LD. Linkage of the major histocompatibility (B) complex and the nucleolar organizer in the chicken. Assignment to a microchromosome. J Hered 1985; 76:146–54; PMID:3998437 [PubMed] [Google Scholar]
- [33].Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 2004; 117:471–82; PMID:15137940; http://dx.doi.org/ 10.1016/S0092-8674(04)00415-5 [DOI] [PubMed] [Google Scholar]
- [34].Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, Aragon L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 2009; 458:219–22; PMID:19158678; http://dx.doi.org/ 10.1038/nature07652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang BD, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 2004; 3:960–7; PMID:15190202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Titos I, Ivanova T, Mendoza M. Chromosome length and perinuclear attachment constrain resolution of DNA intertwines. J Cell Biol 2014; 206:719–33; PMID:25225337; http://dx.doi.org/ 10.1083/jcb.201404039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol 1996; 133:235–46; PMID:8609158; http://dx.doi.org/ 10.1083/jcb.133.2.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011; 2:189–94; PMID:21818412; http://dx.doi.org/ 10.4161/nucl.2.3.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF, et al.. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol 2012; 199:755–70; PMID:23166350; http://dx.doi.org/ 10.1083/jcb.201202155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 2011; 331:1328–32; PMID:21393545; http://dx.doi.org/ 10.1126/science.1201538 [DOI] [PubMed] [Google Scholar]
- [41].Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al.. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011; 13:243–53; PMID:21317883; http://dx.doi.org/ 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- [42].Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 2013; 4:2537; PMID:24088984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gonzalez IL, Sylvester JE. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 1995; 27:320–8; PMID:7557999; http://dx.doi.org/ 10.1006/geno.1995.1049 [DOI] [PubMed] [Google Scholar]
- [44].McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 2008; 24:131–57; PMID:18616426; http://dx.doi.org/ 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- [45].Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res 2005; 15:1079–85; PMID:16024823; http://dx.doi.org/ 10.1101/gr.3970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671–5; PMID:22930834; http://dx.doi.org/ 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell 2010; 18:533–43; PMID:20412769; http://dx.doi.org/ 10.1016/j.devcel.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]